Abstract
The mutation in huntingtin (mHtt) leads to a spectrum of impairments in the developing forebrain of Huntington’s disease (HD) mouse models. Whether these developmental alterations are due to loss- or gain-of-function mechanisms and contribute to HD pathogenesis is unknown. We examined the role of selective loss of huntingtin (Htt) function during development on postnatal vulnerability to cell death. We employed mice expressing very low levels of Htt throughout embryonic life to postnatal day 21 (Hdhd•hyp). We demonstrated that Hdhd•hyp mice exhibit: (1) late-life striatal and cortical neuronal degeneration; (2) neurological and skeletal muscle alterations; and (3) white matter tract impairments and axonal degeneration. Hdhd•hyp embryos also exhibited subpallial heterotopias, aberrant striatal maturation and deregulation of gliogenesis. These results indicate that developmental deficits associated with Htt functions render cells present at discrete neural foci increasingly susceptible to cell death, thus implying the potential existence of a loss-of-function developmental component to HD pathogenesis.
Keywords: Huntingtin, development, degeneration, pathogenesis, neurogenesis, gliogenesis, myelin, loss-of-function, maturation, Huntington’s disease
1. Introduction
Cumulative studies have reported a plethora of early brain abnormalities in Huntington’s disease (HD) that may reflect an impaired developmental substrate. These brain abnormalities may therefore represent the consequences of a developmental diathesis induced by aberrant polyglutamine expansion (polyQ) in huntingtin (Htt). These putative developmental sequelae include impairments in the inhibitory GABAergic system (Philpott et al., 2015), oligodendrocyte abnormalities (Gomez-Tortosa et al., 2001; Myers et al., 1991) and white matter micro-architectural changes (Di Paola et al., 2014; Gregory et al., 2015; Phillips et al., 2014), as well as region-specific brain volumetric alterations (Aylward et al., 2013; Nopoulos et al., 2011; Paulsen et al., 2006). In line with these findings, we and other groups have reported significant developmental abnormalities in striatal and cortical neurogenesis in HD mice (Molero et al., 2009; Molina-Calavita et al., 2014). Further, our in vitro studies using an embryonic stem cell line carrying mutant huntingtin (mHtt) demonstrates that this protein impairs the specification and maturation of organ-specific cellular lineages, as well as those of region-specific neural cellular subtypes (Nguyen et al., 2013a; Nguyen et al., 2013b). Accordingly, recent transcriptomic studies have reported that genes with key developmental and neural functions are preferentially impacted in specimens carrying mHtt (Achour et al., 2015; Jin et al., 2012; Labadorf et al., 2015; Ng et al., 2013).
More recently, we showed that restriction of mHtt expression to the period of neural development (embryonic time to post-natal day [PND] 21) elicit HD-like motoric, electrophysiological and neuropathological features in adult mice (Molero et al., 2016). This provocative finding strongly suggests that mHtt-dependent developmental alterations contribute to HD pathogenesis and progression. Whether HD developmental impairments associated with late life disease hallmarks are a consequence of gain or loss of function mechanisms or a compendium of both pathological processes is unknown. Interestingly, our previous in vitro studies have shown that developmental deficits associated with wild-type Huntingtin (Htt) ablation parallel many of those observed in specimens carrying mHtt (Nguyen et al., 2013a; Nguyen et al., 2013b), which supports a loss of function mechanism. In addition, other studies have shown that loss of huntingtin, via impairments of mitotic spindle orientation, affects neural stem cell (NSC) cycle progression and corresponding cell fate specification potential, further supporting an underlining loss-of-function mechanism (Godin et al., 2010). Collectively, these studies suggest that loss of Htt functional mechanisms play a key role in HD-associated developmental impairments.
Interestingly, cellular (oligodendrocytes and interneurons) and morphometric “developmental sequelae” have shown strong clinical correlates during the HD prodromal phase and can even predict disease progression. For example, myelin breakdown and associated changes in ferritin distribution during the prodromal phase of disease closely mimics topographical profiles of cellular vulnerability to cell death occurring later in HD (Bartzokis et al., 2007). Indeed, the PREDICT-HD and the Huntington Study Group investigations have shown that white matter abnormalities are one of the best predictors of diagnostic proximity to HD (Paulsen et al., 2010), and also correlates with cognitive impairment in pre-manifest cases of HD (Beglinger et al., 2005). On the other hand, selective changes in the complement of interneurons in primary motor cortex and cingulate cortex are strongly linked to symptom heterogeneity in HD, particularly with respect to motoric and mood disorders (Kim et al., 2014). Additional studies have also shown important cognitive correlates with volumetric changes in the nucleus accumbens and the thalamus during the prodromal phase of disease (Aylward et al., 2013). Because of the potential implications for disease pathogenesis, it is essential to examine the role of loss of huntingtin developmental functions for eliciting the postnatal and adult neurological and pathological hallmarks of HD.
A previous study has shown that mice expressing very low levels of huntingtin (HdhneoQ20/null) not only have early brain abnormalities that are suggestive of developmental deficits, but also that these mice develop a progressive lethal neurological phenotype during adult life. Unfortunately, as reduced expression of huntingtin was maintained throughout the life of these mice, it is impossible to predict whether the neurological phenotype was a consequence of developmental impairments or postnatal cellular deficits associated with the continual loss of huntingtin function. Here, we examined the role for disease pathogenesis of the loss of normal Htt developmental functions by severely reducing Htt expression throughout embryonic life until PND21, followed by Htt reconstitution via tamoxifen driven CRE recombination approaches. Accordingly, we found that mice with reduced levels of Htt during neural development (Hdhd•hyp) displayed HD-like motoric abnormalities and neurodegenerative changes. In addition, these mice displayed overt white matter tract (WMT) abnormalities and ultrastructural changes typical of “myelinopathies”. Consistently, Hdhd•hyp embryos displayed impairments in neurogenesis and gliogenesis. These findings support the notion that loss of Htt developmental mechanisms contribute to HD pathogenesis.
2. Materials and Methods
2.1. Animal Models
As a model of selective loss of huntingtin functions, we employed compound heterozygous HdhneoQ20/null mouse models expressing very low levels of a non-pathogenic form of huntingtin (Auerbach et al., 2001; Duyao et al., 1995; White et al., 1997). HdhneoQ20 mice are Swiss Webster (SW) knock-in mice carrying a floxed neomycin cassette within the promoter of the murine HD gene homologous (Hdh), and encoding a normal human sized glutamine tract (20 residues; Q20). The Hdhnull model corresponds to SW mice carrying a pGKneo insertion/replacement inactivation mutation of exons 4/5 in Hdh (Duyao et al., 1995). By mating homozygous HdhneoQ20 mice with mice Hdhnull/+ carrying the CAG-CRE-Esr1 gene (Jax# 004682) and further administering a daily dose of Tamoxifen (1.125 mg/day, oral) for five consecutive days (PND21 to PND26) to the progeny of these mice, we obtained the following experimental groups: (1) compound HdhneoQ20/null;CAG-CRE-Esr1, expressing severely reduced levels (~15%) of huntingtin only during development (thereafter called Hdhd•hyp); and (2) HdhneoQ20/+/CAGCRE-Esr1, expressing higher but still reduced (~65%) levels of huntingtin only during development. As controls, we employed littermate HdhneoQ20/+/CAG-CRE-Esr1 mice, because: (1) previous studies have documented that both Hdhnull/+ mice with targeted mutation of exons 4/5 and HdhneoQ20/+ mice are indistinguishable from wild-type mice (Duyao et al., 1995; White et al., 1997); (2) a recent study has shown that forebrain postnatal ablation of Htt does not lead to brain atrophy or degeneration (Wang et al., 2016); and (3) our studies documented that the absence of detectable developmental and/or postnatal abnormalities in HdhneoQ20/+/CAG-CRE-Esr1 mice. Previous studies have demonstrated highly efficient and widespread levels of recombination using CRE/Esr1 expressed from the CAG promoter (Hayashi and McMahon, 2002). To avoid confusion, we maintained the former genetic nomenclature for the mouse HD gene (Hdh), rather than the more recently adopted, Htt. Both male and female mice were employed unless otherwise specified. All studies were conducted with the approval of the Albert Einstein Animal Institute and were in compliance with all ethical guidelines and regulations.
2.2. Reagents
The following antibodies were used for immunofluorescence staining: mouse anti-APC (1:100; Calbiochem), rat anti-BrdU (1:100; Abcam), mouse anti-Calbindin (1:500; Sigma-Aldrich), goat anti-ChAT (1:250; EMD Millipore), rat anti-CTIP2 (1:500; Abcam); rabbit anti-DARPP32 (1:50; Santa Cruz Biotechnology), rabbit anti-Dystrophin (1:500; Thermo), guinea pig anti-Dlx2 (1:500; gift from Kazuaki Yoshikawa, Osaka University), rabbit anti-GAD65 (1:200; Cell Signaling Technology), mouse anti-GFAP (1:300; BD Biosciences); mouse anti-Mash1 (1:100; BD Biosciences), mouse anti-MBP (1:500; Sternberger Monoclonals); mouse anti-Nestin (1:100; BD Biosciences), mouse anti-NeuN (1:100; EMD Millipore); rabbit anti-NG2 (1:300; EMD Millipore), mouse anti-pNP (1750; Sternberger Monoclonals), mouse anti-Nkx2.1 (1:100; EMD Millipore), mouse anti-O1 (1:700; Sigma-Aldrich), mouse anti-O4 (1:700; Sigma-Aldrich), rabbit anti-μ-opioid receptor (1:500; ImmunoStar), rabbit anti-parvalbumin (1:1000; Sigma-Aldrich), rabbit anti-PDGFRα (1:500 and 1:50 for Cell Signaling Technologies R&D Systems, respectively), mouse anti-somatostatin (1:50; Santa Cruz Biotechnology), and mouse anti-β-tubulin (1:300; Sigma-Aldrich). Alexa-fluor-conjugated secondary antibodies (Invitrogen) were diluted at 1:1500. In our in vitro applications, we employed the NeurocultTM NCFC assay kit from Stem Cell Technologies, CNTF and PDGFα from R&D Systems and bFGF from BD Biosciences.
