Abstract
Intellectual disability encompasses a large set of neurodevelopmental disorders of cognition that are more common in males than females. Although mutations in over 100 X-linked genes linked to intellectual disability have been identified, only a few X-linked intellectual disability proteins have been intensively studied. Hence, the molecular mechanisms underlying the majority of X-linked intellectual disability disorders remain poorly understood. A substantial fraction of X-linked intellectual disability genes encode nuclear proteins, suggesting that elucidating their functions in the regulation of transcription may provide novel insights into the pathogenesis of intellectual disability. Recent studies have uncovered mechanisms by which mutations of the gene encoding Plant Homeodomain (PHD)-like Finger protein 6 (PHF6) contribute to the pathogenesis of the X-linked intellectual disability disorder Börjeson-Forssman-Lehmann syndrome (BFLS). PHF6 plays a critical role in the migration of neurons in the mouse cerebral cortex in vivo, and patient-specific mutations disrupt the ability of PHF6 to promote neuronal migration. Interestingly, PHF6 physically associates with the PAF1 transcriptional elongation complex and thereby drives neuronal migration in the cerebral cortex. PHF6 also interacts with the NuRD chromatin remodeling complex and with the nucleolar transcriptional regulator UBF, though the biological role of these interactions remains to be characterized. In other studies, PHF6 mRNA has been identified as the target of the microRNA miR-128 in the cerebral cortex, providing new insights into regulation of PHF6 function in neuronal migration. Importantly, deregulation of PHF6 function in neuronal migration triggers the formation of white matter heterotopias that harbor neuronal hyperexcitability, which may be relevant to the pathogenesis of intellectual disability and seizures in BFLS. Collectively, these studies are beginning to provide key insights into the molecular pathogenesis of BFLS.
Keywords: X-linked intellectual disability, Börjeson-Forssman-Lehmann syndrome, PHF6, PAF1 complex, Neuronal positioning, Transcription, Heterotopia
X-linked Intellectual disability is a prevalent neurodevelopmental disorder
X-linked intellectual disability (XLID) affects 1 to 3 percent of the population (Bhasin et al., 2006; Larson et al., 2001; van Bokhoven & Kramer, 2010). Clinically, XLID is characterized by a deficit in intellectual function with an intelligence quotient (IQ) of less than 70 before the age of 18 and impairment of adaptive behaviors leading to deficient communication and social interactions (van Bokhoven & Kramer, 2010). Based on IQ score, intellectual disability may be classified as follows: profound (IQ<20), severe (IQ 20–34), moderate (IQ 35–49) and mild (IQ 50–69) (Ropers, 2010). XLID affects more males than females (Ropers, 2010). XLID has been grouped into syndromic and nonsyndromic forms. Nonsyndromic XLID is characterized by intellectual disability in the absence of other symptoms or signs, whereas syndromic XLID patients display intellectual disability together with other developmental abnormalities (Kleine-Kohlbrecher et al., 2010). Gene deletions, duplications, inversions and point mutations on the X chromosome have been associated with XLID. Thus far, mutations in over 100 genes on the X chromosome have been associated with intellectual disability. These mutations affect proteins with functions in transcription, translation, signal transduction and cell cycle regulation (Kleine-Kohlbrecher et al., 2010).
Characterization of XLID-associated proteins has opened up new fields of study in brain development and disease. The X-linked fragile X and Rett syndromes represent two major examples of XLID syndromes that have spawned major lines of research at the intersection of development and cognitive disorders of the brain. Characterized by intellectual disability, autistic features, developmental delay, hyperactivity and attention deficit behavior (Coffee et al., 2008; Huber et al., 2002; Leigh & Hagerman, 2013), fragile X syndrome is caused by expansion of CGG nucleotide repeats in the 5′ untranslated region of the Fmr1 gene, leading to its transcriptional silencing (Verkerk et al., 1991; Yu et al., 1991). A large number of studies have elucidated functions for the Fmr1-encoded protein, FMRP in protein translation and synaptic plasticity (for review, see Leigh & Hagerman, 2013). In mutant mice lacking FMRP, long-term depression triggered by metabotropic glutamate receptors activation is enhanced (Bhakar et al., 2012; Huber et al., 2002). In the cerebral cortex, FMRP knockout mice have increased dendritic spine number with more immature long shaped spines but fewer spines with mature mushroom shape (Irwin et al., 2000). Thus, FMRP regulates both synaptic development and plasticity. At a molecular level, FMRP operates as an RNA-binding protein to control mRNA stability (Zalfa et al., 2007). FMRP may enter the nucleus and assemble with targeted RNAs into mRNP and undergo export to dendrites and regulate local protein synthesis (Eberhart et al., 1996). In fragile X syndrome patients, loss of FMRP function causes deregulation of localized mRNA synthesis, leading to defects in synapse function (Comery et al., 1997; Jin & Warren, 2000).
Rett syndrome patients have normal psychomotor development for the first 6 months after birth but subsequently fail to attain psychomotor milestones and eventually lose language and hand skills (Samaco & Neul, 2011). Mutations in methyl CpG binding protein 2 (MeCP2) cause Rett syndrome (Amir et al., 1999). MeCP2 knockout mice behave normally before weaning age (Guy et al., 2001), but by week eight these mice have unstable gait and reduced spontaneous movement followed by hindlimb clasping (Guy et al., 2001). Induced pluripotent stem cell (iPSC)-derived neurons from patients with Rett syndrome have reduced spine density, soma size and glutamatergic synapses (Marchetto et al., 2010), mimicking neuropathological features of the brain in Rett syndrome patients (Kaufmann & Moser, 2000; Reiss et al., 1993; Samaco & Neul, 2011). MeCP2 binds to methylated cytosine within CpG motifs and has been implicated in the regulation of transcription (Nan et al., 1997). MeCP2 may silence transcription by recruiting co-repressors including histone deacetylases (Hdacs) (Nan et al., 1998; Skene et al., 2010). Remarkably, MeCP2 binds to methylated CA motifs within long genes (>100 kb) to repress transcription, and upon MeCP2 depletion long gene expression is increased (Gabel et al., 2015). Although MeCP2 accumulates in post-mitotic neurons, MeCP2 may also play a role in glial cells and mutations of MeCP2 in glial cells may cause non-cell autonomous defects in neurons (Ballas et al., 2009).
Studies of FMRP and MeCP2 illustrate that significant advances in our understanding of neural development and pathogenesis of intellectual disability can be achieved from systematic studies of individual XLID proteins. Therefore, characterization of Plant Homeodomian (PHD)-like Finger protein 6 (PHF6), which is mutated in the X-linked Börjeson-Forssman-Lehmann syndrome (BFLS), is likely to yield novel insights into our understanding of brain development and intellectual disability.
Börjeson-Forssman-Lehmann syndrome (BFLS)
The Börjeson-Forssman-Lehmann syndrome (BFLS) was first described as an X-linked intellectual disability syndrome by Mats Börjeson and colleagues in 1962 (Börjeson et al., 1962). Since then, additional cases of BFLS patients have been identified. BFLS patients typically have normal birth weight, head circumference and delivery (Lower et al., 2002). However, the main clinical features of BFLS become apparent with age and show considerable variation among different patients (de Winter et al., 2009; Lower et al., 2002). A summary of the clinical features of BFLS is presented in Table 1. Diagnosis of BFLS is typically made clinically based on history and physical examination, and can be confirmed by sequencing of the PHF6 gene. MRI data in patients with BFLS are lacking. In the few BFLS cases where MRI data are available in the literature, enlarged ventricles have been reported in once case (Table 1). Developmental delay usually appears before the first birthday. All BFLS patients have intellectual disability, ranging from mild to severe, as well as small genitalia and short stature (Gécz et al., 2006; Lower et al., 2002). Approximately 75% of the patients have obesity emerging in late childhood; BFLS patients display microcephaly or macrocephaly at a frequency of 6% and 15% respectively; Epileptic seizures have been observed in approximately 8% of patients (Carter et al., 2009; Di Donato et al., 2014; Mangelsdorf et al., 2009; Zweier et al., 2013). Other features of the syndrome include tapered fingers and broad shortened toes. Adult BFLS patients display coarsening of facial features with prominence of the supraorbital ridges and deep-set eyes (Gécz et al., 2006; Lower et al., 2002). Other abnormalities observed uncommonly in BFLS patients include mild polyneuropathy, hearing impairment, cleft lip and hypopituitarism (Gécz et al., 2006). Female carriers of BFLS may have learning problems and show physical manifestations including shortened toes, thickened fleshy ear lobes, pronounced supraorbital ridges and deep-set eyes (Gécz et al., 2006; Lower et al., 2002).
