Summary
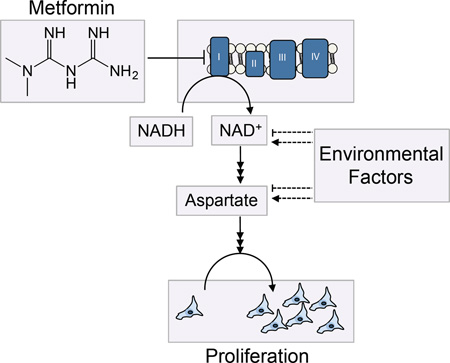
Metformin use is associated with reduced cancer mortality, but how metformin impacts cancer outcomes is controversial. While metformin can act cell autonomously to inhibit tumor growth, the doses of metformin that inhibit proliferation in tissue culture are much higher than what has been described in vivo. Here, we show that environment drastically alters sensitivity to metformin and other complex I inhibitors. We find that complex I supports proliferation by regenerating NAD+, and metformin’s anti-proliferative effect is due to loss of NAD+/NADH homeostasis and inhibition of aspartate biosynthesis. However, complex I is only one of many inputs that determine cellular NAD+/NADH ratio, and dependency on complex I is dictated by the activity of other pathways that affect NAD+ regeneration and aspartate levels. This suggests that cancer drug sensitivity and resistance are not intrinsic properties of cancer cells, and demonstrates that environment can dictate sensitivity to therapies that impact cell metabolism.
Graphical Abstract
Introduction
Metformin is a safe and effective anti-hyperglycemic agent that is commonly used by hundreds of millions of people worldwide to treat type 2 diabetes. Because of its widespread use, an abundance of epidemiological data is available for how metformin might influence other disease states. Retrospective studies have found that taking metformin is associated with improved cancer outcomes, with reductions in cancer incidence and with decreased cancer mortality observed across many tumor types (Evans et al., 2005; Gandini et al., 2014; He et al., 2012; Lee et al., 2012; Zhu et al., 2015). These findings have resulted in studies examining the anti-tumorigenic properties of metformin and other biguanides on cancer cell lines and in mouse models of cancer, as well as clinical trials exploring potential roles for metformin in cancer therapy. Systemic treatment with the biguanides metformin and phenformin can suppress tumor growth in xenograft and autochthonous tumor models (Buzzai et al., 2007; Huang et al., 2008; Shackelford et al., 2013; Wheaton et al., 2014). Metformin may also increase the pathological complete response rate in breast cancer patients receiving neoadjuvant chemotherapy (Jiralerspong et al., 2009). Whether the benefit of metformin in these different settings is attributable to direct action on the tumor is controversial (Birsoy et al., 2012; Foretz et al., 2014). Further, in a recent trial, metformin failed to improve outcomes in patients when added to standard pancreatic cancer therapy (Kordes et al., 2015), highlighting that the factors that determine which tumors are likely to respond to metformin are not known.
The molecular targets of metformin in cells and tissues have only recently come into focus (Luengo et al., 2014; Pernicova and Korbonits, 2014). Metformin can impair respiration by inhibition of the glycerol-phosphate shuttle (Madiraju et al., 2014), and in vitro biochemistry studies have shown that metformin directly inhibits mitochondrial complex I (NADH dehydrogenase), also resulting in decreased mitochondrial respiration (Bridges et al., 2014; El-Mir et al., 2000; Owen et al., 2000; Wheaton et al., 2014). Consistent with these molecular targets, cell culture studies have found that metformin causes increased glucose consumption, increased lactate production, and decreased mitochondrial glucose oxidation (Andrzejewski et al., 2014; Fendt et al., 2013).
Supporting a role for complex I as an important target of metformin in cancer, expression of a metformin resistant yeast analog of complex I (NDI1) makes cells insensitive to metformin in culture and resistant to the anti-tumor growth effects of metformin in xenograft models (Birsoy et al., 2014; Wheaton et al., 2014). Additionally, other complex I inhibitors have shown efficacy as anti-tumor agents (Schockel et al., 2015; Zhang et al., 2014). Together these data support the hypothesis that tumor autonomous inhibition of complex I and respiration play an important role in the anti-tumorigenic effect of metformin.
A major barrier for translating findings pertaining to metformin action in cultured cells to the clinic is a substantial inconsistency in the doses of metformin required to inhibit proliferation in vivo versus in vitro. Inhibition of cancer cell proliferation in vitro generally requires metformin doses that vastly exceed those achievable in vivo (Birsoy et al., 2012; Foretz et al., 2014; Pollak, 2014). Metformin concentrations required to inhibit cancer cell proliferation in vitro range from 1 to 50 mM, while plasma metformin levels in patients and in mice are in the micromolar range (Graham et al., 2011; He and Wondisford, 2015; Memmott et al., 2010). Indeed, even the maximum tolerable plasma concentration of metformin reported for humans, around 400 µM (Dell’Aglio et al., 2009) is still well below the doses used in most studies to decrease proliferation in culture.
In this study, we provide an explanation for the substantial discrepancy between the effective metformin concentration in vivo and in vitro. We find that metabolic environment is a major determinant of complex I dependency, and hence, differences in environment alone can dramatically alter sensitivity to metformin and other complex I inhibitors. Complex I supports proliferation through NAD+ regeneration to maintain cellular NAD+/NADH balance and to allow aspartate synthesis. However, dependency on complex I is directly dictated by the activity of alternative pathways that produce and consume NAD+, which change based on environmental factors. We show that alternative NAD+ utilizing pathways modulates complex I dependency and metformin sensitivity in a predictable manner. Further, we show that perturbing NAD+/NADH balance by modulating complex I activity modulates aspartate levels in a titratable manner both in vitro and in vivo. These data indicate that the anti-proliferative effects of metformin are caused by a decrease in the intracellular NAD+/NADH ratio and aspartate levels and suggest that environment can alter dependency on metabolic drug targets.
Results
Pyruvate suppresses the anti-proliferative effects of complex I inhibition
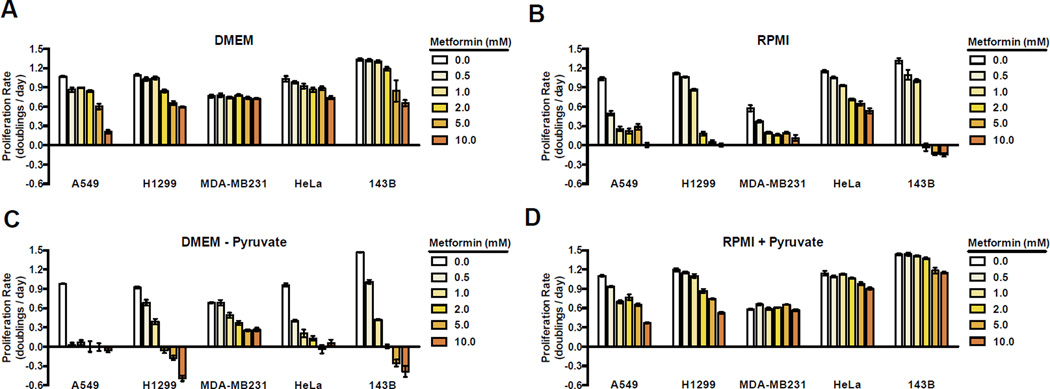
Mirroring the discrepancy between metformin effective concentration in vivo and in vitro, we observed that for all cell lines tested, the choice of culture media dramatically alters metformin sensitivity. We cultured a diverse group of cancer cell lines in two different standard cell culture media, Dulbecco’s Modified Eagle’s Medium (DMEM) and Roswell Park Memorial Institute 1640 medium (RPMI), which both fully support proliferation of the cells examined (Figure 1A, B). Consistent with previous reports, cells cultured in DMEM require up to 10 mM metformin to partially inhibit proliferation (Figure 1A) (Foretz et al., 2014). In contrast, cells cultured in RPMI are much more sensitive to metformin, with lower metformin doses inhibiting proliferation (Figure 1B). While the doses of metformin tested were still higher than what is generally achievable in vivo, these data directly suggest that metabolic environment alone is sufficient to dictate metformin sensitivity.
Figure 1. Pyruvate suppresses the antiproliferative effects of metformin.
