Abstract
The use of Trichoderma isolates with efficient antagonistic activity represents a potentially effective and alternative disease management strategy to replace health hazardous chemical control. In this context, twenty isolates were obtained from tomato rhizosphere and evaluated by their antagonistic activity against four fungal pathogens (Fusarium oxysporum f. sp. lycopersici, Alternaria alternata, Colletotrichum gloeosporoides and Rhizoctonia solani). The production of extracellular cell wall degrading enzymes of tested isolates was also measured. All the isolates significantly reduced the mycelial growth of tested pathogens but the amount of growth reduction varied significantly as well. There was a positive correlation between the antagonistic capacity of Trichoderma isolates towards fungal pathogens and their lytic enzyme production. The Trichoderma isolates were initially sorted according to morphology and based on the translation elongation factor 1-α gene sequence similarity, the isolates were designated as Trichoderma harzianum, T. koningii, T. asperellum, T. virens and T. viride. PCA analysis explained 31.53, 61.95, 62.22 and 60.25% genetic variation among Trichoderma isolates based on RAPD, REP-, ERIC- and BOX element analysis, respectively. ERG-1 gene, encoding a squalene epoxidase has been used for the first time for diversity analysis of antagonistic Trichoderma from tomato rhizosphere. Phylogenetic analysis of ERG-1 gene sequences revealed close relatedness of ERG-1sequences with earlier reported sequences of Hypocrea lixii, T. arundinaceum and T. reesei. However, ERG-1 gene also showed heterogeneity among some antagonistic isolates and indicated the possibility of occurrence of squalene epoxidase driven triterpene biosynthesis as an alternative biocontrol mechanism in Trichoderma species.
Keywords: Antagonism, Biocontrol, Diversity, Ergosterol, Tomato, Trichoderma
Background
The genus Trichoderma has gained immense importance in past several decades due to its antagonistic ability against wide range of plant pathogens and growth promotion in crop plants. Some species of Trichoderma viz., Trichoderma harzianum, T. viride, T. virens and T. koningii are well known antagonists and are being utilized to control plant pathogens under field conditions (Solanki et al. 2011; Srivastava et al. 2012; Galarza et al. 2015). Promising Trichoderma isolates have different mechanisms or combination of direct parasitism, competition for nutrients, stimulators of plant health, or inducers of plant systemic resistance against various pathogens (Harman et al. 2004; Anees et al. 2010; Woo et al. 2014; Jain et al. 2015; Rai et al. 2016). A plethora of antagonistic Trichoderma isolates have been identified by several researchers from different places around the world, having history of varied climate, soil type, cropping system, etc., which differ in their innocuousness and efficacy as biocontrol agents (Sharma et al. 2009; Błaszczyk et al. 2011; Martínez-Medina et al. 2014; Galarza et al. 2015; El_Komy et al. 2015). Therefore, the site specific recommendations are being made according to the fitness potential of a particular isolate for higher efficacy and effectiveness. Despite the commercial successes of these biocontrol agents, the major limitations remain their restricted efficacy and inconsistency under field conditions. Consequently, more efficient Trichoderma isolates with high antagonistic potential capabilities are needed for successful biological control systems.
Due to the ecological importance of Trichoderma spp. and their application as a biocontrol agent in the field, it is important to understand their biodiversity. However, accurate species identification based on morphology is difficult due to the paucity and similarity of morphological characters and increasing numbers of morphologically cryptic species (Kullnig et al. 2001). This has already resulted in incorrect identification. In recent years, the usefulness of molecular markers such as random amplified polymorphic DNA (RAPD) and repetitive-element polymerase chain reaction (REP-PCR) in resolving species differences among microbial species are also well documented (Sharma et al. 2009; Solanki et al. 2013; Srivastava et al. 2014; Singh et al. 2014; Kashyap et al. 2015). RAPD utilized PCR to amplify DNA segments with single primer of arbitrary nucleotide sequence generating fragments by hybridizing with compatible regions of DNA and amplifying the regions where the primers are in correct orientation and appropriately spaced (100–2500 bp). However, REP-PCR uses oligonucelotide primers complementary to repetitive sequences dispersed throughout the genome. Using PCR, this method amplifies diverse regions of DNA flanked by the conserved repetitive sequences, leading to amplicon patterns specific for an individual bacterial and fungal strain. Three different families of repetitive sequences include: the 35–40 bp repetitive extragenic pallindromic (REP) sequence, the 124–127 bp enterobacterial repetitive intergenic consensus (ERIC) sequence and 154 bp BOX (composed of the box A, B and C subunits) element. These sequences appear to be located in distinct, intergenic positions around the genome elements (Mohapatra et al. 2007). Methods based on such repetitive elements have also been used for studying the diversity in the ecosystem, presenting the phylogenetic relationship between strains and discriminating between microorganisms those are genetically close to each other (Rai et al. 2015; Kashyap et al. 2016). Unfortunately, these methods have not been extensively used for the differentiation of Trichoderma spp. Since, species of Trichoderma are reported as the causal agent of green mould disease (Ospina-Giraldo et al. 1998), the understanding of the nature and diversity of Trichoderma is critical for its widespread use against phytopathogenic fungi as there could be the risk of unwanted disease on non-target hosts. Under such situations, it is valuable to establish patterns of gene flow, as well as to develop a fingerprint of Trichoderma isolates. Diversity studies have recently been undertaken to assess its ecological specialization. Several studies reported about a series of new isolates as well as new phylogenetic species of Trichoderma in a series of natural ecosystems (Zachow et al. 2009; Körmöczi et al. 2013). On the other hand, only a few studies were focusing on agricultural environments. However, the results of these studies demonstrated that besides the natural ecosystems, the investigation of agricultural soils also reveals important information about Trichoderma biodiversity. The practical impact of such studies is that the rhizosphere of agricultural soils may be an ideal source of beneficial strains with biocontrol potential. Based on these studies, we speculate that the species composition, distribution, and genetic structure of Trichoderma on the tomato rhizosphere may be different. The confirmation of the differences will help in revealing the biodiversity, origin, and evolutionary processes of Trichoderma under different biological niches.