Table 1.
Summary of PHF6 mutations in BFLS patients in different individuals from different families. Classical features of BFLS includes mild to severe mental retardation, hypogonadism, hypometabolism, obesity with marked gynecomastia, facial dysmorphism, narrow palpebral fissure, large and fleshy ears, tapered fingers.
| F/M | Nucleotide | Amino Acid | Mutation | Reported clinical features | Behavioral disturbances | Ref |
|---|---|---|---|---|---|---|
| M | c. 2T>C | p.M1T | Missense | Classical features of BFLS with no detailed clinical records. | N/A | (Lower et al., 2002) |
| M | c. 2T>C | p.M1T | Missense | Classical features of BFLS. Born at term after uneventful pregnancy, delayed early milestones, walked at age 4, began saying a few words at age 5, height of 140cm (<3rd centile), weight of 45kg (5th centile), and occipital-frontal head circumference of 53.3cm (10th-25th centile) at age 15, short stature, triangular face, hyperplastic supraorbital digits, deep-set eyes, large ears, gynecomastia, lower abdominal obesity, and small penis and testes with no evidence of pubertal change. |
N/A | (Crawford et al., 2006) |
| M | c.134G>A | p.C45Y (familial) | Missense | Classical features of BFLS with no detailed clinical records. | N/A | (Lower et al., 2002) |
| M | c.134G>A | p.C45Y (de novo) | Missense | Classical features of BFLS with no detailed clinical records. | N/A | (Lower et al., 2002) |
| M | c.266G>T | p.G89Y | Missense | Classical features of BFLS. At age 22, height 173cm, weight 108kg, head circumference 61cm. Moderate intellectual disability; delayed development, no epileptic seizures. Brother has similar symptoms with severe intellectual disability, Female carrier in the family has shown mild intellectual disability. |
Good tempered with occasional aggressive attacks | (Mangelsdorf et al., 2009) |
| M | c.296G>T | p.C99F | Missense | Classical features of BFLS. Female carrier has mild clinical features of BFLS including obesity, large ears, amenorrhea, hypothyroidism, epilepsy, and learning difficulties. |
N/A | (Lower et al., 2002) |
| M,F | c.686A>G | p.H229R | Missense | Classical features of BFLS with no detailed clinical records. | N/A | (Lower et al., 2002) |
| M | c.700A>G | p.K234E | Missense | Classical features of BFLS with no detailed clinical records. | N/A | (Lower et al., 2002) |
| M | c.769A>G | p.R257G | Missense | Classical features of BFLS with no detailed clinical records. | N/A | (Lower et al., 2002) |
| M | c.769A>G | p.R257G | Missense | Classical features of BFLS. Severe intellectual disability, self-injurious behavior, stooped hyposcoliotic posture, central obesity, marked gynecomastia, hypogonadism, and hand and feet anomalies. Hodgkin’s lymphoma |
N/A | (Vallée et al., 2004) (Carter et al., 2009) |
| M | c.940A>G | p.I314V | Missense | Classical features of BFLS. Psychomotor developmental delay, language delay, mild intellectual disability (IQ of 61). At age 18, height 171.5 cm, head circumference 55.7cm, moderate obesity, marked gynecomastia. |
Lack of inhibition in sexual contact; limbs showing hypermobility. | (Crawford et al., 2006) |
| M | c.22A>T | p.K8X | Nonsense | Classical features of BFLS. | (Lower et al., 2002) | |
| F | c.955C>T | p.R319X | Nonsense | Classical features of BFLS. Head circumference 34cm (−0.5SD), delayed walking at age 2.5. During first years, frontotemporal alopecia, nystagmus, strabismus, small teeth, hypoplastic labia minora and clitoris. At age 6, head circumference of 49cm (−1.63SD), moderate to severe intellectual disability, facial dimorphism, and tapering fingers. Normal MRI. |
Lack of swallowing coordination; Sociable and happy | (Zweier et al., 2013) |
| M | c.1024C>T (original family) | p.R342X | Nonsense | Classical features of BFLS. Developmental delay, upslanting palpebral fissures, large ears, a high palate, hypotonia, cryptorchidism, and an inguinal hernia. At age 15, talk sentances consisting of three or four words, short stature and obesity, large ears, protruding cheeks,. Female carrier has normal cognitive function. Big hands, ears and feet, with some sensory impairment in the legs. Normal MRI. |
Female carrier is clumsy when running or jumping. | (Gécz, Turner, Nelson, & Partington, 2006; Lower et al., 2004) |
| M | c.1024C>T | p.R342X | Nonsense | Delayed developmental milestones, height on 25th centile, weight over 97th centile, head circumference over 98th centile, narrow bitemporal diameter, upslanting eyes, prominent supraorbiral ridges, hypotelorism, long ears/earlobes, a prominent chin, obese, marked gynecomastia, a hypoplastic scrotum with retractile testes, cubitus valgus, tapered fingers. Female carrier is obese, has deep-set eyes, prominent supraorbital ridges, large ear lobes, tapered fingers, broad feet and short toes. |
N/A | (Lower et al., 2002) |
| M | c.1024C>T | p.R342X | Nonsense | Classical features of BFLS. | N/A | (Lower et al., 2002) |
| M | c.1024C>T | p.R342X | Nonsense | Classical features of BFLS. | N/A | (Crawford et al., 2006) |
| M | c.1024C>T | p.R342X | Nonsense | Classical features of BFLS. | N/A | (Crawford et al., 2006) |
| M | c.1024C>T | p.R342X | Nonsense | Classical features of BFLS and T cell acute lymphoblastic leukemia at age 9 | N/A | (Chao et al., 2010) |
| F | c.27dupA | p.G10fsX21 | Frameshift | Classical features of BFLS. Delayed speech, normal IQ of 87, high level of thyroid stimulating hormone, low level of thyroxin at age 7–8; at age 14, height 180cm, weight 115 kg, shallow forehead, fleshy earlobes, deep set eyes, broad feet, hammertoes, no menstruation at age 14. |
Poor coordination; Tactile defensive and emotionally labile, poor spatial awareness, and startles to noise |
(Crawford et al., 2006) |
| M | IVS2-8A>G | M46fsD exon3 | Frameshift | Classical features of BFLS with mild to moderate intellectual disability, and global developmental delay. | N/A | (Vallée et al., 2004) |
| F | c.677delG | p. G226fsE53 | Frameshift | Classical features of BFLS Diagnosed with Coffin Siris Syndrome in early infancy. At 9 years old, height 1.7SD, weight 30kg, head circumference 54cm (1.4SD). Mild intellectual disability, mild clinodactyly, short toes, and facial dysmorphism. |
Shy and intolerance for being alone; anxiety; sleeping difficulties | (Wieczorek et al., 2013; Zweier et al., 2013) |
| F | c.914G>T | p. C305F | Missense | Classical features of BFLS, previously diagnosed with Coffin Siris Syndrome, unstable gait on stairs, head circumference of −1.0SD at age 11, susceptibility to urinary tract infections, frequent constipation, facial dysmorphism, and tapered fingers. | Aggressive behavior | (Wieczorek et al., 2013; Zweier et al., 2013) |
| F | Dosage gain of exon 4, 5 | Duplication | Classical features of BFLS, delayed milestones, delayed speech, conductive hearing loss, potential retinal dystrophy, and seizures at age 20. At age 22, head circumference 56.5(1.4 SD), facial dysmorphism, tapering fingers. | Unstable gait; behavioral problems including short attention span, hyperactivity, compulsive behavior, sleeping difficulties, and lack of emotional detachment. | (Zweier et al., 2013) | |
| F | Dosage gain of exon 4, 5 | Duplication | Classical features of BFLS. Head circumference of 34cm (1.4 SD) at birth; At age 3–4 month, several single seizures, delayed walking and language, constipation, and neurogenic bladder. At age 32, head circumference 54cm (−0.3 SD). |
