Abstract
H5N2 highly pathogenic avian influenza (HPAI) viruses caused a severe poultry outbreak in the United States (U.S.) during 2015. In order to examine changes in adaptation of this viral lineage, the infectivity, pathogenesis and transmission of poultry H5N2 viruses were investigated in chickens and mallards in comparison to the wild duck 2014 U.S. index H5N2 virus. The four poultry isolates examined had a lower mean bird infectious dose than the index virus but still transmitted poorly to direct contacts. In mallards, two of the H5N2 poultry isolates had similar high infectivity and transmissibility as the index H5N2 virus, the H5N8 U.S. index virus, and a 2005 H5N1 clade 2.2 virus. Mortality occurred with the H5N1 virus and, interestingly, with one of two poultry H5N2 isolates. Increased virus adaptation to chickens was observed with the poultry H5N2 viruses; however these viruses retained high adaptation to mallards but pathogenicity was differently affected.
Keywords: H5N2, H5N8, H5N1, highly pathogenic avian influenza virus, chickens, mallards, infectivity, pathogenicity, transmission, adaptation
Introduction
The natural reservoirs of avian influenza (AI) viruses are wild aquatic birds, with ducks, gulls and shorebirds being the primary hosts (35). Depending on many different factors, the wild bird influenza viruses can adapt to new host species resulting in a virus lineage that can infect, transmit, and persist in the new host population. With few known exceptions, the wild bird adapted viruses appear to cause little disease in the natural host, and these viruses, when experimentally inoculated into chickens, generally cause no clinical disease (28). AI viruses are classified by the disease they cause in chickens, and the wild bird viruses are almost always classified as low pathogenic (LP). Some LPAI viruses, when allowed to replicate in gallinaceous poultry, have mutated to become extremely virulent, and in the standard pathotyping tests kill at least 75% of experimentally inoculated chickens (27). The critical genetic difference determining the LP or the highly pathogenic (HP) phenotype of AI viruses is at the hemagglutinin (HA) cleavage site, and while AI viruses have 16 defined HA subtypes (i.e. H1-16), only some H5 and H7 viruses have the HP phenotype. Few HPAI viruses have become endemic in poultry, but the A/goose/Guangdong/1/96 (Gs/GD) (H5N1) HPAI virus lineage has in the last 20 years spread to over 70 countries and is currently endemic in poultry in at least 8 different countries remaining a constant threat for many countries around the world (19). The HA genes of the virus have diversified into multiple genetic lineages or clades, and specifically subclade 2.3.4.4 has reassorted with different neuraminidase subtypes to generate widely circulating variants including H5N2, H5N3, H5N5, H5N6, and H5N8 subtypes of HPAI viruses (15, 16, 33, 36, 37). In early 2014, outbreaks of H5N8 HPAI were reported in South Korea and Japan in poultry and wild aquatic birds (17), with migratory aquatic birds highly suspected in playing a key role in the spread of the virus (9). In late autumn of 2014 and early 2015, H5N8 HPAI viruses were detected in Russia and several countries in Europe, and in captive falcons, wild birds, and backyard aquatic and gallinaceous poultry in the Western U.S. (2, 8, 15, 33). In addition, another novel reassortant HPAI virus of H5 clade 2.3.4.4, an H5N2, was identified as the cause of an outbreak in poultry farms in British Columbia (24) and was subsequently detected in the U.S. in wild waterfowl and backyard poultry (2, 7, 31). From March through mid-June of 2015, H5N2 viruses caused widespread HPAI infections in commercial poultry flocks in the upper Midwestern U.S. states (10). This represented the worst HPAI event in history for U.S. poultry producers, with more than 49.7 million birds dying or being euthanized (30). The resulting disruption of poultry supply chain, bans on exports of U.S. poultry and poultry products to many countries, and increased costs to the consumer made the economic cost of this outbreak at over 3 billion dollars (30).
The epidemiology of the H5 HPAI virus detections suggested that the initial H5N2 and H5N8 HPAI viruses detected in the U.S. were highly adapted to waterfowl and not yet well adapted to domestic poultry. To better model the outbreak, the pathogenesis and transmission dynamics of representative H5N8 and H5N2 clade 2.3.4.4 HPAI viruses detected early in the U.S. were investigated in chickens (1). Pathobiological features of these isolates were consistent with HPAI virus infection, although the delayed appearance of clinical signs, lesions, and longer mean death times differed from observations with most other Gs/GD lineage H5 HPAI viruses. High mean chicken infectious doses and lack of seroconversion in directly inoculated and contact exposed survivors indicated the viruses were poorly adapted to chickens (1). In contrast, these two index H5 HPAI viruses were highly adapted to mallards and transmitted very well to direct contacts (23). Although these initial U.S. H5 HPAI viruses had reduced adaptation and transmissibility in chickens, multi-generational passage in gallinaceous poultry (chickens or turkeys) could generate chicken adapted viruses with higher infectivity (i.e. lower mean infectious dose) and transmissibility (1). This could also result in changes in adaptation in mallards which could affect the epidemiology of the virus. In order to examine for changes in virus adaptation between the H5N2 wild bird index virus and later poultry isolates, we determined the infectivity, pathogenesis and transmission of H5N2 viruses isolated from the Midwest poultry outbreak in chickens and mallards.
Materials and methods
Viruses
The following HPAI viruses were used in this study; A/Northern pintail/Washington/40964/2014 (H5N2) (A/Np/WA/14), A/gyrfalcon/Washington/40188-6/2014 (H5N8) (A/Gf/WA/14), A/turkey/Minnesota/12582/2015 (H5N2) (A/Tk/MN/15), A/turkey/South Dakota/12511/2015 (H5N2) (A/Tk/SD/15), A/chicken/Iowa/13388/2015 (H5N2) (A/Ck/IA/15), A/turkey/Arkansas/7791/2015 (H5N2) (A/Tk/AR/15), and A/Whooper swan/Mongolia/244/2005 (H5N1) (A/Ws/Mongolia/05). This last virus was included for comparison purposes and belongs to the clade 2.2 H5N1 viruses that spread from Asia into Europe in 2005 via migratory wild waterfowl. The viruses were propagated in specific pathogen free (SPF) embryonating chicken eggs (ECE) according to standard procedures (12). Allantoic fluid was diluted in brain heart infusion (BHI) medium (BD Bioscience, Sparks, MD) in order to obtain an inoculum with 102, 104,106 50% egg infectious dose (EID50) per 0.1 ml/bird. All challenge doses were confirmed by back-titer in ECE’s. All experiments using the HPAI viruses, including work with animals, were conducted according to procedures approved by the institutional biosecurity committee and were performed in biosecurity level-3 enhanced (BSL-3E and ABSL-3E) facilities at the Southeast Poultry Research Laboratory (SEPRL), U.S. National Poultry Research Center, Agricultural Research Service, United States Department of Agriculture (USDA).
Animals and housing
Four week-old specific pathogen free White Leghorn chickens (Gallus gallus domesticus) were obtained from SEPRL’s in-house flocks. Mallard ducks (Anas platyrhynchos) were obtained at 1 day of age from a commercial hatchery and held for 2 weeks at SEPRL. Serum samples were collected from 15 chickens and 15 ducks to confirm that the birds were serologically negative to AIV by blocking ELISA (FlockCheck Avian Influenza MultiS-Screen Antibody Test®, IDEXX Laboratories, Westbrook, ME, USA). Each experimental group was housed in self-contained isolation units ventilated under negative pressure with inlet and exhaust HEPA-filtered air. Feed and water were provided with ad libitum access. Birds were cared for in accordance with an Institutional Animal Care and Use Committee approved animal use protocol.
Experimental design and sampling
The objective of the study was to evaluate the infectivity, transmissibility and pathogenicity of the H5 HPAI viruses in chickens and mallards. The following H5N2 HPAI viruses were evaluated in chickens: A/Tk/MN/15, A/Tk/SD/15, A/Ck/IA/15, and A/Tk/AR/15 (Table 1). The following H5 HPAI viruses were evaluated in mallards: A/Tk/MN/15 (H5N2), A/Ck/IA/15 (H5N2), A/Np/WA/14 (H5N2), A/Gf/WA/14 (H5N8), and A/Ws/Mongolia/05 (H5N1) (Table 2). To evaluate the mean bird infectious dose (BID50) birds were divided into groups of 5-8 birds, and each bird was inoculated intranasally by the choanal cleft with 102, 104, or 106 EID50 in 0.1 ml of the respective viruses. Sham birds were inoculated intranasally with 0.1 ml of sterile allantoic fluid diluted 1:300 in brain heart infusion (BHI) media (Becton, Dickinson and Company, Sparks, MD). To evaluate the transmissibility of each isolate, 2-3 non-inoculated hatch mates were added to each dose group at 1 day post-inoculation (dpi)(contacts). Clinical signs were monitored daily. Body temperatures and weights of mallards inoculated with 106 EID50 of the H5N2 poultry viruses and sham-inoculated controls were taken at 2 and 4 dpi. Oropharyngeal (OP) and cloacal (CL) swabs were collected from chickens at 1, 2, 3 and 4 dpi, and from mallards at 2, 4, 7, 11 and 14 dpi. Swabs were placed in 1.0 ml of BHI with penicillin (2000 units/ml; Sigma Aldrich), gentamicin (200 μg/ml; Sigma Aldrich) and amphotericin B (5 μg/ml; Sigma Aldrich), and stored at −80C. The remaining birds were observed daily for clinical signs over a 14-day period. Birds that were severely lethargic, showed severe neurological signs, stopped eating or drinking or remained recumbent were euthanized. Surviving birds were bled at 14 dpi to evaluate antibody titers and euthanized.