Recent evidences indicated the importance of the sterol biosynthetic pathway in inducing plant defense-related gene expression in both the antagonistic fungus and the plant (Cardoza et al. 2011; Malmierca et al. 2013; Cardoza et al. 2014). The structural and functional analysis of genes involved in the synthesis of ergosterol especially intermediates, such as squalene could provide additional strategies to improve the ability of biocontrol of the Trichoderma strains. To best of the knowledge, there are no reports available on the diversity analysis of ergosterol producing antagonistic Trichoderma species using ERG1 gene, encoding a squalene epoxidase, a key enzyme in the biosynthesis of triterpene derivatives (e.g. ergosterol) from tomato rhizosphere. Thus, to test above mentioned hypothesis, attempts have been made to investigate the species distribution of Trichoderma associated with tomato plants. The comparison of the genetic structure between antagonistic Trichoderma isolates was carried out by molecular (RAPD, REP, ERIC and BOX markers), and biochemical (production of cell wall degrading enzymes) markers. Sequencing based on the characterization of squalene epoxidase (ERG1) gene in antagonistic isolates was performed to get preliminary clues about the role of squalene epoxidase driven triterpene biosynthesis in biocontrol mechanisms of tested isolates.
Methods
Sampling and identification of Trichoderma isolates
Twenty isolates of Trichoderma were obtained from healthy tomato (Solanum lycopersicum cv. VL tamatar 4) rhizosphere (Table 1). Ten healthy plants (~55 days post transplanting) with their roots and rhizospheric soil were randomly sampled and immediately transported to the laboratory. The soil particles attached to roots were carefully collected after uprooting plants, stored at 4 °C and processed within 24 h of collection. Root adhered soil (10 g) was suspended in 90 ml of sterile distilled water and dilution plate technique was used for the isolation of Trichoderma spp. The suspensions from all samples were serially diluted (up to 10−5) and 100 µl of each dilution was added to sterile Petri dishes, in triplicates of each dilution, containing sterile Potato Dextrose Agar (PDA) medium. Streptomycin solution (1%) was added to the medium for preventing bacterial growth, before pouring into Petri plates. The plates were then incubated at 28 ± 1 °C. The isolates were characterized based on the monograph of Gams and Bissett (1998). For morphological analysis, isolates were grown on PDA at 28 ± 1 °C for 5–7 days. Radial growth was measured at 24 h intervals until colony covered the whole Petri dish. Growth rate was calculated as the 7 day average of mean daily growth (mm day−1). All micro morphological data were examined from cultures grown on PDA for 5 days at 28 ± 1 °C. Microscopic observations were done using trinocular microscope (Axio Imager M2 microscope, Carl Zeiss, Germany). For examination of conidial morphology, cultures were washed with sterile water and drops of the suspension were placed on microscope slides and mixed with lactophenol/cotton blue to stain the conidia. Length and width were measured for 30 conidia per isolate. Conidial morphology and size were recorded after 7 days of incubation.
Table 1.
Identification, origin, NCBI Genebank accession numbers, cell wall-degrading enzymes and antagonistic effect of Trichoderma isolates from tomato rhizosphere against fungal plant pathogens
| Code | Isolate (s) | Region | GenBank accession | Cell wall-degrading enzymes | Mycelia inhibition over control (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| EF-1α | ERG1 | Chitinasex | β-1,3 glucanasey | FOL | AA | CG | RS | |||
| UNT60 | Trichoderma harzianum | U.S. Nagar, Uttarakhand | KF360991 | KT989041 | *40.00 ± 1.42h | 83.67 ± 1.95i | 58.54 ± 2.41 g | 58.89 ± 2.14i | 59.80 ± 1.88 g | 53.30 ± 1.99 g |
| UNT64 | T. harzianum | U.S. Nagar, Uttarakhand | KF360992 | KT989042 | 69.00 ± 3.54b | 55.60 ± 0.85l | 62.45 ± 1.56f | 61.50 ± 1.91h | 64.50 ± 2.11cd | 62.78 ± 1.88bcd |
| UNT68 | T. harzianum | U.S. Nagar, Uttarakhand | KF360993 | KT989043 | 76.56 ± 3.95a | 90.56 ± 0.88g | 77.94 ± 2.21a | 79.47 ± 1.88a | 73.94 ± 1.88a | 69.23 ± 1.35a |