2.3. Behavioral Tests
The general behavioral and functional profiles of mice were studied before each motoric examination using the modified SHIRPA protocol (Rogers et al., 1997). Voluntary locomotion and motor coordination were examined in both female and male mice longitudinally at 6 and 12 months of age with the open field and the balance beam tests. All tests were performed after 1 hour of acclimation in the testing room during the light cycle. The 6-month time point corresponds to the upper age limit for mature adult mice (Flurkey et al., 2007), not yet affected by senescence, therefore an appropriated age for the detection of incipient deviations from normal adult behavioral patterns (Semple et al., 2013). In humans, HD onset commonly appears between the third and fourth decade of life. Hence, we chose to examine 12-month mice, as this age falls within the murine middle-age equivalent (Flurkey et al., 2007). Open field test: Mice were placed in an opaque perspex arena (16 square inches) and allowed to freely explore the arena for 6 minutes, during which time voluntary locomotion (total distance traveled in cm) and explorations were recorded and thereafter analyzed using the Viewer software (Biobserve: Bonn, Germany) as previously described (Gulinello et al., 2009). Balance beam test: Motor coordination deficits were measured as the number of slips while traversing a round wooden beam as previously described (Gulinello et al., 2010; Stanley et al., 2005). Before the test, all mice were pre-trained on a wide plank (3 in) to encourage reliable crossing. The start side was brightly illuminated and the end side had a small, darkened chamber that contained palatable food (cereal) to encourage the mice to cross. Immediately after pre-training, missteps (slips, defined by when the paw slipped below the midline of the beam) were assessed on a 1.5 cm diameter beam. The study of gait was conducted by marking the front paws with red, non-toxic paint and the back paws with blue paint. Mice were then placed in a plexiglass “corridor” lined with paper strips. Animals were first habituated to the chamber to encourage stable crossing and to prevent excessive rearing and turning. After drying, the strips were digitized by scanning.
2.4. Tissue processing and staining
Cellular preparations were fixed and processed for immunocytochemistry, as described elsewhere (Molero et al., 2009). Briefly, under deep anesthesia, mice were perfused transcardially with ice-cold 0.25 mg/ml heparin in PBS (10 mL) followed by PFA 4% (50 mL), and their brains thereafter harvested. For histological techniques, the brains were then post-fixed overnight in PFA 4% and incubated in sucrose (20%) until total submersion, subsequently flash frozen in a cryomatrix embedding medium and stored at −80°C until use. For immunostaining procedures, embryo specimens were cryosectioned using 7 μm slices and processed on slides, while postnatal specimens were cryosectioned using 25 μm slices and processed as floating sections. Primary antibody incubations were performed O/N at 4°C. For electron microscopy techniques, perfused brains were sliced (1 mm thick), fixed for an additional hour in a solution composed of 2% PFA/2.5% glutaraldehyde in 0.1M cacodylate buffer and then rinsed in this buffer. Samples were thereafter osmicated with 1% osmium tetroxide for 1 hour, rinsed in buffer, then water, and incubated with 2% uranyl acetate for 1 hour. Specimens were subsequently dehydrated in alcohol mixtures and embedded in Epon LX112. Sections for toluidine blue analyses were sectioned at 1 μm (thick sections).
2.5. Neural cell clonal and differentiation assays
Clonal studies of primary neural stem cell (NSC) cultures were performed with minor modifications to the NeurocultTM NCFC assay kit (Louis et al., 2008). To accomplish this goal, E12.5 timed pregnant mice were sacrificed, respective embryos collected individually, and the lateral ganglionic eminence (LGE) dissected and cultured following our experimental paradigms for clonal expansion (Molero et al., 2009; Yung et al., 2002). After embryo genotyping (24 hours), neurospheres were dissociated with Trypsin-EDTA followed by gentle trituration and thereafter clonally expanded for 10 days following the NeurocultTM NCFC assay kit protocol. Neurosphere size was quantified within the matrix of collagen and grouped according to size as small, medium and large (<0.5, 0.5-1, and >1 mm2, respectively). The clonal differentiation assay was performed with specific modifications of the procedure described elsewhere (Yung et al., 2002). In short, large neurospheres were extracted from the NCFC matrix, dissociated as described above, and plated on 1 cm2 poly-D-lysine coated glass coverslips at a density of 104 cells/well in complete medium supplemented with 10 ng/ml of bFGF and PDGFα. After 48 hours of incubation, the medium was replaced with complete medium supplemented with CNTF, 10 ng/ml, and further cultured for 8 additional days.
2.6. Cell and white matter quantifications
All quantifications were experimentally blinded. Degenerative cells in the external striatum of 12 and 16 month specimens were quantified as the mean ±SEM of cell with ultrastructural morphology of degenerative neurons detected in 8 random grid areas of 10,400 μm2 each (nper genotype = 4 at 12 months, and 3 and 4 for CTL and Hdhd•hyp mice, respectively, at 16 months). All cells within each cortical layer were quantified in a column 150 μm wide within the primary motor cortex in coronal sections obtained at Bregma 1.10 mm. In each case, the cross-sectional column examined corresponded to primary motor cortex immediately juxtaposed to the secondary motor cortex. The number ±SEM of NG2-, O4- and O1-positive cells obtained after 10DIV using the clonal differentiation assay was performed by pooling two distinctive experimental assays (4 embryos per genotype per assay) and thereafter averaging the data obtained from each fields (20 random fields, 0.042 mm2 each). For Sholl analyses, randomly selected traced oligodendrocytes (immunoreactive for GC/O1) were studied by centering nested spheres (spaced 10 μm apart) surrounding the cell body and the radial distribution of arborization patterns framing the cell’s perikaryon automatically quantitated using the Image-J software. G-ratios were calculated in TEM micrographs as the diameter of the axon divided by the diameter of the axon and myelin. A minimum of 100 and 40 axons per mouse, 3 mice/experimental group, were analyzed in striatal and cortex specimens, respectively.
2.7. Quantitative real-time PCR analysis
These studies were performed as described elsewhere (Molero et al., 2009). Specimens were obtained from coronal sections encompassing the whole forebrain (n = 8, 9 and 8 for wild-type, control and Hdhd•hyp specimens, respectively) of 12-month old mice treated with tamoxifen at PND21. Each sample was individually extracted using RNeasy® FFPE kit (Qiagen, Hilden, Germany), and subsequently processed for cDNA synthesis employing standard techniques. QRT-PCR was performed with the TaqMan® Universal Master Mix II per the manufacturer’s instructions on an ABI 7000 Sequence Detector System (Applied Biosystems, Foster City, CA, USA). We employed the TaqMan® primer/probes mm01213820-m1 and mm00446968-m1 to selectively target the Htt and HPRT1 transcripts, respectively (Applied Biosystems, Foster City, CA, USA). Data collection and quality assessment were performed utilizing 1.1 RQ Software (Applied Biosystems, CA, USA). Statistical comparisons were made using the Pair Wise Fixed Reallocation Randomization test (PWFRR) (Pfaffl et al., 2002).
2.8. Statistics
Depending on the data distribution, mean comparisons were performed with unpaired Student t-test or Mann-Whitney-U test. The comparison between proportions was accomplished with two-tailed Chi-Square (χ2) tests. n-values reported correspond to biological replicates. We examined the variance within each group with the Bartlett's and F test and determined whether the data exhibited a Gaussian distribution with the D’Agostino-Pearson omnibus normality test. The slope of myelin g-ratios and the number of neurofilaments by axon diameter were calculated with linear regression analyses and compared with ANCOVA. Comparisons in Sholl analyses were performed with repeated measures ANOVA. Data are presented as either the mean ±SEM or the percentage ±95%CI. All analyses were double-blinded. Unless otherwise indicated, the statistical differences between samples were assigned using a probability of at least < 0.05.
3. Results
3.1. Selective loss of huntingtin developmental functions using the Hdhd•hyp mouse model
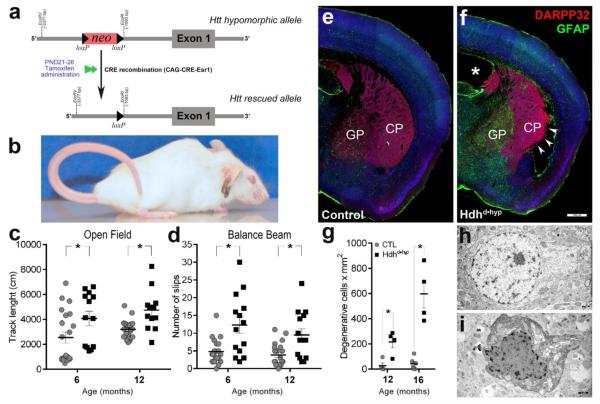
Our experimental model consisted of compound HdhneoQ20/null mice also carrying the CAG-CRE-Esr1 gene. After tamoxifen administration following PND21, the floxed neocassette was excised leading to reconstitution of Htt expression from the HdhneoQ20 allele (Fig.1a). Thus, the targeted experimental mouse expressed very low levels of non-pathogenic Htt (Auerbach et al., 2001) only during the period of neural development (from embryonic life to PND21; hereafter referred as Hdhd•hyp). We choose PND21 as a start point for Htt expression reconstitution, as by this age both striatal neurogenesis and striatal chemo-architecture maturation milestones are finally achieved (Jain et al., 2001; Raghunathan et al., 1996; van der Kooy and Fishell, 1987). Despite reconstitution of Htt expression, Hdhd•hyp mice consistently displayed lower weight during adult life (Fig.A.1d, e). To validate the Hdhd•hyp model, we confirmed the normalization of Htt expression after excisional removal of the neocassette by quantifying via qRT-PCR the level of Htt in 12-month old brain specimens. After tamoxifen administration, the expression Htt from the hypomorph HdhneoQ20 allele was successfully rescued in Hdhd•hyp mice (Hdhd•hyp vs wild type fold change = 0.62, 0.55 – 0.71 95%CI, PWFRR p-value = 0.014). The observed decrease of Htt expression in Hdhd•hyp was anticipated as one of the HTT alleles in Hdhd•hyp mice is null. Conversely, our control group of mice (HdhneoQ20/+;CAG-CRE-Esr1-negative) expressed similar levels of Htt to those displayed by wild-type mice (controls vs wild type fold change = 0.79, 0.67 – 0.91 95%CI, PWFRR p-value = 0.156). Reconstitution of Htt expression from the hypomorph HdhneoQ20 allele was also corroborated in striatal (Hdhd•hyp vs wild type fold change = 0.54, 0.51 – 0.58 95%CI, PWFRR p-value = 0.06) and motor cortex (Hdhd•hyp vs wild type fold change = 0.6, 0.48 – 0.74 95%CI, PWFRR p-value < 0.0001) specimens obtained by tissue microdissection.