Table 1.
Mortality, mean death time, mean bird infectious dose, virus shedding, serology and transmission to direct contacts of H5N2 HPAI poultry viruses in 4 week-old chickens.
| Virus dose (Log 10 EID50) | Mortalitya (MDT) | BID50 | Virus sheddingb (average titer of positive birds) | Serologyc | Mortality contactsd (MDT) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||
| 1 dpi | 2 dpi | 3 dpi | 4 dpi | ||||||||||
|
|
|||||||||||||
| OP | CL | OP | CL | OP | CL | OP | CL | ||||||
| A/Tk/MN/15 | 2 | 0/5 | 3.6 log10 | 1/5 (2.1) | nd | 1/5 (2.0) | nd | 0/5 | nd | 1/5 (2.1) | nd | 0/5 | 0/3 |
| 4 | 3/5 (2.3) | 3/5 (5.2) | nd | 3/5 (5.9) | nd | 0/2 | nd | 1/2 (2.3) | nd | 0/2 | 0/3 | ||
| 6 | 8/8 (2) | 10/10 (5.4) | 10/10 (4.9) | 2/2 (4.2) | 2/2 (4.3) | - | - | - | - | - | 2/2 (4) | ||
| A/Tk/SD/15 | 2 | 0/5 | 3.2 | 0/5 | nd | 0/5 | nd | 0/5 | nd | 0/5 | nd | 0/5 | 0/3 |
| 4 | 4/5 (4) | log10 | 2/5 (3.9) | nd | 1/4 (3.3) | nd | 2/4 (4.0) | nd | 1/3 (3.2) | nd | 0/2 | 0/3 | |
| 6 | 7/7 (2.2) | 8/10 (3.7) | 9/10 (2.9) | 2/3 (4.9) | 2/3 (3.1) | 1/1 (5.1) | 1/10 (4.4) | - | - | - | 0/2 | ||
| A/Ck/IA/15 | 2 | 1/5 (2) | 3.5 log10 | 1/5 (5.4) | nd | 1/4 (2.3) | nd | 1/4 (2.1) | nd | 1/4 (2.2) | nd | 0/4 | 0/3 |
| 4 | 3/5 (3.3) | 3/5 (4.8) | nd | 5/5 (5.9) | nd | 3/3 (6.1) | nd | 2/2 (2.0) | nd | 0/2 | 0/3 | ||
| 6 | 8/8 (2.4) | 10/10 (4.6) | 10/10 (3.5) | 5/5 (5.6) | 5/5 (4.6) | 2/2 (6.1) | 2/2 (6.6) | - | - | - | 0/2 | ||
| A/Tk/AR/15 | 2 | 0/5 | 5.1 | 0/5 | nd | 0/5 | nd | 0/5 | nd | 0/5 | nd | 0/5 | 0/3 |
| 4 | 0/5 | log10 | 0/5 | nd | 0/5 | nd | 0/5 | nd | 0/5 | nd | 0/5 | 0/3 | |
| 6 | 8/9 (2.1) | 5/11 (3.8) | 5/11 (4.5) | 3/5 (4.0) | 4/5 (3.4) | 0/31 | 3/3 (2.7) | 0/1 | 0/1 | 0/1 | 0/2 | ||
EID50, mean egg infectious dose; MDT, mean death time; BID50, mean bird infectious dose; OP, oropharyngeal; CL, cloacal; dpi, days post-inoculation; nd, not done. -, birds dead.
# of inoculated birds dead/total # of birds inoculated
# of virus-positive birds/total # of birds sampled
# of positive birds/total # of birds sampled
# of virus-positive birds/total # of birds sampled
Table 2.
Average distribution of AIV-NP antigen by IHC in tissues from chickens and mallards inoculated with poultry H5N2 HPAI viruses. Tissues were examined at 2 dpi (chickens) and at 3 dpi (mallards) (bird 1/bird 2).
| Species | Virus | Detection of AIV antigen in tissues | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||||
| Nasal epithelium | Eyelid | Trachea | Lung | Heart | Spleen | Brain | Liver | Adrenal gland | Pancreas | Kidney | Cecal tonsils | Thymus | Bursa | Harderian gland | ||
| Chickens | A/Tk/MN/15 | ++/++ | +++/+++ | −/+++ | +++/+++ | +++/++ | ++/+++ | ++/+++ | −/++ | +/+++ | ++/+++ | +/+ | +/+ | +/+++ | −/+++ | ++/+ |
| A/Tk/SD/15 | ++/+ | +++/+ | ++/− | +++/++ | +++/++ | +++/++ | ++/+ | +/+ | ++/+ | ++/+ | +/++ | +/+ | +/+ | +++/− | −/− | |
| A/Ck/IA/15 | ++/++ | +++/+ | +/− | +++/+++ | +++/+++ | +++/+++ | ++/+++ | +/+ | +++/++ | +/++ | ++/++ | +/− | ++/+ | +++/− | +/− | |
| A/Tk/AR/15 | +/+ | −/− | −/− | +/− | +/+ | ++/− | −/− | −/+ | +/− | +/− | −/− | −/− | −/− | −/− | −/− | |
| Mallards | A/Tk/MN/15 | ++/+ | +/− | +/+++ | +/+ | +/++ | +/++ | +/+++ | −/− | −/+ | −/+ | −/− | −/− | ++/++ | −/+ | ++/+++ |
| A/Ck/IA/15 | +/− | −/− | −/− | +/+ | +/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | +/+ | −/− | −/+ | |
= no positive cells;
= single positive cells;
= scattered groups of positive cells;
= widespread positivity.
Two birds were necropsied at 2 dpi (chickens) or 3 dpi (mallards) from the groups inoculated intranasally with 106 EID50 of the H5N2 poultry viruses and from the sham-inoculated control groups. Portions of lung and spleen were collected for virus detection. Tissue samples were collected for microscopic evaluation and included beak, eyelid, trachea, lung, heart, spleen, brain, liver, adrenal gland, pancreas, intestine, thymus, bursa and Harderian gland. Tissues were fixed in 10% neutral buffered formalin solution, sectioned, paraffin embedded, and stained with hematoxylin-and-eosin. Serial sections were also stained by IHC methods to visualize influenza viral antigen distribution in individual tissues as previously described with minor modifications (21).
Viral RNA quantification in swabs and tissues
Viral RNA was extracted from swabs using the MagMAX AI/ND Viral RNA Isolation Kit (Ambion, Austin, TX, USA). Quantitative real time RT-PCR (qRRT-PCR) for AIV detection was performed as previously described (20). qRRT-PCR reactions targeting the influenza virus M gene (25) were conducted using AgPath-ID one-step RT-PCR Kit (Ambion, Austin, TX, USA) and the ABI 7500 Fast Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA). The RT step conditions were 10 min at 45°C and 95°C for 10 min. The cycling conditions were 45 cycles of 15 s, 95°C; 45 s, 60°C. Virus titers in frozen tissue samples were determined by weighing, homogenizing, and diluting tissues in BHI to a 10% (wt/vol) concentration. Viral RNA was extracted using Trizol LS reagent (Invitrogen, Carlsbad, CA) and the Qiagen RNeasy Mini Kit (Qiagen, USA). Equal amounts of RNA extracted from the tissue samples were used in the qRRT-PCR assay (50 ng/μl). For virus quantification, a standard curve was established with RNA extracted from dilutions of the same titrated stock of the challenge virus, and results reported as EID50/ml or EID50/gr equivalents. The calculated qRT-PCR lower detection limit for the viruses varied between 101.5EID50/ml, and 102.5EID50/ml.
Serology
Hemagglutination inhibition (HI) assays were performed to quantify antibody responses to virus infection as previously described (OIE, 2012), with serum collected from surviving birds at 14 dpi. Sera samples were tested by HI assays against antigens specific for the challenge viruses. HI titers were reported as reciprocal log2 titers, with a 3log2 titer or below considered negative.