| NAT69 | T. harzianum | Nanital, Uttarakhand | KF360994 | KT989044 | 53.33 ± 1.44e | 53.33 ± 0.55n | 57.65 ± 1.12g | 53.98 ± 1.34j | 59.50 ± 1.77g | 54.80 ± 1.66g |
| NAT70 | T. harzianum | Nanital, Uttarakhand | KF360995 | KT989045 | 37.00 ± 1.20i | 137.42 ± 0.65e | 62.34 ± 1.21f | 62.50 ± 1.99gh | 60.89 ± 1.55fg | 62.79 ± 1.64bcd |
| ALT73 | T. harzianum | Almora, Uttarakhand | KF360996 | KT989046 | 49.67 ± 2.01f | 47.67 ± 0.44p | 65.78 ± 1.54e | 53.68 ± 1.86j | 64.70 ± 1.66cd | 63.65 ± 1.94b |
| DET89 | T. harzianum | Dehradun, Uttarakhand | KF360997 | KT989047 | 67.00 ± 3.93b | 91.66 ± 1.99g | 61.98 ± 1.34f | 67.87 ± 1.54d | 62.56 ± 1.35e | 58.83 ± 1.51e |
| DET94 | T. harzianum | Dehradun, Uttarakhand | KF360998 | KT989048 | 69.00 ± 4.10b | 171.66 ± 1.01b | 74.35 ± 1.14b | 65.50 ± 1.99e | 68.89 ± 2.01b | 67.89 ± 1.35a |
| HAT96 | T. harzianum | Haridwar, Uttarakhand | KF360999 | KT989049 | 73.60 ± 3.35a | 63.26 ± 0.971 | 68.95 ± 1.21d | 63.45 ± 1.35 fg | 60.54 ± 1.35fg | 61.11 ± 1.91cd |
| UNT38 | T. koningii | U.S. Nagar, Uttarakhand | KF361001 | KT989051 | 74.60 ± 3.55a | 134.61 ± 1.06f | 70.73 ± 1.12c | 69.80 ± 1.36c | 68.90 ± 1.55b | 64.78 ± 1.35b |
| UNS63 | T. koningii | U.S. Nagar, Uttarakhand | KF361002 | KT989052 | 39.00 ± 1.99h | 89.33 ± 1.04h | 52.98 ± 1.33i | 56.89 ± 1.25i | 56.90 ± 1.97h | 59.80 ± 2.11de |
| UNT13 | T. asperellum | U.S. Nagar, Uttarakhand | KF361003 | KT989053 | 56.56 ± 2.44d | 51.26 ± 0.95o | 55.80 ± 1.21h | 61.89 ± 1.44h | 62.80 ± 1.25e | 59.80 ± 1.87de |
| UNT70 | T. asperellum | U.S. Nagar, Uttarakhand | KF361004 | KT989054 | 54.33 ± 1.66e | 34.33 ± 0.65q | 53.90 ± 1.44i | 59.98 ± 0.99h | 60.70 ± 1.35fg | 62.89 ± 1.76bcd |
| UNS28 | T. virens | U.S. Nagar, Uttarakhand | KF361005 | KT989055 | 61.00 ± 1.32c | 78.35 ± 0.78j | 64.30 ± 1.31e | 67.50 ± 1.34d | 65.80 ± 1.97c | 60.80 ± 1.54cde |
| UNS30 | T. virens | U.S. Nagar, Uttarakhand | KF361006 | KT989056 | 73.67 ± 2.12a | 175.1 ± 1.20a | 70.89 ± 1.21c | 75.60 ± 1.65b | 65.70 ± 1.54c | 64.58 ± 1.62b |
| NAS46 | T. virens | Nanital, Uttarakhand | KF361007 | KT989057 | 44.00 ± 1.19g | 54.22 ± 0.55mn | 65.80 ± 1.33e | 60.80 ± 1.21hi | 59.80 ± 1.35g | 57.50 ± 1.31ef |
| ALS47 | T. virens | Almora, Uttarakhand | KF361008 | KT989058 | 57.00 ± 2.37d | 72.52 ± 0.72k | 69.00 ± 1.54d | 65.78 ± 1.35e | 61.90 ± 1.67e | 62.80 ± 1.15bc |
| UNT09 | T. viride | U.S. Nagar, Uttarakhand | KF361011 | KT989061 | 59.90 ± 1.15c | 56.55 ± 0.96l | 67.83 ± 1.34d | 64.45 ± 1.44ef | 60.85 ± 1.88efg | 60.70 ± 1.25cde |
| DET02 | T. viride | Dehradun, Uttarakhand | KF361012 | KT989062 | 31.00 ± 1.25j | 163.33 ± 1.22c | 72.05 ± 1.68c | 68.89 ± 1.55cd | 64.44 ± 1.66cd | 60.35 ± 1.35cde |
| NAT03 | T. viride | Nanital, Uttarakhand | KF361013 | KT989062 | 61.67 ± 2.03c | 151.67 ± 1.10d | 61.80 ± 1.34f | 64.50 ± 1.39ef | 59.48 ± 1.95g | 60.45 ± 1.75cde |
* Within columns, mean ± SE values with a common letter do not differ significantly (P < 0.05), according to DMRT test
xμmol of GlcNAc min−1 mg−1 protein
ynmol of glucose min−1 mg−1 protein
Screening the antagonistic activity of Trichoderma isolates
In vitro antagonistic potential of the biocontrol agent was evaluated against Fusarium oxysporum f. sp. lycopersici (FOL), Alternaria alternata (AA), Colletotrichum gloeosporoides (CG) and Rhizoctonia solani (RS) through dual culture technique. For this, the pathogenic fungi were obtained from National Agriculturally Important Microorganisms Culture Collection (NAIMCC), NBAIM, Mau, Uttar Pradesh. After purification, the culture was maintained on PDA. The isolates were further screened for their antagonistic potential against the pathogen on PDA by measuring the relative growth rates as a function of the incubation period. Five mm mycelial discs taken from the margin of young vigorously growing 5-day-old culture of the antagonists and the pathogen was inoculated at the margin of the Petridish containing 20 ml sterilized PDA medium (opposite to each other). Observations were recorded up to 7 days of incubation (at 28 ± 1 °C). The treatments were replicated five times.