Proliferation rates for A549, H1299, MDA-MB231, HeLa, and 143B cells in media treated with the indicated concentrations of metformin. Cells were cultured in DMEM (A), RPMI 1640 (B), DMEM without pyruvate (C), and RPMI 1640 supplemented with 1 mM pyruvate (D). Values denote mean ± standard error of the mean (SEM). n=3. See also Figure S1.
To better understand the differential effects of metformin in the different culture media, we examined how the media compositions differ. A binary difference is the presence of 1 mM pyruvate in DMEM and the absence of pyruvate in RPMI. To test whether pyruvate concentration could be a determinant of metformin sensitivity, we examined the anti-proliferative effect of metformin on cells cultured in DMEM without pyruvate. Strikingly, cells cultured in DMEM without pyruvate exhibit increased sensitivity to metformin, suggesting that pyruvate suppresses the anti-proliferative effect of metformin (Figure 1C). Consistent with this hypothesis, cells cultured in RPMI supplemented with 1 mM pyruvate (RPMI + Pyruvate) are less sensitive to metformin (Figure 1D). These data indicate that pyruvate availability is a crucial environmental variable that can alter sensitivity to the anti-proliferative effects of metformin.
Interestingly, addition of 1 mM pyruvate to RPMI made cells more resistant to metformin than cells cultured in DMEM with 1 mM pyruvate, further highlighting that environment can influence drug sensitivity. To better understand environmental determinants other than pyruvate that impact metformin sensitivity, we decreased the glucose concentration of DMEM to match RPMI (11 mM), or added aspartate to DMEM at the same concentration as that found in RPMI (150 µM) (Figure S1A, B). Lowering glucose, or addition of aspartate both affected metformin sensitivity to a degree, although the magnitude of effect was much smaller than that observed with pyruvate and was variable across cell lines. Thus, further experiments focused on understanding the influence of pyruvate on metformin sensitivity were performed using DMEM without pyruvate, unless otherwise noted.
Pyruvate modulates complex I dependency by providing an alternative pathway for NAD+ regeneration
It has been classically described that respiration-deficient cells are pyruvate auxotrophs (King and Attardi, 1989). In the absence of respiration, pyruvate can support proliferation by acting as an electron acceptor to regenerate NAD+ and allow aspartate synthesis (Birsoy et al., 2015; Sullivan et al., 2015). We hypothesized that a similar model could explain why pyruvate blocks the anti-proliferative effects of metformin.
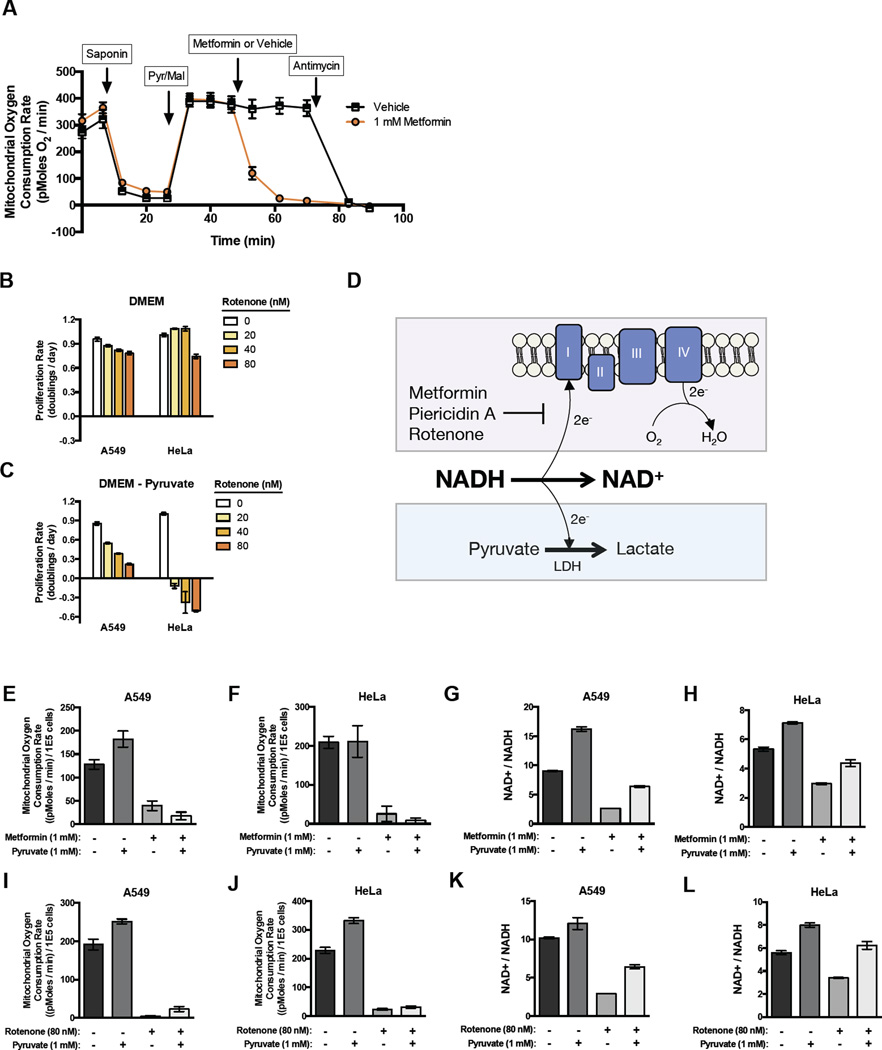
While it is generally accepted that metformin is a mitochondrial inhibitor; the mechanism by which it inhibits mitochondrial activity is controversial. Some reports suggest that metformin acts primarily as an inhibitor of complex I, while recent work has implicated metformin as a mitochondrial glycerol phosphate dehydrogenase (mGPD) inhibitor that disrupts the glycerol phosphate shuttle in the liver (Madiraju et al., 2014). Since both complex I inhibition and mGPD inhibition could lead to NAD+ deficiency, both mechanisms are consistent with pyruvate restoring NAD+ to block metformin’s anti-proliferative effect. Nevertheless, to distinguish between these two mechanisms in the cancer cells studied, we assessed complex I-mediated or glycerol-3-phosphate shuttle-mediated oxygen consumption in saponin-permeabilized cells. Whereas metformin dramatically inhibits oxygen consumption when permeabilized cells are supplied with pyruvate and malate as substrates for complex I, oxygen consumption from glycerol-3-phosphate is not disrupted by metformin (Fig 2A and S2A). These data argue that metformin inhibits electron transport chain from mitochondrial complex I but not from the glycerol-3-phosphate shuttle in these cells.
Figure 2. Complex I activity is dispensable when exogenous pyruvate is available to act as an electron acceptor.
(A) Mitochondrial oxygen consumption of A549 cells following permeabilization by saponin, addition of pyruvate and malate, addition of either metformin or vehicle, and addition of antimycin as indicated. Proliferation rates for A549 and HeLa cells treated with indicated dose of complex I inhibitor rotenone in DMEM (B) or DMEM without pyruvate (C). (D) Schematic illustrating how oxygen allows cells to regenerate NAD+ from NADH via complex I activity and the site of action of complex I inhibitors. Exogenous pyruvate provides an alternate way to regenerate NAD+ through lactate dehydrogenase (LDH) activity. Mitochondrial oxygen consumption rates of A549 (E) and HeLa (F) cells treated with the indicated concentrations of metformin and pyruvate. Intracellular NAD+/NADH ratios of A549 (G) and HeLa (H) cells treated with the indicated concentrations of metformin and pyruvate. Mitochondrial oxygen consumption rates of A549 (I) and HeLa (J) cells treated with the indicated concentrations of rotenone and pyruvate. Intracellular NAD+/NADH ratios of A549 (K) and HeLa (L) cells treated with the indicated concentrations of rotenone and pyruvate. Values denote mean ± SEM. n=5 (A, E-F, I-J) n=3 (B, G-H, K-L). See also Figure S2.
Previous studies have shown that compounds that inhibit respiration by hyperpolarizing the mitochondrial membrane potential, such as the ATP synthase inhibitor oligomycin, also decrease the NAD+/NADH ratio (Sullivan et al., 2015). In contrast to direct electron transport chain inhibition, under these circumstances, proliferation defects caused by these inhibitors can be restored by adding a mitochondrial uncoupler such as the ionophore carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) (Sullivan et al., 2015). While proliferation inhibition by oligomycin is reversed by FCCP, proliferation inhibition due to metformin is not reversed by FCCP, further supporting that metformin acts as an inhibitor of electron transport and not by causing mitochondrial hyperpolarization (Figure S2B).