Statistical analyses
One-way ANOVA with Tukey’s multiple comparison tests was used to analyze body weights, body temperatures, and titers of virus shed, using Prism v.5.01 software (GraphPad Prism™ Version 5 software Inc. La Jolla, CA, USA). A P-value of < 0.05 was considered to be significant. For statistical purposes, all OP and CL swabs and tissues in which virus was not detected were given a numerical value between 101.4 and 102.4 EID50/ml. These values represent the lowest detectable value of virus in these samples based on the methods used.
Sequence analysis
In order to identify genetic changes associated with the changes observed in virus adaptation, full genome sequence analysis of the H5N2 viruses was conducted. Complete genomes of A/Tk/MN/15, A/Tk/SD/15, and A/Ck/IA/15 were sequenced using Ion torrent PGM (Life technologies) and Miseq (Illumina) next-generation sequencer at the National Veterinary Services Laboratories in Ames, Iowa and have been deposited in GenBank under accession no. KX351776-KX351783, KX351768-KX351775, and KX351784-KX351791, respectively. We also retrieved from GenBank the complete genome sequences of A/Np/WA/14 (GenBank accession no. KP307973-KP307980), and A/Tk/AR/15 (GenBank accession no. KR234019-KR234026). The nucleotide sequences for the complete coding regions of H5N2 HPAIV were aligned using MUSCLE (5). Complete coding regions of each segment were aligned and used for subsequent single-nucleotide polymorphisms (SNP) analysis using the Geneious v8.1.2 program (11). The coding sequences discriminating SNPs were classified as either nonsynonymous or synonymous based on whether or not they correspond to differences in encoded amino acid sequences.
Results
Infectivity, pathogenicity and transmission of H5N2 HPAI poultry isolates in chickens
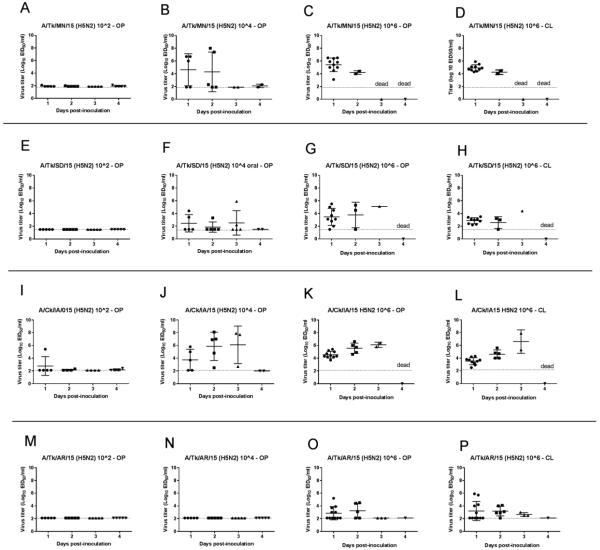
Results for virus infectivity and transmission for chickens are shown in Table 1. Birds, both directly inoculated or contacts, were considered infected if they shed virus, exhibited morbidity, mortality, or seroconverted by 14 dpi. Birds infected with any of the four H5N2 viruses showed similar clinical signs including ruffled feathers, listlessness, infraorbital swelling and prostration. All chickens inoculated with the 106 dose, with the exception of one chicken in the A/Tk/AR/15 group, became infected and died, with mean death times (MDT’s) between 2 and 2.4 days. Three to four chickens inoculated with the 104 virus dose of A/Tk/MN/15, A/Tk/SD/15 and A/Ck/IA/15 became infected and died in less than 4 days. No chickens inoculated with 104 of A/Tk/AR/15 showed clinical signs. Only a single chicken inoculated with A/Ck/IA/15 died at the 102 dose, with chickens in all other 102 groups surviving challenge. The mean bird infectious doses (BID50) for A/Tk/MN/15, A/Tk/SD/15 and A/Ck/IA/15 were similar: 103.6, 103.2 and 103.5 EID50 respectively. The BID50 for A/Tk/AR/15 was higher at 105.1 EID50. The surviving birds did not show evidence of clinical disease and were all serologically negative based on HI data. Only the two contact birds in the group inoculated with 106 of A/Tk/MN/15 became infected and died (Table 1). In the rest of the groups, no contact birds became infected as demonstrated by negative results in virus shed and serology (data not shown).
Chickens inoculated with the lowest dose of the H5N2 viruses shed no or low levels of virus (Table 1, Figure 1), with the exception of the one bird inoculated with A/Ck/IA/15 that died. Three to four birds inoculated with the 104 EID50 dose of A/Tk/MN/15, A/Tk/SD/15 or A/Ck/IA/15, and all birds but one inoculated with the 106 EID50 dose of all four viruses, shed moderate to high amounts of virus. Significantly higher OP titers were shed at 1 dpi by chickens inoculated with A/Tk/MN/15 when compared to A/Tk/SD/15 (P<0.001); and A/Tk/AR/15 (P<0.0001). Higher titers were shed at 1 dpi by chickens inoculated with A/Ck/IA/15 when compared to A/Tk/AR/15 (P<0.0001). Higher CL titers were shed at 1 dpi by chickens inoculated with A/Tk/MN/15 when compared to chickens inoculated with A/Tk/SD/15 (P<0.0001); A/Tk/AR/15 (P<0.0001) and A/Ck/IA/15 (P<0.001).
Figure 1.
Oropharyngeal (OP) and cloacal (CL) viral shed detected by qRRT-PCR from 4-week-old chickens directly inoculated with poultry H5N2 HPAI viruses (bars represent mean and standard deviation). A. A/turkey/Minnesota/12582/2015 (H5N2) (A/Tk/MN/15). B. A/turkey/South Dakota/12511/2015 (H5N2) (A/Tk/SD/15). C. A/chicken/Iowa/13388/2015 (H5N2) (A/Ck/IA/15). D. A/turkey/Arkansas/7791/2015 (H5N2) (A/Tk/AR/15).
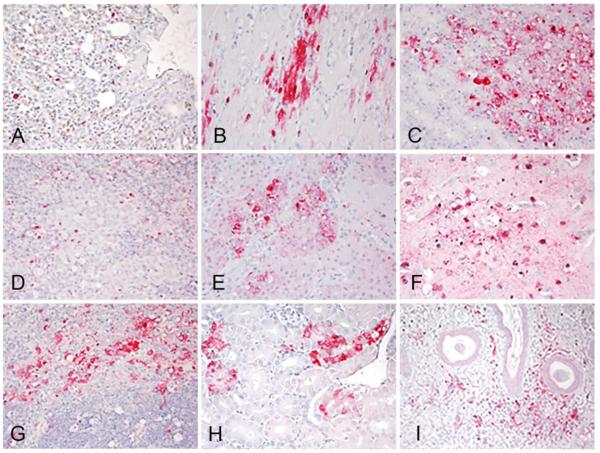
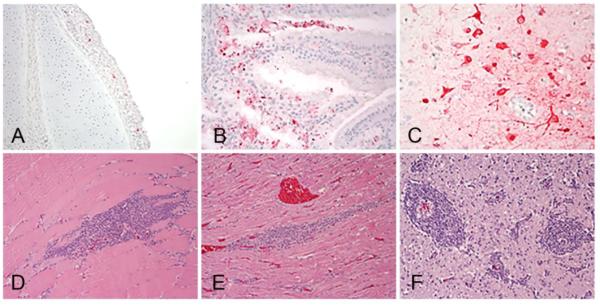
Two birds from the groups of chickens inoculated with 106 of A/Tk/SD/15, A/Ck/IA/15 and A/Tk/AR/15 and two sham inoculated controls were necropsied at 2 dpi. Since there were no survivors in the 106 group inoculated with A/Tk/MN/15 at this time point, two moribund birds from the 104 group were examined. The birds challenged with A/Tk/MN/15, A/Tk/SD/15, and A/Ck/IA/15 were listless, with cyanotic combs and wattles, ruffled feathers, hemorrhages on the shanks and had green watery feces. Similar gross lesions were observed in all six birds and consisted of empty intestines, multifocal necrosis in the pancreas, congested lungs, petechial hemorrhages in the thymus and on skeletal muscle, and splenomegaly with parenchymal mottling. The two chickens necropsied from the A/Tk/AR/15 group had only ruffled feathers and mild gross lesions including empty intestines, congested lungs and splenomegaly. Microscopic lesions and viral antigen staining were similar in severity and distribution among chickens inoculated with A/Tk/MN/15, A/Tk/SD/15, and A/Ck/IA/15, and less prominent in chickens inoculated with A/Tk/AR/15. Microscopic lesions consisted of multifocal necrosis in the parenchyma of several tissues including brain, spleen, adrenal gland, kidney, pancreas, bursa, thymus, cecal tonsils, harderian gland, and liver, and were similar to lesions reported for HPAI viruses (28). Virus antigen was present in parenchymal cells of many organs including cardiac myocytes, hepatocytes, microglial cells and neurons, lung, and kidney tubular epithelial cells (Table 2, Figure 2, A-I). Viral antigen straining in capillary endothelial cells was uncommon, restricted mainly to capillaries in eye lid and air capillaries of the lungs. Sham-inoculated birds were clinically healthy throughout the experiment, and showed no lesions or virus antigen in tissues.