Molecular characterization of antagonists
Total genomic DNA from fungus was extracted with cetyl-trimethylammonium bromide (CTAB) as described by Kumar et al. (2013a). Briefly, for each fungal isolates, fresh mycelium (~5 g) was dried on sterile blotter paper and was ground in liquid nitrogen to make a fine powder. This powder was taken in a centrifuge tube and 2× CTAB (hexadecyltrimethyl ammonium bromide) buffer (15 ml) was added in each tube separately. Extraction buffer contained (per 1 l) 2 g CTAB, 1 M Tris pH-8 (10 ml), 5 M NaCl (28 ml), 0.5 M EDTA (4 ml) with sterile distilled water (57 ml) and 1 ml β-mercaptoethanol. This was incubated in water bath at 65 °C for 30 min with intermittent shaking. The mixture was centrifuged at 13,000 rpm for 15 min at 4 °C to pellet the mycelium. Supernatant was taken into another Oakridge tube and an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added with slow inversion. The mixture was again centrifuged at 13,000 rpm for 15 min at 4 °C. The aqueous supernatant was taken in a fresh tube and added 0.6 volume isopropanol and was incubated at −20 °C overnight. After incubation, it was again centrifuged at 13,000 rpm for 20 min at 4 °C temperature. The supernatant was discarded and pellet was washed with 70% ethanol. The pellet was dissolved in 500 μl of TE buffer for the use in PCR and stored at −20 °C.
For the molecular identification of ergosterol producing isolates, ERG1F (5ʹ-CGCTCCGTGCTTCTTCTTCTC-3ʹ) and EGR1R (5ʹ-CTTCTTCTCTCCCGTCTCC-3ʹ) primers were used. The PCR reaction was carried out in a 25-μl reaction mixture containing the following: 10× PCR buffer, 50 ng DNA template, 2 mM MgCl2, 0.25 mM dNTP mixture and 0.25 μM each of primer, and one unit of Taq Polymerase (Bangalore Genie, India). Amplifications were performed in Thermal Cycler (G Storm GS4, Somerset, UK) under the following conditions: initial denaturation 5 min at 94 °C, 35 cycles of 45 s at 94 °C, 45 s at 58 °C, 1 min at 72 °C, with the final extension of 10 min at 72 °C.
Polymerase chain reaction (PCR) assay for translation elongation factor (TEF-1a) gene was conducted using primers TEF1-728 F and TEF1-986R (Al-Sadi et al. 2015). The PCR reactions were carried out in 25 μl reaction mixture containing 10× PCR buffer, 50 ng DNA template, 2 mM MgCl2, 0.25 mM dNTP mixture and 0.25 μM each of primer, and one unit of Taq Polymerase (Bangalore Genie, India). Thermocycling was run with the following settings: heating at 94 °C (5 min); then 35 cycles of 94 °C (30 s), 60 °C (30 s) and 72 °C (90 s). The final extension was done at 72 °C for 10 min.
Molecular characterization of Trichoderma isolates was assessed by rep-PCR using the BOXA1R, Rep1R-I, Rep2-I, ERIC-1R and ERIC-2F primers (Srivastava et al. 2014). All the PCR reactions were carried out in 25 μl reaction mixture containing 5× Gitschier buffer, 50 ng DNA template, 2 mM MgCl2, 0.25 mM dNTP mixture and 0.25 μM each of primer, and one unit of Taq Polymerase (Bangalore Genie, India). Thermal Cycler (G Storm GS4, Somerset, UK) was programmed as an initial denaturation at 94 °C for 5 min, 40 cycles of 94 °C for 1 min, 36 °C for 1 min and 72 °C for 2 min and a final extension at 72 °C for 10 min.
For RAPD assay, the DNA extracted from tested isolates was amplified with the RAPD primers using the five RAPD primer set (Bangalore Genei, India). The thermal profile used was initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation step (94 °C for 1 min), annealing (47 °C, 1 min), extension (72 °C for 1.5 min), and a final extension step (72 °C for 7 min).
Amplified products were resolved in 2.0% agarose gels using 1× TAE buffer on a gel electrophoresis apparatus. Ethidium bromide (0.25 mg ml−1) was used as an intercalating agent. The gel was run at 2 V cm−1 of the length of gel till the bands resolved. The amplified bands, after separation on the gel, were visualized and documented using a gel documentation imaging system (Bio-Rad, USA).
Statistical analysis
Experimental data for conidial morphology and growth rate were analyzed using Duncan’s multiple range test (DMRT). Standard errors were calculated for all mean values. All RAPD, ERIC, REP and BOX-PCR reactions were repeated to ensure validity of results. The presence or absence of individual, distinct, and reproducible bands was scored as ‘1’ for presence and ‘0’ for absence. Principle component analysis (PCA) was performed using XLSTAT software.