To determine if the ability of pyruvate to restore proliferation is specific to metformin or generalizable to other complex I inhibitors, we treated cells in the presence or absence of pyruvate with the complex I inhibitors rotenone, phenformin, and piericidin A. While cells were generally resistant to these inhibitors in the presence of pyruvate, in the absence of pyruvate, cells exhibited increased sensitivity to the anti-proliferative effects of complex I inhibition (Figure 2B-C, S2C). Importantly, the presence or absence of pyruvate minimally alters proliferation rate in the absence of complex I inhibitors. Taken together, these data suggest that exogenous pyruvate does not alter the intrinsic proliferation rate of cells in culture, but rather, pyruvate decreases cellular dependence on complex I activity to support proliferation.
When complex I is inhibited, electrons from NADH cannot be transferred to molecular oxygen as an electron acceptor to form water and regenerate NAD+. However, in the presence of exogenous pyruvate, cells can regenerate NAD+ via conversion of pyruvate to lactate (Figure 2D). Thus, when excess pyruvate is available in the environment, cells can utilize this orthogonal pathway to maintain the NAD+/NADH ratio in the absence of complex I activity. Consistent with this idea, addition of the lactate dehydrogenase (LDH) inhibitor GSK2837808A, which partially inhibits pyruvate to lactate conversion, restores metformin sensitivity to cells cultured in pyruvate containing media (Figure S2D). Additionally, we cultured cells in media with a range of lactate to pyruvate ratios. In the absence of metformin, cells grew at a similar rate regardless of the lactate to pyruvate ratios supplied. Conversely, in the presence of metformin, increasing the lactate to pyruvate ratio significantly decreased proliferation rate (Figure S2E). These data support the hypothesis that net pyruvate to lactate conversion is required to suppress metformin effects on proliferation.
To further test this hypothesis, we treated cells with metformin and other complex I inhibitors at doses that inhibited proliferation in pyruvate-free media but did not markedly affect proliferation in pyruvate containing media. Under these conditions, we measured mitochondrial oxygen consumption and found that exposure to metformin decreases mitochondrial oxygen consumption (Figure 2E-F). Importantly, while the presence of pyruvate suppresses the anti-proliferative effect of these drugs, it does not restore mitochondrial oxygen consumption (Figure 2E-F). In contrast, while treatment with metformin decreases NAD+/NADH ratio in the absence of pyruvate, supplementation with pyruvate restores the NAD+/NADH ratio under these conditions (Figure 2G-H). We repeated these experiments with other complex I inhibitors including rotenone and piercidin A, and obtained similar results (Fig 2I-L, S2F-G). These results are consistent with a model where complex I activity is dispensable in environmental contexts in which NAD+ can be regenerated through orthogonal pathways. Of note, proliferation rate in such contexts is not correlated with oxygen consumption or, by extension, with mitochondrial ATP production.
Modulating pathways that alter NAD+/NADH homeostasis changes complex I dependency
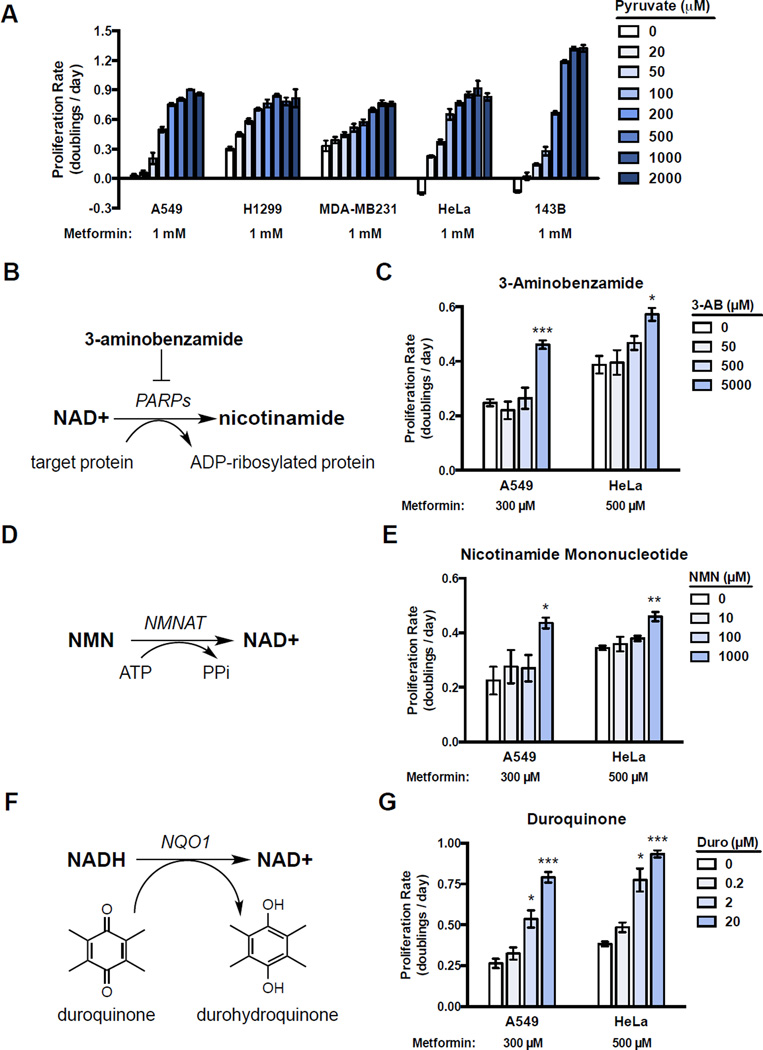
The observation that pyruvate can maintain the NAD+/NADH ratio despite complex I inhibition demonstrates that orthogonal pathways can influence the NAD+/NADH ratio. Further, the activity of orthogonal NAD+ modulating pathways should alter complex I dependency and, thus, modify metformin sensitivity in a titratable way. Consistent with this notion, we show that increasing pyruvate to lactate conversion by titrating the concentration of exogenous pyruvate in the environment decreases metformin sensitivity in a dose dependent manner (Figure 3A). To generalize this finding, we tested if changing the activity of other orthogonal NAD+/NADH modulating pathways could also alter complex I dependence and metformin sensitivity. Poly(ADP-ribose) polymerases (PARPs) are a family of proteins that use NAD+ to transfer ADP-ribose groups onto proteins (Figure 3B), and PARP activity is reported to be a major consumption pathway for NAD+ (Pillai et al., 2005). Blocking PARP activity increases intracellular NAD+ (Bai et al., 2011; Pirinen et al., 2014); thus, we tested if altering PARP activity could change complex I dependence. A549 and HeLa cells were treated with increasing amounts of the PARP inhibitor 3-aminobenzamide (Fang et al., 2014) in the presence of metformin. Metformin was added at approximately the IC50 dose for each cell line in DMEM in the absence of pyruvate. Since blocking NAD+ consumption will decrease the need for NAD+ regeneration, a PARP inhibitor is expected to decrease complex I dependency, and as predicted, addition of 3-aminobenzamide decreased metformin sensitivity (Figure 3C).
Figure 3. Altering cellular NAD+/NADH ratio titrates dependency on complex I activity.
(A) Proliferation rates for A549, H1299, MDA-MB231, HeLa, and 143B cells treated with 1 mM metformin and supplemented with the indicated amounts of pyruvate. (B) Schematic illustrating how poly(ADP-ribose) polymerases (PARPs) consume NAD+. (C) Proliferation rates of A549 and HeLa cells treated with metformin at the indicated concentration in media supplemented with the indicated concentration PARP inhibitor 3-aminobenzamide (3-AB). (D) Schematic illustrating how nicotinamide mononucleotide (NMN) supplementation can increase NAD+ synthesis by nicotinamide mononucleotide adenylyltransferase (NMNAT) activity. (E) Proliferation rates of metformin treated A549 and HeLa cells supplemented with the indicated concentration of NMN. (F) Schematic illustrating how duroquinone can oxidize NADH to yield NAD+ and durohydroquinone by the activity of NAD(P)H dehydrogenase, quinone 1 (NQO1). (G) Proliferation rates of metformin treated A549 and HeLa cells supplemented with the indicated concentration of duroquinone. Values denote mean ± SEM. n=3. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S3.