Figure 1. Selective loss of Huntingtin function during development in Hdhd•hyp mice leads to late-life motor abnormalities, gliosis and striatal neuronal degeneration.
Compound heterozygous mice carrying both hypomorphic and null Htt alleles received tamoxifen between PND21 to 26, which rescue the expression Htt from the hypomorphic gene (a). Mice selectively expressing low levels of Htt during development (Hdhd•hyp) exhibit Straub tail, atrophy of hindlimb muscles and lordokyphosis (b). 6- and 12-month old Hdhd•hyp mice traversed significantly longer track lengths within the Open Field arena (c) and made more slips in the Balance Beam test (d) as compared to controls (CTL). e-f depicts 12-month of age brain sections immunostained with GFAP and DARPP32. GP and CP stand for globus pallidum and caudate-putamen, respectively. *asterisk and arrowheads in f indicates lateral ventricle hypertrophy and lateral CP degenerative changes in Hdhd•hyp, respectively. Electron microscopic analyses of the lateral striatum revealed significantly larger numbers of degenerating neurons in 12- and 16-months Hdhd•hyp specimens as compared with CTL (g-i). Scale bars in f and h-i correspond to 500 and 1 μm, respectively. Data in c-d is presented as mean ± SEM. * in c, d and g corresponds to p-values < 0.05.
3.2. Hdhd•hyp mice exhibited deficits in motor coordination and progressive neurological abnormalities
We next examined whether the selective loss of Htt function during neural development impairs sensorimotor function. At six months of age, Hdhd•hyp mice exhibited a hyperkinetic phenotype and abnormalities of motor coordination (Fig.1, c-d). These two behavioral traits did not display progression over the course of the next six months. Like in HdhneoQ20/null mice, Hdhd•hyp mice also exhibited clasping and handling induced seizures, the later characterized by alternating clonic movement of forelimbs, clonic and versive movements of the head, chewing movement and facial myoclonus, flagpole tail dorsiflexion, followed by intermittent eye blinking, immobility, head nodding and clonic mouth jerks (Fig.A.2, A.3 [videos]). There was also frequent back arching and tonic posturing of the hind limbs. The clinical features of these seizure-like events suggest a forebrain origin, possibly generalizing to deeper structures.
One year old Hdhd•hyp mice, but not controls, showed Straub tail (46.2%; 23-71 95%CI, Fig.1b) and gait disturbances, including flattened pelvic elevation, abnormally wide hindlimb stance and occasional hopping and stiffness (Fig.A.1). Gait abnormalities qualitatively worsened over time, to the extent that 12-month Hdhd•hyp mice lacked the typical uniformity of step alternation (loss of fore- /hind-paw overlap in the paw print test) in addition to exhibiting smaller gait strides (Fig.A3). Finally, 16-month Hdhd•hyp mice, but not controls, displayed lordokyphosis (convex curvature of the thoracic spine) in association with atrophy of hindlimb muscles (Fig.1b, Fig.A.1k-o), the later, a typical trait documented not only in HD patients but also in the R6/2 mouse model of HD (Ribchester et al., 2004). Interestingly, the majority of Hdhd•hyp mice exhibited significantly smaller cerebellar lobules as compared with controls (6.9 ±0.2 vs 8.5 ±0.5, respectively; t(15) = 2.82, two-tailed p = 0.013, n = 10 and 7 for Hdhd•hyp and CTL, respectively; Fig.A1h-i), a difference that remained significant even after normalization of cerebellar dimensions by forebrain size (0.53 ±0.008 vs 0.6147 ±0.04, respectively; t(15) = 2.256, two-tailed p = 0.039, n = 10 and 7 for Hdhd•hyp and CTL, respectively). Cerebellar changes may in part, explain late-life muscle atrophic changes that are observed in these mice. Overall these findings demonstrate the existence of progressive neurological abnormalities in Hdhd•hyp mice.
3.3. Hdhd•hyp mice developed striatal neurodegenerative pathology and WMT abnormalities
We next examined whether the developmental loss of huntingtin function is associated with HD-like striatal neuropathological profiles. To accomplish this goal, we first examined for striatal astrogliosis in 12-month old mice by staining coronal sections with the astrocyte marker, GFAP. Accordingly, 83.3% of 12-month Hdhd•hyp striata exhibited reactive gliosis as compared with 20% in controls (Χ2(1)= 4.412; p = 0.035; n = 6 and 5 for Hdhd•hyp and controls, respectively, Fig.1e, f). Reactive gliosis in Hdhd•hyp mice was exceedingly severe in the globus pallidum (GP; Fig.1f) and observed in 100% of Hdhd•hyp specimens examined. In addition, 27.3% of Hdhd•hyp specimens (3/11) showed degenerative changes in the region corresponding to the posterior ventrolateral striatum and the endopiriform claustrum (arrowheads; Fig.1f). Moreover, 12-month old Hdhd•hyp mice displayed an aberrant number of neurons with degenerative features as assessed by ultrastructural analysis as compared with controls (216.1 ±46 vs 27.4 ±23 cells x mm2; nper genotype = 4; two-tailed Mann-Whitney-U exact p < 0.05; Fig.1g-i). By 16 months of age, the number of striatal degenerative neurons markedly increased in Hdhd•hyp specimens (540.1 ±73.4 vs 41.2.01 ±19.4 cells x mm2; n = 4 and 6, respectively; two-tailed Mann-Whitney-U exact p = 0.009). These data support the occurrence of progressive neuropathological changes in the Hdhd•hyp striatum.
3.4. Hdhd•hyp embryos exhibited heterotopias within the germinative zone of the striatal anlagen and impairments in the maturation of striatal compartmentalization
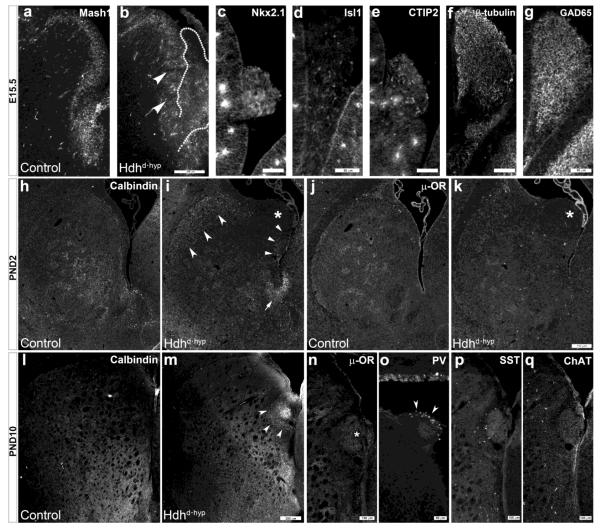
Previous studies have documented the role of huntingtin in forebrain neurogenesis (Godin et al., 2010; Nguyen et al., 2013a; White et al., 1997). Further, MacDonald et al. reported that embryos with hypomorphic expression of mutant HTT (HdhneoQ50/neoQ50) exhibit ectopic tissue masses in the developing lateral ventricles and striatal subventricular zone (White et al., 1997). Our studies now demonstrate that all embryos (three E14.5, seven E15.5, and six E17.5), and early postnatal specimens (two PND21, two PND10, and eleven PND21) expressing low levels of the non-pathogenic form of huntingtin also displayed analogous masses (Fig.2b-g). We further defined the nature of these structures: (1) they constituted heterotopic evaginations of germinative zone-derived cells from the posterior ventromedial aspect of the lateral ganglionic eminence (LGE; Fig.2b, outline), the developing anlage giving rise to striatal medium spiny neurons (MSNs); (2) cells within these heterotopic masses expressed the medial ganglionic (MGE) patterning marker, Nkx2.1 (Fig.2c), as well as the MSN progenitor markers, Isl1 and CTIP2 (Fig.2d & e), thus signifying that they encompass an amalgam of MGE and LGE telencephalic origin, respectively; (3) the broad expression of β–tubulin and GAD65 indicates the GABAergic neuronal identity of cells contained within these structures (Fig.2f, g); (4) the expression of both calbindin (Fig.2m, arrowheads) and μ-opioid receptor (μ-OR; Fig.2n) indicates that these heterotopias share both striatal MSN striosome and matrix subtype compartmental identity; and (5) the presence of parvalbumin (PV; Fig.2o) but not somatostatin (SST; Fig.2p) or choline acetyltransferase (ChAT; Fig.2q) within these structures indicates that Nkx2.1 precursors within these masses selectively give rise to PV+ GABAergic interneurons. Another interesting finding was the loss of the typical palisade appearance of pseudostratified neuroepithelium encompassing Mash1+ neuronal progenitors within the E15.5 Hdhd•hyp LGE (Fig.2b, arrowheads). These findings indicate the presence of deficits in early neural lineage specification, migration and regional organization as a consequence of the loss of Htt function within the embryonic striatal anlagen.