Results
Antagonistic activity of Trichoderma isolates
Antagonistic capabilities of the Trichoderma isolates were assessed by the growth inhibition of four fungal pathogens (FOL, AA, CG and RS) through the dual culture assay. In general, all the antagonistic isolates grew faster than pathogen. The interaction of biological control agents versus four different fungal pathogens showed significant differences in growth inhibition of the pathogen isolates (Table 1). Isolate UNT68 showed highest inhibition effect on the percent mycelia growth of FOL (77.94%), AA (79.47%), CG (73.94%) and RS (69.23). Contrarily, isolate UNT70, ALT73, UNS63 and UNT60 showed least percent mycelia growth of FOL (53.90%), AA (53.68%), CG (56.90%) and RS (53.30%), respectively. Most of the isolates showed per cent mycelium inhibition values ranged between 60 and 70% against pathogens. The interaction between pathogens and Trichoderma isolates were determined and illustrated by a biplot (Fig. 1). The first two principal component axis of the biplot accounted for 25.54% (PC1) and 27.36% (PC2) of the total variation of the pathogen–antagonist interaction. In this biplot, all the Trichoderma isolates were located very far from the origin of biplot, indicating strong antagonism of mycoparasitic isolates towards fungal plant pathogens. Eigen values of the first and second components were 10.508 and 5.471, respectively.
Fig. 1.
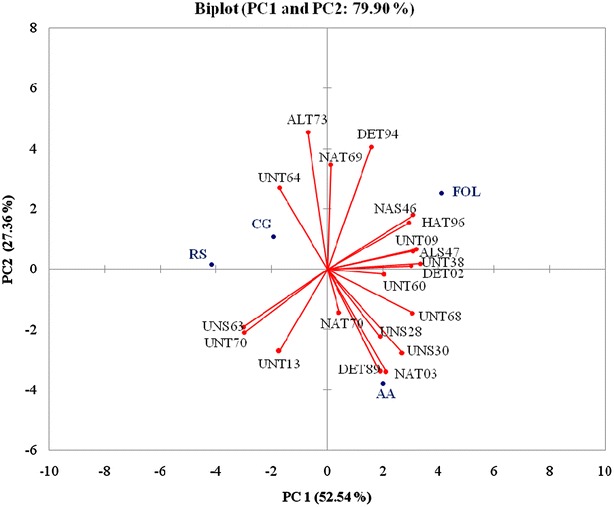
PCA biplot of in vitro dual culture assay showing antagonistic effect of twenty Trichoderma isolates against four fungal plant pathogens viz., Fusarium oxysporum f. sp. lycopersici (FOL), Alternaria alternata (AA), Colletotrichum gloeosporoides (CG) and Rhizoctonia solani (RS)
Production of hydrolytic enzymes
All Trichoderma isolates used in the present study produced cell wall-degrading enzymes (chitinase and β-1, 3 glucanase). Data presented in Table 1 showed that all the mycoparasitic strains produced chitinase and β-1,3 glucanase in the range of 31.0–76.56 μmol GlcNAc min−1mg−1 protein and 47.67–175.1 nmol glucose min−1 mg−1 protein, respectively. Among all the isolates, maximum chitinase was produced by UNT68, HAT96, UNT38 and UNS30. Similarly, maximum β-1,3 glucanase production was observed in UNS30 followed by DET94 (Table 1). The lowest activities of chitinase (31.00 μmol GlcNAc min−1 mg−1 protein) and β-1,3-glucanases (51.56 nmol glucose min−1 mg−1 protein) were obtained for isolates DET02 and UNT13, respectively. However, most of the isolates showed moderate activities of both lytic enzymes (Table 1).
Identification of antagonists
Distinct morphological differences were observed in 5 days old cultures of tested antagonistic isolates grown on PDA (Table 2). A perusal of data indicated that there was a significant difference in growth rate among isolates. Isolates UNT60, UNT68, NAT70, DET89, HAT96, UNT38, UNT13 and UNS30 grew faster (13.3 mm day−1) than other isolates. Less growth rate (11.4 mm day−1) was recorded in case of ALT73 and ALS47 isolates (Table 2). Ellipsoidal and sub-globose to globose conidia were noticed in thirteen isolates (UNT60, UNT64, UNT68, NAT69, NAT70, ALT73, DET89, DET94, HAT96, UNT38, UNS63, UNT09 and DET02). However, it was ellipsoidal and obovoid in rest of the eight isolates (UNT13, UNT70, UNS28, UNS30, NAS46, ALS47 and NAT03). Conidia colour varied from white to watery in all tested isolates. Fourteen isolates (UNT60, UNT64, UNT68, NAT70, DET89, DET94, HAT96, UNS63, UNT13, UNS28, UNS30, NAS46, UNT09 and DET02) showed conidiation concentric zone, while rings were also recorded in six isolates (NAT69, ALT73, UNT38, UNT70, ALS47 and NAT03). Phialides of most of the isolates were tending clustered in 2–3 whorls, but four isolate (NAT69, DET89, DET94 and NAT03) showed solitary disposition (Table 2). The phialides were nine-pin shaped and their size varied between 3.9–13.7 × 1.7–2.9 to 7.0–15.0 × 2.0–3.0 µm in seventeen isolates. However, globose and sigmoid or hooked phialides were also observed in two (UNT68 and ALT73) and one isolate (NAT03), respectively (Table 2).
Table 2.