Another input into cellular NAD+/NADH homeostasis is NAD+ synthesis. Recent data have shown that the NAD+ salvage pathway is frequently upregulated in cancer and providing nicotinamide mononucleotide (NMN) increases NAD+ biosynthesis (Canto et al., 2012; Gomes et al., 2013; Wang et al., 2011) (Figure 3D). This predicts that NMN treatment may decrease complex I dependency, and indeed, we found that treating cells with NMN also decreased metformin sensitivity (Figure 3E).
Next, we reasoned that other redox reaction pairs that use NAD+/NADH as cofactors should also modulate cellular NAD+/NADH balance. When cells are exposed to duroquinone, the intracellular reductase NQO1 converts duroquinone to reduced durohydroquinone and, in the process, consumes NADH to produce NAD+ (Merker et al., 2006) (Figure 3F). Thus, providing cells with duroquinone allows for an orthogonal source of NAD+ regeneration and is predicted to decrease complex I dependency. Consistent with this, we found that addition of duroquinone dose-dependently decreases metformin sensitivity (Figure 3G).
To verify that these orthogonal pathways modulate NAD+/NADH in the expected manner, NAD+/NADH was measured after the respective treatments, and indeed, all treatments restored the NAD+/NADH ratio as predicted (Figure S3A). Next, we investigated whether the orthogonal NAD+/NADH modulators affected NAD+ and NADH levels as would be predicted by their mechanism of action. We found that, as expected, 3-aminobenzamide, NMN, and duroquinone all raised NAD+ levels (Figure S3B). Additionally, in HeLa cells, consistent with the expected effects of modulating enzymes that influence the production or consumption of NAD+ but not net cycling NADH back to NAD+, neither 3-aminobenzamide nor NMN significantly changed NADH levels, while duroquinone, which can regenerate NAD+ from NADH, decreased NADH levels (Figure S3C). Similar results were observed in A549 cells, although there were small decreases in NADH in NMN treated cells (Figure S3C). These data are consistent with NMN having an outsized effect on NAD+/NADH ratio relative to its effect on proliferation. How NMN causes decreased NADH levels in A549 cells is unknown, however it suggests that other feedback regulation of NAD+ synthesis may exist in cells.
Changes in NAD+/NADH ratio correlate with the anti-proliferative effects of metformin
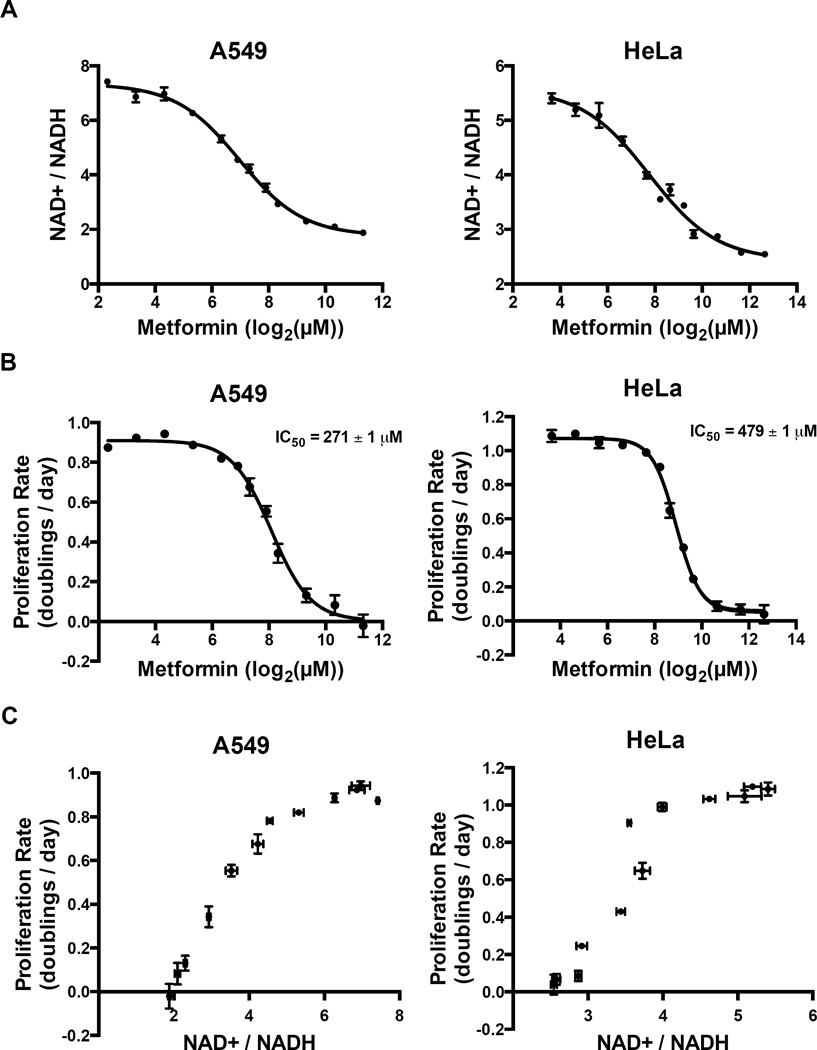
Our data indicate that while complex I activity is dispensable in the presence of pyruvate, complex I is important for maintenance of cellular NAD+/NADH ratio in the absence of pyruvate. We treated cells with a large range of metformin doses and found that in pyruvate-free media, decreasing complex I activity by increasing metformin concentration decreases both the NAD+/NADH ratio (Figure 4A) and proliferation rate (Figure 4B) in a dose dependent manner. Plotting the effects of metformin on NAD+/NADH ratio and proliferation rate reveals a striking correlation between the two phenotypes (Figure 4C). In metformin-treated A549 cells, an NAD+/NADH ratio less than three is associated with a dramatic decrease in cell proliferation, whereas an NAD+/NADH ratio greater than six is associated with maximal rates of cell proliferation. Interestingly, this relationship between the NAD+/NADH ratio and proliferation rate is also observed in A549 cells treated with other complex I inhibitors either in the presence or absence of pyruvate. In HeLa cells, although the absolute NAD+/NADH ratio that supports proliferation differs from that of A549 cells, a similar correlation between NAD+/NADH ratio and proliferation is observed that is also generalizable to other complex I inhibitors.
Figure 4. NAD+/NADH ratio tracks with proliferation rate in metformin treated cells.
Intracellular NAD+/NADH ratios (A) and proliferation rates (B) were measured after treatment with metformin at the indicated doses in A549 cells and HeLa cells. Proliferation rates and NAD+/NADH ratios were plotted independent of metformin concentrations to determine the relationship between NAD+/NADH ratio and proliferation rate in A549 cells and HeLa cells, respectively (C). Values denote mean ± SEM. n=3. See also Figure S4.
We next investigated the effect of metformin on the ATP/AMP ratio as this has also been proposed to be a mechanism by which metformin affects cell proliferation. Again, we treated cells with a large range of metformin doses and found that in pyruvate-free media, decreasing complex I activity also decreases the ATP/AMP ratio (Figure S4A). Interestingly, this decrease in the ATP/AMP ratio is also reversed by pyruvate (Figure S4B). Previous work has shown that maintaining NAD+/NADH balance is required for proliferation in order to support de novo aspartate biosynthesis (Birsoy et al., 2015; Sullivan et al., 2015); however, an abundance of literature implicates activation of AMP-activated protein kinase (AMPK) from a decreased ATP/AMP ratio as being a key modulator of metformin’s anti-proliferative effect (Foretz et al., 2014; Hardie, 2015; Li et al., 2015; Pernicova and Korbonits, 2014). Thus, we sought to distinguish whether the NAD+/NADH correlation with proliferation inhibition under metformin treatment was downstream of effects on aspartate or the ATP/AMP ratio.