Figure 2.
Immunohistochemical detection of viral antigen in 4-week-old chickens intranasally inoculated with106 EID50 of poultry H5N2 viruses. A/turkey/Minnesota/12582/2015 (A, B, D, G, I), and A/chicken/Iowa/13388/2015 (C, E, F, H). A. Viral antigen (in red) in epithelium of air capillaries and mononuclear cells in the lung. B. Viral antigen in cardiac myocytes. C. Viral antigen in acinar cells in pancreas. D. Viral antigen in mononuclear cells in the spleen. E. Viral staining in adrenal corticotropic cells. F. Viral antigen in neurons and ependymal cells in the brain. G. Viral staining in histiocytes in the thymus. H. Viral staining in tubular epithelial cells in kidney. I. Viral staining in vascular endothelial cells and infiltrating mononuclear cells in eyelid. Magnification 40X.
Virus replication was also examined at 2 dpi in lung and spleen following infection with the HPAI viruses (Table. 3). Similar high virus titers were found in tissues of chickens infected with all four viruses, with the exception of one chicken inoculated with A/Tk/AR/15 which had lower titers in spleen and lung than the rest.
Table 3.
Virus titers in tissues collected from chickens inoculated with poultry H5N2 HPAI viruses at 2 dpi (bird1/bird2).
| Virus | Log 10 dose | Spleen | Lung |
|---|---|---|---|
| A/Tk/MN/15 | 4 | 6.2/8.0a | 7.1/8.2 |
| A/Tk/SD/15 | 6 | 7.7/6.5 | 8.1/7.6 |
| A/Ck/IA/15 | 6 | 6.5/7.0 | 7.2/7.9 |
| A/Tk/AR/15 | 6 | 6.7/3.1 | 5.7/2.7 |
EID50/g
Infectivity, pathogenicity and transmission of H5N2 HPAI poultry isolates in mallards and comparison with other H5 HPAI viruses
Results for virus infectivity and transmission of the H5 HPAI viruses in mallards are shown in Table 4. Based on virus shed and seroconversion, all mallards, even those given the low virus doses and all contacts became infected. Therefore, the mean bird infectious dose (BID50) for all viruses in mallards was less than 102 EID50. However mortality was limited, only observed in two mallards inoculated with 104 or 106 of A/Tk/MN/15 (H5N2) and one of the contacts in the same 106 dose group, and in all mallards inoculated with A/Ws/Mongolia/05 (H5N1) and contacts, regardless of the dose given. Mallards inoculated with A/Ws/Mongolia/05 and the contacts, presented severe clinical disease with listlessness, anorexia and mild to severe neurological signs beginning at 2 dpi and characterized by tremors, lack of coordination, head tilt, seizures, and paralysis. Directly inoculated and contacts ducks in these groups died in less than five days, with the exception of one contact mallard from the 102 group which died at 6 dpi. The two ducks inoculated with A/Tk/MN/15 (H5N2) and the contact duck presented with neurological signs and were euthanized at 9 dpi. No clinical signs were observed in the rest of the mallards.
Table 4.
Mortality, virus shedding, serology and transmission to direct contacts of H5N2, H5N8, and H5N1 HPAI viruses in mallards.
| Virus | Log 10 Dose | Mortalitya (MDT) | Virus sheddingb (average titer of positive birds) | Serologyc (Log2 titers) | Contacts infectedd | Contacts mortality (MDT) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| 2 dpi | 4 dpi | 7 dpi | 11 dpi | 14 dpi | |||||||||||
|
| |||||||||||||||
| OP | CL | OP | CL | OP | CL | OP | CL | OP | CL | ||||||
| A/Tk/MN/15 (H5N2) | 2 | 0/5 | 5/5 (5.9) | 5/5 (3.7) | 5/5 (7.6) | 5/5 (5.5) | 5/5 (5.6) | 5/5 (5.3) | 5/5 (4.3) | 5/5 (4.6) | 5/5 (4.1) | 5/5 (4.0) | 5/5 (5.4) | 3/3 | 0/3 |
| 4 | 1/5 (9) | 5/5 (6.6) | 5/5 (5.0) | 5/5 (7.3) | 5/5 (4.8) | 5/5 (4.6) | 5/5 (2.9) | 3/4 (2.9) | 4/4 (3.1) | 3/4 (2.4) | 4/4 (2.9) | 4/4 (5.6) | 3/3 | 0/3 | |
| 6 | 1/8 (9) | 10/10 (7.4) | 10/10 (3.6) | 8/8 (6.8) | 8/8 (4.3) | 8/8 (4.8) | 8/8 (3.2) | 7/7 (2.9) | 7/7 (3.2) | 4/7 (2.6) | 4/7 (2.7) | 7/7 (5.6) | 3/3 | 1/3 (9) | |
| A/Ck/IA/15 (H5N2) | 2 | 0/5 | 1/5 (4.8) | 1/5 (3.3) | 4/5 (3.8) | 2/5 (2.8) | 5/5 (4.8) | 5/5 (2.6) | 1/5 (3.0) | 0/5 | 0/5 | 0/5 | 5/5 (4.6) | 3/3 | 0/3 |
| 4 | 0/5 | 5/5 (4.6) | 0/5 | 5/5 (5.9) | 4/5 (3.6) | 3/5 (3.4) | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 5/5 (6.4) | 3/3 | 0/3 | |
| 6 | 0/8 | 10/10 (4.7) | 3/8 (3.3) | 7/8 (4.9) | 6/8 (3.1) | 1/8 (2.8) | 0/8 | 1/8 (2.7) | 0/8 | 0/8 | 0/8 | 8/8 (5.5) | 3/3 | 0/3 | |
| A/Np/WA/14 (H5N2) | 2 | 0/5 | 2/5 (4.7) | 1/5 (2.9) | 5/5 (4.5) | 4/5 (3.5) | 5/5 (4.2) | 5/5 (3.6) | 5/5 (4.1) | 0/5 | 0/5 | 0/5 | 4/5 (3.2) | 3/3 | 0/3 |
| 4 | 0/5 | 5/5 (5.7) | 5/5 (4.1) | 5/5 (5.6) | 5/5 (4.1) | 5/5 (3.4) | 4/5 (5.0) | 3/5 (2.8) | 4/5 (4.6) | 0/5 | 0/5 | 5/5 (4.4) | 3/3 | 0/3 | |
| 6 | 0/5 | 5/5 (5.5) | 5/5 (3.0) | 5/5 (5.5) | 5/5 (3.5) | 5/5 (3.4) | 5/5 (4.3) | 5/5 (4.2) | 5/5 (3.7) | 0/5 | 0/5 | 5/5 (3.6) | 3/3 | 0/3 | |
| A/Gf/WA/14 (H5N8) | 2 | 0/5 | 4/5 (3.6) | 3/5 (2.7) | 5/5 (6.2) | 5/5 (2.8) | 5/5 (4.3) | 4/5 (2.9) | 2/5 (3.4) | 4/5 (2.5) | 0/5 | 0/5 | 3/5 (3.8) | 3/3 | 0/3 |
| 4 | 0/5 | 5/5 (5.9) | 5/5 (3.1) | 5/5 (5.4) | 5/5 (3.7) | 5/5 (3.3) | 4/5 (3.3) | 3/5 (2.8) | 3/5 (3.0) | 0/5 | 0/5 | 5/5 (4.3) | 3/3 | 0/3 | |
| 6 | 0/5 | 5/5 (5.8) | 5/5 (2.8) | 5/5 (5.2) | 4/5 (3.3) | 5/5 (3.3) | 4/5 (3.1) | 5/5 (2.6) | 2/5 (2.0) | 0/5 | 0/5 | 5/5 (4.2) | 3/3 | 0/3 | |
| A/Ws/Mongolia/05 (H5N1) | 2 | 5/5 (4) | 5/5 (5.5) | 5/5 (3.3) | 2/5 (4.7) | 2/5 (3.0) | - | - | - | - | - | - | - | 3/3 | 3/3 (5) |
| 4 | 5/5 (3.5) | 5/5 (5.6) | 5/5 (3.3) | 2/5 (5.2) | 2/5 (3.2) | - | - | - | - | - | - | - | 3/3 | 3/3 (4.5) | |
| 6 | 5/5 (3.5) | 5/5 (5.5) | 5/5 (3.1) | 2/5 (4.5) | 2/5 (2.8) | - | - | - | - | - | - | - | 3/3 | 3/3 (4.5) | |
EID50, mean egg infectious dose; MDT, mean death time; OP, oropharyngeal; CL, cloacal; dpi, days post-inoculation; na, not applicable.