Morphological characteristics of Trichoderma isolates
| Isolate | Colony | Mycelial | Conidia | Phialides | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Growth rate (mm day−1) | Colour | Reverse colour | Edge | Form | Colour | Conidiation | Branching | Shape | Size (µm) | Colour | Shape | Size (µm) | Disposition | |
| UNT60 | *1.33 ± 0.17 | Dark green | Creamish | Wavy | Floccose to Arachnoid | White | Concentric zones | Branched | ellipsodal, subglobose | 1.5–3.4 | Green | Nine-Pin shape | 6–14 × 1.4–2.6 | Tending clustered, 2–3 whorls |
| UNT64 | 1.32 ± 0.17 | Dark green | Creamish | Wavy | Floccose to Arachnoid | Watery white | Concentric zones | Highly branched | ellipsodal, globose | 1.3–3.6 | Green | Nine-Pin shape | 7–15 × 2–3 | Tending clustered, 2–3 whorls |
| UNT68 | 1.33 ± 0.17 | Light green | Light yellow | Wavy | Floccose to Arachnoid | Watery white | Concentric zones | Branched | ellipsodal, subglobose | 1.6–3.0 | Light Green | Globose | 8–14 × 2–3 | Tending clustered, 2–3 whorls |
| NAT69 | 1.16 ± 0.09 | Dark green | Colourless | Wavy | Arachnoid | Watery white | Ring like zones | Highly branched | ellipsodal, subglobose | 1.7–4.1 | Green | Nine-Pin shape | 6–14 × 1.4–3 | Solitary |
| NAT70 | 1.33 ± 0.17 | Dark green | Colourless | Smooth | Arachnoid | Watery white | Concentric zones | Highly branched | ellipsodal, subglobose | 1.4–3.7 | Green | Nine-Pin shape | 5.6–14.8 × 2–3 | Tending clustered, 2–3 whorls |
| ALT73 | 1.14 ± 0.12 | Light green | Light yellow | Smooth | Floccose to Arachnoid | White | Ring like zones | Branched | ellipsodal, globose | 1.3–3.3 | Green | Globose | 4.9–11.2 × 1.9–3 | Tending clustered, 2–3 whorls |
| DET89 | 1.33 ± 0.17 | Yellowish green | Light yellow | Wavy | Arachnoid | Watery white | Concentric zones | Branched | ellipsodal, globose | 1.5–3.4 | Light Green | Nine-Pin shape | 6–15 × 1.4–2.8 | Solitary |
| DET94 | 1.15 ± 0.09 | Light green | Creamish | Smooth | Arachnoid | Watery white | Concentric zones | Highly branched | ellipsodal, subglobose | 1.5–3.4 | Light Green | Nine-Pin shape | 5.9–15.2 × 1.9–2.8 | Solitary |
| HAT96 | 1.33 ± 0.17 | Dark green | Creamish | Smooth | Floccose to Arachnoid | White | Concentric zones | Moderately branched | ellipsodal, globose | 1.5–3.6 | Green | Nine-Pin shape | 7–14.8 × 1.9–2.6 | Tending clustered, 2–3 whorls |
| UNT38 | 1.33 ± 0.17 | Light green | Light yellow | Wavy | Arachnoid | Watery white | Ring like zones | Highly branched | ellipsodal, globose | 1.4–3.8 | Green | Nine-Pin shape | 6.2–10.2 × 2.2–2.9 | Tending clustered, 2–3 whorls |
| UNS63 | 1.31 ± 0.16 | Light green | Creamish | Wavy | Floccose | White | Concentric zones | Highly branched | ellipsodal, globose | 1.5–3.9 | Green | Nine-Pin shape | 5.8–12.4 × 2.7–3.2 | Tending clustered, 2–3 whorls |
| UNT13 | 1.33 ± 0.17 | White to green | Light yellow | Wavy | Arachnoid | Watery white | Concentric zones | Highly branched | ellipsodal, obovoid | 1.4–3.9 | Dark Green | Nine-Pin shape | 6.5–11.7 × 2.7–3.5 | Tending clustered, 2–3 whorls |
| UNT70 | 1.32 ± 0.16 | White to green | Light yellow | Smooth | Floccose to Arachnoid | White | Ring like zones | Moderately branched | ellipsodal, obovoid | 1.5–3.8 | Light Green | Nine-Pin shape | 6.1–12.5 × 2.7–3 | Tending clustered, 2–3 whorls |
| UNS28 | 1.31 ± 0.17 | Light green | Creamish | Wavy | Floccose | Watery white | Concentric zones | Highly branched | ellipsodal, obovoid | 1.4–3.6 | Light Green | Nine-Pin shape | 5.6–15 × 1.4–3 | Tending clustered, 2–3 whorls |
| UNS30 | 1.33 ± 0.17 | Light green | Light yellow | Wavy | Floccose | Watery white | Concentric zones | Highly branched | ellipsodal, obovoid | 1.3–3.6 | Green | Nine-Pin shape | 6.8–14.4 × 2.2–3.2 | Tending clustered, 2–3 whorls |