Sociable behavior | (Zweier et al., 2013) | |
| F | 6kb deletion | Affecting exon 4, 5 | Deletion | Classical features of BFLS with moderate intellectual disability. Head circumference 53cm (2.2SD), linear skin hyperpigmentation, and facial dimorphism. | Sociable behavior, | (Zweier et al., 2013) |
| F | 100kb deletion | Deletion of last 5 coding exons | Deletion | Classical features of BFLS. Delayed developmental milestone, feeding difficulties, muscular hypotonia, single horseshoe kidney, obstructive uropathy, recurrent pyelonephritis, and multiple febrile seizures. | N/A | (Di Donato et al., 2014) |
| F | 15kb deletion | Deletion of last 3 exons | Deletion | Classical features of BFLS with mild intellectual disability. At age 7, mild hearing loss, and recurrent middle ear infections; At age 16, head circumference 56.6cm (75th centile), slightly widened ventricles and increased white matter signal abnormality on MRI. | N/A | (Berland et al., 2011) |
| M | c.999-1001 delTGA | p.D333del | Deletion | Classical features of BFLS with delayed milestones. | N/A | (Baumstark et al., 2003) |
| M | c.999-1001 delTGA | p.D333del | Deletion | First patient description. Delayed milestones, severe mental deficiency with IQ of 20, seizures, large ears, short stature, small genitalia, and small pituitary gland. Coarse gyri, widened ventricular system, cortical dysplasia with indistinct lamination, and heterotopias. |
Hyperactive and aggressive behaviors | (Börjeson, Forssman, & Lehmann, 1962; Gécz et al., 2006) |
| F | Entire gene deleted | Deletion | Classical features of BFLS. Head circumference 35.5 (0.5SD) at birth, delayed motor milestones and facial dysmorphism. muscular hypotonia, ectopic left kidney, strabismus, hyperopia, bilateral sensorineural hearing loss, enlarged ventricles on MRI. | Compulsive eating behavior; Severe sleeping difficulties, | (Zweier et al., 2013) | |
| F | 270kb deletion | Deletion | Classical features of BFLS. Mild muscular hypotonia, mild speech delay, narrow external auditory canal, mild learning disability; IQ within borderline to normal range. | N/A | (Di Donato et al., 2014) |
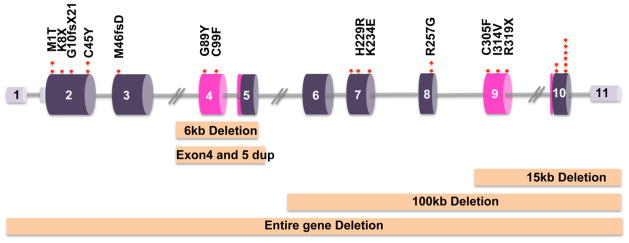
Molecular genetics studies have led to the identification of multiple mutations in the PHF6 gene on the X chromosome in BFLS patients (Berland et al., 2011; Carter et al., 2009; Lower et al., 2002; Turner et al., 2004). Most PHF6 mutations in BFLS are either missense or lead to truncation of PHF6 protein. Five PHF6 mutations in BFLS are recurrent including c.2T>C/p.M1T, c134G>A/p.C45Y, c769A>G/p.R257G, c.999-1001delTGA/p.D333del and c1024C>T/p.R342X, found in 21% of patients. The truncation mutation c.1024C>T/p.R342X is found in five different BFLS families (Chao et al., 2010; Crawford et al., 2006; Lower et al., 2002, 2004). The majority of PHF6 mutations occur in exon 2, which encodes the N-terminus of PHF6 protein. A fundamental question that remains to be addressed is how PHF6 mutations lead to BFLS.
Mechanisms of PHF6 function
The PHF6 gene, which is located on Xq26-27 between the markers DXS425 and DXS105, is comprised of 11 exons that is transcribed into a 4.5 kb mRNA and translated into a 41 kDa protein with 365 amino acids. Two major isoforms of PHF6 mRNA, depending on whether intron 10 is spliced out, give the same protein product (Gécz et al., 2006). PHF6 protein is highly conserved in vertebrates. Structurally, PHF6 protein contains two plant homeodomain (PHD)-like zinc fingers, two nuclear localization sequences and a nucleolar localization sequence (Liu et al., 2014), suggesting that PHF6 may play a role in the regulation of transcription. PHD zinc fingers are structurally conserved modules that interact with chromatin or mediate protein-protein interactions (Sanchez & Zhou, 2011). The Cys4-His-Cys3 motifs within the PHD domain of PHF6 coordinate two Zn2+ ions and stabilize PHF6 protein structure (Liu et al., 2014). The evolutionary conserved PHF6 cysteine residues C45, C99 and C305 are targeted by missense mutations in BFLS. PHD fingers also recognize specific histone modifications (Aasland et al., 1995; Sanchez & Zhou, 2011; Wysocka et al., 2006), and regulate the function of epigenome readers in transcription via recruitment of multi-protein complexes. However, unlike other PHD domain proteins, PHF6 contains two imperfect PHD domains (Liu et al., 2014). Therefore, it is unclear whether the PHD domains in PHF6 function similarly as other PHD domains to recognize specific chromatin marks.
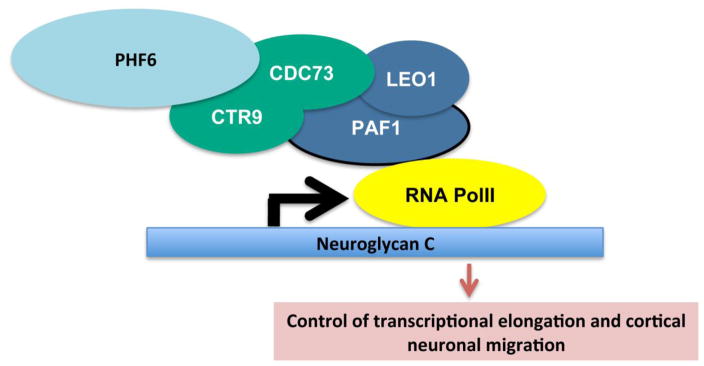
Employing an unbiased computation-assisted proteomics approach, Zhang et al. discovered that PHF6 associates with subunits of the PAF1 transcription elongation complex including PAF1, LEO1, CTR9, and CDC73 (Zhang et al., 2013). The interaction of PHF6 with the PAF1 complex plays a critical role in PHF6 function in neuronal migration in the cerebral cortex. Knockdown of PAF1 mimics the PHF6 knockdown-induced phenotype of impaired neuronal migration in the mouse cerebral cortex (Zhang et al., 2013). The PAF1 complex occupies the promoter and gene body of actively transcribed genes and associates with RNA polymerase II (PolII) to promote transcriptional elongation and transcription-coupled histone modifications (Chen et al., 2009; Krogan et al., 2002; Marton & Desiderio, 2008; Rondon et al., 2004; Squazzo et al., 2002). The C-terminal domain (CTD) phosphorylation of PolII is thought to mediate PAF1 recruitment to PolII. The PAF1 complex may directly stimulate RNA PolII transcription or act synergistically with other factors to promote transcriptional elongation (Jaehning et al., 2010). During transcriptional elongation, the PAF1 complex may recruit the histone methyltransferase Set2 to induce trimethylation of H3K36 and the ATPase Chd1 to remodel chromatin structure (Tomson & Arndt, 2013). In yeast, the PAF1 complex also promotes H2B K123 monoubiquitylation by recruiting the ubiquitin-ligase Rad6-Bre1. The H2B moniubiquitination facilitates dimethylation and trimethylation of H3K4 and H3K79 by the histone methyltransferases Set1 and Dot1 (Tomson & Arndt, 2013; Wood et al., 2003). Recent studies have shown that PAF1 may also regulate promoter-proximal pausing of PolII (Chen et al., 2015). PAF1 regulates PolII pausing approximately 50 nucleotides downstream of the transcription start site (TSS) where significant PolII occupancy is found. Upon PAF1 knockdown, PolII is released to the gene body (Chen et al., 2015). Together, these studies indicate the PAF1 complex regulates multiple aspects of transcription. By interacting with the PAF1 complex, PHF6 is thus poised to influence transcription. Interestingly, both of PHF6 and the PAF1 complex were shown to induce the expression of the gene encoding Neuroglycan C, which in turn mediates the ability of PHF6 and PAF1 complex to promote neuronal migration (Zhang et al., 2013). Knockdown of Neuroglycan C phenocopies the effect of PHF6 knockdown and PAF1 knockdown on neuronal migration, and expression of Neuroglycan C in PHF6-depleted progenitors rescues the defects in neuron migration (Zhang et al., 2013). These observations suggest that PAF1 and PHF6 may share a core network of target genes.