# of inoculated birds dead/total # of birds inoculated
# of virus-positive birds/total # of birds sampled
# of positive birds/total # of birds sampled
# of virus-positive contact birds/total # of contact birds
Body temperatures and weights were taken at 2 and 4 dpi from mallards inoculated with 106 of A/Tk/MN/15 and A/Ck/IA/15 and the sham-inoculated controls. At 2 dpi, but not at 4dpi, mallards inoculated with A/Tk/MN/15 and A/Ck/IA/15 had significantly higher body temperatures (109±0.5°C, 108.4±0.9°C), than the controls (107.2±0.5°C) (P<0.0001 and 0.001, respectively), but there were no differences between the virus-inoculated groups. No differences in body weights were observed at 2 dpi among all three groups, but at 4 dpi mallards inoculated with A/Tk/MN/15 and A/Ck/IA/15 had significantly lower body weights (290±3g and 340±2g) than controls (398± 3g) (P<0.0001 and 0.001, respectively), and body weights were significantly lower in the A/Tk/MN/15 group compared to the A/Ck/IA/15 group (P<0.001).
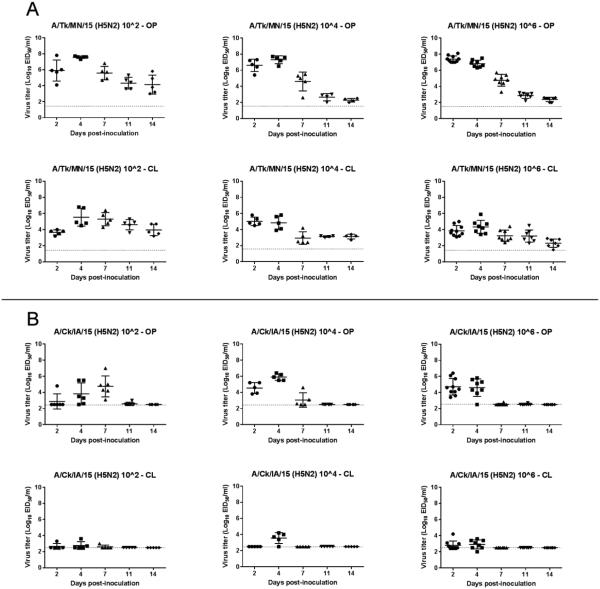
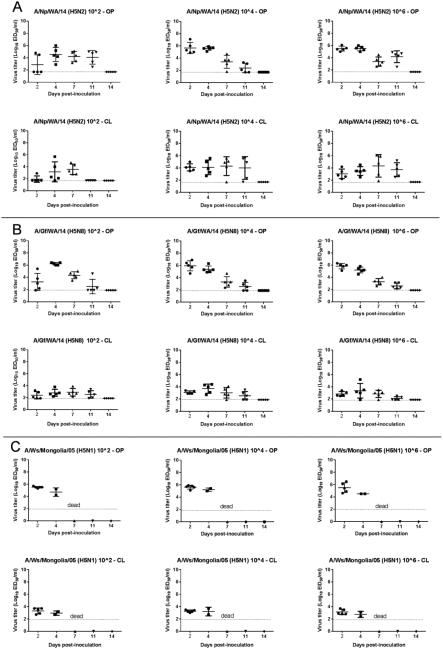
Viral RNA was detected in both OP and CL swabs from all mallards inoculated with A/Tk/MN/15 at all sampling time points regardless of the dose given (Table 4, Figure 3). Titers were higher in OP swabs. On the contrary, mallards inoculated with A/Ck/IA/15 shed minimal amounts of virus by the CL route, and stopped shedding virus before 11 dpi. Mallards inoculated with A/Np/WA/14 (H5N2) and A/Gf/WA/14 (H5N8) showed similar patterns of virus shedding, with higher titers in OP swabs and shedding detected until 11 dpi (Table 4, Figure 4). Mallards inoculated with A/Ws/Mongolia/05 (H5N1) shed high amount of virus by the OP route before dying (Table 4, Figure 4). Mallards inoculated with A/Tk/MN/15 shed significantly higher amount of virus at 2 dpi by the OP route than mallards inoculated with A/Ck/IA/15, A/Np/WA/14, and A/Gf/WA/14 (P<0.00001, 0.0001, and 0.001, respectively). This difference in virus shed was also observed at 4 dpi. Similar patterns of virus shed were observed in the contact ducks when compared to the virus-inoculated ducks (Supplemental Figure 1 and 2). Contact ducks seroconverted at 13 dpi with titers also similar to virus-inoculated ducks.
Figure 3.
Oropharyngeal (OP) and cloacal (CL) viral shed detected by qRRT-PCR from 2-week-old mallards directly inoculated with poultry H5N2 HPAI viruses (bars represent mean and standard deviation). A. A/turkey/Minnesota/12582/2015 (H5N2) (A/Tk/MN/15). B. A/chicken/Iowa/13388/2015 (H5N2) (A/Ck/IA/15).
Figure 4.
Oropharyngeal (OP) and cloacal (CL) viral shed detected by qRRT-PCR from 2-week-old mallards directly inoculated with the H5N2 and H5N8 HPAI U.S. index viruses and a H5N1 Goose/Guangdong lineage virus (bars represent mean and standard deviation). A, A/Northern pintail/Washington/40964/2014 (H5N2) (A/Np/WA/14). B. A/gyrfalcon/Washington/40188-6/2014 (H5N8) (A/Gf/WA/14). C A/Whooper swan/Mongolia/244/2005 (H5N1) (A/Ws/Mongolia/05).
Two mallards from each of the groups inoculated with 106 of A/Tk/MN/15, A/Ck/IA/15 and the sham-inoculated control group were necropsied at 3 dpi, as well as the ducks that had to be euthanized during the course of the experiment (3 ducks from the A/Tk/MN/15 group that were euthanized at 9 dpi). Control ducks and ducks inoculated with A/Ck/IA/15 lacked gross lesions. Gross lesions in the ducks inoculated with A/Tk/MN/15 included empty intestines, splenomegaly, and thymus atrophy. More severe microscopic lesions and more widespread viral staining were also present in tissues of A/Tk/MN/15-inoculated ducks compared to ducks inoculated with A/Ck/IA/15 (Table 2). Lesions included mild to moderate rhinitis and tracheitis, mild focal degeneration of pancreatic acinar cells and splenic macrophages, mild lymphocyte depletion in the thymus and bursa, and mild lymphocyte infiltration in the liver. The lesions present in the lung consisted of mild congestion and interstitial inflammation with mixed mononuclear cells. In the brain, randomly scattered foci of malacia with gliosis were observed. In the heart, mild focal myocardial degeneration to necrosis and minimal to mild mononuclear cell inflammation was present. The corticotrophic cells of the adrenal gland had mild focal vacuolar degeneration to necrosis. The intestinal epithelium was only minimally affected, with mild inflammatory changes in the lamina propria. Mild to moderate necrosis of hepatocytes with sinusoidal histiocytosis was observed in the liver. The spleen, thymus, bursa, and mucosa-associated lymphoid tissue had moderate lymphoid depletion.
Viral antigen staining in A/Tk/MN/15 inoculated ducks was present in lymphoid organs within resident and infiltrating phagocytes (Table 2, Figure 5 A-C). Vascular endothelium was consistently negative for the presence of viral antigen. Viral antigen was detected in the nasal, trachea, and Harderian gland epithelium and infiltrating mononuclear cells, in phagocytes in the lung, in neurons and glial cells of the brain, in cardiac myocytes, and in pancreatic acinar epithelium. No viral staining was found in tissues collected from the duck euthanized at 9 dpi, however lymphoplasmacytic perivascular cuffs in the brain and lymphoplasmacytic infiltration in the heart and muscle were present (Figure 5 D-F). No or very mild lesions and viral staining was detected in tissues of mallards inoculated with A/Ck/IA/15.
Figure 5.
Histological lesions and immunohistochemical detection of viral antigen in 2-week-old mallards intranasally inoculated with 106 EID50 of A/Tk/MN/15 H5N2 HPAI virus. Tissues were collected at 3 dpi (A, B, C) and at 9 dpi (D, E, F). Viral antigen (in red) in epithelial cells and infiltrating mononuclear cells in trachea (A) and Harderian gland (B) and in neurons and glial cells of the cerebrum (C). Lymphoplasmacytic cell infiltration in skeletal muscle (D), heart (F), and forming perivascular cuffs in the cerebrum (E). Magnification 40X).
Virus replication in spleens and lungs collected at 3 dpi from necropsied birds or at 9 dpi from one of the ducks that had to be euthanized because of severe neurological signs, was also examined (Table. 5). Higher virus titers were found in both spleens and one lung at 3 dpi in mallards infected with A/Tk/MN/15 compared to mallards infected with A/Ck/IA/15. Virus was still detected at 9 dpi in the lung and spleen of the euthanized duck.
