Abstract
Radiotherapy is the standard therapy for nasopharyngeal carcinoma (NPC); however, radioresistance can hinder successful treatment. Here, we report that miR-24 acts as a tumor suppressor and radiosensitizer in NPC cells and xenografts by targeting Jab1/CSN5. Although accumulating evidence has shown that Jab1/CSN5 functions as an oncoprotein in human cancers, its regulation through miRs has not been described. In this study, we found that Jab1/CSN5 functioned in a manner opposite that of miR-24 in NPC tumorigenesis and radioresistance. We demonstrated that miR-24 inhibits Jab1/CSN5 translation via direct binding to its 3’UTR and 5’UTR, leading to tumor growth inhibition, and sensitizes NPC tumors to radiation in vivo. Furthermore, silencing Jab1/CSN5 phenocopied the function of miR-24 in NPC cells after ionizing radiation treatment, resulting in increased apoptosis. Finally, we analyzed 50 paired samples of primary and matched recurrent NPC tissues from 25 NPC patients and subjected them to high-throughput genomic quantitative nuclease protection assay for quantifying simultaneously miR and mRNA levels. Our results showed that miR-24 levels were significantly decreased in recurrent NPC and that levels of Jab1/CSN5, as its target, were higher than those in primary NPC. Together, our findings indicate that miR-24 inhibits NPC tumor growth and increases NPC radiosensitivity by directly regulating Jab1/CSN5 and that both miR-24 and Jab1/CSN5 can serve as prognostic markers for NPC recurrence; this, in turn, may provide a promising therapeutic strategy for reversing NPC radioresistance.
Keywords: nasopharyngeal carcinoma, radioresistance, microRNA, recurrence, tumorigenesis
Introduction
Nasopharyngeal carcinoma (NPC) is a head and neck malignancy with local invasion and early distant metastasis. It is endemic to Southern China, Southeast Asia, Northern Africa, and Alaska. Epstein-Barr virus infection, genetic susceptibility, and endemic environmental factors are believed to be the main etiologic factors of NPC 9, 41, 48. Owing to the distinct anatomical location (pharyngeal recess) and relative high radiosensitivity of NPC, radiotherapy is the mainstay treatment for primary NPC presenting at early stages. In recent decades, concurrent chemotherapy and radiotherapy have been recommended as a standard treatment for NPC patients at late stages 5. Nevertheless, local failure or regional failure presenting as persistent or recurrent tumor still occurs and radioresistance remains the main cause of NPC treatment failure 24. Therefore, it’s urgent to elucidate the mechanism of NPC radioresistance to identify biomarkers for clinical therapy optimization and to develop novel effective treatment strategies.
MicroRNAs (miRs) are a class of small non-coding RNAs (of about 22 nucleotides in length) that regulate levels of multiple proteins, primarily by binding to the 3’ untranslated region (UTR) of their target mRNAs and suppressing protein translation 3, 21, 25, 51. Mature miRs act as trans-acting factors capable of degrading or suppressing the expression of mRNA targets, and around 60% of genes may be regulated by miRs 2. miRs have emerged as important epigenetic regulators of gene encoding proteins, and aberrant miR expression has been found in many cancers including NPC 6. However, the role of miR expression in radioresistance remains to be characterized.
MiR-24 is clustered with miR-23 and miR-27 at two genomic loci: 1) the miR-24-1 gene cluster (chromosome 9q22.32), encoding miR-23b, −27b, and −24-1; and 2) the miR-24-2 gene cluster (chromosome 19p13.13), encoding miR-23a, −27a, and −24-2 5. Studies have shown that miR-24 can inhibit cancer cell proliferation 29 and promote cancer cell apoptosis 50, indicating its role as a tumor suppressor. However, miR-24 has been found to play different roles in several cancers. For example, it inhibits HeLa cell proliferation 7 and represses cell death in gastric cancers 45, showing its role as an onco-miR. It has been documented that miRs can be valuable in manipulating the tumor radiation response in the clinic to enhance susceptibility to radiation 28. Therefore in this study we investigated the potential role of miR-24 as a novel therapeutic target to reverse NPC radioresistance.
Jab1/CSN5 (c-Jun activation domain-binding protein-1, Jab1 hereafter) was originally identified as a c-Jun co-activator by our group 10 and subsequently discovered by other groups as the fifth member and an integral component of the constitutive photomorphogenic-9 (COP9) signalosome complex (CSN5) 47. As one of the eight core subunits (CSN1-CSN8 in order of descending size), it not only harbors the catalytic center of CSN isopeptidase activity but also stably exists independently of the CSN complex in vivo 47. It participates in multiple cellular processes, both as part of the CSN-associated complex and as free forms independently of the CSN complex 44.
Accumulating evidence has shown that Jab1 functions as an oncoprotein in human cancers. Jab1 is overexpressed in multiple human cancers, including hepatocellular carcinoma 14 and NPC 32 and its overexpression is positively correlated with poor survival rate in various human malignancies 40. Our group recently found that Jab1 contributes to NPC tumorigenesis and resistance to radiotherapy by promoting NPC cell growth, dysregulating NPC cell cycle progression, and inhibiting radiation-induced apoptosis 32, 33, 40. Previous investigation by our group showed that Jab1 deletion sensitized both primary embryonic fibroblasts and osteosarcoma cells to γ-radiation–induced apoptosis and increased spontaneous DNA damage and homologous recombination defects 43. These findings indicate the critical role of Jab1 in cancer development and radiotherapy failure.
Ionizing radiation (IR) damages cells by producing intermediate ions and free radicals, leading to increased DNA damage including DNA double-strand breaks and genomic instability 28. Increased DNA damage response has been reported to be one of the mechanisms of radioresistance 42. Jab1 is critically involved in DDR and is linked to the maintenance of genome integrity 31, 42, 43. Loss of Jab1 leads to spontaneous DNA breaks that are associated with increased expression of the histone γ-H2AX, initiating the recruitment of DDR proteins 43. In NPC cells, Jab1 deletion could reduce the capacity of NPC cells to repair DNA lesions by reducing Rad51 (a key protein in the HR repair pathway) expression 34. These findings suggest that Jab1 plays a crucial role in radioresistance via increasing DNA repair ability and decreasing IR-induced cell apoptosis, indicating that blocking Jab1 might be a feasible and appealing approach to sensitize NPC cells to radiotherapy.
To date, no study has explained why Jab1 is overexpressed or regulated in NPC. In this current study, we first demonstrated that miR-24 functioned as an NPC cell growth inhibitor and a radiosensitizer for NPC radiotherapy both in vitro and in vivo, which is the opposite of the effect of Jab1 in NPC. Therefore, we hypothesized that miR-24 might be the molecule upstream of Jab1 leading to its overexpression during radiotherapy and therefore to NPC radioresistance. To test our hypothesis, we investigated the correlation between miR-24 and Jab1 in vitro and in vivo and used the luciferase assay to validate the binding sites of miR-24 to Jab1. The inverse correlation between miR-24 and Jab1 was then confirmed by the high-throughput genomic quantitative nuclease protection (qNPA) in NPC primary and recurrent patient samples. In addition, miR-24 downregulation or Jab1 upregulation in NPC recurrent tissues compared with primary paired tissues is closely associated with a shorter interval from initial treatment to NPC recurrence, indicating potential roles of miR-24 and Jab1 as prognostic markers for NPC recurrence and in novel therapeutic strategies for treating NPC patients. However, whether miR-24 contributes to the progression of NPC or radioresistance and underlying mechanism remains poorly understood. In the present study, we showed that miR-24 inhibits NPC and increases NPC radiosensitivity by directly targeting Jab1, suggesting a tumor suppressor role for miR24 in NPC. Our finding suggested that the restoration of miR-24 levels in tumors provide a promising therapeutic strategy for NPC treatment.