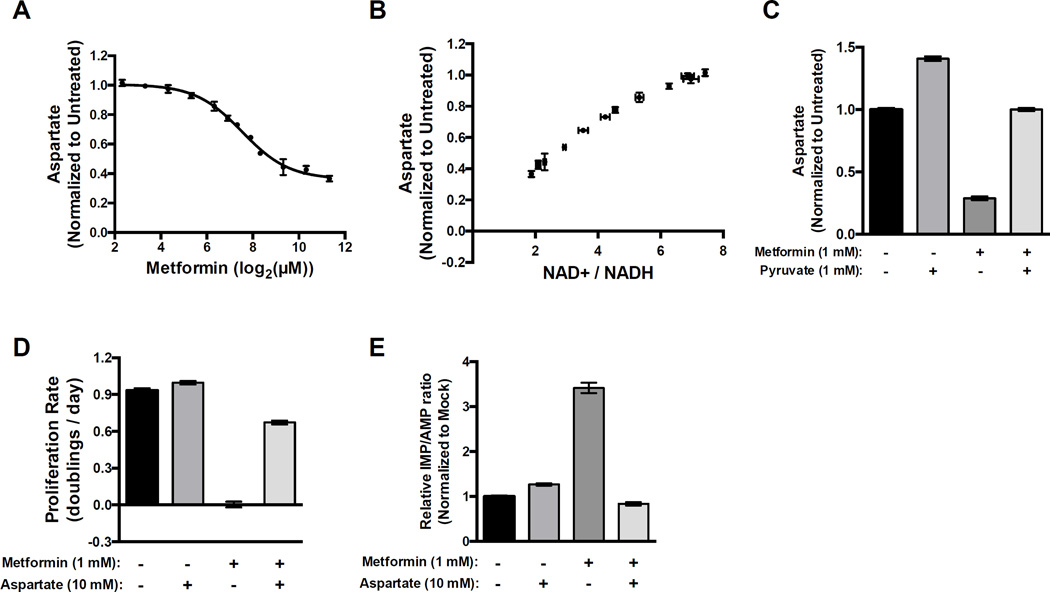
Treating cells with a titration of metformin showed that metformin dose-dependently decreases intracellular aspartate levels (Figure 5A). The decrease in aspartate levels strongly correlates with metformin’s effect on cellular NAD+/NADH ratio (Figure 5B). Addition of pyruvate, which increases cellular NAD+/NADH ratio, restores cellular aspartate levels, and addition of aspartate alone is sufficient to reverse the anti-proliferative effects of metformin (Figure 5C and D). An important anabolic role of aspartate is to support synthesis of purines, including the conversion of IMP to AMP. Consistent with metformin causing nucleotide insufficiency downstream of decreased aspartate levels, we found that in the presence of metformin the IMP/AMP ratio is dramatically elevated and that this elevation is reversed by addition of aspartate (Figure 5E).
Figure 5. The antiproliferative effects of metformin are suppressed by exogenous aspartate.
(A) Intracellular aspartate levels were measured after treatment with metformin at the indicated doses in A549 cells. (B) Aspartate levels and NAD+/NADH ratios were plotted independent of metformin concentrations to determine the relationship between NAD+/NADH ratio and aspartate levels in A549 cells. (C) Intracellular aspartate levels in A549 cells treated with the indicated doses of metformin and pyruvate. (D) Proliferation rates of A549 treated with the indicated doses of metformin and exogenous aspartate. (E) Normalized IMP/AMP ratio in A549 treated with the indicated doses of metformin and exogenous aspartate. Values denote mean ± SEM. n=3. See also Figure S5.
To show that the ability of aspartate to rescue metformin toxicity is downstream of the effects of metformin on NAD+/NADH, we verified that addition of aspartate does not change the cellular NAD+/NADH ratio (Figure S5A). Interestingly, we observed that ATP/AMP levels are also partially restored by aspartate (Figure S5B). However, comparing the proliferation rate with ATP/AMP ratio we observe that the degree to which aspartate restores proliferation rate is outsized relative to its effect on ATP/AMP ratio, suggesting that the effect of metformin on aspartate via changing NAD+/NADH is at least, in part, orthogonal to its effect on ATP/AMP ratio (Figure S5C). Indeed, given that there are no known pathways to generate ATP from aspartate in the absence of complex I activity, it is mechanistically unclear how aspartate can directly alter the ATP/AMP ratio, suggesting that perhaps ATP/AMP ratio changes are downstream of changes in proliferation rate, rather than a direct effect of changes in NAD+/NADH.
We next explored the relationship between the anti-proliferative effects of metformin, and AMPK signaling and mechanistic target of rapamycin complex I (mTORC1) signaling. We studied a panel of cell lines including A549 cells that are null for the upstream AMPK activator liver kinase B1 (LKB1). In H1299 and MDA-MB231 with intact LKB1, an increase in phosphorylation of the AMPK substrate acetyl-CoA carboxylase (ACC) upon metformin treatment is not reversed with pyruvate or aspartate supplementation (Figure S5D), further suggesting that changes in AMPK signaling and ATP/AMP ratio cannot fully explain the growth inhibitory effects of metformin (Griss et al., 2015). We did not observe any consistent trends in AMPK or mTORC1 signaling across cell lines with respect to metformin treatment, and pyruvate or aspartate restoration of proliferation in the presence of metformin (Figure S5D). These data suggest that effects on these signaling pathways cannot account for all of the anti-proliferative effects of metformin in these cells.
Metformin treatment decreases NAD+ and aspartate levels in tumors and slows tumor growth
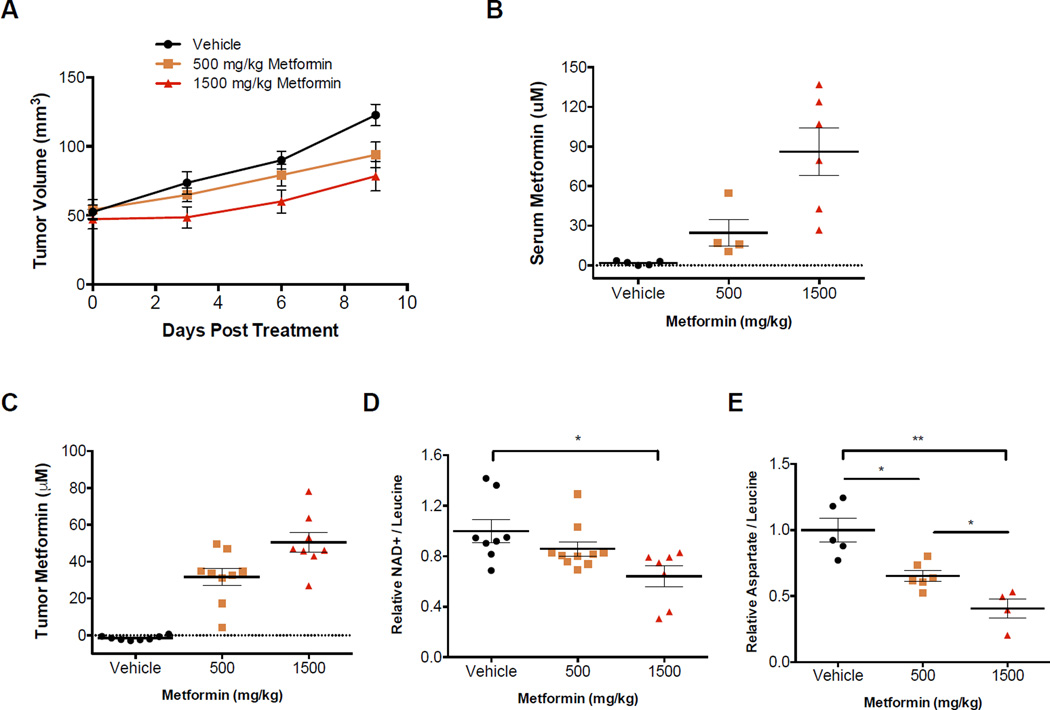
Finally, we sought to explore whether our findings regarding the anti-proliferative effects of metformin were relevant in vivo. For these studies A549 cells were xenografted into nude mice. When the tumors reached 50 mm3 the mice were randomized into three groups and treated with once a day oral gavage with vehicle, metformin at 500 mg/kg, or metformin at 1500 mg/kg. Increasing dosages of metformin dose-dependently inhibited tumor growth (Figure 6A). Measurement of plasma metformin concentration confirmed that the levels achieved with these doses are therapeutically relevant and expected to be tolerable in humans (Dell’Aglio et al., 2009) (Figure 6B). Further, we showed that escalating doses of metformin resulted in an increase in the tumor concentration of metformin (Figure 6C). NADH levels were too low to detect in tumors; however, NAD+ levels dose-dependently decreased in tumors with increasing doses of metformin (Figure 6D). We also assessed whether intratumoral ATP/AMP ratios were effected by metformin and found that while metformin appeared to decrease the ATP/AMP ratio in some tumors, the effect was neither statistically significant nor dose dependent. (Figure S6A). Finally, intratumoral aspartate levels decreased with increasing doses of metformin (Figure 6E). Taken together these data suggest that there is a correlation between metformin treatment at therapeutically relevant doses and tumor growth inhibition. In addition, the degree of tumor growth inhibition correlates with both intratumoral NAD+ and aspartate levels.