Table 5.
Virus titers in tissues collected from mallards inoculated with poultry H5N2 HPAI viruses.
| Virus | Log 10 dose | dpi | Spleen | Lung |
|---|---|---|---|---|
| A/Tk/MN/15 | 6 | 3 | 4.3/5.8a | 4.2/6.7 |
| A/Ck/IA/15 | 6 | 3 | 2.9/2.9 | 4.4/4.0 |
| A/Tk/MN/15 | 4 | 9 | 3.1 | 3.0 |
EID50/g
Sequence analysis
All of the H5N2 viruses tested in this study were consistent with HPAI virus on the basis of the amino acid sequence at the hemagglutinin cleavage site (A/Tk/AR/15, A/Tk/MN/15, and A/Tk/SD/15: PLRERRRKR/G; A/Ck/IA/15: PQRERRRKR/G), and phylogenetic analysis of the HA gene corroborated that the H5N2 viruses are descendants of the Gs/GD lineage H5 clade 2.3.4.4 virus that spread from East Asia to North America in late 2014 (15). All of the poultry H5N2 viruses used in this study belonged to H5N2 Midwestern U.S. cluster. Single nucleotide polymorphism (SNP) analysis revealed that multiple non-synonymous mutations occurred in all genes for the poultry H5N2 isolates when compared to the index A/Np/WA/14 virus. A total of 30 non-synonymous mutations were identified (Table 6). A total of 15 non-synonymous mutations were identified from A/Tk/AR/15, 16 from A/Tk/MN/15, 15 from A/Tk/SD/15, and 21 from A/Ck/IA/15 virus. We identified common substitutions R215K in PB1, A337V in PA, K217T in NS1, and N60H in NEP protein from A/Tk/MN/15, A/Tk/SD/15, and A/Ck/IA/15 viruses which had higher infectivity and pathogenicity than index H5N2 virus and A/Tk/AR/15 in chickens. In addition, we found unique substitutions H15P in PB1-F2 and R723L in PB1 protein from A/Tk/MN/15 which had higher pathogenicity than other Midwest H5N2 viruses in mallards.
Table 6.
Non-Synonymous substitutions found in the poultry H5N2 HPAI isolates when compared to A/Northern pintail/Washington/40964/2014 (H5N2) (A/Np/WA/14).
| Protein | Codon Number |
Amino Acid Change |
Nucleotide Position |
Nucleotide Change |
Codon Change |
A/Tk/AR/15 | A/Tk/MN/15 | A/Tk/SD/15 | A/Ck/IA/15 |
|---|---|---|---|---|---|---|---|---|---|
| HA | 7 | L -> P | 20 | T -> C | CTT -> CCT | X | |||
| HA | 8 | L -> F | 22 | C -> T | CTT -> TTT | X | X | X | X |
| HA | 82 | M -> I | 246 | G -> A | ATG -> ATA | X | |||
| HA | 130 | N -> T | 389 | A -> C | AAT -> ACT | X | X | X | X |
| HA | 157 | S -> P | 469 | T -> C | TCC -> CCC | X | X | X | X |
| HA | 338 | L -> Q | 1,013 | T -> A | CTA -> CAA | X | |||
| NA | 150 | H -> N | 448 | C -> A | CAT -> AAT | X | |||
| NA | 253 | R -> K | 758 | G -> A | AGA -> AAA | X | X | X | X |
| NA | 368 | E -> K | 1,102 | G -> A | GAA -> AAA | X | X | X | X |
| NA | 412 | V -> A | 1,235 | T -> C | GTT -> GCT | X | X | X | |
| NA | 416 | S -> G | 1,246 | A -> G | AGC -> GGC | X | |||
| PB2 | 386 | L -> V | 1,156 | T -> G | TTA -> GTA | X | X | X | X |
| PB2 | 649 | V -> I | 1,945 | G -> A | GTA -> ATA | X | X | X | X |
| PB1-F2 | 15 | H -> P | 44 | A -> C | CAC -> CCC | X | |||
| PB1 | 180 | E -> D | 540 | A -> C | GAA -> GAC | X | |||
| PB1 | 215 | R -> K | 644 | G -> A | AGG -> AAG | X | X | X | |
| PB1 | 317 | M -> V | 949 | A -> G | ATG -> GTG | X | |||
| PB1 | 531 | K -> R | 1,592 | A -> G | AAG -> AGG | X | |||
| PB1 | 667 | I -> T | 2,000 | T -> C | ATC -> ACC | X | |||
| PB1 | 723 | R -> L | 2,168 | G -> T | CGA -> CTA | X | |||
| PA | 337 | A -> V | 1,010 | C -> T | GCT -> GTT | X | X | X | |
| PA | 475 | A -> T | 1,423 | G -> A | GCA -> ACA | X | |||
| NP | 109 | I -> T | 326 | T -> C | ATC -> ACC | X | |||
| NP | 347 | I -> L | 1,039 | A -> C | ATC -> CTC | X | |||
| M2 | 78 | Q -> R | 233 | A -> G | CAG -> CGG | X | X | X | X |
| NS1 | 64 | I -> K | 191 | T -> A | ATA -> AAA | X | |||
| NS1 | 176 | I -> T | 527 | T -> C | ATT -> ACT | X | X | X | X |
| NS1 | 203 | W -> R | 607 | T -> C | TGG -> CGG | X | |||
| NS1 | 217 | K -> T | 650 | A -> C | AAA -> ACA | X | X | X | |
| NEP | 60 | N -> H | 178 | A -> C | AAC -> | X | X | X |
Discussion
In this study we examined the infectivity, pathogenicity and transmission of H5N2 HPAI viruses isolated from commercial turkeys and chickens from the Midwestern U.S. in 2015, in chickens, the primary gallinaceous poultry species, and mallards, the principal migratory waterfowl species. Our goal was to characterize changes in host adaptation of these viruses after circulation in gallinaceous poultry. The H5N2 HPAI virus is a reassortant that contains the Eurasian clade 2.3.4.4 H5 gene plus four other Eurasian genes (polymerase acidic protein subunit [PA], matrix protein [M], polymerase basic protein subunit 2 [PB2], nonstructural protein [NS]) and three North American wild bird lineage LPAI viral genes (neuraminidase [NA], nucleoprotein [NP], polymerase basic protein 1 [PB1] (8). In late 2014 and early 2015, this H5N2 virus caused outbreaks in turkey and chicken commercial operations in British Columbia, Canada (24). In the Pacific Flyway, this virus was commonly detected in wild waterfowl species, mostly in mallards and American wigeons (Anas Americana), but also in Northern pintails (Anas acuta), Wood ducks (Aix sponsa), Northern shovelers (Anas clypeata), Canada geese (Anas clypeata), American green-winged teal (Anas carolinensis), Gadwall (Anas stepera), and Cinnamon teal (Anas Cyanoptera) (32, 2, 6), and was also detected in backyard poultry in the U.S. (7, 30). Subsequently, the H5N2 virus was detected in Midwestern U.S., causing the devastating outbreak in commercial poultry in from March to June of 2015. Continued passage of the H5N2 virus in poultry could have increased adaptation of the virus to gallinaceous species rendering them more infectious.
The mean infectious dose of AI virus isolates could be considered a measure of the infectivity and adaptation of a virus to a specific host, serving as a quantitative predictor for which strains of AI virus, given the right conditions, would be more likely transmitted to and maintained in a given species (26). When looking at AI viruses under the parasite-host perspective, LPAI viruses in wild ducks, gulls, and shorebirds appear to have highly co-evolved, with the virus replicating to high titers, but causing minimal to no disease in these birds. It is expected that the duck adapted H5N2 virus would quickly adapt to chickens and turkeys if the initial infectious dose and transmission rate allowed for sustained infection. A previous study found that the early wild bird H5N2 HPAI virus from the initial case within the Pacific flyway was not yet optimally adapted to chickens (1). This conclusion was based on experimental findings of a chicken infectious dose50 greater than 104.4 EID50/bird, long MDT (4 days), and lack of transmission to contact chickens (1). On the other hand, mallards challenged with 106 EID50 of the same H5N2 virus (A/Np/WA/14) did not show mortality and shed high titers of virus for more than 11 days, which would favor dissemination and transmission of this virus (23).
In the current study we found that, based on the lower BID50 and higher levels of virus shedding, three of the poultry isolates examined were better adapted to chickens compared to the wild duck origin 2014 U.S. index H5N2 virus. It was anticipated that the lower BID50 would also translate to improved contact transmission in the experimental model used. Improved transmission was observed in the A/TK/MN/15 group, but not with the other viruses. This lack of transmission in the experimental model is at odds with the strong epidemiologic data showing farm-to-farm spread of the virus (31). Likely reasons for the higher susceptibility in chickens observed in the field include negative health impact from environmental and management conditions, secondary infections, immunosuppression, and the physiologic stress of egg laying in the field birds. So although we see indications of increased adaptation to chickens, the viruses are still in transition to being fully chicken adapted-viruses.