Results
Low Expression of miR-24 Promotes NPC Cell Growth, and Resists NPC Cells to Ionizing Radiation
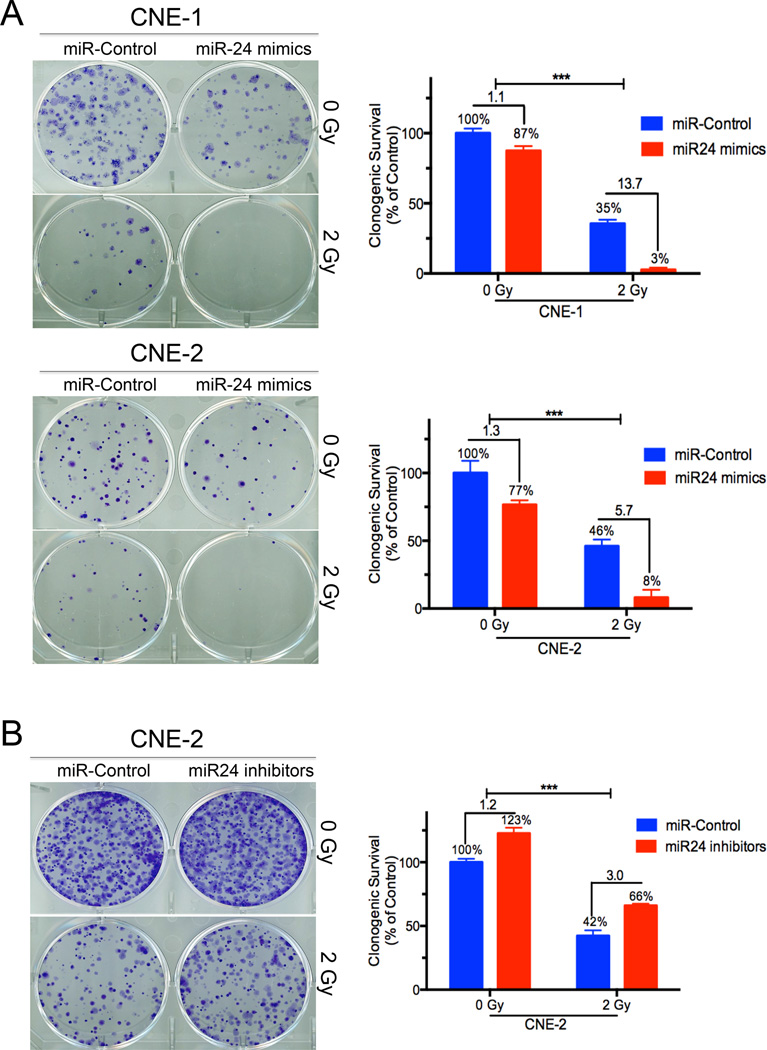
Our previous study has shown that miR-24 played an important role in NPC 46. To further identify the precise role of miR-24 in NPC, we overexpressed miR-24 by transfecting NPC cells (CNE-2, CNE-2R, CNE-1, HONE-1 and C666.1) with miR-24 mimics (Supplementary Figure 1A). NPC cell growth was inhibited (over 50%) (Supplementary Fig. 1B) and cell apoptosis was induced (about 6-fold) (Supplementary Fig. 1C) in a dose-dependent manner by ectopic miR-24 expression. Furthermore, the colony-forming ability of NPC cells was suppressed by miR-24 overexpression and was further inhibited when miR-24-overexpressing cells were treated with IR (Fig. 1A, p<0.001). Conversely, miR-24 inhibition with miR-24 inhibitors increased the colony-forming ability of NPC cells and significantly reduced NPC cell response to irradiation (Supplementary Fig. 1D, Fig. 1B). Taken together, our results showed that miR-24 functions as a tumor-suppressive miR in NPC and could sensitize NPC cells to irradiation in vitro.
Figure 1.
High expression of miR-24 inhibited NPC cell growth and Low expression of miR-24 promoted NPC cell growth in vitro. (A) Effects of overexpression of miR-24 on the colonogenic ability of CNE-1 and CNE-2 cells when combined with IR. Shown are percentages of colonies treated with miR-24 mimics (10 nM) in combination with IR relative to colonies treated with miR control (right panel). (B) Effects of inhibition of miR-24 on the clonogenic ability of CNE-2 cells when combined with IR. Shown are percentages of colonies treated with miR-24 inhibitors (10 nM) in combination with IR relative to colonies treated with miR control (right panel). Data represent means ± s.d. of three independent experiments. ***P<0.001, Student’s t-test.
MiR-24 Inhibits NPC Xenografts and Sensitizes NPC Tumors to Radiation in vivo
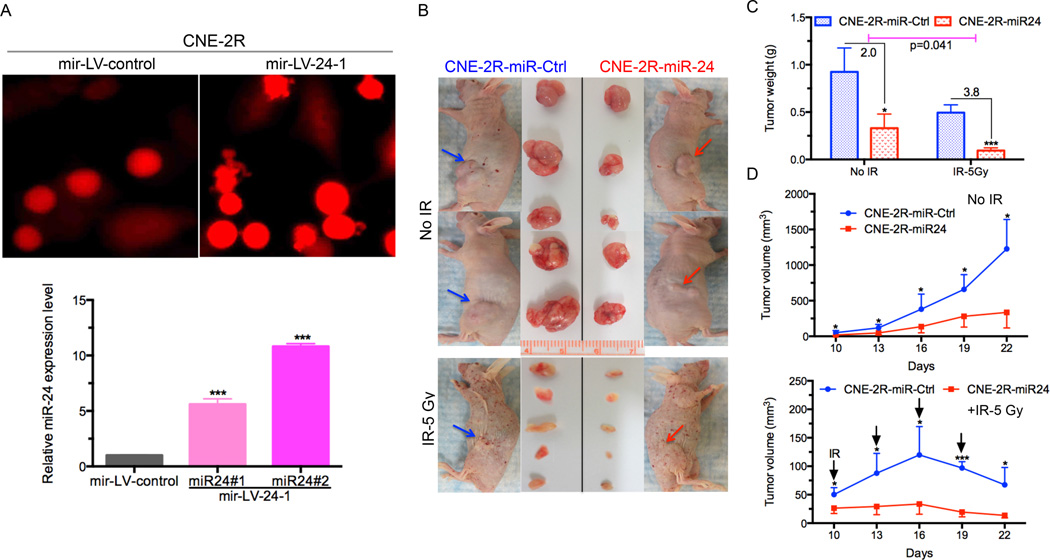
The in vitro data above showed the critical role of miR-24 in NPC cells. To further confirm the function of miR-24 in NPC, we established NPC cells stably overexpressing miR-24 by infecting CNE-2R with a lentivirus mir-LV-24-1 or mir-LV-control. Colonies were selected to confirm the miR-24 expression level by qRT-PCR analysis. Our results showed about a ten-fold increase in miR-24 levels in colony #2 and a six-fold increase in colony #1 compared with the cells expressing mir-LV-ctrl (Fig. 2A).
Figure 2.
MiR-24 inhibits NPC tumor growth and sensitize NPC tumor to radiation in vivo. (A) Representative photos of successful establishment of NPC stable cells (CNE-2R) overexpressing miR-24 by transducing with mir-LV-24-1 or mir-LV-control as control group (upper panel). qRT-PCR analysis of miR-24 expression level in two colonies 31 and #2 of CNE-2R cells that were transduced with a lentivirus mir-LV-24-1 for overexpressing miR-24. Results are presented as the mean ± s.d. of three independent experiments. Raw data were analyzed with 2−ΔΔCt. U6 snRNA was used as an internal control. ***P<0.001, Student’s t-test. (B) BALB/c athymic nu/nu mice with tumors and resected tumors are shown (n=5 per group). Red arrow: miR-24–overexpressing group injected in right flank. Blue arrow: miR-ctrl group injected in left flank. The upper panel is without IR treatment and the bottom panel is treated with IR (at a dose of 5Gy). (C) Weight of xenograft tumors is significantly reduced by miR-24 overexpressing xenografts than control, combined with IR or not. Tumors were weighed after resection. Results are expressed as the mean ± s.e.m. *, P <0.05; **, P <0.0001, Student’s t-test. (D) miR-24 expressing NPC xenografts induced tumor regression and slow the rate of tumor growth compared with miR-Control, throughout the course of the treatment. Growth curves of xenograft tumors after the injection with miR-24 overexpressing cells and mir-LV-ctrl cells combined with IR (bottom panel) or not (top panel). Tumor volume is reported as mean tumor volume ± s.d for each group of five mice. *, P <0.05; ***, P <0.0001, Student’s t-test.
We then transplanted the colony #2 cells stably overexpressing miR-24 into nude mice and treated the mice with IR when tumors were palpable (Fig. 2B). As shown in Fig. 2C, tumor weight was significantly lower (2-fold) compared with tumors from cells infected with mir-LV-control. Consistently, the xenograft mouse model also showed the suppressive effects of miR-24 on tumor growth (around 60% and 80% inhibition at day 19 and 22, respectively). When mice were treated with IR, tumor weight was significantly lower (3.8-fold) in the miR-24-overexpressing group than in the control group without IR. Moreover, tumor growth was reduced significantly (around 3-fold at day 16) when compared with cells infected with a control lentivirus (Fig. 2D, top panel). As expected, when combined with IR treatment, there was a significant difference (around 6-fold at day 16) between the miR-24–overexpressing group and controls (Fig. 2D, bottom panel). Our findings suggest that miR-24 functions as a tumor-suppressor in NPC tumorigenicity and may improve the efficacy of NPC radiotherapy.