Figure 6. Metformin slows tumor growth and decreases intratumoral NAD+ and aspartate levels.
(A) A549 xenografts in nude mice were treated with vehicle or the indicated doses of metformin, once a day by oral gavage. (B) Serum and (C) tumor metformin concentrations measured in material from mice treated for 10 days at the indicated dose of metformin, with tissue harvested 2 hours after the last dose. Relative intratumoral (D) NAD+ and (E) aspartate levels after 10 days of vehicle or metformin treatment at the indicated dose. Relative NAD+ and aspartate levels are shown as NAD+ and aspartate total ion counts (TIC) normalized to leucine TIC. Values denote mean ± SEM. *p < 0.05, **p < 0.01. Related to Figure S6.
Discussion
Recent data suggests that a primary function of respiration in support of proliferation is to provide electron acceptors and regenerate oxidized cofactors (Birsoy et al., 2015; Sullivan et al., 2015). Extending this idea, we show that the anti-proliferative effect of metformin tracks with its effect on cellular NAD+/NADH ratio. Further, while there are correlations between ATP/AMP ratio and the cellular NAD+/NADH ratio, we find the downstream anti-proliferative effects of metformin track more consistently with its effect on NAD+/NADH ratio and intracellular aspartate. Importantly, we find that altering the metabolic environment in a way that influences NAD+/NADH homeostasis is sufficient to change cellular dependency on complex I and, by extension, alter metformin sensitivity. In contexts where cells are allowed to regenerate NAD+ by alternative pathways, complex I dependency is low. However, in other environmental contexts, including those found in vivo with A549 xenografts, complex I activity appears to be important for maintaining cellular NAD+/NADH homeostasis. Treatments with metformin at therapeutically relevant doses are sufficient to alter intratumoral NAD+ levels and intratumoral aspartate levels. While our data does not rule out the possibility of other whole body metabolic anti-tumor effects of metformin, our findings show that the cell autonomous anti-proliferative effects of metformin are, at least in part, mediated by its effect on NAD+/NADH homeostasis and aspartate levels and further support that complex I itself can be a cancer target.
The sensitivity of cancer cells to a drug is typically considered to be a cell intrinsic property, with efficacy matched directly to ability to inhibit a protein target in a specific genetic context. In many cases, the degree to which the direct drug target is inhibited is an effective proxy and predictor for downstream effects on proliferation. In contrast to this paradigm, we find that for metformin and other complex I inhibitors, the environmental context can decouple the biological effect of decreased proliferation from inhibition of the drug target. We show that inhibition of complex I activity, as measured by oxygen consumption, is a poor predictor of the anti-proliferative effects of metformin. Indeed, while media supplementation of pyruvate does not change metformin action as a complex I inhibitor, it is sufficient to alter the effective anti-proliferative dose of metformin by approximately two orders of magnitude. While variable sensitivity to biguanides has been attributed to specific genetic events (Buzzai et al., 2007; Cuyas et al., 2015; Shackelford et al., 2013) or to factors that alter drug uptake (Chen et al., 2015; Chen et al., 2010; Goswami et al., 2014; Madera et al., 2015; Nies et al., 2011; Shu et al., 2007), our data suggest that the metabolic environment is an additional determinant of drug sensitivity independent of cell intrinsic factors.
These findings indicate that considering environmental context is critical for in vitro screening of drugs targeting metabolism. Indeed, had metformin not been identified to promote cancer survival in patients taking this drug for diabetes, in vitro studies in DMEM would have concluded that clinically unachievable doses are required for metformin to have an effect on cancer cells. These results highlight the importance of considering environmental context for determining the potential efficacy of newly developed drugs.
A major barrier to extrapolating preclinical data on the anti-tumorigenic effects of metformin has been the large inconsistency in dosing between tissue culture and tumor models. Showing that metabolic environment is a crucial determinant of complex I dependency begins to explain this discrepancy. Indeed, cellular sensitivity to metformin in DMEM without pyruvate is within a few fold of what has been observed to have clinical benefit in patients. Additionally, due to dose limiting toxicity in animals, it is impossible to achieve high enough doses to determine the in vivo tumor autonomous IC50 and because other environmental factors also influence complex I sensitivity, comparing the effects of metformin in DMEM without pyruvate to the effects in tumors may not be the best comparison.
Standard in vitro tissue culture is a poor mimic for the metabolic environment of tumors (Davidson et al., 2016; Mayers and Vander Heiden, 2015). Given the dramatic differences in oxygen and nutrient availability in culture versus tumors, electron acceptors are likely to be more limited in vivo, thus increasing dependence on pathways that regenerate NAD+ (Sullivan et al., 2015). One notable example regarding metformin and nutrient availability involves recent work showing that serine limitation increases in vivo metformin sensitivity (Gravel et al., 2014). Serine levels in vivo are much lower than what is found in vitro and de novo serine biosynthesis requires multiple NAD+ consuming redox reactions, likely impinging on cellular NAD+/NADH homeostasis and potentially partially explaining its interaction with metformin sensitivity. Additionally, given other differences in waste disposal and carbon dioxide levels, there are other environmental influences on the cellular NAD+/NADH ratio that are not accounted for in vitro. A better understanding of NAD+/NADH homeostasis in tumors will be critical for identifying contexts in which metformin therapy may be most impactful and for developing combination therapies that increase the efficacy of metformin in cancer. Developing culture systems that better mimic the in vivo metabolic environment could also be important for identifying cancer targets that might otherwise be missed. Regardless, our findings highlight that consideration of the metabolic environment is critical for the development of cancer therapies that target metabolism.
Experimental Procedures
Cell Culture
A549, H1299, MDA-MB231, HeLa, and 143B cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Corning) with or without pyruvate or in Roswell Park Memorial Institute 1640 Media (RPMI) (Corning) as indicated. In all cases media was supplemented with 10% dialyzed fetal bovine serum (Sigma) and penicillin-streptomycin (Corning). Cells were cultured at 37°C with 5% CO2.
Proliferation Rates
Cell were plated in replicate in 6 well dishes (Corning), with an initial seeding density of 20,000 cells per well for A549, H1299, HeLa, and 143B and an initial seeding density of 40,000 cells for MDA-MB231. After seeding, cells were allowed to settle overnight and one 6 well dish was counted to determine starting cell number prior to treatment. For the remaining dishes, cells were washed twice in phosphate buffered saline (PBS), and 4 mL of media containing the indicated treatment was added. For pyruvate titration experiments, media was changed daily to maintain a constant pyruvate concentration. Cell counts were determined four days after initial treatment using a Cellometer Auto T4 Plus Cell Counter (Nexcelom Bioscience) (Sullivan et al., 2015). Proliferation rate was calculated based on the following formula:
Proliferation Rate (Doublings per day) = (Log2(Final cell count (day 5)/ Initial cell count (day 1))/ 4 (days)
Mitochondrial Oxygen Consumption
Oxygen consumption rates (OCR) were measured using a Seahorse Bioscience Extracellular Flux Analyzer (XF24). Cells were plated in Seahorse Bioscience 24-well plates at a seeding density of 50,000 cells per well in 100 µl of DMEM without pyruvate. After a 1 hour incubation, an additional 500 µl of DMEM was added with or without metformin and/or pyruvate before overnight incubation. The next day, cells were washed twice in DMEM without phenol red containing 0.5% dialyzed FBS at pH 7.4 containing treatments, and were then incubated in 500 µl of the same media with the corresponding treatments. OCR measurements were determined 24 hours after metformin and/or pyruvate treatment. Mitochondrial OCR was calculated by taking the total OCR and subtracting the residual OCR following addition of high dose rotenone and antimycin (2 µM each). Following the OCR measurements, cells from each well were counted using a Cellometer Auto T4 Plus Cell Counter (Nexcelom Bioscience) and used to normalized OCR per 100,000 cells. For determining complex I and mGPD dependent oxygen consumption, cells were seeded at 100,000 cells per well, incubated overnight, and prior to measurement media was changed to mitochondrial assay buffer (70 mM sucrose, 220 mM mannitol, 10 mM KH2PO4, 5 mM MgCl2, 2 mM HEPES, 1 mM EGTA, 0.2 % (w/v) fatty acid free BSA, pH 7.2) supplemented with 10 mM ADP. The first injection contained saponin (50 µg/mL final) to permeabilize cells and OCR was monitored until intracellular metabolites diffused out and OCR stabilized. Respiration substrates of complex I (10 mM pyruvic acid and 2 mM malic acid) or mGPD (10 mM glycerol-3-phosphate) were then injected until respiration was induced and stabilized followed by injection of metformin (1 mM final) or vehicle. Mitochondrial OCR was determined by subtracting OCR after addition of 2 µM antimycin from the experimental OCR measurements.