Figure 2. Loss of Huntingtin function leads to subpallial ventricular heterotopias and abnormalities in striatal chemo-architectural maturation.
a depicts immunoreactivity to Mash1 within the E15.5 subpallial germinative zone. Arrowheads in b point to disorganized regions within the Hdhd•hyp SVZ where the pseudo-stratified palisades of Mash1 progenitor cells are lost. A heterotopic mass of cells emerging from the most ventral aspect of the LGE is outlined in b. c-g depicts immunoreactivity to Nkx2.1 (c), Isl1 (d), CTIP2 (e), β-tubulin (f) and GAD65 (g) of E15.5 Hdhd•hyp subpallial heterotopias. h-k depicts representative PND2 striatal specimens immunostained against calbindin (h-i) and μ-opioid receptor (μ-OR; j-k). Concave-base arrowheads and standard arrowheads in i point to the ectopic expression of calbindin+ cells in the dorso-lateral and medial Hdhd•hyp striatum, respectively. Arrow in i points to aberrant calbindin immunoreactivity in the ventral end of the lateral ventricle. *asterisk in i and k point to Hdhd•hyp striatal ventricular heterotopias lacking immunoreactivity to calbindin and μ-OR. By contrast, both calbindin (arrowheads, l) and μ-OR (asterisk, m) are expressed in Hdhd•hyp heterotopias by PND10. Immunoreactivity to PV (o), but not to SST (p) or ChAT (q), is detected in these heterotopias. Scale bar in b corresponds to 200 μm, in c-g and o to 50 μm, in k and m to 250 μm, and in p, q to 100 μm.
In studies employing a knock-in model of HD, we previously observed maturational changes in the acquisition of the striosomal and matrix striatal chemo-architecture. Similarly, E17.5 Hdhd•hyp striata did not acquired the typical honeycomb immunoreactive pattern of calbindin indicative of this embryonic time (Fig.2h-i). Instead, calbindin was expressed ectopically in the external aspect of the striatum (Fig.2i, concave-base arrowheads), around ventricular heterotopias (arrowheads) and in the ventral tip of the lateral ventricle (arrow). By contrast, PND10 immunoreactivity to calbindin was similar between Hdhd•hyp and control specimens (Fig.2l-m). Conversely, although μ-OR immunoreactivity appears decreased in E17.5 striatal Hdhd•hyp specimens, this MSN marker exhibited the typical patchy distribution of striosomes at this time (Fig.2m-n). Overall, these findings suggest that loss of huntingtin function during embryonic life also leads to maturational impairments in the acquisition of the striatal chemo-architecture.
3.5. Hdhd•hyp mice exhibited cortical deep layer degeneration and myelination abnormalities
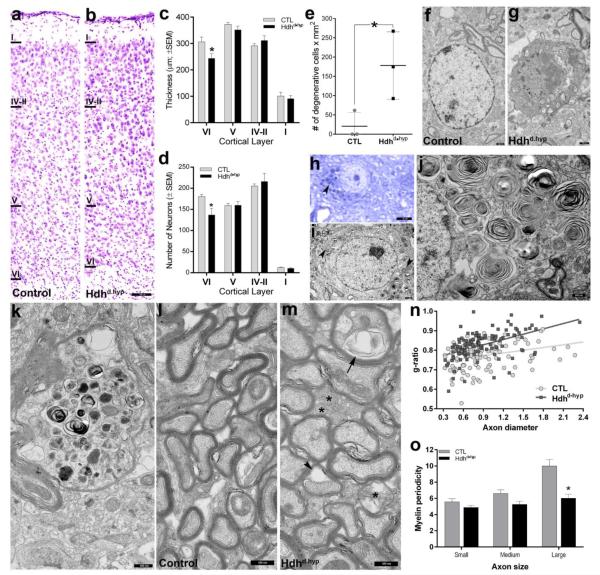
Cortical degeneration is one of the main hallmarks of HD (Waldvogel et al., 2015). In motor cortex, degeneration particularly affects deep layers (Hedreen et al., 1991). Thus, we next examined the motor cortex of 12-month old specimens. At this age, layer VI thickness (244.1 ±17 vs 306.4 ±18 μm) and numbers of neurons (137.2 ±14.9 vs 180.6 ±5.2) was significantly reduced in Hdhd•hyp mice compared to controls (t(11) = 2.4, two-tailed p = 0.035; and t(11) = 2.21, p = 0.04, respectively; Fig.3a-d). Consistently, the number of neurons with degenerative-like ultrastructural features in deep layer motor cortex was significantly higher in Hdhd•hyp specimens than in controls (178 ±50.5 vs 20.6 ±20 cell x mm2; Mann-Whitney-U one-tailed exact p = 0.05; Fig.3e-g). We extended our screening for cellular degeneration to outer cortical layers by examining 12-month old thick sections stained with toluidine blue. Degenerating cells appeared dark when stained with toluidine blue. We detected a significantly higher number of darkly stained neuronal cells lacking their typical morphology, especially in layer III/IV as compared with controls (9.9% 9-10.9 95%CI vs 0.4% 0.22-0.62 95%CI, respectively, Χ2(1) = 407; p < 0.0001). This complementary approach further confirmed our electron microscopy findings: 8.9% (7.9-9.8 95%CI) of neuronal species appeared with degenerative changes in Hdhd•hyp layer III/VI as compared with 0.03% (0.001-0.2 95%CI) in controls (Χ2(1) = 284, p < 0.0001). Unexpectedly, a number of non-degenerative cortical neurons, particularly those in layer III/IV, exhibited darkly stained intracytoplasmic inclusions (Fig.3h). Ultrastructural examination revealed that these structures corresponded to concentric membrane-bound laminated inclusions of zebra bodies (Fig.3i, j). We also observed the existence of axons of larger caliber displaying Wallerian-like degenerative morphology (Fig.3k). Overall these findings are indicative of HD-like late-life cortical neuropathology in Hdhd•hyp specimens.
Figure 3. 12-month of age motor cortex of Hdhd•hyp mice exhibit neuronal loss and myelination abnormalities.
a-b depicts Nissl stained cross-sections of 12-month of age motor cortices. Compared to CTL (n = 5), Hdhd•hyp (n = 8) cortical thickness (μm; c) and numbers of neurons (d) of layer VI are reduced, whereas the numbers of degenerating cells is increased (nper genotype = 3; e). Representative TEM micrographs of normal and degenerating neurons from 12-month of age CTL and Hdhd•hyp are shown in f and g, respectively. h-j shows Hdhd•hyp neurons containing intracytoplasmic zebra-bodies (h-i arrowheads). k depicts Hdhd•hyp axonal processes displaying Wallerian-like degenerative morphology. l-m corresponds to TEM micrographs showing layer VI myelinated axonal processes. *asterisk in m points to under-myelinated axons. Arrowhead and arrow in m point to a myelin ballon and tubulovesicular structures, respectively. n depicts g-ratios of myelinated axons. Compared with CTL, the periodicity of myelin lamellae of large axons located in Hdhd•hyp cortical layer VI is reduced (n = 37 axons [pool of 3 biological replicas per genotype]; o). Error bars in c-e and o represent ±SEM. Scale bars in b, f-g, h, i and j-m correspond to 150, 0.75, 4, 1 and 0.5 μm, respectively.
Numerous studies have also reported extensive alterations in white matter tract (WMT) architecture in HD cases and animal models (Bohanna et al., 2011; Jin et al., 2015; Paulsen et al., 2006; Phillips et al., 2014). Thus, we next examined the status of myelination of cortical deep layers at 12-months of age. Ultrastructural analyses revealed that Hdhd•hyp specimens, compared with controls, frequently exhibited hypomyelinated axons (Fig.3m, asterisks), as well as of axons with myelin balloons and tuberovesicular structures (Fig.3m, arrowhead and arrow, respectively). Consistently, Hdhd•hyp specimens displayed significantly higher g-ratios as compared to controls (F(1, 224) = 4.6247, p = 0.032; Fig.3n). Interestingly, myelination deficits in Hdhd•hyp specimens appeared to preferentially affect axons of larger caliber, as is indicated by the reduced periodicity of myelin lamellae exhibited by large axons (t(73) = 4.084, p = 0.0001, sample was obtained from the pool of 3 biological replicates per genotype; Fig.3o). Indeed, similar myelinopathic profiles were also observed on white matter tracts containing axonal bundles traveling across the striatum (Fig. A4).
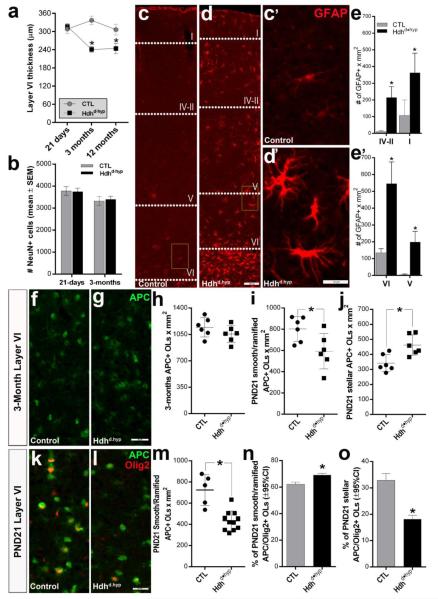
We next defined the onset of changes in cortical thickness and neuronal numbers in motor cortex layer VI (Fig.4a, b). Accordingly, the reduction in Hdhd•hyp layer VI was detected as early as 3-months of age (241 ±9.8 vs 337 ±13.8 for Hdhd•hyp [n = 6] and CTL [n = 5], respectively, t(9) = 5.8; two-tailed p = 0.0003), but not at PND21 (316.8 ±8.5 vs 308.3 ±14.3 for Hdhd•hyp [n = 10] and CTL [n = 6], respectively, t(14) = 0.54; two-tailed p = 0.594). Interestingly, despite the differences in thickness, at 3-months of age the number of neurons in layer VI was comparable between Hdhd•hyp and control specimens (t(9) = 0.28; two-tailed p = 0.79, n = 6 and 4, respectively). To examine whether these early changes occurred in response to intercurrent insults, we examined for the presence of reactive astrogliosis. Consistently, compared with controls, Hdhd•hyp specimens exhibited a significant increase in the number of GFAP+ astrocytes in both, superficial (t(8) = 7.01, p < 0.0001, and t(8) = 3.833, p = 0.0049, for layers IV-II and I, respectively, nper genotype = 5) and deep (t(8) = 7.016, p < 0.0001 and t(8) = 6.873, p < 0.0001, for layers VI and V, respectively, nper genotype = 5) cortical layers (Fig.4e, e’, respectively), although they were clearly more prominent in layer VI. Further, astrocytes in Hdhd•hyp specimens were clearly reactive, as indicated by the substantial enhancement of GFAP immunoreactivity and the complexity of cellular processes (Fig.4c’ and d’). The existence of reactive astrogliosis at this early time suggests the active presence of cellular insults or an increase in susceptibility to normal physiological stressors at this time.