Jab1 is a Direct Target of miR-24
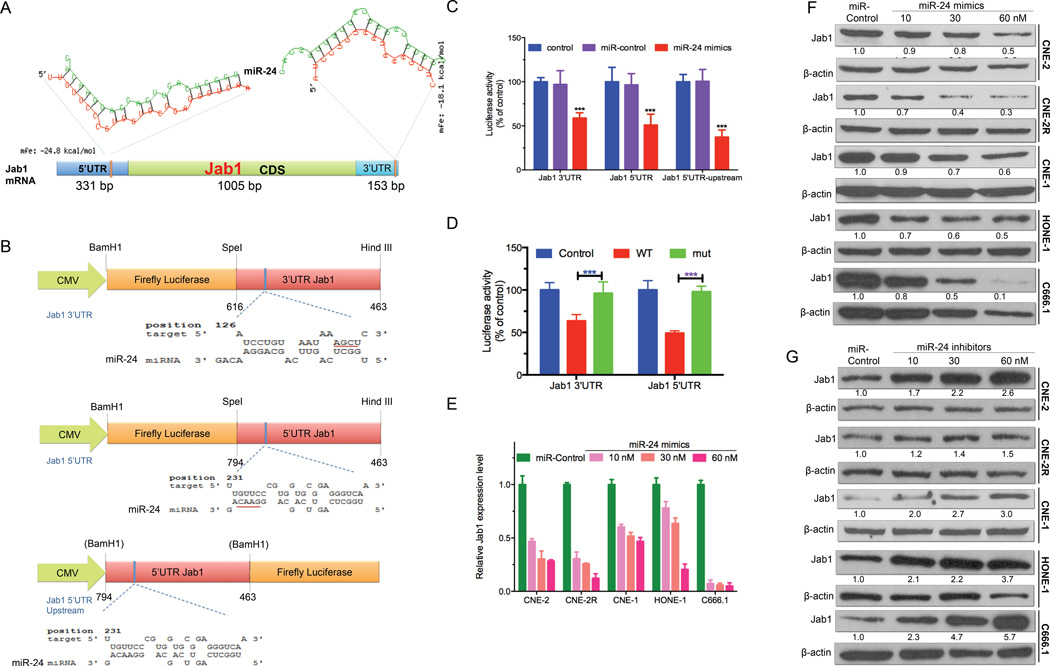
In vitro and in vivo studies demonstrated that ectopic restoration of miR-24 could significantly inhibit NPC cell growth and growth of NPC xenografts and could enhance the sensitivity of both NPC cells and xenografts to IR. How it functions in NPC, however, remains to be elucidated. Our previous findings indicated that Jab1 is a key regulator of several signaling pathways. It acts as a putative oncogene in NPC and increases NPC radioresistance 32, 33, 40. Interestingly, Jab1 has the opposite function of miR-24 in NPC. We therefore hypothesized that Jab1 might be a target of miR-24. To investigate this hypothesis, we used a computational target prediction algorithm, RNAhybrid, to identify the binding motifs of miR-24 to the 3’UTR or 5’UTR of Jab1 18, 38. MFEs (Minimum Free Energies) are determined from the optimization of duplexes between a miR and a putative target sequence. In our case, as shown in A, both 3’UTR [153 bp; MFE: −18.1 kcal/mol] and 5’UTR [331 bp; MFE: −24.8 kcal/mol] of Jab1 harbored a targeting sequence by miR-24 with rational and applicable MFEs.
Next, to validate that Jab1 is a direct target of miR-24, we used three miR luciferase constructs containing the 3’UTR or 5’UTR of Jab1, pMIR-luc-Jab1 3’UTR and pMIR-luc-Jab1 5’UTR, located at 1462–1479 nt and 231–255 nt, respectively, and also test the Jab1 5’UTR inserted upstream of the luciferase coding region, pMIR-luc-Jab1 5’UTR Upstream (Fig. 3B). Treatment of co-transfected CNE-2 cells with miR-24 mimics markedly reduced luciferase activity in cells with miR reporter pMIR-luc-Jab1 3’UTR (approximately 60%, Fig. 3C) or pMIR-luc-Jab1 5’UTR (approximately 50%, Fig. 3C), indicating that both the 3’UTR and 5’UTR are functional binding sites of miR-24. Repeating the experiments with the Jab1 5’UTR inserted upstream of the luciferase coding region gave similar results (approximately 50% inhibition, Fig. 3C). To further confirm the specific nature of the miR-24-mRNA interaction, the sequences within the pMIR-luc-Jab1 3’UTR (AGCU deletion) or pMIR-luc-Jab1 5’UTR (GUUC deletion) binding sites were mutated, resulting in the total loss of miR-24–mediated repression of luciferase activity (Fig. 3D). These findings support the notion that miR-24 directly targets both binding sites within the 3’UTR and 5’UTR of Jab1 mRNA to exert an inhibitory effect on Jab1 expression.
Figure 3.
MiR-24 directly targets Jab1 mRNA. (A) In silico prediction of the binding sites of miR-24 to Jab1 at 3’UTR and 5’UTR by RNAhybrid was shown. Mfe: minimum free energy. 5’UTR: 5’ untranslated region; CDS: coding sequence; 3’UTR: 3’ untranslated region. (B) Schematic map showing the luciferase reporter constructs pMIR-Luc-Jab1 3’UTR, pMIR-Luc-Jab1 5’UTR and pMIR-Luc-Jab1 5’UTR upstream and miR-24 binding sites position. Bam H1, SpeI and HindIII: restriction enzymes; blue bar: the exact predicted binding sites of miR-24 to Jab1. Red underline: deleted sequences for mutation. (C) Assay of luciferase activities when co-transfected with pMIR-Luc-Jab1 3’UTR, pMIR-Luc-Jab1 5’UTR or pMIR-Luc-Jab1 5’UTR upstream and miR-24 mimics or miRNA control. The control group was transfected with pMIR-Luc-Jab1 3’UTR, pMIR-Luc-Jab1 5’UTR or pMIR-Luc-Jab1 5’UTR upstream only. Data were normalized to control. ***P <0.0001, Student’s t-test. (D) Assay of luciferase activities when co-transfected with pMIR-Luc-Jab1 3’UTR, or -mut or pMIR-Luc-Jab1 5’UTR, or -mut and miR-24 mimics. The control group was transfected with pMIR-Luc-Jab1 3’UTR or pMIR-Luc-Jab1 5’UTR only. Data were normalized to control. ***P <0.0001, Student’s t-test. (E) qRT-PCR analysis of Jab1 expression after miR-24 overexpression in a dose-dependent manner was shown. Results are presented as the means ± s.d. of three independent experiments. Raw data were analyzed with 2−ΔΔCt. GAPDH was used as an internal control. (F) Western blot results of Jab1 protein level by transfecting with miR-24 mimics in a dose-dependent manner. β-Actin was used as an internal control. Data were quantified using ImageJ software. (G) Western blot results of Jab1 protein level by transfecting with miR-24 inhibitors in a dose-dependent manner. β-Actin was used as an internal control. Data were quantified using ImageJ software.
The functional significance of miR-24 was further evaluated under conditions of miR-24 overexpression and attenuation in multiple NPC cell lines. Forced expression of miR-24 mimics led to a robust increase in mature miR-24 levels (Supplementary Fig. 1A). Accordingly, western blot analysis confirmed that Jab1 expression decreased dramatically (up to 90%) at both the mRNA (Fig. 3E) and protein levels (Fig. 3F, Supplementary Fig. 2A and 2B) in a dose- and time-dependent manner, indicating that miR-24–mediated reduction of Jab1 is mediated by mRNA degradation. Conversely, there was a significant increase in Jab1 protein levels (Fig. 3G) following miR-24 inhibition (Supplementary Fig. 1E) in a dose-dependent manner. Together, these data indicate that miR-24 targets Jab1 by binding to its 3’UTR and 5’UTR, leading to Jab1 mRNA degradation and translational repression.
MiR-24 Expression Inversely Correlates with Jab1 Levels Both In Vitro and In Vivo
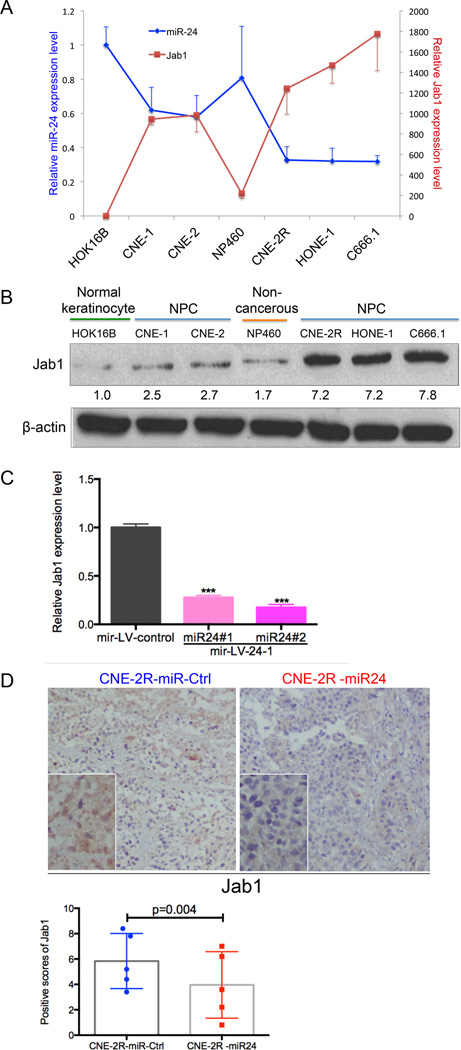
To further confirm the correlation between miR-24 and Jab1, quantitative real-time PCR (qRT-PCR) and western blot analysis were performed to measure Jab1 mRNA and protein expression levels in a panel of cell lines, including the normal keratinocyte cell line HOK16B, non-cancerous human immortalized nasopharyngeal epithelial cell line NP460, and five NPC cell lines (CNE-1, CNE-2, CNE2R, HONE-1 and C666.1). As shown in Fig. 4A and 4B, the expression levels of miR-24 and Jab1 were correlated negatively. The cells with the relatively low endogenous miR-24 expression level exhibited high levels of Jab1, and inversely. These data showed an in vitro inverse correlation between miR-24 and Jab1.
Figure 4.