Measurement of NAD+/NADH
Cells were plated in 6-well dishes at 20,000 cells per well, allowed to adhere overnight, washed twice in PBS, and cultured in 4 ml of the indicated treatment media for 24 hours. Cells were then washed three times in ice cold PBS and extracted in 100 µL of ice cold lysis buffer (1% Dodecyltrimethylammonium bromide (DTAB) in 0.2 N NaOH diluted 1:1 with PBS), and immediately frozen at −80°C. NAD+/NADH ratio was measured using a modified version of instructions provided with the NAD/NADH Glo Assay Kit (Promega). For NADH measurement, 20 µL of the freshly thawed lysate was transferred to PCR tubes and incubated at 75°C for 30 minutes which allows for base-mediated degradation of NAD+. For NAD+ measurement, 20 µL of the freshly thawed lysate was transferred to PCR tubes containing 20 µL lysis buffer and 20 µL 0.4 N HCl. NAD+ samples were incubated at 60°C for 15 minutes, where acidic conditions result in selective degradation of NADH. Following respective incubations, samples were allowed to equilibrate to room temperature for 8 minutes and then quenched with the respective neutralizing solution with 20 µL 0.25 M Tris in 0.2 N HCl (NADH) or 20 µL 0.5 M Tris base (NAD+). Following sample prep, manufacturer instructions were used for enzyme-linked luminescence based NAD+ and NADH measurement (Sullivan et al., 2015).
Aspartate measurement by GCMS
Cells were seeded in 6 well dishes and incubated overnight to allow the cells to adhere. Following incubation cells were washed twice in PBS and media containing the indicated treatments was added. After overnight treatment, aspartate was extracted using 80% methanol in water with 1 µg norvaline standard added per sample. The extracted content was dried under nitrogen gas, derivatized, and measured as detailed in (Lewis et al., 2014).
Polar Metabolite Quantification by LCMS
Measurements were as described previously (Sullivan et al., 2015). Briefly, metabolites were first extracted using ice cold 80% methanol and chloroform. Relative metabolites abundances were measured using a Dionex UltiMate 3000 ultra-high performance liquid chromatography system connected to a Q Exactive benchtop Orbitrap mass spectrometer, equipped with an Ion Max source and a HESI II probe (Thermo Fisher Scientific). To quantify metabolite abundance from resulting chromatogram XCalibur QuanBroswer 2.2 (Thermo Fisher Scientific) was used in conjunction with in-house retention time library of chemical standards.
Immunoblotting
Cells were treated with metformin, pyruvate, and aspartate for 24 hrs, washed with cold PBS, and lysed with cold RIPA buffer containing cOmplete Mini protease inhibitors (Roche) and phosSTOP phosphatase inibitors (Roche). Protein concentration was quantified by BCA Protein Assay (Pierce) using BSA as a standard. Samples were resolved by SDS-PAGE using standard techniques, and protein was detected with the following antibodies: 4EBP1 (total and S65), ACC (total and pS79), Raptor (pS792) and S6K (total and S6K-pT389) from Cell Signalling Technologies, and Raptor (total) from Millipore.
Xenografts
Two million A549 cells were injected into flanks of nu/nu mice (088; Charles River Laboratories). Tumor volume was measured by caliper in two dimensions and volumes were estimated using the equation V = (π/6)(L*W2). The tumors were permitted to grow to 50mm3, after which the animals were randomly assigned to a treatment or vehicle group. Vehicle, 500 mg/kg, or 1500 mg/kg metformin were dosed via oral gavage daily for 10 days. The vehicle consisted of 0.1% (w/v) Tween 80 and 0.5% (w/v) methylcellulose. The tumors and plasma samples were collected 2 hours after the final dosage on day 10, and metformin and metabolite concentrations were quantified by LCMS.
Statistical Analysis
Data are presented as the mean ± standard error of the mean (SEM). Sample size (n) indicates experimental replicates from a single representative experiment; the results of experiments were validated by independent repetitions. Statistical significance was determined using an unpaired two-tailed t test with Welch’s correction where significance was determined as p ≤ 0.05.
Supplementary Material
Acknowledgments
This work was supported by the Burroughs Wellcome Fund, SU2C, the Ludwig Center at MIT and the NIH (P30CA1405141, GG006413, R01 CA168653, and R01 CA201276) to M.G.V.H., NIH (T32 GM007753) to D.Y.G., a postdoctoral fellowship (PF-15-096-01-TBE) from the American Cancer Society to L.B.S., NSF (GRFP DGE-1122374) to A.L. and S.M.D. We thank members of the Vander Heiden lab for thoughtful discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
D.Y.G. and L.B.S. performed experiments to determine proliferation rates, oxygen consumption measurements, NAD+/NADH ratio measurements, and metabolite measurements. A.L. performed xenograft experiments and assisted with writing the manuscript. A.M.H. performed immunoblotting experiments. L.N.B. and N.G. performed proliferation rate experiments. S.M.D. provided preliminary data and technical expertise. E.F. performed LCMS experiments. C.J.T. provided critical supplies and experimental design. D.Y.G., L.B.S., and M.G.V.H. designed the study and wrote the manuscript.