PHF6 also interacts with the nucleosome remodeling and deacetylation (NuRD) chromatin remodeling complex (Todd & Picketts, 2012). The NuRD complex is composed of the histone deacetylase Hdac1 or Hdac2 and ATPase Chd3 or Chd4, with combinatorial assembly of other subunits, including histone-binding protein RbAp46 and RbAp48, scaffold protein Mta1/2, DNA-binding protein Mbd3, and GATA zinc finger domain-containing protein 2A Gatad2a and Gatad2b (Zhang et al., 1998, 1999). The NuRD complex has been widely studied in cancer and embryonic stem (ES) cells (Lai & Wade, 2011). In acute promyelocytic leukemia (APL), the NuRD complex alters DNA methylation levels leading to gene repression. Upon removing the NuRD subunit Mbd3 in leukemia cells, DNA methylation levels at the RARb2 gene are significantly reduced (Morey et al., 2008). In addition, depletion of Mbd3 allows reprogramming of somatic cells to induced pluripotent stem (iPS) cells (Rais et al., 2013). The Mbd3-NuRD complex is thought to be recruited to the OSKM (Oct4, Sox2, Klf4, and Myc) target genes and inhibit robust reprogramming. Upon Mbd3 depletion, OSKM proteins interact with other pluoripotency-inducing chromatin regulators to drive efficient reprogramming (Rais et al., 2013). Recently, two de novo mutations in the NuRD subunit Gatad2a have been described in patients with severe intellectual disability (Willemsen et al., 2013), suggesting that the NuRD complex plays a critical role in brain development.
The NuRD complex is highly expressed in the developing brain. Recent studies by Yamada, Yang et al. have identified a novel function for the NuRD complex in neuronal connectivity in the brain (Yamada et al., 2014). The NuRD complex decommissions a subset of gene promoters by removing H3K9/14 acetylation marks and thereby represses the expression of developmentally regulated genes to trigger granule neuron parallel fiber/Purkinje cell synaptic connectivity in the mammalian brain. Upon depletion of Chd4, the core ATPase subunit of the NuRD complex, these developmental genes are derepressed, leading to defects in granule neuron presynaptic bouton formation (Yamada et al., 2014). Whether the interaction of PHF6 with the NuRD complex influences gene expression and establishment of neuronal connectivity remains unexplored.
PHF6 also interacts with the nucleolar transcription factor upstream binding factor (UBF) in cells (Wang et al., 2013). UBF recruits RNA polymerase I to rDNA promoter regions. Upon PHF6 depletion, HeLa cells arrest at the G2/M phase of the cell cycle and have increased levels of UBF and ribosomal RNA synthesis. PHF6 interacts with UBF and is recruited to rDNA promoters to repress rDNA expression. Knockdown of PHF6 derepresses the rDNA promoter, leading to increased ribosomal RNA levels (Wang et al., 2013). However, the mechanism of rDNA repression by the PHF6-UBF interaction localized to the rDNA promoter remains unclear.
In situ RNA hybridization and expression analyses have shown that PHF6 is highly expressed in the developing central nervous system (CNS) and declines along the course of development (Voss et al., 2007). These observations suggest that PHF6 expression is tightly regulated during brain development. Recent studies by Franzoni et al provide insights into the mechanisms underlying the regulation of PHF6 expression (Franzoni et al., 2015). Downregulation of PHF6 expression is correlated with an increase in the expression levels of miR-128, suggesting that miR-128 may regulate PHF6 expression during brain development. Consistent with this interpretation, expression of miR-128 in cortical neurons triggers the downregulation of PHF6, an effect that is mediated by miR-128 binding sites within the 3′UTR of PHF6 mRNA (Franzoni et al., 2015). A prediction of these results is that miR-128 might influence neuronal migration in the cerebral cortex. Consistent with this prediction, expression of miR-128 by in utero electroporation in the mouse brain arrests the migration of cortical neurons in layers III and IV–VI (Franzoni et al., 2015), which phenocopies the migration defects observed upon PHF6 knockdown (Zhang et al., 2013). Importantly, expression of PHF6 reverses the miR-128-induced migration defects (Franzoni et al., 2015), suggesting that PHF6 operates downstream of miR-128 in the regulation of cortical migration. Whether miR-128 regulation of PHF6 expression influences other aspects of brain development remains to be explored.
PHF6 in BFLS Pathogenesis
Identification of a critical role for PHF6 in radial neuronal migration in the cerebral cortex raises the important question of whether deregulation of PHF6 function in migration contributes to BFLS pathogenesis. Proper migration and positioning of neurons is a prerequisite for correct neuronal connectivity (Ayala et al., 2007; Morgan-Smith et al., 2014; Nadarajah & Parnavelas, 2002). Radially migrating neurons in the cerebral cortex are born in the ventricular zone (VZ) and sub-ventricular zone (SVZ), migrate along the radial glia scaffold to reach the cortical plate (Ayala et al., 2007; Kriegstein & Noctor, 2004; Vaillend et al., 2008). The oldest neurons reside in the deep layer of the cortical plate, and newly generated neurons pass the deep layers to reside in more superficial layers (Greig et al., 2013). Upon PHF6 knockdown, neurons arrest in the intermediate zone, and the ectopic cells have neuronal hyperexcitability (Zhang et al., 2013). Introduction of an RNAi-resistant form of wild type-PHF6 into PHF6 knockdown mice rescues the migration defects, suggesting that the RNAi-induced phenotype is the result of specific knockdown of PHF6 rather than off target effects (Zhang et al., 2013). In vivo structure-function analyses of PHF6 demonstrate that BFLS patient-specific mutations impair the ability of PHF6 to promote neuronal migration (Zhang et al., 2013). Strikingly, neuropathological studies performed in the first identified BFLS patient have revealed defects of neuronal migration in the cortical plate including coarse gyri, cortical dysplasia with indistinct lamination and heterotopic neurons observed in the subcortical white matter (Brun et al., 1974).
PHF6 knockdown in the mouse cerebral cortex also triggers the formation of white matter heterotopias with aberrant neuronal activity (Zhang et al., 2013). Importantly, electrophysiological recordings reveal hyperexcitability of heterotopic neurons. Whole-cell patch clamp recordings reveal an aberrant pattern of activity in heterotopic neurons in PHF6 knockdown animals (Zhang et al., 2013). The membrane potential of heterotopic neurons oscillates, leading to frequent firing of action potentials. Further, knockdown of Neuroglycan C, a PHF6 target gene, also leads to formation of heterotopias that harbor neuronal hyperexcitability (Zhang et al., 2013). Heterotopias have been postulated to contribute to seizure susceptibility (Ackman et al., 2009). Heterotopias may serve as loci that initiate seizure activity. Studies in both genetically induced animal models and depth electrode recordings from patients have suggested that heterotopic loci may function to generate epilepsy (Aghakhani et al., 2005; Kothare et al., 1998, Manent et al., 2009). Together, these data suggest that inhibition of the PHF6 pathway triggers the formation of heterotopias in which neurons are hyperexcitable. These findings may be relevant to reduced seizure threshold in BFLS patients.
Although recent studies of PHF6 function have focused on its role in the regulation of neuronal migration and positioning in the cerebral cortex, it is likely that PHF6 has additional functions in the developing brain owing to its expression pattern (Voss et al., 2007; Zhang et al., 2013). PHF6 is expressed throughout the cerebral cortex and although levels of PHF6 drop with maturation, PHF6 is poised to regulate other aspects of neuronal development beyond neuronal positioning. In addition, PHF6 may have developmental roles outside of the cerebral cortex that may be relevant to BFLS pathogenesis.
Perspectives
Recent studies have advanced our understanding of the underlying molecular mechanisms of PHF6 function and its relevance to BFLS pathogenesis. These studies suggest that PHF6 is required for proper neuronal migration and that deregulation of PHF6 function in neuronal positioning contributes to the pathogenesis of BFLS. Analyses of PHF6 signaling in cells, primary neurons, and in the developing mouse brain suggest a role for PHF6 in transcription. Although progress has been made in identifying PHF6 binding partners through proteomic analyses of cells in which PHF6 is expressed, comprehensive proteomic analyses of PHF6 partners during neuronal development or in BFLS-derived iPSC cells should advance our understanding of the mechanisms by which PHF6 regulates brain development as well as provide novel insights into BFLS pathogenesis. In addition, mapping genome wide targets of PHF6 in the developing brain should provide insights on the transcriptional roles of PHF6 specifically in the brain. Therefore, further research is required to gain a better understanding of PHF6 functions in brain development and pathogenic mechanisms underlying BFLS.