In contrast to chickens, when examined in mallards, two of these poultry H5N2 isolates had a similar high infectivity as the U.S. index H5N2 virus, the U.S. index Eurasian H5N8 virus, and the 2005 H5N1 Gs/GD lineage virus, and transmitted efficiently to direct contacts, although infection with one of the viruses (A/Ck/IA/15) resulted in lower virus replication levels as measured by OP and CL swab titers. Interestingly, different from the index virus, one of the poultry H5N2 viruses (A/Tk/MN/15) caused occasional mortality when given at high doses. This increased virulence in mallards was unexpected since this virus showed increased adaptation to chickens. However, previous experiments have shown great variability in virulence in ducks of Gs/GD H5N1 HPAI lineage viruses, which are well adapted and virulent to chickens, from failing to produce illness to producing high mortality in mallards and domestic ducks (3, 14, 22, 35).
During the initial phase of the U.S. HPAI outbreak in December 2014-February 2015, transmission of H5N2 virus from wild birds to poultry was confined primarily to small backyard flocks in the Pacific flyway. Through a combination of poor transmission rates and relatively small numbers of exposed birds, the virus did not change fast enough to develop a sustained gallinaceous poultry transmission chain. One of the first outbreak cases in the Midwest was in Arkansas and was contained to the index flock. This virus (A/Tk/AR/15) was the least chicken-adapted of the four Midwest strains tested (BID50 = 105.1 versus 103.2-3.6). The other three viruses had been circulating in densely populated poultry flocks for several weeks. Minnesota, the original epicenter of the Midwest outbreak, is a major turkey producing state with a high density of farms and large number of birds on each farm. This higher density, with the increased opportunities for replication in more hosts and increased opportunities for spread between farms, favored the maintenance of the virus while it was still poorly adapted, thus giving the virus the time and opportunity to increase adaptation in gallinaceous poultry. The outbreak in the egg laying areas of Iowa, where farm complexes of over 1 million chickens are common, also provided unique opportunities for the virus to adapt.
The phenotype of a HPAI virus, with its extreme virulence for gallinaceous poultry, means that with the increased replication seen with the later viruses, the practical outcome was a shorter mean death time (MDT) than the 2014 original wild bird-adapted H5N8 or H5N2 HPAI strains. The clade 2.3.4.4 virus lineage evolved after 18 years of replication in the poultry population primarily in China, and the virus was identified in the wild waterfowl population serving as a vector for intermediate to long distance dispersion. In the last 10 years, three major occurrences of Gs/GD lineage virus jumping between poultry and wild birds have been identified and each has persisted for at least a few years in wild waterfowl populations. The A/Ws/Mongolia/05 virus was a representative of the clade 2.2 viruses that was part of the first long distance wild waterfowl dispersion seen in 2005 across western Asia and into Europe. The clade 2.2 viruses were unusual in that at least some of them were not only highly pathogenic in chickens, but it also caused mortality in some wild duck species (4, 6, 18). The A/Ws/Mongolia/05 strain, as was shown in this study, was both highly infectious and deadly for mallard ducks. It has been hypothesized that the clade 2.2 viruses did not spread to North America in part because this higher virulence in wild birds made it less probable for wild birds to disseminate the virus from Asia to Alaska across the Bering Sea since sick birds were unlikely to attempt a long distance migration. The clade 2.3.4.4 Gs/GD lineage viruses appeared to be the result of reassortant events that created viruses that were able to replicate to high titers in ducks, increasing the opportunities for transmission, but with a phenotype of mild clinical disease. This combination would presumable be an ideal combination to allow long distance movement of the virus and subsequent transmission.
An interesting result of this study was the increased virulence, and consequently some mortality, observed when infecting mallards with one of the more poultry adapted viruses (A/Tk/MN/15). The gallinaceous poultry adaptation process in this virus resulted in changes that likely also affected pathogenesis in mallards, i.e. phenotypic effect of a mild increase in virulence. Another aspect of virus adaptation in poultry that needs to be taken into account is that not all gallinaceous species are the same species, and adaptation changes occurring in, for example, turkeys might not be the same as changes in chickens. We do not know how the virus passed in the field and could have passed both in turkeys and in chickens. SNP analysis revealed that 30 non-synonymous mutations occurred in genes for the poultry H5N2 isolates when compared to the index A/Np/WA/14 virus. The Midwest H5N2 outbreak appears to have started at multiple point sources, although with closely related viruses from the wild waterfowl population, and the evolutionary path may be slightly different. The examination of these different sub-lineages by SNP analysis showed some convergent evolution where the same mutations were positively selected enhancing gallinaceous adaptation. As the SNP changes are further investigated using reverse genetics techniques, it will be possible to determine specific amino acids that are correlated with interorder adaptation between Anseriformes and Galliformes, but also intraorder changes that might point to any adaptation specific to chickens or turkeys.
Fortunately, because of the immense efforts of Canadian and U.S. veterinary forces, the H5N2 HPAI outbreak was contained and the virus was eradicated from the poultry population. The 2.3.4.4 Gs/GD lineage of HPAI virus was not detected from wild waterfowl in North America during later 2015 (13, 32), but the potential for the virus to persist in wild birds with fresh introductions into U.S. poultry remains an ongoing concern. Enhanced surveillance in wild birds and poultry is advisable for the foreseeable future to limit the impact of AI in the future.
Supplementary Material
Highlights.
Poultry H5N2 clade 2.3.4.4 HPAI viruses were investigated in chickens and mallards
The viruses were better adapted to chickens than the initial U.S. H5N2 virus
The viruses retained high adaptation to mallards but pathogenicity differed
Acknowledgments
This research was supported by the USDA/ ARS CRIS Project 6612-32000-063-00D and by CRIP (Center of Research in Influenza Pathogenesis) an NIAID funded Center of Excellence in Influenza Research and Surveillance (CEIRS, contract HHSN272201400008C). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the USDA or NIH. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The authors appreciate the technical assistance provided by Kira Moresco and Nicolai Lee, and the animal care provided by Keith Crawford, Gerald Damron, and Roger Brock in conducting these studies. USDA is an equal opportunity provider and employer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare they have no conflict of interest.
References
- 1.Bertran K, Swayne DE, Pantin-Jackwood MJ, Kapczynski DR, Spackman E, Suarez DL. Lack of chicken adaptation of newly emergent Eurasian H5N8 and reassortant H5N2 high pathogenicity avian influenza viruses in the U.S. is consistent with restricted poultry outbreaks in the Pacific flyway during 2014-2015. Virology. 2016;494:190–197. doi: 10.1016/j.virol.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 2.Bevins SN, Dusek RJ, White CL, Gidlewski T, Bodenstein B, Mansfield KG, DeBruyn P, Kraege D, Rowan E, Gillin C, Thomas B, Chandler S, Baroch J, Schmit B, Grady MJ, Miller RS, Drew ML, Stopak S, Zscheile B, Bennett J, Sengl J, Brady C, Ip HS, Spackman E, Killian ML, Torchetti MK, Sleeman JM, Deliberto TJ. Widespread detection of highly pathogenic H5 influenza viruses in wild birds from the Pacific Flyway of the United States. Scientific reports. 2016;6:28980. doi: 10.1038/srep28980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown JD, Stallknecht DE, Beck JR, Suarez DL, Swayne DE. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg Infect Dis. 2006;12:1663–1670. doi: 10.3201/eid1211.060652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Smith GDJ, Zhang SY, Qin K, Wang J, Li KS, Webster RG, Peiris JSM, Guan Y. H5N1 virus outbreak in migratory waterfowl: a worrying development could help to spread this dangerous virus beyond its stronghold in southeast Asia. Nature (London) 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- 5.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert M, Jambal L, Karesh WB, Fine A, Shiilegdamba E, Dulam P, Sodnomdarjaa R, Ganzorig K, Batchuluun D, Tseveenmyadag N, Bolortuya P, Cardona CJ, Leung CY, Peiris JS, Spackman E, Swayne DE, Joly DO. Highly Pathogenic Avian Influenza Virus among Wild Birds in Mongolia. PLoS One. 2012;7:e44097. doi: 10.1371/journal.pone.0044097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greene JL. Update on the highly-pathogenic avian influenza outbreak of 2014-2015. Congresional Research Service. 2015 https://fas.org/sgp/crs/misc/R44114.pdf [Online.]