References
- Andrzejewski S, Gravel SP, Pollak M, St-Pierre J. Metformin directly acts on mitochondria to alter cellular bioenergetics. Cancer & metabolism. 2014;2:12. doi: 10.1186/2049-3002-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B, Wang T, Chen WW, Clish CB, Sabatini DM. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508:108–112. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Sabatini DM, Possemato R. Untuning the tumor metabolic machine: Targeting cancer metabolism: a bedside lesson. Nature medicine. 2012;18:1022–1023. doi: 10.1038/nm.2870. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges HR, Jones AJ, Pollak MN, Hirst J. Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. The Biochemical journal. 2014;462:475–487. doi: 10.1042/BJ20140620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, Viollet B, Thompson CB. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer research. 2007;67:6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EC, Liang X, Yee SW, Geier EG, Stocker SL, Chen L, Giacomini KM. Targeted disruption of organic cation transporter 3 attenuates the pharmacologic response to metformin. Mol Pharmacol. 2015;88:75–83. doi: 10.1124/mol.114.096776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Pawlikowski B, Schlessinger A, More SS, Stryke D, Johns SJ, Portman MA, Chen E, Ferrin TE, Sali A, et al. Role of organic cation transporter 3 (SLC22A3) and its missense variants in the pharmacologic action of metformin. Pharmacogenet Genomics. 2010;20:687–699. doi: 10.1097/FPC.0b013e32833fe789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuyas E, Fernandez-Arroyo S, Corominas-Faja B, Rodriguez-Gallego E, Bosch-Barrera J, Martin-Castillo B, De Llorens R, Joven J, Menendez JA. Oncometabolic mutation IDH1 R132H confers a metformin-hypersensitive phenotype. Oncotarget. 2015;6:12279–12296. doi: 10.18632/oncotarget.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell metabolism. 2016 doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Aglio DM, Perino LJ, Kazzi Z, Abramson J, Schwartz MD, Morgan BW. Acute metformin overdose: examining serum pH, lactate level, and metformin concentrations in survivors versus nonsurvivors: a systematic review of the literature. Annals of emergency medicine. 2009;54:818–823. doi: 10.1016/j.annemergmed.2009.04.023. [DOI] [PubMed] [Google Scholar]
- El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. The Journal of biological chemistry. 2000;275:223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. Bmj. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157:882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt SM, Bell EL, Keibler MA, Davidson SM, Wirth GJ, Fiske B, Mayers JR, Schwab M, Bellinger G, Csibi A, et al. Metformin decreases glucose oxidation and increases the dependency of prostate cancer cells on reductive glutamine metabolism. Cancer research. 2013;73:4429–4438. doi: 10.1158/0008-5472.CAN-13-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretz M, Guigas B, Bertrand L, Pollak M, Viollet B. Metformin: from mechanisms of action to therapies. Cell Metab. 2014;20:953–966. doi: 10.1016/j.cmet.2014.09.018. [DOI] [PubMed] [Google Scholar]
- Gandini S, Puntoni M, Heckman-Stoddard BM, Dunn BK, Ford L, DeCensi A, Szabo E. Metformin and cancer risk and mortality: a systematic review and meta-analysis taking into account biases and confounders. Cancer prevention research. 2014;7:867–885. doi: 10.1158/1940-6207.CAPR-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami S, Yee SW, Stocker S, Mosley JD, Kubo M, Castro R, Mefford JA, Wen C, Liang X, Witte J, et al. Genetic variants in transcription factors are associated with the pharmacokinetics and pharmacodynamics of metformin. Clin Pharmacol Ther. 2014;96:370–379. doi: 10.1038/clpt.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham GG, Punt J, Arora M, Day RO, Doogue MP, Duong JK, Furlong TJ, Greenfield JR, Greenup LC, Kirkpatrick CM, et al. Clinical pharmacokinetics of metformin. Clinical pharmacokinetics. 2011;50:81–98. doi: 10.2165/11534750-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Gravel SP, Hulea L, Toban N, Birman E, Blouin MJ, Zakikhani M, Zhao Y, Topisirovic I, St-Pierre J, Pollak M. Serine deprivation enhances antineoplastic activity of biguanides. Cancer research. 2014;74:7521–7533. doi: 10.1158/0008-5472.CAN-14-2643-T. [DOI] [PubMed] [Google Scholar]
- Griss T, Vincent EE, Egnatchik R, Chen J, Ma EH, Faubert B, Viollet B, DeBerardinis RJ, Jones RG. Metformin Antagonizes Cancer Cell Proliferation by Suppressing Mitochondrial-Dependent Biosynthesis. PLoS Biol. 2015;13:e1002309. doi: 10.1371/journal.pbio.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. Molecular Pathways: Is AMPK a Friend or a Foe in Cancer? Clin Cancer Res. 2015;21:3836–3840. doi: 10.1158/1078-0432.CCR-14-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Wondisford FE. Metformin action: concentrations matter. Cell Metab. 2015;21:159–162. doi: 10.1016/j.cmet.2015.01.003. [DOI] [PubMed] [Google Scholar]
- He X, Esteva FJ, Ensor J, Hortobagyi GN, Lee MH, Yeung SC. Metformin and thiazolidinediones are associated with improved breast cancer-specific survival of diabetic women with HER2+ breast cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2012;23:1771–1780. doi: 10.1093/annonc/mdr534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wullschleger S, Shpiro N, McGuire VA, Sakamoto K, Woods YL, McBurnie W, Fleming S, Alessi DR. Important role of the LKB1-AMPK pathway in suppressing tumorigenesis in PTEN-deficient mice. The Biochemical journal. 2008;412:211–221. doi: 10.1042/BJ20080557. [DOI] [PubMed] [Google Scholar]
- Jiralerspong S, Palla SL, Giordano SH, Meric-Bernstam F, Liedtke C, Barnett CM, Hsu L, Hung MC, Hortobagyi GN, Gonzalez-Angulo AM. Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:3297–3302. doi: 10.1200/JCO.2009.19.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- Kordes S, Pollak MN, Zwinderman AH, Mathot RA, Weterman MJ, Beeker A, Punt CJ, Richel DJ, Wilmink JW. Metformin in patients with advanced pancreatic cancer: a double-blind, randomised, placebo-controlled phase 2 trial. The Lancet. Oncology. 2015;16:839–847. doi: 10.1016/S1470-2045(15)00027-3. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim TI, Jeon SM, Hong SP, Cheon JH, Kim WH. The effects of metformin on the survival of colorectal cancer patients with diabetes mellitus. International journal of cancer. Journal international du cancer. 2012;131:752–759. doi: 10.1002/ijc.26421. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Saud SM, Young MR, Chen G, Hua B. Targeting AMPK for cancer prevention and treatment. Oncotarget. 2015;6:7365–7378. doi: 10.18632/oncotarget.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luengo A, Sullivan LB, Heiden MG. Understanding the complex-I-ty of metformin action: limiting mitochondrial respiration to improve cancer therapy. BMC biology. 2014;12:82. doi: 10.1186/s12915-014-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madera D, Vitale-Cross L, Martin D, Schneider A, Molinolo AA, Gangane N, Carey TE, McHugh JB, Komarck CM, Walline HM, et al. Prevention of tumor growth driven by PIK3CA and HPV oncogenes by targeting mTOR signaling with metformin in oral squamous carcinomas expressing OCT3. Cancer prevention research. 2015;8:197–207. doi: 10.1158/1940-6207.CAPR-14-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, Prigaro BJ, Wood JL, Bhanot S, MacDonald MJ, et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2014;510:542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Vander Heiden MG. Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem Sci. 2015;40:130–140. doi: 10.1016/j.tibs.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memmott RM, Mercado JR, Maier CR, Kawabata S, Fox SD, Dennis PA. Metformin prevents tobacco carcinogen--induced lung tumorigenesis. Cancer prevention research. 2010;3:1066–1076. doi: 10.1158/1940-6207.CAPR-10-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker MP, Audi SH, Bongard RD, Lindemer BJ, Krenz GS. Influence of pulmonary arterial endothelial cells on quinone redox status: effect of hyperoxia-induced NAD(P)H:quinone oxidoreductase 1. American journal of physiology. Lung cellular and molecular physiology. 2006;290:L607–L619. doi: 10.1152/ajplung.00302.2005. [DOI] [PubMed] [Google Scholar]
- Nies AT, Hofmann U, Resch C, Schaeffeler E, Rius M, Schwab M. Proton pump inhibitors inhibit metformin uptake by organic cation transporters (OCTs) PloS one. 2011;6:e22163. doi: 10.1371/journal.pone.0022163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. The Biochemical journal. 2000;(348 Pt 3):607–614. [PMC free article] [PubMed] [Google Scholar]
- Pernicova I, Korbonits M. Metformin--mode of action and clinical implications for diabetes and cancer. Nature reviews. Endocrinology. 2014;10:143–156. doi: 10.1038/nrendo.2013.256. [DOI] [PubMed] [Google Scholar]
- Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. The Journal of biological chemistry. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- Pirinen E, Canto C, Jo YS, Morato L, Zhang H, Menzies KJ, Williams EG, Mouchiroud L, Moullan N, Hagberg C, et al. Pharmacological Inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell Metab. 2014;19:1034–1041. doi: 10.1016/j.cmet.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak M. Overcoming Drug Development Bottlenecks With Repurposing: Repurposing biguanides to target energy metabolism for cancer treatment. Nature medicine. 2014;20:591–593. doi: 10.1038/nm.3596. [DOI] [PubMed] [Google Scholar]
- Schockel L, Glasauer A, Basit F, Bitschar K, Truong H, Erdmann G, Algire C, Hagebarth A, Willems PH, Kopitz C, et al. Targeting mitochondrial complex I using BAY 87-2243 reduces melanoma tumor growth. Cancer & metabolism. 2015;3:11. doi: 10.1186/s40170-015-0138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer cell. 2013;23:143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, Castro RA, Ianculescu AG, Yue L, Lo JC, Burchard EG, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. The Journal of clinical investigation. 2007;117:1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Hasan MK, Alvarado E, Yuan H, Wu H, Chen WY. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011;30:907–921. doi: 10.1038/onc.2010.468. [DOI] [PubMed] [Google Scholar]
- Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budinger GR, et al. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife. 2014;3 doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Fryknas M, Hernlund E, Fayad W, De Milito A, Olofsson MH, Gogvadze V, Dang L, Pahlman S, Schughart LA, et al. Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments. Nat Commun. 2014;5:3295. doi: 10.1038/ncomms4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang Y, Gong YI, He J, Chen X. Metformin and lung cancer risk of patients with type 2 diabetes mellitus: A meta-analysis. Biomedical reports. 2015;3:235–241. doi: 10.3892/br.2015.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.