Figure 4. Cortical cell loss in Hdhd•hyp mice is long preceded by reductions in cortical thickness, reactive gliosis, and changes in the composition of myelinating oligodendrocytes.
Graph in a and b depict layer VI cortical thickness and neuronal number, respectively, determined from coronal section stained with Nissl at PND21, 3 months and 12 months. c and d depict coronal sections of 3 month cortices immunostained against GFAP. c’ and d’ correspond to higher magnification of insets in c and d, respectively. e and e’ correspond to graphs where GFAP+ astrocytes were quantified in the superficial and deep motor cortical layers, respectively (nper genotype = 5). h-j graphs the overall number (h), smooth- and ramified-type (i) and stellar-type (j) APC+ oligodendrocytes in the 3-month old motor cortex layer VI (n per genotype = 6). k-l depict representative specimens at PND21 co-stained with APC and Olig2. The overall numbers of APC+ smooth- and ramified-type oligodendrocytes from layer VI PND21 Hdhd•hyp specimens are depicted in m. The fraction of APC+ smooth- and ramified-type and stellar-type oligodendrocytes also immunoreactive against Olig2 (k, i) are represented in n and o, respectively. Each point in graphs h-j and m represent a biological replicate. Error bars in a, b, e, e’, h-j, and m represent ±SEM. Error bars in n and o represent ±95%CI. Scale bars in d, d’, and g/l correspond to 100, 20 and 25 μm, respectively. *asterisk in all graphs correspond to p-values < 0.05.
Considering our findings of myelin deficits at 12-months of age and the fact that myelinated axons traversing layer VI substantially contribute to the volume of the neuropil in this region (Tomassy et al., 2014), it is possible that reductions in the thickness of this layer in Hdhd•hyp mice are a result of early myelination deficits. To properly assess this issue, we examined the number of myelinating APC+ oligodendrocytes in layer VI of 3-month old motor cortex (Fig.4f-h). Although at this time, the overall number of APC+ oligodendrocytes was similar between CTL and Hdhd•hyp mice (t(10) = 1.29, p = 0.224; f-g), when defined according to morphology, Hdhd•hyp mice showed a targeted reduction of smooth- and ramified-type oligodendrocytes (t(10) = 2.52, p = 0.034), with an increase of stellar-type oligodendrocytes (t(10) = 3.26, p = 0.0086) as compared with CTLs (nper genotype = 6; i). Likewise, PND21 Hdhd•hyp specimens also displayed significantly fewer APC+ smooth- and ramified-type oligodendrocytes as compared with controls (434.4 ±27.8 vs 724.8 ±65.5, respectively, t(14) = 4.87, p = 0.0002, n = 10 [Hdhd•hyp] and 5 [CTL]; Fig.4k-m), which may explain our previous observations of preferential deficits in the myelination of larger caliber axons, as these are differently myelinated by smooth- and ramified-type oligodendrocytes (Murtie et al., 2007). Interestingly, we also detected a mild but significant increase in the number of PND21 APC+ smooth- and ramified oligodendrocytes expressing the myelinogenic transcription factor, Olig2 (Χ2(1) = 44.85, p < 0.0001, n = the pool of 6 and 10 CTL and Hdhd•hyp biological replicas, respectively; Fig.4n). Changes in Olig2 may be a consequence of compensatory increases of myelinogenesis in remaining smooth- and ramified-type oligodendrocytes. By contrast, fewer APC+ stellar-types oligodendrocytes expressed Olig2 in Hdhd•hyp mice as compared with CTLs (Χ2(1) = 109.9, p < 0.0001, n = 6 and 10, respectively). Hence, it is possible that any compensatory increase in the number of stellar-type oligodendrocytes in Hdhd•hyp mice may not be sufficient to adequately sustain axonal myelination.
3.6 Oligodendrocyte progenitors derived from Hdhd•hyp primary embryonic neural stem cells display progressive maturation abnormalities
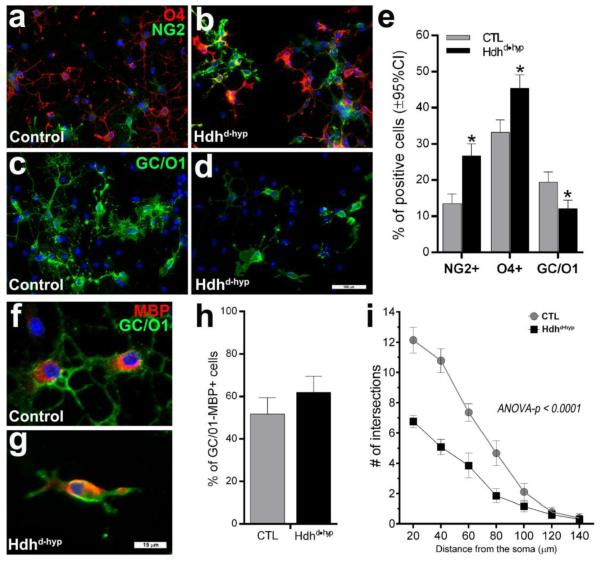
Some groups have suggested that various morphological types of oligodendrocytes may also represent different stages of oligodendrocyte maturation (Vinet et al., 2010), progressing from smooth to ramified and stellate oligodendrocytes according to maturity level. To further define impairments in gliogenesis and myelinogenesis in Hdhd•hyp mice, embryonic day 12.5 (E12.5) neural stem cells (NSCs) of the LGE were clonally expanded as neurospheres, dissociated, plated and differentiated into oligodendrocytes. Although the germinative zone of the LGE gives rise to the striatum, it is also the source of an important subpopulation of myelinating forebrain oligodendrocytes (Kessaris et al., 2006). After 10 days in vitro (DIV) of differentiation, cellular species were stained with the early oligodendrocyte precursor marker, NG2, the more lineage-committed oligodendrocyte progenitor marker, O4, and the oligodendrocyte post-mitotic differentiation marker, GC/01. Interestingly, the number of proliferating NG2 and O4 oligodendrocyte species were significantly higher in Hdhd•hyp preparations than in controls (Χ2(1) = 40.4, p < 0.0001 and Χ2(1) = 22.9, p < 0.0001, for NG2 and O4 preparations, respectively; Fig.5a-e). By contrast, Hdhd•hyp specimens not only exhibited fewer GC/O1+ oligodendrocytes (Χ2(1) = 16.97, p < 0.0001), but these post-mitotic species appeared morphologically more immature as compared with those observed in controls, namely the number, length and branching of cellular processes were substantially reduced. GC/O1 immunoreactivity was used to assess process branching using Sholl analyses, a quantitative method for radial distribution of dendritic morphology and complexity (Fig.5i). There was a significant reduction in the number of intersections of processes in Hdhd•hyp GC/O1+ oligodendrocytes compared to controls (two-way ANOVA F(13, 952) = 6.104, p < 0.0001, n = 743 and 1376 Hdhd•hyp and CTL oligodendrocytes, respectively). Normally, after 10 DIV, morphologically mature oligodendrocytes begin to express the myelin protein, MBP. Although the fraction of GC/O1+ oligodendrocytes expressing MBP was comparable between experimental mice (Χ2(1) = 3.47, p = 0.06, h), MBP expression was unexpectedly observed in very immature Hdhd•hyp oligodendrocytes species (bipolar precursors with very short processes; f vs g). Overall, these findings reveal important deficits in Hdhd•hyp oligodendrocytes, from impairments in the transition from proliferating progenitors to post-mitotic precursors, as well as in subsequent morphological maturation and myelination.
Figure 5. Loss of Htt function leads to abnormalities in oligodendrocyte maturation.
The lateral ganglionic eminences (LGE) of E12.5 embryos were dissected, and associated NSCs clonally expanded under mitogenic conditions, and induced to undergo oligodendrocyte differentiation on plated cells by bFGF withdrawal and CNTF addition. Preparations represent the pool of two independent biological replicates per experimental group. a-d depicts representative oligodendrocytes species after 10 DIV in differentiation conditions and stained against O4 and NG2 (a and b) and GC/O1 (c and d). e depicts the fraction of immunoreactive NG2, O4 and GC/O1 oligodendrocyte positive species. f-h depicts the fraction of GC/O1+ oligodendrocytes co-expressing MBP after 10 DIV. i depicts the Sholl profiles of oligodendrocytes: compared with CTL, the number of intersecting processes (indicative of oligodendrocyte branching and length) is significantly reduced in Hdhd•hyp specimens. Error bars in e/h and i correspond to ±95%CI and ±SEM, respectively. Blue staining in a-d and f-g correspond to DAPI. Scale bars in d and g correspond to 100 and 15 μm, respectively.