| NAS46 | 1.16 ± 0.09 | Yellow to green | Light yellow | Wavy | Floccose to Arachnoid | Watery white | Concentric zones | Branched | ellipsodal, obovoid | 1.4–3.5 | Green | Nine-Pin shape | 5.5–13.7 × 1.7–3.2 | Tending clustered, 2–3 whorls |
| ALS47 | 1.14 ± 0.09 | White to green | Light yellow | Wavy | Floccose | Watery white | Ring like zones | Branched | ellipsodal, obovoid | 1.5–3.9 | Light Green | Nine-Pin shape | 4.5–12.0 × 1.7–3.0 | Tending clustered, 2–3 whorls |
| UNT09 | 1.31 ± 0.16 | Dark green | Creamish | Wavy | Arachnoid | White | Concentric zones | Moderately branched | ellipsodal, subglobose | 1.3–3.9 | Dark Green | Nine-Pin shape | 3.9–13.7.0 × 1.7–2.9 | Tending clustered, 2–3 whorls |
| DET02 | 1.32 ± 0.15 | Yellow to green | Light yellow | Wavy | Floccose | White | Concentric zones | Moderately branched | ellipsodal, subglobose | 1.5–3.7 | Green | Nine-Pin shape | 4.5–11.9 × 1.7–2.7 | Tending clustered, 2–3 whorls |
| NAT03 | 1.16 ± 0.11 | Light green | Light yellow | Smooth | Floccose | White | Ring like zones | Branched | ellipsodal, obovoid | 1.5–3.7 | Green | Sigmoid or hooked | 5–12 × 2.2–2.7 | Solitary |
* Within columns, mean ± SE values with a common letter do not differ significantly (P < 0.05), according to DMRT test
Molecular identification based on sequences of Tef1gene confirmed that the isolates belonged to five different species viz., T. harzianum (UNT60, UNT64, UNT68, NAT69, NAT70, ALT73, DET89, DET94 and HAT96), T. koningii (UNT38 and UNS63), T. asperellum (UNT13 and UNT70), T. virens (UNS28, UNS30, NAS46 and ALS47) and T. viride (UNT09, DET02 and NAT03) (Table 1). The result of the phylogenetic analysis based on the Tef1 gene sequences of 20 Trichoderma isolates is shown in Fig. 2.
Fig. 2.

Neighbor joining tree (Kimura two-parameter distance) of twenty Tef-1a sequences of Trichoderma isolates from tomato rhizosphere. The numbers given over branches indicate bootstrap coefficient
RAPD-PCR analysis
Five primers viz., OPA-2 (TGCCGAGCTG), OPA-3 (AGTCAGCCAC), OPA-13 (CAGCACCCAC), OPA-15 (TTCCGAACCC) and OPA-18 (AGGTGACCGT) produced a total of 641 fragments among all the 20 isolates (Fig. 3). The size of RAPD fragments ranged 250–2500 bp. Principle component analysis (PCA) showed that RAPD markers explained 31.53% variation among Trichoderma isolates at genetic level (Fig. 3). PCA divided the 20 Trichoderma isolates in four clusters with pronounced separation of isolates. The first (PCA1) and second (PCA2) principal components were accounted for 20.47 and 11.06%, respectively. Two isolates occupied distinct position, UNT13 was far from the origin while HAT96 was near to the origin of biplot. Cluster I consisted of five isolates (ALT73, NAT03, UNS30, NAS46 and UNT70). However, cluster II comprised nine isolates (NAT70, UNT64, NAT69, ALS47, DET94, UNT38, UNT68, UNT60 and UNS28). Cluster III and IV contained two isolates each.
Fig. 3.
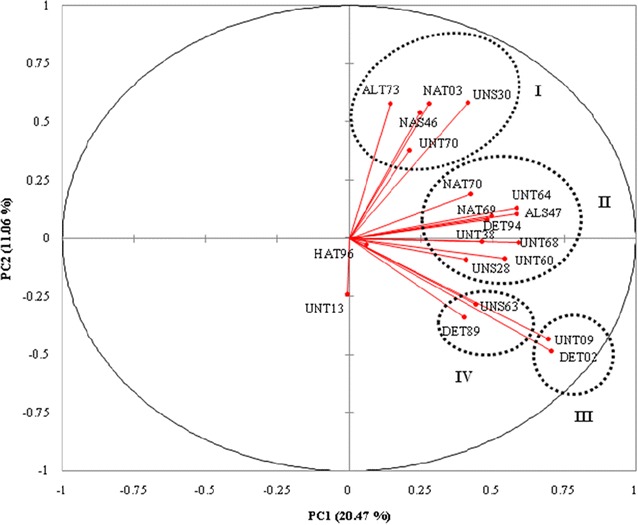
Principal component analysis score plot of twenty Trichoderma isolates based on RAPD-PCR data
BOX-PCR analysis
BOX-PCR banding pattern showed a total of 200 fragments in the range of 250–4000 bp. The results of PCA analysis based on first and second coordinates showed a maximum Eigen value of 9.306 and minimum value of 0.012 with a percentage variation of 46.53 and 13.72%, respectively (Fig. 4). PCA analysis revealed that nine isolates (UNS28, UNT38, UNT09, DET94, NAT03, NAT69, UNS63, UNT60 and UNT68) formed a major cluster (cluster IV), while three isolates (DET02, UNT70 and UNT13) were grouped in cluster II and two isolates were grouped in Cluster-I (UNS30 and NAS46), VI (DET89 and HAT96) and VII (ALS47 and NAT70).
Fig. 4.