Inverse correlation between the expression level of Jab1 and miR-24 both in vitro and in vivo. (A) Double y-axis diagram represented the relative miR-24 and Jab1 mRNA expression levels in seven cell lines: HOK16B, NP460, CNE-1, CNE-2, CNE-2R, HONE-1, and C666.1. U6 snRNA was used as an endogenous control for miR-24. Results are presented as the means ± s.d. of three independent experiments. (B) Western blotting assay of Jab1 protein level in seven cell lines: HOK16B, NP460, CNE-1, CNE-2, CNE-2R, HONE-1, and C666.1. Protein levels were quantified using ImageJ software. (C) qRT-PCR results for the Jab1 mRNA levels in the cells stably overexpressing miR-24. Data represent means ± s.d. of three independent experiments. ***P < 0.0001, Student’s t-test. (D) Representative pictures of Jab1 expression in the tissues resected from mice injected with cells overexpressing miR-24 or not (top panel). The bottom panel shows positive scores of Jab1 expression in the tissues resected from mice injected with CNE-2R overexpressing miR-24 or not. P = 0.004, Student’s t-test.
To confirm this inverse correlation in vivo, we tested the Jab1 level in NPC cells infected with lentivirus expressing miR-24. Western blot analysis confirmed that stable expression of miR-24 downregulated Jab1 levels compared to control (Fig. 4C). We next evaluated the impact of miR-24 stable expression on xenograft tumor formation. Xenograft tumors overexpressing miR-24 showed a significant reduction in Jab1 levels compared to control-miR by immunohistochemical analysis (Fig. 2, Fig. 4D; P = 0.004), providing evidence of the inverse correlation between miR-24 and Jab1 in vivo.
Jab1 Expression is Induced by IR, and Silencing Jab1 Replicates the Effects of miR-24 Under IR, Whereas Exogenous Expression of Jab1 without its 3’UTR and 5’UTR can Rescue the Inhibition Effect by miR-24
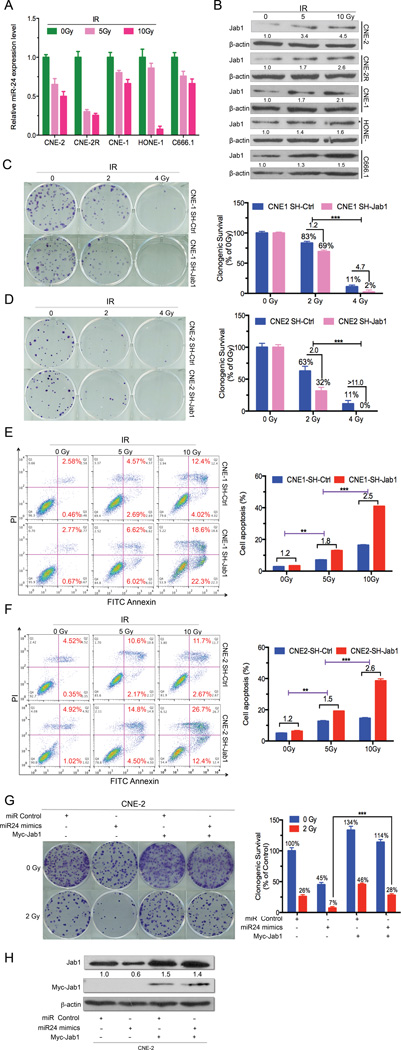
To explore whether Jab1 could mediate the function of miR-24 in NPC as a tumor suppressor and radiosensitizer, we detected both miR-24 and Jab1 expression after irradiation of NPC cells. MiR-24 was downregulated (over 2-fold) after irradiation in a dose-dependent manner, as shown by qRT-PCR analysis (Fig. 5A), whereas Jab1 was upregulated (over 2-fold) accordingly at the protein level (Fig. 5B). This finding suggests that the Jab1 increase under IR might be due to the inhibition of miR-24 post-irradiation. To further identify the role of Jab1 in mediating miR-24 function under IR, NPC cells (CNE-1 and CNE-2) were infected with a lentivirus expressing an shRNA targeting Jab1 (Supplementary Fig. 2C). After treatment with IR, loss of Jab1 increased NPC cell response to radiation in both CNE-1 and CNE-2 cells (Fig. 5C and 5D, p<0.001), consistent with the function of miR-24 (Fig. 1B). Additionally, cell apoptosis was further increased (around 2-fold) in Jab1-deleted cells (Fig. 5E and 5F) and miR-24–overexpressing cells (Fig. 1C). Interestingly, our colony formation assay data revealed that the inhibition effect of Jab1 by miR-24 (under IR or not) could be rescued significantly (0Gy: 1.2 fold vs 2.2 fold; 2Gy: 1.6 fold vs 3.7 fold. Fig 5G, p<0.001) when overexpressed exogenous Jab1 without its 3’UTR and 5’UTR (Fig. 5H). Taken together, our present evidences showed that miR-24 exerts its effect on NPC tumorigenesis and radioresistance by targeting Jab1.
Figure 5.
Jab1 expression is induced and its deletion, or exogenous expression increases NPC radiosensitivity, or rescue under ionizing radiation (IR). (A) MiR-24 expression is downregulated under IR treatment. qRT-PCR results for miR-24 expression after treatment with ionizing radiation (IR) (0, 5, or 10 Gy). Raw data were analyzed with 2−ΔΔCt. Data represent means ± s.d. of three independent experiments. U6 snRNA was used as an internal control. (B) Western blot results of Jab1 expression when cells were treated with IR in a dose-dependent manner. β-Actin was used as an internal control. Data were quantified using ImageJ software. (C and D) Effects of Jab1 deletion on the clonogenic ability of CNE-1 and CNE-2 cells when treated with IR in a dose-dependent manner. Shown are representative examples of colony-formation assays in CNE-1 and CNE-2 cells (left panel) with Jab1 deletion or not when cells were treated with IR. Percentage of colonies treated with IR (2 and 4 Gy) relative to colonies without IR treatment (right panel). Data are means ± s.d. of three independent experiments. ***P<0.001, Student’s t-test. (E and F) Representative results of cell apoptosis assay by flow cytometry in NPC cells with Jab1 deletion or not (left panel) treated with IR (0, 5, and 10 Gy). Red numbers indicate percentages of apoptosis. Quantification of the percentage of apoptotic cells (right panel) is shown. Data represent means ± s.d. of three independent experiments. **P<0.01, ***P<0.001, Student’s t-test. (G) Rescue experiment. Effects of exogenous Jab1 on the clonogenic ability of CNE-2 cells. Shown are representative examples of colony-formation assays with CNE-2 cells (left panel) for the rescue effect of exogenous Jab1 with or without IR. Percentage of colonies treated with IR (2 Gy) relative to colonies without IR treatment (right panel). Data are means ± s.d. of three independent experiments. ***P<0.001, Student’s t-test. (H) Westernblot results for exogenous (Myc-Jab1) and endogenous Jab1 protein levels following transfection with miR-24 mimics or Myc-Jab1 plasmids only, or both in NPC cells (CNE-2). β-Actin was used as an internal control. Data were quantified using ImageJ software.
Jab1 Expression in NPC Patients Inversely Correlated with miR-24, and Both Associated with NPC Recurrence
To reduce genetic variation between NPC patients, 50 samples from 25 NPC patients (paired primary and matched recurrent NPC tissues from the same patients) were collected and subjected to a novel microarray analysis, the high-throughput genomic quantitative nuclease protection assay (qNPA, from HTG Molecular) for quantifying simultaneously miR and mRNA levels from the same patient sample, to further validate the miR-24–Jab1 pathway in NPC. qNPA is a well-established approach for quantifying gene expression (including mRNAs and miRs) 35. Due to the scant biopsy material from NPC patients, qNPA is an optimal approach for measuring the miRs and gene expression levels on the same sample using FFPE tissues.