4. Discussion
This study demonstrates that loss of Htt function during neural development leads to HD-like neurological abnormalities in mice. The evidence is that mice with reduced expression of Htt during embryonic development until PND21 exhibit early myelination abnormalities and reactive gliosis, and late cortical and striatal neuropathological abnormalities paralleling those described in HD. This study also shows that dysfunction of Htt causes discrete abnormalities in the developmental program of striatal neurogenesis and forebrain myelinogenesis.
By using a compound HdhneoQ20/null mouse carrying the CAG-CRE-Esr1, and administering tamoxifen at PND21, we were able to reconstitute the normal expression levels of Htt from the HdhneoQ20 allele, thus allowing us to examine the postnatal effects of the selective loss of Htt develop mental functions. This experimental approach allowed us to demonstrate that previous observations of neurological abnormalities in the HdhneoQ20/null hypomorph model are mostly the consequence of developmental impairments and not due to the ongoing reduction of Htt expression throughout life (Auerbach et al., 2001; White et al., 1997). This is also consistent with a previous study showing that full ablation of Htt in adult mice does not lead to neurological abnormalities (Huang et al., 2015). Further, our use of electron microscopic techniques and immunostaining approaches at discrete ages furnished several new observations that provide a more complete understanding of the role of loss-of-Htt functions during development for the occurrence of late-life cell death and WMT pathology. However, our findings of progressive neurological abnormalities in a loss of Htt developmental function model are at apparent odds with the absence of progression that was reported by Auerbach et al. 2001 (Auerbach et al., 2001). This discrepancy may stem from the fact that degeneration was not directly interrogated in their study; rather, it was indirectly examined by histological techniques (cresyl violet cell quantification). Further, one possibility is that the ongoing but reduced Htt expression present throughout life might have accounted for this disparity.
Previous studies have shown that ablation of normal Htt in mouse models is embryonically lethal (Duyao et al., 1995; Nasir et al., 1995), which highlight the seminal roles of this protein during development. However, the expression of Htt from a single non-pathogenic allele not only does not adversely affect normal development, but also is sufficient to sustain the steady state throughout postnatal life (Duyao et al., 1995). Conversely, although embryos expressing ~10% of the normal levels of wild-type Htt are viable, this reduction leads to developmental abnormalities that are associated with postnatal motoric disorders. Remarkably, similar to nullizygous mice, homozygosity for the mutant HdhneoQ111 hypomorphic allele, as well as compound heterozygosity for the mutant HdhneoQ111/null allele is embryonically lethal (Auerbach et al., 2001). This lethality can be explained by two non-mutually exclusive mechanisms: (1) gain of novel toxic functions by the presence of aberrant polyQ expansion, and (2) polyQ-dependent abrogation of key developmental functions of Htt. The fact that lethality of mutant hypomorph mice can be overcome by the presence of a single non-pathogenic allele, even at reduced expression levels (HdhneoQ20), strongly indicates that the polyQ expansion is interfering with an important developmental function(s) of Htt. This possible dominant-negative mechanism is also supported by the following facts: (1) the array of developmental and postnatal neurological deficits exhibited by Hdhd•hyp, HdhneoQ20/null, HdhneoQ111/neoQ20 and HdhneoQ50/neoQ20 are analogous; (2) the phenotype and course of neurological disorders in HdhneoQ50/neoQ20 and HdhneoQ111/neoQ20 mice are more aggressive than in HdhneoQ20/null, possibly reflecting a more severe loss of Htt developmental functions (Auerbach et al., 2001; White et al., 1997); (3) developmental abnormalities in Hdhd•hyp, HdhneoQ20/null and HdhneoQ111/neoQ20 parallel those described in a HD knock-in model (Auerbach et al., 2001; Molero et al., 2009; White et al., 1997); and (4) many stem cell-mediated developmental alterations associated with mHtt are recapitulated by wild-type Htt ablation (Nguyen et al., 2013a; Nguyen et al., 2013b). This putative dominant-negative mechanism may be secondary to polyQ-dependent aberrant splicing of Htt or aberrant cleavage of the protein resulting in a short amino-terminal fragment (Sathasivam et al., 2013; Zheng and Diamond, 2012). This may explain why R6/2 mice, a model expressing the exon 1 carrying the pathogenic expansion, develops HD-like symptoms most rapidly, even though the expression of this fragment occurs at a lower levels as compared with the endogenous gene (Carter et al., 1999; Li et al., 2005). This is further supported by the fact that heterozygous mice expressing a truncated form of endogenous Htt (Exons 1-4, 6), also display motor and neuropathological abnormalities (Nasir et al., 1995). Overall, these results indicate that the loss of normal Htt developmental functions, either by direct ablation or by means of the polyQ expansion, may disrupt the genesis of the cortico-basal system, ultimately leading to its dysfunction and cell death during postnatal life.
Cumulative investigations have uncovered multiple HD-associated cell-autonomous impairments in striatal MSNs that are detrimental for these cells, including transcriptional deregulation, neuronal excitotoxicity, mitochondrial dysfunction, activation of proteases, protein aggregation and deregulation of intracellular trafficking (Bithell et al., 2009; Cha, 2007; Chen, 2011; Cowan et al., 2008; Graham et al., 2009; Leavitt et al., 2006; McGuire et al., 2006; Mehta et al., 2013; Metzler et al., 2007; Qin and Gu, 2004; Rona-Voros and Weydt, 2010; Rosenstock et al., 2010; Sun et al., 2001; Yuen et al., 2012). However, none of these putative pathogenic mechanisms adequately explains the selective profiles of cellular vulnerability in HD, the defining hallmark of all neurodegenerative diseases. On the other hand, our study shows that, even in the absence of the pathogenic protein, Hdhd•hyp mice develop analogous profile of neurological and pathological abnormalities to those observed in HD during adult life. As an explanation, we suggest that the loss of Htt functions results in disruption of developmental neurogenesis and gliogenesis at discrete niches throughout the neuraxis leading to region-specific foci of stressed but metastable cells. Consequently, these cells may not be capable of properly responding to normal physiological stressors at a later time, which would make them more vulnerable to cell death (Kerschbamer and Biagioli, 2015; Mehler and Gokhan, 2001; Molero et al., 2009). Indeed, our previous findings of late-life motor, neurophysiological and pathological abnormalities in a mouse model that expresses mutant Htt only during development seem to lend support for this possibility (Molero et al., 2016). Indeed, this putative pathogenic mechanism associated with development impairments may not be unique to HD, as related findings has also been reported by manipulating the developmental expression of mutant ATXN1 and LRRK2 in SCA and Parkinson’s disease, respectively (Matikainen-Ankney et al., 2016; Serra et al., 2006). However, it is also feasible that additional pathological stress resulting from the ongoing expression of the mutant protein may further accelerate the demise of those “developmentally” vulnerable cells. Certainly, it is likely that brain regions displaying the earliest and greatest vulnerabilities to cell death result from the synergy of both developmental and postnatal insults.
Our findings may also have implications for juvenile Huntington’s disease (JHD), where this developmental component may play a key role. Individuals with JHD usually exhibit very large polyQ expansions (>60 repeats) (Ribai et al., 2007; Squitieri et al., 2006). It is possible that the expansion directly disrupts some developmental functions by altering the ability of Htt to associate with key protein partners (Li and Li, 2004). In this context, the longer the expansion, the greater the disruption of Htt when associated with a significant subset of its protein partners. Another possibility is that the polyQ expansion not only alters the function of its protein partners, but may also turn mHtt into a dominant-negative factor, further disrupting the developmental functions of the non-pathogenic allele (Feero and Hoffman, 1995; Rubinsztein, 2003). Therefore, both the HdhneoQ20/null model and its derivative, Hdhd•hyp, by depriving neural structures of adequate levels of Htt during development, may be imitating the severe loss-of-Htt function of the long polyQ expansions associated with JHD. Indeed, the Hdhd•hyp model exhibits several characteristic features of JHD, including reduced body mass index (Tereshchenko et al., 2015), seizures (Cloud et al., 2012), hindlimb stiffness and muscle atrophy (Aubeeluck and Brewer, 2008; Nicolas et al., 2011), extreme gliosis of the globus pallidum (Byers et al., 1973), and cerebellar involvement (Gonzalez-Alegre and Afifi, 2006; Ho et al., 1995).
Another interesting finding of this study was the detection of myelination abnormalities in Hdhd•hyp mice. Our data revealed myelination abnormalities in both motor cortex and striatal WMTs. Further, we demonstrated the presence of deficits in the elaboration of oligodendrocyte species tasked with myelinating axons of larger caliber within deep cortical layers. In addition, we observed that reduced expression of Htt within developing oligodendrocytes leads to stage-specific maturational abnormalities. These results have important implications for HD, as oligodendrocytes and WMT abnormalities are amongst the earliest, most stable and robust findings in HD. Although is not yet clear the exact role of these abnormalities in HD pathogenesis, they represent one of the best predictors of disease onset and are significantly correlated with cognitive impairments during the HD prodromal phase (Beglinger et al., 2005; Bohanna et al., 2011; Ciarmiello et al., 2006; Halliday et al., 1998; Stoffers et al., 2010). Indeed, oligodendrocyte dysfunction may affect axonal integrity, transport, structure, metabolism and ultimately, neuronal survival (Huang et al., 2015). Further, myelin breakdown may lead to failure of neurotransmission and associated excitotoxicity secondary to corresponding overstimulation by afferent feedback (Bartzokis et al., 2007). It is important to consider that WMT abnormalities in HD have been observed in association with an increase in the number of oligodendrocyte species (Gomez-Tortosa et al., 2001; Myers et al., 1991). In this regard, we did not detect changes in the overall number, but in the complement of post-mitotic oligodendrocytes subtypes. This apparent discrepancy may, in part, be due to differences in study design. In our study, the analyses interrogated post-mitotic oligodendrocytes using the lineage marker APC, whereas previous neuropathological studies have examined oligodendrocytes with cresyl-violet histological techniques (Gomez-Tortosa et al., 2001; Myers et al., 1991). Alternatively, it is also possible that intrinsic human vs mouse evolutionary differences in gliogenesis may also have accounted for this discrepancy (Freeman and Rowitch, 2013). Overall, our findings suggest that loss-of-Htt function during development results in impairments in gliogenesis that are associated with early deficits in myelinogenesis, and possibly in myelin breakdown and axonal pathology late in life. Indeed, it is highly likely that these deficits are contributing to neuronal death in Hdhd•hyp mice.