The finding that migration arrest in the PHF6 knockdown animals leads to the formation of heterotopias has important implications in the pathogenesis of BFLS. Clinically, heterotopia formation is associated with epileptic activity in the brain, which has been documented in BFLS patients (Robinson et al., 1983). Additionally, heterotopic neurons are hyperexcitable (Ackman et al., 2009). In human neuropathological studies, in addition to evidence of whiter matter heterotopias, the cerebral cortex displays dysplasia with indistinct or absent lamination (Brun et al., 1974). Whether cortical dysplasia due to impairment of PHF6 function is relevant to BFLS requires further study.
The identification of a novel transcriptional PHF6/PAF1 pathway in the developing brain has opened new avenues for the study of the molecular mechanisms that underlie XLID, and has also brought new insights into the regulation of transcription. The association of PHF6 with the PAF1 complex suggests an important role for PHF6 in the regulation of transcriptional elongation during neuronal development. Intriguingly, nascent transcriptional initiation is widespread across the genome, and the transition from transcriptional initiation to elongation represents a critical step of gene regulation (Guenther et al., 2007; Krumm et al., 1995; Muse et al., 2007; Nechaev & Adelman, 2012). Previous studies have revealed that the PAF1 complex cooperates with DSIF and the elongation factor SII/TFIIS, and thereby promotes elongation of PolII along the gene body (Chen et al., 2009; Zhu et al., 2005). In view of the findings that PHF6 may function to negatively or positively regulate gene expression (Todd & Picketts, 2012; Wang et al., 2013; Zhang et al., 2013) and PAF1’s critical role in gene pausing (Chen et al., 2015), it would be interesting to investigate whether PHF6 regulates gene pausing, pause release, or both. The PAF1 complex appears to be critical for histone modifications including H3K4 tri-methylation (H3K4me3) or histone H2B (ubH2B) ubiquitination (Krogan et al., 2003; Wood et al., 2003). Whether PHF6 promotes modifications of histones via interactions with the PAF1 complex remains an open question.
The identification of Neuroglycan C as a target gene of PHF6 and the PAF1 complex provides new insights in neuronal migration and intellectual disability (Zhang et al., 2013). The Neuroglycan C gene, a member of the neuregulin family, encodes a transmembrane chondroitin sulfate glycoprotein, which has been implicated in the regulation of cortical radial migration (Anton et al., 1997; Brandt et al., 2007; Kinugasa et al., 2004; Rio et al., 1997). The Neuroglycan C gene might represent a locus associated with schizophrenia, a neuropsychiatric disorder in which impaired neuronal migration may play a pathogenic role (Impagnatiello et al., 1998; Lee et al., 2011; So et al., 2010; Tomita et al., 2011). These observations raise the possibility that mutations of PHF6 may contribute to the pathogenesis of neuropsychiatric disorders beyond intellectual disability. It will be also interesting to determine whether deregulation of Neuroglycan C might play a role in intellectual disability and epilepsy.
Beyond the role of the PHF6-PAF1 interaction in neuronal positioning, it will be important to characterize the significance of the PHF6 interaction with the NuRD complex in brain development. Recently, two de novo mutations in the NuRD subunit Gatad2a have been reported in patients with severe intellectual disability (Willemsen et al., 2013). The NuRD complex represses transcription of developmentally regulated genes and thereby drives synaptogenesis in the mammalian brain (Yamada et al., 2014). Whether the NuRD complex regulates neuronal migration in the cerebral cortex remains unclear. In granule neurons of the cerebellum, the NuRD complex removes H3K9/14 acetylation marks at a subset of genes during development to drive synaptogenesis (Yamada et al., 2014). However, how the NuRD complex and PHF6 might regulate the epigenetic landscape of the genome in the cerebral cortex remains unknown.
Identification of the PHF6/UBF interaction in HeLa cells suggests that PHF6 may function in the nucleolus to regulate rDNA transcription (Wang et al., 2013). The nucleolus is a prominent subnuclear domain implicated in ribosomal RNA (rRNA) transcription, which accounts for up to 80% of RNA synthesis in eukaryotic cells (Nemeth & Langst, 2011). Transcription of rDNA is essential during the cell cycle (Roussel et al., 1996); however, its importance in post-mitotic neurons remains to be elucidated. Recently, intellectual disability-linked proteins including FMRP and the RNA methyltransferase NSUN2 have been shown to be transported to nucleoli (Khan et al., 2012; Nemeth & Langst, 2011). Whether PHF6 may harbor regulatory roles in the nucleolus has been unexplored.
Future studies of PHF6 should focus on elucidation of the biochemical functions of PHF6 including the role of its PHD domains and pathological relevance. Mouse models of the disease will assist in understanding the pathological consequences of PHF6 inactivation and their impact on clinical features including intellectual disability and seizures. In addition, it will be important to explore whether PHF6 harbors additional biological functions in brain development including neural progenitor proliferation, differentiation, synaptic development and circuit integration, and determine whether deregulation of such functions contributes to the pathogenesis of BFLS.
Figure 1.
PHF6 gene with patient mutations. The small blocks in light purple represent the 3′ and 5′ UTR of PHF6 gene. The blocks in dark purple represent the coding exons with PHD domains shown in pink. Point mutations in BFLS are shown as red dots. Five large domain deletions in BFLS are also shown.
Figure 2.
A proposed model for PHF6-PAF1 complex in regulation of neuronal migration during cortical development. During critical period of neuronal migration, PHF6 interacts with PAF1 transcriptional elongation complex to regulate downstream targets including Neuroglycan C to regulate neuronal migration and positioning.
Highlights.
Mutations of the PHF6 gene are linked with Börjeson-Forssman-Lehmann syndrome (BFLS).
PHF6 regulates transcription via interactions with the PAF1 complex, NuRD complex, and UBF.
miR-128 controls neuronal positioning by inhibiting PHF6 in the cerebral cortex.
Deregulation of the PHF6/PAF1 complex triggers formation of white matter heterotopia.