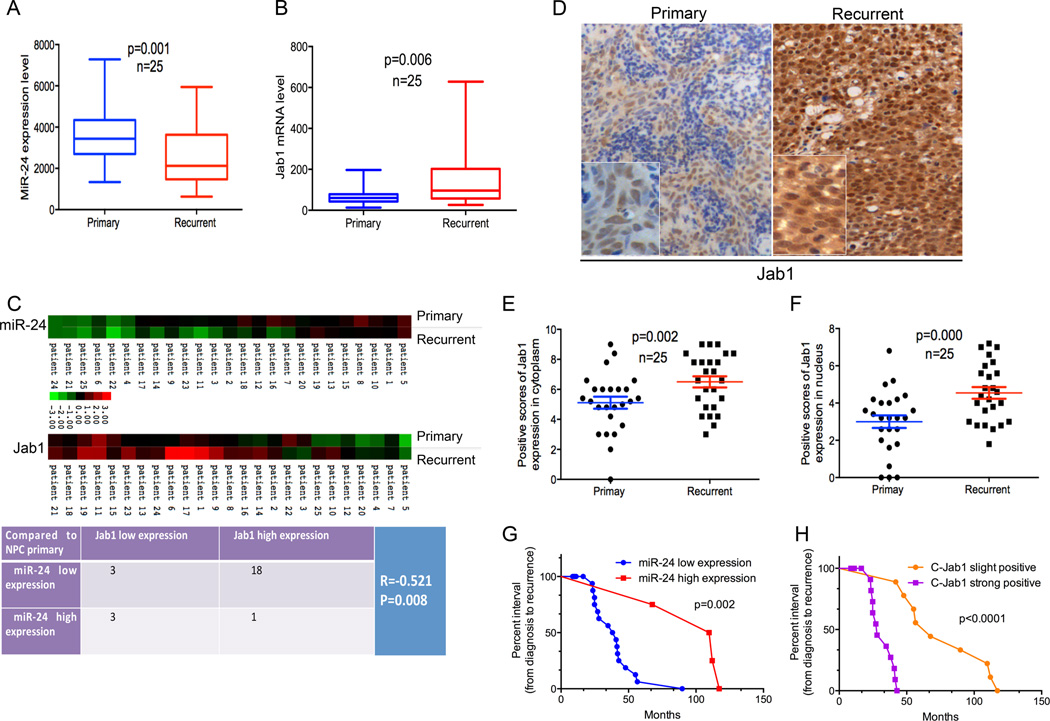
Of importance, the expression level of miR-24 was found to be significantly lower in recurrent NPC tissues than in the paired primary tissues from the same patients (Fig. 6A, P = 0.001), with a lower expression level observed in 84% (N=21/25) of recurrent NPC tissues compared with matched primary tissues (Supplementary Fig. 3A). Also, the Jab1 mRNA expression level was found to be significantly higher in recurrent NPC tissues than in the paired primary tissues (Fig. 6B, P = 0.006), with a higher level seen in 88% (N=22/25) of recurrent NPC tissues compared with matched primary tissues (Supplementary Fig. 3B, 22/25), thus, revealing an inverse correlation between miR-24 and Jab1 in NPC patients. To further correlate the expression of miR-24 and Jab1 mRNA from the same samples, we utilized a heat-map to show the association more clearly (Fig. 6C). The Spearman correlation test results indicated a strong inverse correlation between miR-24 and Jab1 mRNA levels in NPC patient samples (Fig. 6C, R = −0.521, P = 0.008). The detailed inverse correlation between miR-24 and Jab1 from NPC samples is also shown in Supplementary Fig. 3C. All together, these data provide solid evidence that elevated Jab1 expression negatively correlated with reduced miR-24 expression.
Figure 6.
Inverse correlation between miR-24 and Jab1 in NPC patient samples and closely associated with a shorter interval from initial treatment to NPC recurrence. (A) qNPA analysis of miR-24 levels in both primary and matched recurrent NPC samples from 25 patients was shown. Experiments were performed in triplicate. The paired Student’s t-test was used for statistical analysis. (B) qNPA analysis of Jab1mRNA levels in primary and matched recurrent NPC samples from 25 patients. Experiments were performed in triplicate. The paired Student’s t-test was used for statistical analysis. (C) Heat-map showing the expression of miR-24 and Jab1 in 25 patient samples, comparing primary and recurrent NPC tissues (upper panel). The lowest expression is represented by green and the highest expression by red. Data were normalized to the median value by log2 before mapping using software programs Cluster and TreeView. The Spearman correlation test was used to validate the correlation between miR-24 and Jab1 expression in NPC primary and recurrent samples (bottom panel). (D) Representative immunohistochemistry photos of Jab1 protein level in primary and recurrent NPC tissues from patient samples. (E and F) The positive scores of Jab1 expression in cytoplasm and nucleus are shown. The paired Student’s t-test was used for statistical analysis. (G) Kaplan-Meier analysis of miR-24 expression, with the interval from initial treatment to NPC recurrence. MiR-24 high level is consistent with long interval. The log-rank test was used to perform statistical analysis. (H) Kaplan-Meier analysis of Jab1 expression, with the interval from initial treatment to NPC recurrence. Jab1 low protein level in cytoplasm is consistent with long interval. P-value is from log-rank analysis.
In terms of the Jab1 mRNA degradation and translational inhibition by miR-24 from in vitro data, we next detected the Jab1 protein level in NPC primary and recurrent samples from the 25 patients by immunohistochemical analysis (same patient set as qNPA patient set). Jab1 was expressed to a higher degree in both nucleus and cytoplasm in the recurrent NPC samples than in the primary NPC samples (Fig. 6D, 6E and 6F). This result is consistent with Jab1 mRNA expression levels, which were also inversely correlated with miR-24 expression in NPC patient samples, further confirming the translational repression of Jab1 by miR-24 in NPC patient samples.
To identify the crucial role of miR-24 and Jab1 in NPC recurrence, we performed Kaplan-Meier analyses of miR-24 and Jab1 expression, using the interval from initial treatment to NPC recurrence. MiR-24 was positively correlated with the interval from initial treatment to recurrence (Fig. 6G, Supplementary Table 1, P = 0.002). Although low Jab1 mRNA levels correlated positively with a longer interval to recurrence, the P-value was not significant (Supplementary Fig. 3D, Table 2). However, both Jab1 protein expression in nucleus (Supplementary Fig. 3E, Table 3, P = 0.000) and in cytoplasm (Fig. 6H, Supplementary Table 4, P = 0.000) correlated negatively with the interval. Herein we report for the first time that miR-24 downregulation or Jab1 upregulation at the protein level in NPC recurrent tissues is closely associated with a shorter interval from initial treatment to NPC recurrence, indicating the potential roles of miR-24 and Jab1 as prognostic markers for NPC recurrence. Together, our findings define a central role for miR-24 as a tumor-suppressive miR in NPC and suggest its use in novel strategies for treatment of this cancer.
Discussion
NPC radioresistance following radiotherapy is the major cause for NPC treatment failure 24. Studies have reported that miR expression changes upon IR treatment in different cell types. Investigation of the use of miRs as drugs or drug targets against tumors is under way 13, 16. A few molecular-targeting drugs have shown clinical activity against advanced NPC 23, and use of miR as a therapeutic strategy is a promising strategy. It has been reported that lentiviral vectors have been applied successfully to deliver miRs for gene therapy of NPC. Silencing the radioresistance manganese superoxide dismutase (SOD2) by miR-155 could reduce NPC resistance to IR 37, and lenti-miR-26a inhibited the tumorigenicity of NPC cells in nude mice 26. These studies indicate the potential application of miRs in increasing NPC sensitivity to radiotherapy and inhibiting NPC development.
Sufficient evidence shows that a subset of tumors, such as colorectal and gastric cancer, has reduced levels of miR-24, indicating its potential role as a tumor-suppressive miR 8, which is consistent with our results in NPC model. MiR-24 overexpression could induce apoptosis in endothelial cells 12, facilitating TRAIL-induced apoptosis via restored function of the canonical apoptosis pathway by targeting the 3’UTR of XIAP, leading to the activation of intracellular caspase-8 50. In addition, miR-24 can also regulate cell cycle progression by inhibiting the cell cycle 19. Another group previously documented that overexpression of miR-24 led to higher chromosomal breaks and sensitivity to IR 20, supporting our findings in this NPC model. Certain miRs are decreased in response to IR, and overexpressing these miRs makes cells more sensitive to radiation, and vice versa. For example, let-7b is markedly downregulated following IR, and overexpression of let-7b increases radiosensitivity while inhibiting let-7b reduces radiosensitivity 49. Therefore, it is possible to develop miR-24 as an anti-tumor agent to enhance radiosensitivity, and improve the efficacy of radiotherapy.
On the other hand, we observed a lower expression level of miR-24 in recurrent NPC samples compared with matched primary samples from the same patients. Low expression of miR-24 was associated with a short interval from initial treatment to NPC recurrence, indicating the possible use of miR-24 as a biomarker for NPC recurrence. A lower miR-24 expression level in recurrent NPC samples compared with primary samples indicates a greater chance of reoccurrence. Detecting circulating cell-free miRs released from secretion of cancer cells would avoid the need for tumor tissue biopsies 39. Therefore, measurement of miR-24 expression in the patient’s body fluids may be a feasible method to predict reoccurrence in patients with primary NPC.
Jab1 is an oncoprotein and prognostic marker for multiple cancers. It plays an important role in cell proliferation, apoptosis, and the regulation of genomic stability and DNA repair. Dysregulation of Jab1 contributes to oncogenesis by functionally inactivating key negative regulatory proteins such as p27 17, leading to their degradation. Knockdown of Jab1 inhibits proliferation and induces apoptosis in hepatocellular carcinoma cells 22. In NPC, high expression of Jab1 32 contributed to an advanced stage of NPC and a low overall survival rate. Systemic delivery of the modified CSN5siRNA encapsulated in SNALP remarkably inhibited hepatic tumor growth in an orthotopic xenograft model of human liver cancer 22, indicating Jab1 to be a promising novel target for anti-cancer therapy.
Jab1 can also mediate the nuclear export and degradation of DNA damage and repair proteins. Double-strand breaks arising during meiotic recombination cannot be efficiently repaired in Jab1-mutant cells 11. DNA-damaging agents are commonly used as important cancer therapies, triggering DNA repair and then rendering the cells resistant to therapy. NPC is one of the typical examples where Jab1 is involved in NPC radioresistance 32.