5. Conclusion
In summary, we have shown that the loss of Htt function during neural development leads to progressive HD-like striatal and cortical impairments during later life. These findings suggest that disruption of key functions of Htt at discrete developing neural niches may result in selective foci of enhanced vulnerability to cell death throughout the neuraxis. It is possible that additional postnatal physiological stress in Hdhd•hyp mice, or pathological stressors associated with the ongoing expression of mHtt, may further affect a developmentally metastable cell population, which would ultimately lead to its demise.
Supplementary Material
Highlights.
Postnatal effects of developmental loss-of-huntingtin (Htt) function were studied.
Developmental dysfunction of Htt leads to later striatal and cortical degeneration.
Neuropathological changes also included white matter tract impairments.
Disruption of Htt at developing neural niches enhances vulnerability to cell death.
ACKNOWLEDGMENTS
This work was supported by US National Institutes of Health grant NS073758 to A.E.M, grant NS071571 to M.F.M, grant HD071593 to A.E.M. and M.F.M., and the F. M. Kirby, Alpern Family, Harold and Isabel Feld and Roslyn and Leslie Goldstein Foundations to M.F.M. We are grateful to Dr. Marcy E. MacDonald (Center for Human Genetic Research, Massachusetts General Hospital) for sharing the Hdhnull and HdhneoQ20 mouse models utilized in these research studies. We thank Dr. Kazuaki Yoshikawa (Osaka University) for sharing the anti-Dlx2 antibody.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Achour M, et al. Neuronal identity genes regulated by super-enhancers are preferentially downregulated in the striatum of Huntington's disease mice. Hum Mol Genet. 2015;24:3481–96. doi: 10.1093/hmg/ddv099. [DOI] [PubMed] [Google Scholar]
- Aubeeluck A, Brewer H. Huntington's disease. Part 2: treatment and management issues in juvenile HD. Br J Nurs. 2008;17:260–3. doi: 10.12968/bjon.2008.17.4.28715. [DOI] [PubMed] [Google Scholar]
- Auerbach W, et al. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Human Molecular Genetics. 2001;10:2515–23. doi: 10.1093/hmg/10.22.2515. [DOI] [PubMed] [Google Scholar]
- Aylward EH, et al. Regional atrophy associated with cognitive and motor function in prodromal Huntington disease. J Huntingtons Dis. 2013;2:477–89. doi: 10.3233/JHD-130076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartzokis G, et al. Myelin breakdown and iron changes in Huntington's disease: pathogenesis and treatment implications. Neurochem Res. 2007;32:1655–64. doi: 10.1007/s11064-007-9352-7. [DOI] [PubMed] [Google Scholar]
- Beglinger LJ, et al. White matter volume and cognitive dysfunction in early Huntington's disease. Cognitive & Behavioral Neurology. 2005;18:102–7. doi: 10.1097/01.wnn.0000152205.79033.73. [DOI] [PubMed] [Google Scholar]
- Bithell A, et al. Transcriptional dysregulation of coding and non-coding genes in cellular models of Huntington's disease. Biochem Soc Trans. 2009;37:1270–5. doi: 10.1042/BST0371270. [DOI] [PubMed] [Google Scholar]
- Bohanna I, et al. Diffusion tensor imaging in Huntington's disease reveals distinct patterns of white matter degeneration associated with motor and cognitive deficits. Brain Imaging Behav. 2011;5:171–80. doi: 10.1007/s11682-011-9121-8. [DOI] [PubMed] [Google Scholar]
- Byers RK, et al. Huntington's disease in children. Neuropathologic study of four cases. Neurology. 1973;23:561–9. doi: 10.1212/wnl.23.6.561. [DOI] [PubMed] [Google Scholar]
- Carter RJ, et al. Characterization of progressive motor deficits in mice transgenic for the human Huntington's disease mutation. Journal of Neuroscience. 1999;19:3248–57. doi: 10.1523/JNEUROSCI.19-08-03248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JH. Transcriptional signatures in Huntington's disease. Prog Neurobiol. 2007;83:228–48. doi: 10.1016/j.pneurobio.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM. Mitochondrial dysfunction, metabolic deficits, and increased oxidative stress in Huntington's disease. Chang Gung Med J. 2011;34:135–52. [PubMed] [Google Scholar]
- Ciarmiello A, et al. Brain white-matter volume loss and glucose hypometabolism precede the clinical symptoms of Huntington's disease. Journal of Nuclear Medicine. 2006;47:215–22. [PubMed] [Google Scholar]
- Cloud LJ, et al. Seizures in juvenile Huntington's disease: frequency and characterization in a multicenter cohort. Mov Disord. 2012;27:1797–800. doi: 10.1002/mds.25237. [DOI] [PubMed] [Google Scholar]
- Cowan CM, et al. Polyglutamine-modulated striatal calpain activity in YAC transgenic huntington disease mouse model: impact on NMDA receptor function and toxicity. J Neurosci. 2008;28:12725–35. doi: 10.1523/JNEUROSCI.4619-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paola M, et al. MRI measures of corpus callosum iron and myelin in early Huntington's disease. Hum Brain Mapp. 2014;35:3143–51. doi: 10.1002/hbm.22391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao MP, et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–10. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- Feero W, Hoffman EP. Huntington's disease. Their loss is our gain? Curr Biol. 1995;5:1229–31. doi: 10.1016/s0960-9822(95)00244-2. [DOI] [PubMed] [Google Scholar]
- Flurkey K, et al. Mouse Models in Aging Research. 2007;III:637–672. [Google Scholar]
- Freeman MR, Rowitch DH. Evolving concepts of gliogenesis: a look way back and ahead to the next 25 years. Neuron. 2013;80:613–23. doi: 10.1016/j.neuron.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin JD, et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67:392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- Gomez-Tortosa E, et al. Quantitative neuropathological changes in presymptomatic Huntington's disease. Annals of Neurology. 2001;49:29–34. [PubMed] [Google Scholar]
- Gonzalez-Alegre P, Afifi AK. Clinical characteristics of childhood-onset (juvenile) Huntington disease: report of 12 patients and review of the literature. J Child Neurol. 2006;21:223–9. doi: 10.2310/7010.2006.00055. [DOI] [PubMed] [Google Scholar]
- Graham RK, et al. Differential susceptibility to excitotoxic stress in YAC128 mouse models of Huntington disease between initiation and progression of disease. J Neurosci. 2009;29:2193–204. doi: 10.1523/JNEUROSCI.5473-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory S, et al. Neuropsychiatry and White Matter Microstructure in Huntington's Disease. J Huntingtons Dis. 2015;4:239–49. doi: 10.3233/JHD-150160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulinello M, et al. Acquired infection with Toxoplasma gondii in adult mice results in sensorimotor deficits but normal cognitive behavior despite widespread brain pathology. Microbes Infect. 2010;12:528–37. doi: 10.1016/j.micinf.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulinello M, et al. Validation of a 2-day water maze protocol in mice. Behav Brain Res. 2009;196:220–7. doi: 10.1016/j.bbr.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday GM, et al. Regional specificity of brain atrophy in Huntington's disease. Experimental Neurology. 1998;154:663–72. doi: 10.1006/exnr.1998.6919. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–18. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Hedreen JC, et al. Neuronal loss in layers V and VI of cerebral cortex in Huntington's disease. Neuroscience Letters. 1991;133:257–61. doi: 10.1016/0304-3940(91)90583-f. [DOI] [PubMed] [Google Scholar]
- Ho VB, et al. Juvenile Huntington disease: CT and MR features. AJNR Am J Neuroradiol. 1995;16:1405–12. [PMC free article] [PubMed] [Google Scholar]
- Huang B, et al. Mutant huntingtin downregulates myelin regulatory factor-mediated myelin gene expression and affects mature oligodendrocytes. Neuron. 2015;85:1212–26. doi: 10.1016/j.neuron.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, et al. Cellular and molecular aspects of striatal development. Brain Res Bull. 2001;55:533–40. doi: 10.1016/s0361-9230(01)00555-x. [DOI] [PubMed] [Google Scholar]
- Jin J, et al. Interrogation of brain miRNA and mRNA expression profiles reveals a molecular regulatory network that is perturbed by mutant huntingtin. J Neurochem. 2012;123:477–90. doi: 10.1111/j.1471-4159.2012.07925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, et al. Early white matter abnormalities, progressive brain pathology and motor deficits in a novel knock-in mouse model of Huntington's disease. Hum Mol Genet. 2015;24:2508–27. doi: 10.1093/hmg/ddv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschbamer E, Biagioli M. Huntington's Disease as Neurodevelopmental Disorder: Altered Chromatin Regulation, Coding, and Non-Coding RNA Transcription. Front Neurosci. 2015;9:509. doi: 10.3389/fnins.2015.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N, et al. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nature Neuroscience. 2006;9:173–9. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EH, et al. Cortical interneuron loss and symptom heterogeneity in Huntington disease. Ann Neurol. 2014;75:717–27. doi: 10.1002/ana.24162. [DOI] [PubMed] [Google Scholar]
- Labadorf A, et al. RNA Sequence Analysis of Human Huntington Disease Brain Reveals an Extensive Increase in Inflammatory and Developmental Gene Expression. PLoS One. 2015;10:e0143563. doi: 10.1371/journal.pone.0143563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt BR, et al. Wild-type huntingtin protects neurons from excitotoxicity. Journal of Neurochemistry. 2006;96:1121–9. doi: 10.1111/j.1471-4159.2005.03605.x. [DOI] [PubMed] [Google Scholar]