Acknowledgments
We thank the members of the Bonni laboratory for critical reading of the manuscript. The authors are supported by National Institutes of Health grant NS088378 (to A.B.), the Mathers Foundation (to A.B.), Natural Sciences and Engineering Research Council of Canada (NSERC) grant # RGPIN-2016-06605 (to A.J.-A).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aasland R, Gibson TJ, Stewart aF. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends in Biochemical Sciences. 1995;20(2):56–59. doi: 10.1016/s0968-0004(00)88957-4. http://doi.org/10.1016/S0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Ackman JB, Aniksztejn L, Crepel V, Becq H, Pellegrino C, Cardoso C, Ben-Ari Y, Represa A. Abnormal Network Activity in a Targeted Genetic Model of Human Double Cortex. Journal of Neuroscience. 2009;29(2):313–327. doi: 10.1523/JNEUROSCI.4093-08.2009. http://doi.org/10.1523/JNEUROSCI.4093-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghakhani Y, Kinay D, Gotman J, Soualmi L, Andermann F, Olivier A, Dubeau F. The role of periventricular nodular heterotopia in epileptogenesis. Brain. 2005;128(3):641–651. doi: 10.1093/brain/awh388. http://doi.org/10.1093/brain/awh388. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genetics. 1999;23(2):185–188. doi: 10.1038/13810. http://doi.org/10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Anton ES, Marchionni Ma, Lee KF, Rakic P. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development. 1997;124(18):3501–3510. doi: 10.1242/dev.124.18.3501. [DOI] [PubMed] [Google Scholar]
- Ayala R, Shu T, Tsai LH. Trekking across the Brain: The Journey of Neuronal Migration. Cell. 2007;128(1):29–43. doi: 10.1016/j.cell.2006.12.021. http://doi.org/10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Ballas N, Lioy DT, Grunseich C, Mandel G. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nature Neuroscience. 2009;12(3):311–7. doi: 10.1038/nn.2275. http://doi.org/10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berland S, Alme K, Brendehaug a, Houge G, Hovland R. PHF6 Deletions May Cause Borjeson-Forssman-Lehmann Syndrome in Females. Molecular Syndromology. 2011;1(6):294–300. doi: 10.1159/000330111. http://doi.org/10.1159/000330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakar AL, Dölen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annual Review of Neuroscience. 2012;35:417–43. doi: 10.1146/annurev-neuro-060909-153138. http://doi.org/10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin T, Brocksen S, Avchen R, Van Naarden Braun K. Morbidity and Mortality Weekly Report: Prevalence of four developmental disabilities among children aged 8 Years — Metropolitan Atlanta Developmental Disabilities Surveillance Program, 1996 and 2000. 2006;55 [PubMed] [Google Scholar]
- Börjeson M, Forssman H, Lehmann O. An X-linked, Recessively Inherited Syndrome Characterized by Grave Mental Deficiency, Epilepsy, and Endocrine Disorder. Acta Medica Scandinavica. 1962;171(1):13–22. doi: 10.1111/j.0954-6820.1962.tb04162.x. [DOI] [PubMed] [Google Scholar]
- Brandt N, Franke K, Rasin MR, Baumgart J, Vogt J, Khrulev S, Hassel B, Pohl E, Sestan N, Nitsch R, Schumacher S. The neural EGF family member CALEB/NGC mediates dendritic tree and spine complexity. The EMBO Journal. 2007;26(9):2371–86. doi: 10.1038/sj.emboj.7601680. http://doi.org/10.1038/sj.emboj.7601680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun A, Borjeson M, Forssman H. An inherited syndrome with mental deficiency and endocrine disorder. A patho-anatomical study. Journal of Intellectual Disability Research: JIDR. 1974;18(4):317–325. doi: 10.1111/j.1365-2788.1974.tb01243.x. [DOI] [PubMed] [Google Scholar]
- Carter MT, Picketts DJ, Hunter AG, Graham GE. Further clinical delineation of the Börjeson-Forssman-Lehmann syndrome in patients with PHF6 mutations. American Journal of Medical Genetics. Part A. 2009;149A(2):246–50. doi: 10.1002/ajmg.a.32624. http://doi.org/10.1002/ajmg.a.32624. [DOI] [PubMed] [Google Scholar]
- Chao MM, Todd Ma, Kontny U, Neas K, Sullivan MJ, Hunter AG, Picketts D, Kratz CP. T-cell acute lymphoblastic leukemia in association with Börjeson-Forssman-Lehmann syndrome due to a mutation in PHF6. Pediatric Blood & Cancer. 2010;55(4):722–4. doi: 10.1002/pbc.22574. http://doi.org/10.1002/pbc.22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Woodfin AR, Gardini A, Rickels RA, Marshall SA, Smith ER, Shiekhattar R, Shilatifard A. PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell. 2015;162(5):1003–1015. doi: 10.1016/j.cell.2015.07.042. http://doi.org/10.1016/j.cell.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes & Development. 2009;23(23):2765–77. doi: 10.1101/gad.1834709. http://doi.org/10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Ikeda M, Budimirovic DB, Hjelm LN, Kaufmann WE, Warren ST. Mosaic FMR1 deletion causes fragile X syndrome and can lead to molecular misdiagnosis: a case report and review of the literature. American Journal of Medical Genetics. Part A. 2008;146A(10):1358–67. doi: 10.1002/ajmg.a.32261. http://doi.org/10.1002/ajmg.a.32261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery Ta, Harris JB, Willems PJ, Oostra Ba, Irwin Sa, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(May):5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford J, Lower KM, Hennekam RCM, Van Esch H, Mégarbané a, Lynch S, Turner G, Gécz J. Mutation screening in Borjeson-Forssman-Lehmann syndrome: identification of a novel de novo PHF6 mutation in a female patient. Journal of Medical Genetics. 2006;43(3):238–43. doi: 10.1136/jmg.2005.033084. http://doi.org/10.1136/jmg.2005.033084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter CF, van Dijk F, Stolker JJ, Hennekam RCM. Behavioural phenotype in Börjeson-Forssman-Lehmann syndrome. Journal of Intellectual Disability Research: JIDR. 2009;53(4):319–28. doi: 10.1111/j.1365-2788.2009.01156.x. http://doi.org/10.1111/j.1365-2788.2009.01156.x. [DOI] [PubMed] [Google Scholar]
- Di Donato N, Isidor B, Lopez Cazaux S, Le Caignec C, Klink B, Kraus C, Schrock E, Hackmann K. Distinct phenotype of PHF6 deletions in females. European Journal of Medical Genetics. 2014;57(2–3):85–9. doi: 10.1016/j.ejmg.2013.12.003. http://doi.org/10.1016/j.ejmg.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Human Molecular Genetics. 1996;5(8):1083–1091. doi: 10.1093/hmg/5.8.1083. http://doi.org/10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- Franzoni E, Booker SA, Parthasarathy S, Rehfeld F, Grosser S, Srivatsa S, Fuchs RH, Tarabykin V, Vida I, Wulczyn FG. miR-128 regulates neuronal migration, outgrowth and intrinsic excitability via the intellectual disability gene Phf6. eLife. 2015;2015(4):1–23. doi: 10.7554/eLife.04263. http://doi.org/10.7554/eLife.04263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, Hemberg M, Ebert DH, Greenberg ME. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522(7554):89–93. doi: 10.1038/nature14319. http://doi.org/10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gécz J, Turner G, Nelson J, Partington M. The Börjeson-Forssman-Lehman syndrome (BFLS, MIM #301900) European Journal of Human Genetics: EJHG. 2006;14(12):1233–7. doi: 10.1038/sj.ejhg.5201639. http://doi.org/10.1038/sj.ejhg.5201639. [DOI] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nature Reviews. Neuroscience. 2013;14(11):755–69. doi: 10.1038/nrn3586. http://doi.org/10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A Chromatin Landmark and Transcription Initiation at Most Promoters in Human Cells. Cell. 2007;130(1):77–88. doi: 10.1016/j.cell.2007.05.042. http://doi.org/10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird a. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nature Genetics. 2001;27(3):322–6. doi: 10.1038/85899. http://doi.org/10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(11):7746–50. doi: 10.1073/pnas.122205699. http://doi.org/10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagnatiello F, Guidotti AR, Pesold C, Dwivedi Y, Caruncho H, Pisu MG, Uzunov DP, Smalheiser NR, Davis JM, Pandey GN, Pappas GD, Tuetinh P, Sharma PR, Costa E. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(26):15718–23. doi: 10.1073/pnas.95.26.15718. http://doi.org/10.1073/pnas.95.26.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin Sa, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cerebral Cortex (New York, NY: 1991) 2000;10(10):1038–1044. doi: 10.1093/cercor/10.10.1038. http://doi.org/10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- Jaehning JA. The Paf1 complex: Platform or player in RNA polymerase II transcription? Biochimica et Biophysica Acta - Gene Regulatory Mechanisms. 2010;1799(5–6):379–388. doi: 10.1016/j.bbagrm.2010.01.001. http://doi.org/10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Warren ST. Understanding the molecular basis of fragile X syndrome. Human Molecular Genetics. 2000;9(6):901–908. doi: 10.1093/hmg/9.6.901. http://doi.org/10.1093/hmg/9.6.901. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cerebral Cortex. 2000;10(10):981–991. doi: 10.1093/cercor/10.10.981. http://doi.org/10.1207/S15326942DN1603_18. [DOI] [PubMed] [Google Scholar]
- Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, Vincent AK, Malli R, Ali G, Khan FS, Ishak GE, Doherty D, Weksberg R, Ayub M, Windpassinger C, Ibrahim S, Frye M, Ansar M, Vincent JB. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. American Journal of Human Genetics. 2012;90(5):856–863. doi: 10.1016/j.ajhg.2012.03.023. http://doi.org/10.1016/j.ajhg.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinugasa Y, Ishiguro H, Tokita Y, Oohira A, Ohmoto H, Higashiyama S. Neuroglycan C, a novel member of the neuregulin family. Biochemical and Biophysical Research Communications. 2004;321(4):1045–1049. doi: 10.1016/j.bbrc.2004.07.066. http://doi.org/10.1016/j.bbrc.2004.07.066. [DOI] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, … Helin K. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Molecular Cell. 2010;38(2):165–78. doi: 10.1016/j.molcel.2010.03.002. http://doi.org/10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothare SV, VanLandingham K, Armon C, Luther JS, Friedman A, Radtke RA. Seizure onset from periventricular nodular heterotopias: depth-electrode study. Neurology. 1998;51(6):1723–7. doi: 10.1212/wnl.51.6.1723. http://doi.org/10.1212/WNL.51.6.1723. [DOI] [PubMed] [Google Scholar]
- Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends in Neurosciences. 2004;27(7):392–399. doi: 10.1016/j.tins.2004.05.001. http://doi.org/10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Molecular Cell. 2003;11(3):721–729. doi: 10.1016/s1097-2765(03)00091-1. http://doi.org/10.1016/S1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA Polymerase II Elongation Factors of Saccharomyces cerevisiae: a Targeted Proteomics Approach. Society. 2002;22(20):6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. http://doi.org/10.1128/MCB.22.20.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Hickey LB, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes and Development. 1995;9(5):559–572. doi: 10.1101/gad.9.5.559. http://doi.org/10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- Lai AY, Wade Pa. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nature Reviews. Cancer. 2011;11(8):588–96. doi: 10.1038/nrc3091. http://doi.org/10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson S, KCLLANKJHL DA. Prevalence of mental retardation and developmental disabilities: Estimates from the 1994/1995 National Health Interview Survey Disability Supplements. American Journal on Mental Retardation. 2001;106(3):231–295. doi: 10.1352/0895-8017(2001)106<0231:POMRAD>2.0.CO;2. http://doi.org/10.1352/0895-8017(2001) [DOI] [PubMed] [Google Scholar]
- Lee SCS, Cowgill EJ, Al-Nabulsi A, Quinn EJ, Evans SM, Reese BE. Homotypic regulation of neuronal morphology and connectivity in the mouse retina. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2011;31(40):14126–33. doi: 10.1523/JNEUROSCI.2844-11.2011. http://doi.org/10.1523/JNEUROSCI.2844-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh MJ, Hagerman RJ. Fragile X Clinical Features and Neurobiology. Neural Circuit Development and Function in the Heathy and Diseased Brain. Chapter 33. Elsevier Inc; 2013. http://doi.org/10.1016/B978-0-12-397267-5.00044-3. [Google Scholar]
- Li J, Pelletier MR, Perez Velazquez JL, Carlen PL. Reduced cortical synaptic plasticity and GluR1 expression associated with fragile X mental retardation protein deficiency. Molecular and Cellular Neurosciences. 2002;19(2):138–151. doi: 10.1006/mcne.2001.1085. http://doi.org/10.1006/mcne.2001.1085. [DOI] [PubMed] [Google Scholar]
- Liu Z, Li F, Ruan K, Zhang J, Mei Y, Wu J, Shi Y. Structural and functional insights into the human Börjeson-Forssman-Lehmann syndrome-associated protein PHF6. The Journal of Biological Chemistry. 2014;289(14):10069–83. doi: 10.1074/jbc.M113.535351. http://doi.org/10.1074/jbc.M113.535351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower KM, Solders G, Bondeson ML, Nelson J, Brun A, Crawford J, Malm G, Borjeson M, Turner G, Partington M, Gécz J. 1024C> T (R342X) is a recurrent PHF6 mutation also found in the original Börjeson-Forssman-Lehmann syndrome family. European Journal of Human Genetics: EJHG. 2004;12(10):787–9. doi: 10.1038/sj.ejhg.5201228. http://doi.org/10.1038/sj.ejhg.5201228. [DOI] [PubMed] [Google Scholar]
- Lower KM, Turner G, Kerr Ba, Mathews KD, Shaw Ma, Gedeon AK, Schelley S, Hoyme E, White SM, Delatycki MB, Lampe AK, Clayton-Smith J, Stewart H, van Ravenswaay CMA, de Vries BBA, Cox B, Grompe M, Ross S, Thomas P, Mulley JC, Gécz J. Mutations in PHF6 are associated with Börjeson-Forssman-Lehmann syndrome. Nature Genetics. 2002;32(4):661–5. doi: 10.1038/ng1040. http://doi.org/10.1038/ng1040. [DOI] [PubMed] [Google Scholar]
- Manent JB, Wang Y, Chang Y, Paramasivam M, LoTurco JJ. Dcx reexpression reduces subcortical band heterotopia and seizure threshold in an animal model of neuronal migration disorder. Nature Medicine. 2009;15(1):84–90. doi: 10.1038/nm.1897. http://doi.org/10.1038/nm1111-1521c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf M, Chevrier E, Mustonen A, Picketts DJ. Börjeson-Forssman-Lehmann Syndrome due to a novel plant homeodomain zinc finger mutation in the PHF6 gene. Journal of Child Neurology. 2009;24(5):610–4. doi: 10.1177/0883073808327830. http://doi.org/10.1177/0883073808327830. [DOI] [PubMed] [Google Scholar]
- Marchetto MCN, Carromeu C, Acab A, Yu D, Yeo G, Mu Y, Chen G, Gage FH, Muotri Alysson R. A model for neural development and treatment of Rett Syndrome using human induced pluripotent stem cells. Cell. 2010;143(4):527–539. doi: 10.1016/j.cell.2010.10.016. http://doi.org/doi:10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton HA, Desiderio S. The Paf1 complex promotes displacement of histones upon rapid induction of transcription by RNA polymerase II. BMC Molecular Biology. 2008;9:4. doi: 10.1186/1471-2199-9-4. http://doi.org/10.1186/1471-2199-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Brenner C, Fazi F, Villa R, Gutierrez A, Buschbeck M, Nervi C, Minucci S, Fuks F, Di Croce L. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Molecular and Cellular Biology. 2008;28(19):5912–23. doi: 10.1128/MCB.00467-08. http://doi.org/10.1128/MCB.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan-Smith M, Wu Y, Zhu X, Pringle J, Snider WD. GSK-3 signaling in developing cortical neurons is essential for radial migration and dendritic orientation. eLife. 2014;3:e02663. doi: 10.7554/eLife.02663. http://doi.org/10.7554/eLife.02663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nature Genetics. 2007;39(12):1507–11. doi: 10.1038/ng.2007.21. http://doi.org/10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah B, Parnavelas JG. Modes of neuronal migration in the developing cerebral cortex. Nature Reviews. Neuroscience. 2002;3(6):423–32. doi: 10.1038/nrn845. http://doi.org/10.1038/nrn845. [DOI] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88(4):471–481. doi: 10.1016/s0092-8674(00)81887-5. http://doi.org/10.1016/S0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–389. doi: 10.1038/30764. http://doi.org/10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Transcription Initiation Into Productive Elongation. 2012;1809(1):34–45. doi: 10.1016/j.bbagrm.2010.11.001. http://doi.org/10.1016/j.bbagrm.2010.11.001.Pol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A, Langst G. Genome organization in and around the nucleolus. Trends in Genetics. 2011;27(4):149–156. doi: 10.1016/j.tig.2011.01.002. http://doi.org/10.1016/j.tig.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Paradee W, Melikian HE, Rasmussen DL, Kenneson A, Conn PJ, Warren ST. Fragile X mouse: Strain effects of knockout phenotype and evidence suggesting deficient amygdala function. Neuroscience. 1999;94(1):185–192. doi: 10.1016/s0306-4522(99)00285-7. http://doi.org/10.1016/S0306-4522(99)00285-7. [DOI] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, Maza I, Mor N, Baran D, Weinberger L, Jaitin DA, Lara-Astiaso D, Blecher-Gonen R, Shipny Z, Mukamel Z, Hagai T, Gilad S, Amann-Zalcenstein D, Tanay A, Amit I, Novershtern N, Hanna JH. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502(7469):65–70. doi: 10.1038/nature12587. http://doi.org/10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Reiss AL, Faruque F, Naidu S, Abrams M, Beaty T, Bryan RN, Moser H. Neuroanatomy of Rett Syndrome: A VolumetIric Imagmg Study. 1993:227–234. doi: 10.1002/ana.410340220. [DOI] [PubMed] [Google Scholar]