In this study, we demonstrated for the first time that miR-24 binds to both the 3’UTR and 5’UTR of Jab1, leading to Jab1 mRNA degradation and translational repression. Although it is common for miRs to target 3’UTR of its targets, resulting to translational repression 19, we reported here that both the 3’UTR and 5’UTR of Jab1could be targeted by miR-24, and overexpression of Jab1 without both 3’UTR and 5’UTR could rescue the inhibition effect mediated by miR-24 with or without IR treatment. Translational inhibition is often a consequence of mRNA deadenylation and degradation 1, 4. The effects of miRs targeting 5’UTR of mRNA are contradictory. Jopling et al first reported in 2005 that miR-122 could bind to the 5’UTR of its targets, leading to translational inhibition 15. However, in 2008, miR-10a was reported to bind ribosomal protein mRNAs at the 5’UTR downstream of the conserved 5’TOP motif, enhancing their translation 30. This may be due to the fact that some miRs can induce translation of target mRNAs during cell cycle arrest.
Furthermore, observed miR-24 levels correlated inversely with Jab1 mRNA and protein levels in both NPC cells and NPC patients. MiR-24 expression was reduced while Jab1 was induced under IR treatment, and knockdown of Jab1 could largely imitate the function of miR-24. Additionally, a higher expression level of Jab1 was observed in recurrent NPC samples compared with the matched primary samples from the same patients, and Jab1 is associated with a short interval from initial treatment to NPC recurrence, representing a possible biomarker for NPC recurrence. All in all, we determined that Jab1 as an oncoprotein in NPC can be blocked by miR-24, presenting a novel approach for treating NPC and reversing NPC radioresistance. Other targets of miR-24, in addition to Jab1, are to be discovered to achieve its full activity. Our elucidation of the intricate network and roles of miR-24 in NPC development and therapy failure will allow for specific therapeutic approaches for NPC patients.
Materials and Methods
Patient Samples
Fifty formalin-fixed, paraffin-embedded (FFPE) human primary and matched recurrent NPC tissues were obtained from 25 NPC patients between 2001 and 2012 from First People’s Hospital of Foshan (30 samples) and Shantou Central Hospital (20 samples), both affiliated with Sun Yat-Sen University. Patients with both primary and recurrent NPC after radiotherapy were chosen to participate in this study. Ethics approval was given by these two hospitals, and fully informed consent was obtained from all patients before sample collection. In this study, primary NPC indicates a patient with a diagnosis of NPC for the first time, prior to any therapy. Recurrent NPC indicates the same patient who has NPC reoccurrence in the local region after radiotherapy. The clinicopathologic characteristics of the NPC patients are listed in Supplementary Table 5.
Cell Lines and Cell Culture
Cells of the normal keratinocyte cell line HOK16B has been described previously 32 and were cultured in keratinocyte-SFM basal medium containing 30 µg/mL bovine pituitary extract, 0.2 ng/mL epidermal growth factor (EGF), 5% fetal bovine serum (FBS), and 0.5% penicillin-streptomycin sulfate. Cells of the non-cancerous human immortalized nasopharyngeal epithelial cell line NP460 (a generous gift from Prof. George Sai Wah Tsao, Department of Anatomy, Faculty of Medicine, The University of Hong Kong) were cultured in keratinocyte-SFM basal medium combined with Epilife medium at a 1:1 ratio, supplemented with growth factors, 2% FBS and 0.5% penicillin-streptomycin sulfate. The human NPC cell lines CNE-1 (well-differentiated), CNE-2 (poorly differentiated), HONE-1 (poorly differentiated), and C666.1 (undifferentiated) have been described previously. CNE-2R, a radioresistant cells, was established by exposing CNE-2 cells to an increasing does of IR and has been described previously 36. All the NPC cells were cultured in RPMI 1640 medium containing 10% FBS and 0.5% penicillin-streptomycin sulfate. NPC cells (CNE-2R) stably overexpressing miR-24 or not by transduction with lentivirus (LV) expressing mir-LV-24-1 or mir-LV-control were maintained in medium containing puromycin at a concentration of 4 µg/mL. NPC cells (CNE-1, CNE-2) with Jab1 knockdown or not by infection with a lentivirus expressing a small hairpin RNA (shRNA) targeting sh-Jab1 or sh-Ctrl were maintained in medium containing puromycin at a concentration of 0.2 µg/mL puromycin. All cell lines were digested with 0.25% trypsin except for NP460 and C666.1, which were digested with 0.05% trypsin. Cells were stained with trypan blue and counted using a Countess automated cell counter (Invitrogen).
Reagents and Chemicals
All cell culture media were purchased from Mediatech and Gibco. Phosphate buffered saline (PBS) and chemiluminescent western blotting substrate were from ThermoFisher Scientific (Grand Island, NY, USA). Trypsin, hematoxylin, β-actin antibody (Cat# A5441) and the primers for reporter pMIR-Luc-Jab1 and mutation were from Sigma-Aldrich (St. Louis, MO, USA). Trizol, mirVana miR-24 mimics (Cat# 4464066) and mirVana mimic negative control Cat# 446458), and miRNA inhibitors (Cat# 17000) were from Ambion, Thermo Fisher Scientific. TaqMan microRNA assays of U6 snRNA (Cat# 00193), and hsa-miR-24 (Cat# 000402), TaqMan microRNA reverse transcription kit, TaqMan universal master mix II, and SYBR green PCR master mix were from Applied Biosystems (Foster City, CA, USA). The Miniprep kit, Plus regent, and Lipofectamine 2000 were from Invitrogen, Life Technologies (Carlsbad, CA, USA). The FITC annexin V apoptosis detection kit I was from BD Biosciences (San Jose, CA, USA). Lentiviral miRNA Expression system was from Biosettia (San Diego, CA, USA), and Matrigel matrix was from BD. The pMIR-REPORT System was from ThermoFisher Scientific. Endogenous restriction enzymes HindIII and SpeI were purchased from New England BioLabs (Ipswich, MA, USA). Purelink Genomic DNA mini kit, and Purelink Quick Plasmid Gel Extraction Kit were from Qiagen (Valencia, CA, USA). The QuickChange Multi Site-Directed Mutagenesis Kit was from Agilent Technologies (Santa Clara, CA, USA), the Rapid DNA Ligation Kit was from Roche (Indianapolis, IN, USA), and the Improm-II reverse transcription system and Dual-luciferase Reporter Assay System were from Promega (Madison, WI, USA). Jab1/Csn5 antibody for Western blot and immunohistochemistry studies was from Santa Cruz, Dallas, TX, USA (Cat# sc-13157). Myc antibody was from Roche (Cat#11667149001), and the Liquid DAB Substrate Chromogen System was from Dako (Carpinteria, CA, USA).
MiR-24 Mimic and miR-24 Inhibitor Transfection Studies
Exponentially growing NPC cells were plated onto six-well plates using medium without antibiotics 24 hours before transfection. mirVana miR-24 mimics, miRNA inhibitor, or miR control (Ambion) was transfected using Lipofectamine 2000 as a carrier at a 1:1 ratio to overexpress the mature miR-24 level, to inhibit the level, or as a control. Opti-MEM medium (Gibco) was replaced with regular culture medium without antibiotics after transfection for 6 hours. Cells were harvested to extract protein and RNA.
Quantitative Real-Time Polymerase Chain Reaction
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect miR-24 and Jab1 mRNA levels. Cells were harvested using Trizol (Ambion). For detection of mature miR-24, reverse transcription was performed to convert RNAs to cDNAs using a TaqMan miRNA reverse-transcription kit. For Jab1 detection, reverse transcription was performed using the ImProm-II reverse transcription system. PCR reaction mixtures containing TaqMan universal master mix II (miR-24) or SYBR Green PCR master mix (Jab1) and TaqMan miRNA or gene expression assays (Applied Biosystems) were used according to the manufacturer’s protocols (gradient S, Mastercycler, Eppendorf). Cycling variables were as follows: 50°C for 2 minutes and 95°C for 10 minutes followed by 40 cycles at 95°C (15 s) and annealing/extension at 60°C (1 minute). Data were analyzed with 2−ΔΔCt for relative changes in miR or gene expression. U6 snRNA and GAPDH were used to normalize miR-24 and Jab1 values, respectively.
Colony Formation Assay
The colony-formation assay was used to analyze cell growth after treatment with miR-24 mimics or IR. After approximately 10 days, cell colonies were washed with PBS, fixed with methanol, stained by 0.1% crystal violet, and scored by counting the number of colonies with an inverted microscope, using the standard definition that a colony consists of 50 or more cells. The inhibition ratios (%) of colony formation were calculated as the ratio of the indicated treatment group to the control group as follows: % inhibition ratio = 100% × Nt/Nc, where Nt is the number of the treatment group colonies and Nc is the number of control group colonies.
Irradiation Studies
In vitro irradiation was performed using a Gammacell 1000 irradiator (Nordion) with iodine-131 at MD Anderson Cancer Center. Cells were suspended in cell medium before irradiation at a rate of 283 rads/min. For in vivo irradiation, mice were irradiated in animal chambers using a Mark I irradiator (J. L. Shepherd and Associates) at MD Anderson Cancer Center, with an irradiation rate of 1059 rads/min.
Flow Cytometric Analysis of Cell Apoptosis
NPC cells were collected after treatment with miR-24 mimics or IR. Cells were stained with the annexin V-FITC apoptosis detection kit I (BD Biosciences) for cell apoptosis detection according to the manufacturer’s recommendations. Samples were measured with a FACScan flow cytometer (Becton Dickinson), and results were analyzed using FlowJo software.