- Li JY, et al. The use of the R6 transgenic mouse models of Huntington's disease in attempts to develop novel therapeutic strategies. NeuroRx. 2005;2:447–64. doi: 10.1602/neurorx.2.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 2004;20:146–54. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Louis SA, et al. Enumeration of neural stem and progenitor cells in the neural colony-forming cell assay. Stem Cells. 2008;26:988–96. doi: 10.1634/stemcells.2007-0867. [DOI] [PubMed] [Google Scholar]
- Matikainen-Ankney BA, et al. Altered Development of Synapse Structure and Function in Striatum Caused by Parkinson's Disease-Linked LRRK2-G2019S Mutation. J Neurosci. 2016;36:7128–41. doi: 10.1523/JNEUROSCI.3314-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire JR, et al. Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. Journal of Biological Chemistry. 2006;281:3552–9. doi: 10.1074/jbc.M509806200. [DOI] [PubMed] [Google Scholar]
- Mehler MF, Gokhan S. Developmental mechanisms in the pathogenesis of neurodegenerative diseases. Progress in Neurobiology. 2001;63:337–63. doi: 10.1016/s0301-0082(00)00052-6. [DOI] [PubMed] [Google Scholar]
- Mehta A, et al. Excitotoxicity: bridge to various triggers in neurodegenerative disorders. Eur J Pharmacol. 2013;698:6–18. doi: 10.1016/j.ejphar.2012.10.032. [DOI] [PubMed] [Google Scholar]
- Metzler M, et al. NMDA receptor function and NMDA receptor-dependent phosphorylation of huntingtin is altered by the endocytic protein HIP1. Journal of Neuroscience. 2007;27:2298–308. doi: 10.1523/JNEUROSCI.5175-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molero AE, et al. Selective expression of mutant huntingtin during development recapitulates characteristic features of Huntington's disease. Proc Natl Acad Sci U S A. 2016;113:5736–41. doi: 10.1073/pnas.1603871113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molero AE, et al. Impairment of developmental stem cell-mediated striatal neurogenesis and pluripotency genes in a knock-in model of Huntington's disease. Proc Natl Acad Sci U S A. 2009;106:21900–5. doi: 10.1073/pnas.0912171106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Calavita M, et al. Mutant huntingtin affects cortical progenitor cell division and development of the mouse neocortex. J Neurosci. 2014;34:10034–40. doi: 10.1523/JNEUROSCI.0715-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtie JC, et al. Morphometric analysis of oligodendrocytes in the adult mouse frontal cortex. J Neurosci Res. 2007;85:2080–6. doi: 10.1002/jnr.21339. [DOI] [PubMed] [Google Scholar]
- Myers RH, et al. Decreased neuronal and increased oligodendroglial densities in Huntington's disease caudate nucleus. Journal of Neuropathology & Experimental Neurology. 1991;50:729–42. doi: 10.1097/00005072-199111000-00005. [DOI] [PubMed] [Google Scholar]
- Nasir J, et al. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–23. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- Ng CW, et al. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc Natl Acad Sci U S A. 2013;110:2354–9. doi: 10.1073/pnas.1221292110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen GD, et al. Selective roles of normal and mutant huntingtin in neural induction and early neurogenesis. PLoS One. 2013a;8:e64368. doi: 10.1371/journal.pone.0064368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen GD, et al. Functions of huntingtin in germ layer specification and organogenesis. PLoS One. 2013b;8:e72698. doi: 10.1371/journal.pone.0072698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas G, et al. Juvenile Huntington disease in an 18-month-old boy revealed by global developmental delay and reduced cerebellar volume. Am J Med Genet A. 2011;155A:815–8. doi: 10.1002/ajmg.a.33911. [DOI] [PubMed] [Google Scholar]
- Nopoulos PC, et al. Smaller intracranial volume in prodromal Huntington's disease: evidence for abnormal neurodevelopment. Brain. 2011;134:137–42. doi: 10.1093/brain/awq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, et al. Brain structure in preclinical Huntington's disease. Biol Psychiatry. 2006;59:57–63. doi: 10.1016/j.biopsych.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Paulsen JS, et al. Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain Res Bull. 2010;82:201–7. doi: 10.1016/j.brainresbull.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, et al. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips O, et al. Deep white matter in Huntington's disease. PLoS One. 2014;9:e109676. doi: 10.1371/journal.pone.0109676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott AL, et al. Cortical inhibitory deficits in premanifest and early Huntington's disease. Behav Brain Res. 2015;296:311–317. doi: 10.1016/j.bbr.2015.09.030. [DOI] [PubMed] [Google Scholar]
- Qin ZH, Gu ZL. Huntingtin processing in pathogenesis of Huntington disease. Acta Pharmacologica Sinica. 2004;25:1243–9. [PubMed] [Google Scholar]
- Raghunathan A, et al. Transient compartmental expression of a family of protein tyrosine phosphatases in the developing striatum. Brain Res Dev Brain Res. 1996;91:190–9. doi: 10.1016/0165-3806(95)00176-x. [DOI] [PubMed] [Google Scholar]
- Ribai P, et al. Psychiatric and cognitive difficulties as indicators of juvenile huntington disease onset in 29 patients. Arch Neurol. 2007;64:813–9. doi: 10.1001/archneur.64.6.813. [DOI] [PubMed] [Google Scholar]
- Ribchester RR, et al. Progressive abnormalities in skeletal muscle and neuromuscular junctions of transgenic mice expressing the Huntington's disease mutation. Eur J Neurosci. 2004;20:3092–114. doi: 10.1111/j.1460-9568.2004.03783.x. [DOI] [PubMed] [Google Scholar]
- Rogers DC, et al. Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm Genome. 1997;8:711–3. doi: 10.1007/s003359900551. [DOI] [PubMed] [Google Scholar]
- Rona-Voros K, Weydt P. The role of PGC-1alpha in the pathogenesis of neurodegenerative disorders. Current Drug Targets. 2010;11:1262–9. doi: 10.2174/1389450111007011262. [DOI] [PubMed] [Google Scholar]
- Rosenstock TR, et al. Mitochondrial-Associated Metabolic Changes and Neurodegeneration in Huntington's Disease - from Clinical Features to the Bench. Current Drug Targets. 2010;11:1218–1236. doi: 10.2174/1389450111007011218. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. How does the Huntington's disease mutation damage cells? Sci Aging Knowledge Environ. 2003;2003:PE26. doi: 10.1126/sageke.2003.37.pe26. [DOI] [PubMed] [Google Scholar]
- Sathasivam K, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci U S A. 2013;110:2366–70. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple BD, et al. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol. 2013;106-107:1–16. doi: 10.1016/j.pneurobio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra HG, et al. RORalpha-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell. 2006;127:697–708. doi: 10.1016/j.cell.2006.09.036. [DOI] [PubMed] [Google Scholar]
- Squitieri F, et al. Juvenile Huntington's disease: does a dosage-effect pathogenic mechanism differ from the classical adult disease? Mechanisms of Ageing & Development. 2006;127:208–12. doi: 10.1016/j.mad.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Stanley JL, et al. The mouse beam walking assay offers improved sensitivity over the mouse rotarod in determining motor coordination deficits induced by benzodiazepines. J Psychopharmacol. 2005;19:221–7. doi: 10.1177/0269881105051524. [DOI] [PubMed] [Google Scholar]
- Stoffers D, et al. Contrasting gray and white matter changes in preclinical Huntington disease: an MRI study. Neurology. 2010;74:1208–16. doi: 10.1212/WNL.0b013e3181d8c20a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, et al. Polyglutamine-expanded huntingtin promotes sensitization of N-methyl-Daspartate receptors via post-synaptic density 95. Journal of Biological Chemistry. 2001;276:24713–8. doi: 10.1074/jbc.M103501200. [DOI] [PubMed] [Google Scholar]
- Tereshchenko A, et al. Abnormal Weight and Body Mass Index in Children with Juvenile Huntington's Disease. J Huntingtons Dis. 2015;4:231–8. doi: 10.3233/JHD-150152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassy GS, et al. Distinct profiles of myelin distribution along single axons of pyramidal neurons in the neocortex. Science. 2014;344:319–24. doi: 10.1126/science.1249766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooy D, Fishell G. Neuronal birthdate underlies the development of striatal compartments. Brain Research. 1987;401:155–61. doi: 10.1016/0006-8993(87)91176-0. [DOI] [PubMed] [Google Scholar]
- Vinet J, et al. Subclasses of oligodendrocytes populate the mouse hippocampus. Eur J Neurosci. 2010;31:425–38. doi: 10.1111/j.1460-9568.2010.07082.x. [DOI] [PubMed] [Google Scholar]
- Waldvogel HJ, et al. The Neuropathology of Huntington's Disease. Curr Top Behav Neurosci. 2015;22:33–80. doi: 10.1007/7854_2014_354. [DOI] [PubMed] [Google Scholar]
- Wang G, et al. Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis. Proc Natl Acad Sci U S A. 2016;113:3359–64. doi: 10.1073/pnas.1524575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, et al. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nature Genetics. 1997;17:404–10. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- Yuen EY, et al. Disrupted GABAAR trafficking and synaptic inhibition in a mouse model of Huntington's disease. Neurobiol Dis. 2012;46:497–502. doi: 10.1016/j.nbd.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung SY, et al. Differential modulation of BMP signaling promotes the elaboration of cerebral cortical GABAergic neurons or oligodendrocytes from a common sonic hedgehog-responsive ventral forebrain progenitor species. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16273–8. doi: 10.1073/pnas.232586699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Diamond MI. Huntington disease and the huntingtin protein. Prog Mol Biol Transl Sci. 2012;107:189–214. doi: 10.1016/B978-0-12-385883-2.00010-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.