- Rio C, Rieff HI, Qi P, Khurana TS, Corfas G. Neuregulin and erbB receptors play a critical role in neuronal migration. Neuron. 1997;19:39–50. doi: 10.1016/s0896-6273(00)80346-3. http://doi.org/S0896-6273(00)80346-3 [pii] [DOI] [PubMed] [Google Scholar]
- Robinson LK, Jones KL, Culler F, Nyhan WL, Sakati N, Jones KL. The Borjeson-Forssman-Lehmann Syndrome. 1983;468:457–468. doi: 10.1002/ajmg.1320150311. [DOI] [PubMed] [Google Scholar]
- Rondon A, Gallardo M, Garcia-Rubio M, Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 2004;5(1):47–53. doi: 10.1038/sj.embor.7400045. http://doi.org/10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers HH. Genetics of early onset cognitive impairment. Annual Review of Genomics and Human Genetics. 2010;11(X):161–87. doi: 10.1146/annurev-genom-082509-141640. http://doi.org/10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- Roussel P, André C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. Journal of Cell Biology. 1996;133(2):235–246. doi: 10.1083/jcb.133.2.235. http://doi.org/10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaco RC, Neul JL. Complexities of Rett Syndrome and MeCP2. Journal of Neuroscience. 2011;31(22):7951–7959. doi: 10.1523/JNEUROSCI.0169-11.2011. http://doi.org/10.1523/JNEUROSCI.0169-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. The PHD finger: A versatile epigenome reader. Trends in Biochemical Sciences. 2011;36(7):364–372. doi: 10.1016/j.tibs.2011.03.005. http://doi.org/10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr ARW, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 Is Expressed at Near Histone-Octamer Levels and Globally Alters the Chromatin State. Molecular Cell. 2010;37(4):457–468. doi: 10.1016/j.molcel.2010.01.030. http://doi.org/10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So HC, Fong PY, Chen RYL, Hui TCK, Ng MYM, Cherny SS, Mak WWM, Cheung EFC, Chan RCK, Chen EYH, Li T, Sham PC. Identification of neuroglycan C and interacting partners as potential susceptibility genes for schizophrenia in a Southern Chinese population. American Journal of Medical Genetics, Part B: Neuropsychiatric Genetics. 2010;153(1):103–113. doi: 10.1002/ajmg.b.30961. http://doi.org/10.1002/ajmg.b.30961. [DOI] [PubMed] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO Journal. 2002;21(7):1764–1774. doi: 10.1093/emboj/21.7.1764. http://doi.org/10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd MaM, Picketts DJ. PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. Journal of Proteome Research. 2012;11(8):4326–37. doi: 10.1021/pr3004369. http://doi.org/10.1021/pr3004369. [DOI] [PubMed] [Google Scholar]
- Tomita K, Kubo K ichiro, Ishii K, Nakajima K. Disrupted-in-schizophrenia-1 (Disc1) is necessary for migration of the pyramidal neurons during mouse hippocampal development. Human Molecular Genetics. 2011;20(14):2834–2845. doi: 10.1093/hmg/ddr194. http://doi.org/10.1093/hmg/ddr194. [DOI] [PubMed] [Google Scholar]
- Tomson BN, Arndt KM. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochimica et Biophysica Acta - Gene Regulatory Mechanisms. 2013;1829(1):166–126. doi: 10.1016/j.bbagrm.2012.08.011. http://doi.org/10.1016/j.bbagrm.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G, Lower KM, White SM, Delatycki M, Lampe aK, Wright M, Clayton-Smith J, Kerr B, Schelley S, Hoyme HE, De Vries BBA, Kleefstra T, Grompe M, Cox B, Gecz J, Partington M. The clinical picture of the Börjeson-Forssman-Lehmann syndrome in males and heterozygous females with PHF6 mutations. Clinical Genetics. 2004;65(3):226–32. doi: 10.1111/j.0009-9163.2004.00215.x. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/14756673. [DOI] [PubMed] [Google Scholar]
- Vaillend C, Poirier R, Laroche S. Genes, plasticity and mental retardation. Behavioural Brain Research. 2008;192(1):88–105. doi: 10.1016/j.bbr.2008.01.009. http://doi.org/10.1016/j.bbr.2008.01.009. [DOI] [PubMed] [Google Scholar]
- van Bokhoven H, Kramer JM. Disruption of the epigenetic code: an emerging mechanism in mental retardation. Neurobiology of Disease. 2010;39(1):3–12. doi: 10.1016/j.nbd.2010.03.010. http://doi.org/10.1016/j.nbd.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Verkerk AJM, Pieretti M, Sutcliffe JS, Fu Y, Kuhl DPA, Pixxuti A, Reiner O, Richards S, Victoria MF, Zhang F, Eussen BE, van Ommen G, Blonden LAJ, Riggins GJ, Chastain JL, Kunst CB, Galjaard H, Caskey CT, Nelson DL, Oostra BA, Warren ST. Identification of a Gene ( FMR-1 ) Containing a CGG Repeat Coincident with a Breakpoint Cluster Region Exhibiting length Variation in Fragile X Syndrome. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Voss AK, Gamble R, Collin C, Shoubridge C, Corbett M, Gécz J, Thomas T. Protein and gene expression analysis of Phf6, the gene mutated in the Börjeson-Forssman-Lehmann Syndrome of intellectual disability and obesity. Gene Expression Patterns: GEP. 2007;7(8):858–71. doi: 10.1016/j.modgep.2007.06.007. http://doi.org/10.1016/j.modgep.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Wang J, Leung JW, Gong Z, Feng L, Shi X, Chen J. PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis. The Journal of Biological Chemistry. 2013;288(5):3174–83. doi: 10.1074/jbc.M112.414839. http://doi.org/10.1074/jbc.M112.414839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen MH, Nijhof B, Fenckova M, Nillesen WM, Bongers EMHF, Castells-Nobau A, … Kleefstra T. GATAD2B loss-of-function mutations cause a recognisable syndrome with intellectual disability and are associated with learning deficits and synaptic undergrowth in Drosophila. Journal of Medical Genetics. 2013;50(8):507–14. doi: 10.1136/jmedgenet-2012-101490. http://doi.org/10.1136/jmedgenet-2012-101490. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. Journal of Biological Chemistry. 2003;278(37):34739–34742. doi: 10.1074/jbc.C300269200. http://doi.org/10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442(7098):86–90. doi: 10.1038/nature04815. http://doi.org/10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yang Y, Hemberg M, Yoshida T, Cho HY, Murphy JP, Fioravante D, Regehr WG, Gygi SP, Georgopoulos K, Bonni A. Promoter decommissioning by the NuRD chromatin remodeling complex triggers synaptic connectivity in the mammalian brain. Neuron. 2014;83(1):122–34. doi: 10.1016/j.neuron.2014.05.039. http://doi.org/10.1016/j.neuron.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AS, Pritchard M, Kremer E, Lynch M, Nancarrow J, Baker E, Holman K, Mulley JC, Warren ST, Schlessinger D, Sutherland GR, Richards RI. Fragile X Genotype Characterized by an Unstable Region of DNA. 1991;252(5009):1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SGN, Bagni C. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nature Neuroscience. 2007;10(5):578–87. doi: 10.1038/nn1893. http://doi.org/10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Mejia La, Huang J, Valnegri P, Bennett EJ, Anckar J, Jahani-Asl A, Gallardo G, Ikeuchi Y, Yamada T, Rudnicki M, Harper JW, Bonni A. The X-linked intellectual disability protein PHF6 associates with the PAF1 complex and regulates neuronal migration in the mammalian brain. Neuron. 2013;78(6):986–93. doi: 10.1016/j.neuron.2013.04.021. http://doi.org/10.1016/j.neuron.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The Dermatomyositis-Specific Autoantigen Mi2 Is a Component of a Complex Containing Histone Deacetylase and Nucleosome Remodeling Activities. Cell. 1998;95(2):279–289. doi: 10.1016/s0092-8674(00)81758-4. http://doi.org/10.1016/S0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng H, Erdjument-bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation 1924–1935. 1999 doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes and Development. 2005;19(14):1668–1673. doi: 10.1101/gad.1292105. http://doi.org/10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier C, Kraus C, Brueton L, Cole T, Degenhardt F, Engels H, Gillessen-Kaesbach G, Graul-Neumann L, Horn D, Hoyer J, Just W, Rauch A, Reis A, Wollnik B, Zeschnigk M, Ludecke HJ, Wieczorek D. A new face of Borjeson-Forssman-Lehmann syndrome? De novo mutations in PHF6 in seven females with a distinct phenotype. Journal of Medical Genetics. 2013;50(12):838–47. doi: 10.1136/jmedgenet-2013-101918. http://doi.org/10.1136/jmedgenet-2013-101918. [DOI] [PubMed] [Google Scholar]