Lentivirus Infection, Constitutive Expression and miR Stable Expression
CNE-2R cells were seeded (1.5 × 104 per well) onto a 12-well plate 1 day before transduction with hsa-mir-LV-24-1 or mir-LV-control. Medium was replaced by new medium containing 6 µg/mL Polybrene, and 100 µL of mir-LV-24-1 or mir-LV-control was added, followed by centrifugation at 1.000 × g for 40 minutes at room temperature. When cell confluence was greater than 50%, transduced cells were selected by puromycin at a concentration of 4 µg/mL. Transduction efficiency was measured under a microscope, and a single colony was chosen to determine the differential transduction efficiency of the stable cells overexpressing miR-24 or not.
Mouse Model
All experimental procedures using mice were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Texas MD Anderson Cancer Center. For the orthotopic xenograft tumor model, both flanks of 4- to 6-week-old male BALB/c athymic nu/nu mice were subcutaneously injected with 50 µL of 1.0 × 106 NPC CNE-2R cells stably overexpressing miR-24 or miR-Control (in PBS) combined with 50 µL of Matrigel. When the tumor mass became palpable (at day 10 after injection), mice were randomly divided into a control group and an IR-treated group (five mice per group). The investigator was blinded to the group allocation during experiment. Mice in the IR-treated group were irradiated with a dose of 5 Gy and re-irradiated every two other day. Experiments were repeated twice. Tumors were measured every 2 days with digital calipers, and the tumor volume (in mm3) was calculated using the formula: Volume = (L × W2)/2. Mice were killed on day 22 after injection, when some of the tumors reached the size limit set by the institutional animal care and use committee. Mice were killed by CO2 asphyxiation, and tumors were weighed after careful resection (two mice in the IR-treated group died 1 day before CO2 asphyxiation).
Western Blotting Analysis
Western blot analysis was performed as described previously 32. Cell Lysates were immunoblotted with purified mouse anti-human Jab1 antibody and β-actin antibody was used as the internal positive control. Finally, the signals were developed with chemiluminescent western blotting substrate (ThermoFisher Scientific), and ImageJ64 software (NIH) was used to quantify signal intensity.
Immunohistochemical Analysis
Jab1 expression at the protein level was detected immunohistochemically on 50 paraffin-embedded NPC tissues from the 25 patients. Briefly, deparaffinized sections were treated with 10 mM sodium citrate buffer (pH 6.0) for heat-induced retrieval of the antigen and then immersed in 3% hydrogen peroxide solution to inhibit endogenous peroxidase activity, followed by incubation of the sections in 5% bovine serum albumin to block nonspecific binding. The sections were then incubated with primary antibodies against Jab1 (1:150) at 4°C overnight and then incubated with biotinylated secondary antibody followed by the Liquid DAB Substrate Chromogen System according to the manufacturer’s instructions. Jab1 expression level was evaluated by counting at least 500 tumor cells in at least five representative high-power fields. The Jab1 staining in the nucleus and cytoplasm of the tumor cells were scored separately and scores combined for each case as described previously 27.
Computational Analysis of miR-24 Binding to Jab1
RNAhybrid can be used in a very simple version to find binding sites in a gene of interest that is posited to be regulated in the miRNA pathway. We therefore used RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html; offered by BiBiServ, Bielefeld University Bioinformatics Server), which is unique in offering a flexible online prediction, to predict the binding sites of miR-24 to Jab1 with the secondary structure and minimum free energies (MFEs). MFEs result from the optimization of duplexes between a miRNA and a putative target sequence 18, 38.
Vector Construction and Mutagenesis of Predicted Binding Sites of Jab1
To generate a luciferase reporter for evaluating miR activity, we amplified two fragments of Jab1 mRNA (3’UTR and 5’UTR) by PCR from genomic DNA isolated from NPC cell CNE-2. Inserts were retrieved with HindIII and SpeI and cloned into the same sites of pMIR-REPORT Lucirefase vector (Applied Biosystems) downstream of the firefly open reading frame (ORF) for the Jab1 3’UTR and 5’UTR and in the BamH1 site (pMIR-REPORT) upstream of the luciferase ORF for the Jab1 5’UTR Upstream. The primers for PCR amplification were as follows:
-
3’UTR: Sense: 5’ CGCACTAGTACAGTCTCTGAGAAGTACTTTACCTG 3’;
Anti-sense: 5’ GGTAAGCTTTTCATTTTAAAGAGCTTTATTACAGG 3’.
-
5’UTR: Sense: 5’ CGCACTAGTGACTATACCACTCCCATACCCT 3’;
Anti-sense: 5’ AATAAGCTTCGCCGAGGAAGCGGAGAA 3’.
-
5’UTR-Upstream: Sense: 5’ CGGGATCCGACTATACCACTC CCATACCCTA 3’;
Anti-sense: 5’ TCCGTGGATCCCGCCGAGGAAGCGGAGAAGTTG 3’.
The QuikChange Multi Site-Directed Mutagenesis Kit was used to generate the mutation in the predicted binding sites. Four nucleotides (AGCT) in the 3’UTR of the Jab1 gene or GGTC in the 5’UTR of Jab1 were deleted by PCR from the wild-type Jab1 gene. Both wild-type and mutant inserts were confirmed by sequencing (DeWalch Technologies). The primers for mutation were as follows:
3’UTR mutant: AGCT deletion:
Forward: 5’ CAAGGATTTATAATTATAGGACATTATTGAAATTTTAC 3’;
Reverse: 5’ GTAAAATTTCAATAATGTCCTATAATTATAAATCCTGTG 3’.
5’UTR mutant: GGTC deletion:
Forward: 5’ AAACAAGGGCACCACGCCTCTTCCGGGTGTGGGCCTT 3’;
Reverse: 5’ AAAGGCCCACACCCGGAAGAGGCGTGGTGCCCTTGTTT 3’.
Dual Luciferase Reporter Assay
Luciferase plasmids (pMIR) containing 5’-UTR or 3’-UTR of Jab1 were transiently transfected with miR-24 mimics into CNE-2 cells. Cells were seeded onto 24-well plates and co-transfected 24 hours later with pMIR-5’-UTR or 3’-UTR, 30 nmol/L miR-24 mimics, or miR control and a pRL plasmid control using Plus reagent following the manufacturer’s protocol. After 48 hours, cells were washed and resuspended in the lysis buffer, and the luciferase activity was assayed with a luminometer using the Dual-Light system (Promega, Madison, WI, USA). All experiments were performed in triplicate, and the results are presented as the means of these separate experiments.
Establishment of shRNA Stable Cells
To knockdown Jab1 in NPC cells, CNE1 and CNE2 cells were stably transfected with Jab1 shRNA (sh-Jab1) or control shRNA (sh-Cont) as described previously 33. Briefly, Jab1 shRNA oligonucleotides were cloned into a retrovirus pSIREN-RetroQ system (Clontech, Mountain View, CA, USA). The packaging cell line 293T was co-transfected with Jab1 shRNA-vector DNA along with the vectors pCGP and pVSVG using the Lipofectamine PLUS reagent. Stable clones were selected following treatment with puromycin at 0.8 µg/mL for 2 weeks. Positive clones were further confirmed by immunoblot analysis and maintained in 0.2 µg/mL puromycin.
Quantitative Nuclease-Protection Assay (qNPA)
Fifty total FFPE NPC samples (25 primary, 25 recurrent) were analyzed for miR and gene expression profiling (HTG Molecular qNPA service, Tucson, AZ, USA). Briefly, section areas were measured and scraped from the slides. Tissues were then lysed in HTG lysis buffer to a concentration of approximately 0.1 cm2 per well. Nuclease protection probes were added first to hybridize the samples, and S1 nuclease was then added to degrade the non-hybridized single-strand molecules. Samples were then transferred to the reader plate, followed by adding programming linkers for capturing specific nuclease protection probes (HTG Molecular). Detection linkers and biotinylated detection probes were then added one after the other to bind to the probes and detect the linkers. Avidin-horseradish peroxidase as substrate was finally added for the chemiluminescence detection. All samples were analyzed in triplicate, and Universal Human Reference RNA was analyzed in triplicate (100 ng/well) on each plate as a qNPA array plate run control.
Statistical Data Analysis
Statistical analysis was performed using the SPSS statistical software. Statistical evaluation for data analysis used Student’s t-test when there were only two groups (two sided). The paired t-test was used to calculate the P-value of miR-24 and Jab1 expression between primary and recurrent NPC tissues (two sided). The Spearman correlation test was used to calculate the correlation coefficient (r) and P-value for the correlation between miR-24 expression and Jab1 expression. Kaplan-Meier curves were drawn to show the association of miR-24 or Jab1 expression and the interval from initial treatment to recurrence, with P-values from log-rank analysis. All data are reported as means ± standard deviation. Differences between groups were considered significant statistically when P ≤ 0.05.
Supplementary Material
Acknowledgments
We thank Michael Worley and the Department of Scientific Publications at MD Anderson Cancer Center for editing the manuscript.
Funding: This study was supported by a fellowship from the China Scholarship Council (201206380043 to SW), the National Natural Science Foundation of China (81071837, 81372410, and 30670627 to HY), and the Scientific and Technological Project of Guangdong, China (2008A030201009 and 2010B050700016 to HY) and by grants from the National Cancer Institute (R01-CA90853 to FXC), the Sister Institution Network Fund (FXC), and The University of Texas MD Anderson Functional Proteomics Core Facility (NCI Cancer Center Support Grant CA16672).