- 8.Ip HS, Torchetti MK, Crespo R, Kohrs P, DeBruyn P, Mansfield KG, Baszler T, Badcoe L, Bodenstein B, Shearn-Bochsler V, Killian ML, Pedersen JC, Hines N, Gidlewski T, DeLiberto T, Sleeman JM. Novel eurasian highly pathogenic avian influenza a h5 viruses in wild birds, washington, USA, 2014. Emerg Infect Dis. 2015;21:886–890. doi: 10.3201/eid2105.142020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong J, Kang HM, Lee EK, Song BM, Kwon YK, Kim HR, Choi KS, Kim JY, Lee HJ, Moon OK, Jeong W, Choi J, Baek JH, Joo YS, Park YH, Lee HS, Lee YJ. Highly pathogenic avian influenza virus (H5N8) in domestic poultry and its relationship with migratory birds in South Korea during 2014. Vet Microbiol. 2014;173:249–257. doi: 10.1016/j.vetmic.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Jhung MA, Nelson DI, Centers for Disease Control and Prevention (CDC) Outbreaks of avian influenza A (H5N2), (H5N8), and (H5N1) among birds-United States, December 2014-January 2015. MMWR Morb. Mortal. Wkly. Rep. 2015;64:111. [PMC free article] [PubMed] [Google Scholar]
- 11.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Briefings in bioinformatics. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 12.Killian ML, Spackman E. Avian Influenza Virus Isolation, Propagation, and Titration in Embryonated Chicken Eggs. In: Spackman E, editor. Animal Influenza virus. Methods in Molecular Biology 1161. Humana Press; 2014. p. 125. [DOI] [PubMed] [Google Scholar]
- 13.Krauss S, Stallknecht DE, Slemons RD, Bowman AS, Poulson RL, Nolting JM, Knowles JP, Webster RG. The enigma of the apparent disappearance of Eurasian highly pathogenic H5 clade 2.3.4.4 influenza A viruses in North American waterfowl. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1608853113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon YK, Thomas C, Swayne DE. Variability in pathobiology of South Korean H5N1 high-pathogenicity avian influenza virus infection for 5 species of migratory waterfowl. Vet Pathol. 2010;47:495–506. doi: 10.1177/0300985809359602. [DOI] [PubMed] [Google Scholar]
- 15.Lee DH, Torchetti MK, Winker K, Ip HS, Song CS, Swayne DE. Intercontinental Spread of Asian-Origin H5N8 to North America through Beringia by Migratory Birds. J Virol. 2015;89:6521–6524. doi: 10.1128/JVI.00728-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee MS, Chen LH, Chen YP, Liu YP, Li WC, Lin YL, Lee F. Highly pathogenic avian influenza viruses H5N2, H5N3, and H5N8 in Taiwan in 2015. Vet Microbiol. 2016;187:50–57. doi: 10.1016/j.vetmic.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Lee YJ, Kang HM, Lee EK, Song BM, Jeong J, Kwon YK, Kim HR, Lee KJ, Hong MS, Jang I, Choi KS, Kim JY, Lee HJ, Kang MS, Jeong OM, Baek JH, Joo YS, Park YH, Lee HS. Novel reassortant influenza A(H5N8) viruses, South Korea, 2014. Emerg Infect Dis. 2014;20:1087–1089. doi: 10.3201/eid2006.140233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Xiao H, Lei F, Zhu Q, Qin K, Zhang X-W, Zhang X-L, Zhao D, Wang G, Feng Y, Ma J, Liu W, Wang J, Gao GF. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science. 2005;309:1206. doi: 10.1126/science.1115273. [DOI] [PubMed] [Google Scholar]
- 19.OIE-WAHIS 2015, posting date World Animal Health Information Database (WAHIS) Interface. http://www.oie.int/wahis_2/public/wahid.php/Wahidhome/Home [Online.]
- 20.Pantin-Jackwood M, Swayne DE, Smith D, Shepherd E. Effect of species, breed and route of virus inoculation on the pathogenicity of H5N1 highly pathogenic influenza (HPAI) viruses in domestic ducks. Vet Res. 2013;44:62. doi: 10.1186/1297-9716-44-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantin-Jackwood MJ. Immunohistochemical staining of influenza virus in tissues. Methods Mol Biol. 2014;1161:51–58. doi: 10.1007/978-1-4939-0758-8_5. [DOI] [PubMed] [Google Scholar]
- 22.Pantin-Jackwood MJ, Swayne DE. Pathobiology of Asian highly pathogenic avian influenza H5N1 virus infections in ducks. Avian Dis. 2007;51:250–259. doi: 10.1637/7710-090606R.1. [DOI] [PubMed] [Google Scholar]
- 23.Pantin-Jackwood MJ, Costa-Hurtado M, Shepherd E, DeJesus E, Smith D, Spackman E, Kapczynski DR, Suarez DL, Stallknecht D, Swayne DE. Pathogenicity and transmission of H5 and H7 highly pathogenic avian influenza viruses in mallards. J Virology. doi: 10.1128/JVI.01165-16. Accepted JV 8-19-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasick J, Berhane Y, Joseph T, Bowes V, Hisanaga T, Handel K, Alexandersen S. Reassortant highly pathogenic influenza A H5N2 virus containing gene segments related to Eurasian H5N8 in British Columbia, Canada, 2014. Scientific reports. 2015;5:9484. doi: 10.1038/srep09484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT, Suarez DL. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swayne DE, Slemons RD. Using mean infectious dose of wild duck- and poultry-origin high- and low-pathogenicity avian infleunza viruses as one measure of infectivity and adaptation to poultry. Avian Dis. 2008;52:455–460. doi: 10.1637/8229-012508-Reg.1. [DOI] [PubMed] [Google Scholar]
- 27.Swayne DE, Suarez DL, Sims LD. Influenza. In: Wiley-Blackwell A, Swayne DE, Glisson JR, McDougald LR, Nair V, Nolan LK, Suarez Dl, editors. Diseases of Poultry. 2013. [Google Scholar]
- 28.Swayne DE, Pantin-Jackwood M. Pathobiology of avian influenza virus infections in birds and mammals. In: Swayne DE, editor. Avian Influenza. Blackwell Publishing; Ames, Iowa: 2008. pp. 87–122. [Google Scholar]
- 29.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.United States Department of Agriculture, Animal and Plant Health Inspection Service Highly Pathogenic Avian Influenza Infected Premises 2014-2015. 2015 posting date. https://www.aphis.usda.gov/animal_health/animal_dis_spec/poultry/downloads/hpai-positive-premises-2014-2015.pdf. [Online.]
- 31.United States Department of Agriculture, Animal and Plant Health Inspection Service USDA-APHIS Veterinary Services Epidemiologic and Other Analyses of HPAI-Affected Poultry Flocks: September 9, 2015 Report. 2015 posting date. https://www.aphis.usda.gov/animal_health/animal_dis_spec/poultry/downloads/Epidemiologic-Analysis-Sept-2015.pdf [Online.]
- 32.United States Department of Agriculture, Animal and Plant Health Inspection Service Wild bird highly pathogenic avian influenza cases in the United States. 2016 https://www.aphis.usda.gov/wildlife_damage/downloads/DEC%202014%20-%20JUNE%202015%20WILD%20BIRD%20POSITIVE%20HIGHLY%20PATHOGENIC %20AVIAN%20INFLUENZA%20CASES%20IN%20THE%20UNITED%20STATES.pdf. [Online.]
- 33.Verhagen JH, Herfst S, Fouchier RA. Infectious disease. How a virus travels the world. Science. 2015;347:616–617. doi: 10.1126/science.aaa6724. [DOI] [PubMed] [Google Scholar]
- 34.Wasilenko JL, Arafa AM, Selim AA, Hassan MK, Aly MM, Ali A, Nassif S, Elebiary E, Balish A, Klimov A, Suarez DL, Swayne DE, Pantin-Jackwood MJ. Pathogenicity of two Egyptian H5N1 highly pathogenic avian influenza viruses in domestic ducks. Arch Virol. 2008;156:37–51. doi: 10.1007/s00705-010-0813-y. [DOI] [PubMed] [Google Scholar]
- 35.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong FY, Phommachanh P, Kalpravidh W, Chanthavisouk C, Gilbert J, Bingham J, Davies KR, Cooke J, Eagles D, Phiphakhavong S, Shan S, Stevens V, Williams DT, Bounma P, Khambounheuang B, Morrissy C, Douangngeun B, Morzaria S. Reassortant Highly Pathogenic Influenza A(H5N6) Virus in Laos. Emerg Infect Dis. 2015;21:511–516. doi: 10.3201/eid2103.141488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao K, Gu M, Zhong L, Duan Z, Zhang Y, Zhu Y, Zhao G, Zhao M, Chen Z, Hu S, Liu W, Liu X, Peng D, Liu X. Characterization of three H5N5 and one H5N8 highly pathogenic avian influenza viruses in China. Vet Microbiol. 2013;163:351–357. doi: 10.1016/j.vetmic.2012.12.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.