Footnotes
Conflict of interest statement: The authors have no conflicts of interest to declare.
Authors’ Contributions
Conception and design: S. Wang, Y. Pan, H. Yang, F.X. Claret
Development of methodology: S. Wang, Y. Pan, H. Zhang R, Yang, F.X. Claret
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S. Wang, Y. Pan, R. Zhang, T. Xu, C. Wu, W. Wang, H. Huang, H. Yang, F.X. Claret
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S. Wang, F.X. Claret
Writing, review, and/or revision of the manuscript: S. Wang, Y. Pan, F.X. Claret
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): H. Yang, F.X. Claret
Study supervision: F.X. Claret
Other (helped with discussion and comments on the project): G. Calin
Supplementary information is available at Oncogene’s website.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. The New England journal of medicine. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 4.Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochemical Society transactions. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- 5.Chan AT, Gregoire V, Lefebvre JL, Licitra L, Felip E, Group E-E-EGW. Nasopharyngeal cancer: EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2010;(21 Suppl 5):v187–v189. doi: 10.1093/annonc/mdq186. [DOI] [PubMed] [Google Scholar]
- 6.Chen HC, Chen GH, Chen YH, Liao WL, Liu CY, Chang KP, et al. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. British journal of cancer. 2009;100:1002–1011. doi: 10.1038/sj.bjc.6604948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic acids research. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Molecular cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien YC, Chen JY, Liu MY, Yang HI, Hsu MM, Chen CJ, et al. Serologic markers of Epstein-Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. The New England journal of medicine. 2001;345:1877–1882. doi: 10.1056/NEJMoa011610. [DOI] [PubMed] [Google Scholar]
- 10.Claret FX, Hibi M, Dhut S, Toda T, Karin M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature. 1996;383:453–457. doi: 10.1038/383453a0. [DOI] [PubMed] [Google Scholar]
- 11.Doronkin S, Djagaeva I, Beckendorf SK. CSN5/Jab1 mutations affect axis formation in the Drosophila oocyte by activating a meiotic checkpoint. Development. 2002;129:5053–5064. doi: 10.1242/dev.129.21.5053. [DOI] [PubMed] [Google Scholar]
- 12.Fiedler J, Jazbutyte V, Kirchmaier BC, Gupta SK, Lorenzen J, Hartmann D, et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 2011;124:720–730. doi: 10.1161/CIRCULATIONAHA.111.039008. [DOI] [PubMed] [Google Scholar]
- 13.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature reviews Drug discovery. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu MC, Huang CC, Chang HC, Hu TH, Hung WC. Overexpression of Jab1 in hepatocellular carcinoma and its inhibition by peroxisome proliferator-activated receptor{gamma} ligands in vitro and in vivo. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:4045–4052. doi: 10.1158/1078-0432.CCR-07-5040. [DOI] [PubMed] [Google Scholar]
- 15.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 16.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouvaraki MA, Korapati AL, Rassidakis GZ, Tian L, Zhang Q, Chiao P, et al. Potential role of Jun activation domain-binding protein 1 as a negative regulator of p27kip1 in pancreatic adenocarcinoma. Cancer research. 2006;66:8581–8589. doi: 10.1158/0008-5472.CAN-06-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic acids research. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Molecular cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nature structural & molecular biology. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee RC, Feinbaum RL, Ambros V, The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 22.Lee YH, Judge AD, Seo D, Kitade M, Gomez-Quiroz LE, Ishikawa T, et al. Molecular targeting of CSN5 in human hepatocellular carcinoma: a mechanism of therapeutic response. Oncogene. 2011;30:4175–4184. doi: 10.1038/onc.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim WT, Ng QS, Ivy P, Leong SS, Singh O, Chowbay B, et al. A Phase II study of pazopanib in Asian patients with recurrent/metastatic nasopharyngeal carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:5481–5489. doi: 10.1158/1078-0432.CCR-10-3409. [DOI] [PubMed] [Google Scholar]
- 24.Liu SC, Tsang NM, Chiang WC, Chang KP, Hsueh C, Liang Y, et al. Leukemia inhibitory factor promotes nasopharyngeal carcinoma progression and radioresistance. The Journal of clinical investigation. 2013;123:5269–5283. doi: 10.1172/JCI63428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 26.Lu JA, He ML, Wang L, Chen Y, Liu XO, Dong Q, et al. MiR-26a Inhibits Cell Growth and Tumorigenesis of Nasopharyngeal Carcinoma through Repression of EZH2. Cancer research. 2011;71:225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 27.McCarty KS, Jr, Szabo E, Flowers JL, Cox EB, Leight GS, Miller L, et al. Use of a monoclonal anti-estrogen receptor antibody in the immunohistochemical evaluation of human tumors. Cancer research. 1986;46:4244s–4248s. [PubMed] [Google Scholar]
- 28.Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Current opinion in genetics & development. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13513–13518. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5 ‘ UTR of ribosomal protein mRNAs and enhances their translation. Molecular cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Pan Y, Claret FX. Targeting Jab1/CSN5 in nasopharyngeal carcinoma. Cancer letters. 2012;326:155–160. doi: 10.1016/j.canlet.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan Y, Zhang Q, Tian L, Wang X, Fan X, Zhang H, et al. Jab1/CSN5 negatively regulates p27 and plays a role in the pathogenesis of nasopharyngeal carcinoma. Cancer research. 2012;72:1890–1900. doi: 10.1158/0008-5472.CAN-11-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan Y, Zhang Q, Atsaves V, Yang H, Claret FX. Suppression of Jab1/CSN5 induces radio- and chemo-sensitivity in nasopharyngeal carcinoma through changes to the DNA damage and repair pathways. Oncogene. 2013;32:2756–2766. doi: 10.1038/onc.2012.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan Y, Yang H, Claret FX. Emerging roles of Jab1/CSN5 in DNA damage response, DNA repair, and cancer. Cancer biology & therapy. 2014;15:256–262. doi: 10.4161/cbt.27823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pechhold S, Stouffer M, Walker G, Martel R, Seligmann B, Hang Y, et al. Transcriptional analysis of intracytoplasmically stained, FACS-purified cells by high-throughput, quantitative nuclease protection. Nature biotechnology. 2009;27:1038–1042. doi: 10.1038/nbt.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu CJ, Liang ZH, Huang JL, Zhao RY, Su CH, Wang SM, et al. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell cycle. 2012;11:785–796. doi: 10.4161/cc.11.4.19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu YM, Zhao SP, Hong JD, Tang S. Radiosensitive gene therapy through imRNA expression for silencing manganese superoxide dismutase. J Cancer Res Clin. 2010;136:953–959. doi: 10.1007/s00432-009-0739-5. [DOI] [PubMed] [Google Scholar]
- 38.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature Reviews Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 40.Shackleford TJ, Claret FX. JAB1/CSN5: a new player in cell cycle control and cancer. Cell division. 2010;5:26. doi: 10.1186/1747-1028-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sham JS, Wei WI, Zong YS, Choy D, Guo YQ, Luo Y, et al. Detection of subclinical nasopharyngeal carcinoma by fibreoptic endoscopy and multiple biopsy. Lancet. 1990;335:371–374. doi: 10.1016/0140-6736(90)90206-k. [DOI] [PubMed] [Google Scholar]
- 42.Sorrells S, Carbonneau S, Harrington E, Chen AT, Hast B, Milash B, et al. Ccdc94 protects cells from ionizing radiation by inhibiting the expression of p53. PLoS genetics. 2012;8:e1002922. doi: 10.1371/journal.pgen.1002922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian L, Peng G, Parant JM, Leventaki V, Drakos E, Zhang Q, et al. Essential roles of Jab1 in cell survival, spontaneous DNA damage and DNA repair. Oncogene. 2010;29:6125–6137. doi: 10.1038/onc.2010.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomoda K, Kubota Y, Arata Y, Mori S, Maeda M, Tanaka T, et al. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. The Journal of biological chemistry. 2002;277:2302–2310. doi: 10.1074/jbc.M104431200. [DOI] [PubMed] [Google Scholar]
- 45.Walker JC, Harland RM. microRNA-24a is required to repress apoptosis in the developing neural retina. Genes & development. 2009;23:1046–1051. doi: 10.1101/gad.1777709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, Zhang R, Claret FX, Yang H. Involvement of microRNA-24 and DNA Methylation in Resistance of Nasopharyngeal Carcinoma to Ionizing Radiation. Molecular cancer therapeutics. 2014;13:3163–3174. doi: 10.1158/1535-7163.MCT-14-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei N, Deng XW. The COP9 signalosome. Annual review of cell and developmental biology. 2003;19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- 48.Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365:2041–2054. doi: 10.1016/S0140-6736(05)66698-6. [DOI] [PubMed] [Google Scholar]
- 49.Weidhaas JB, Babar L, Nallur SM, Trang P, Roush S, Boehm N, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer research. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Y, Tobin LA, Camps J, Wangsa D, Yang J, Rao M, et al. MicroRNA-24 regulates XIAP to reduce the apoptosis threshold in cancer cells. Oncogene. 2013;32:2442–2451. doi: 10.1038/onc.2012.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.