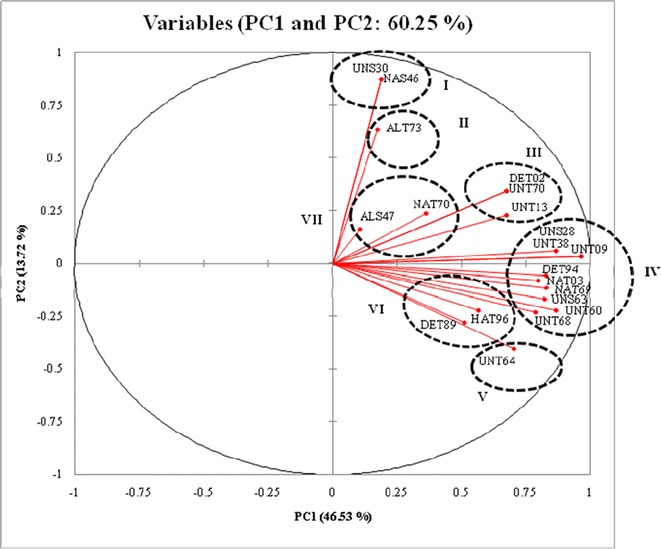
Principal component analysis score plot of twenty Trichoderma isolates based on BOX-PCR data
ERIC-PCR analysis
The genetic discrimination among the 20 isolates was assessed using ERIC-PCR and a high level of variability in the banding pattern was obtained (Fig. 5). The number of bands in the amplification profile was 182, and their size was found to vary from 250 to 3000 bp among these isolates (Fig. 5). Principal component analysis (PCA) based on first and second coordinates showed a maximum Eigen value of 10.027 and minimum value of 0.01 with a percentage variation of 50.13 and 12.09, respectively (Fig. 5). A perusal of the PCA analysis revealed that eight isolates (HAT96, UNT68, DET94, UNT60, UNT64, NAT69, DET02, DET89 and UNT09) formed a major cluster (cluster IV), while three isolates were grouped in cluster II (UNS28, UNS63 and UNT38) and IV (UNT70, NAT03 and ALT73).
Fig. 5.
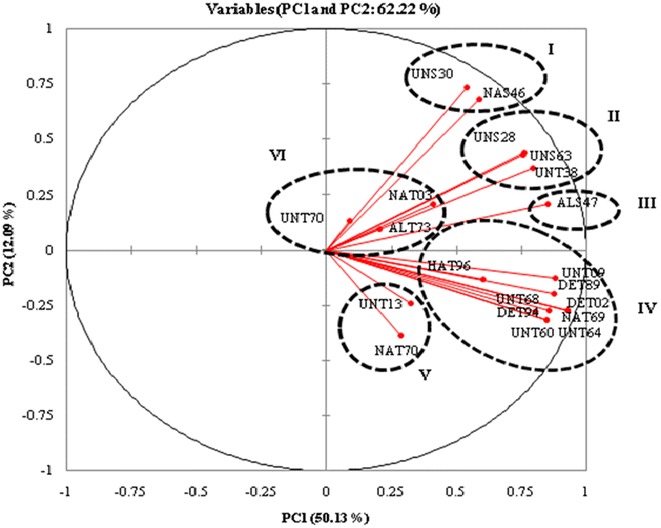
Principal component analysis score plot of twenty Trichoderma isolates based on ERIC-PCR data
REP-PCR analysis
The genetic discrimination among the 20 isolates was assessed using REP-PCR and a high level of variability in the banding pattern was obtained (Fig. 6). The number of bands in the amplification profile was 350, and their size was found to vary from 270 to 3000 bp among these isolates (Fig. 6). Principal component analysis (PCA) based on first and second coordinates showed a maximum Eigen value of 9.758 and minimum value of 0.017 with a percentage variation of 48.71 and 13.16, respectively (Fig. 6). A perusal of the PCA analysis revealed that six isolates (UNS30, NAS46, NAT03, ALS47, UNS28 and UNT70) formed a major cluster (cluster V), while four isolates were grouped in cluster VI (UNT13, DET02, UNT09 and UNS63).
Fig. 6.
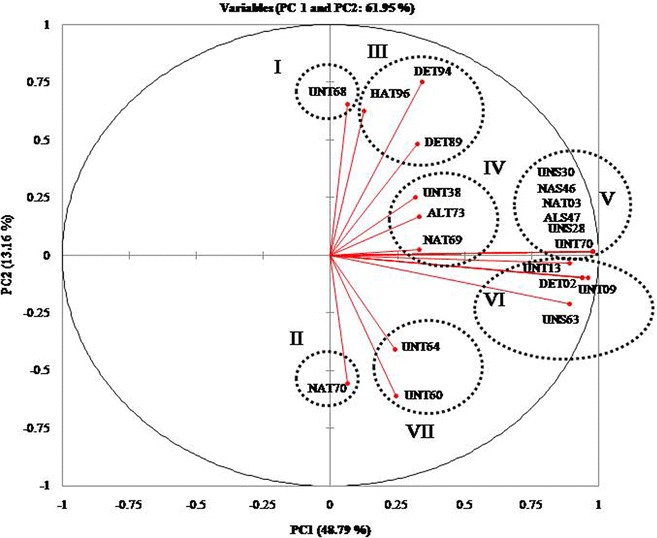
Principal component analysis score plot of twenty Trichoderma isolates based on REP-PCR data
ERG1 sequencing and phylogenetic analysis
Detection of squalene epoxidase (ERG-1) gene in Trichoderma isolates was shown in Fig. 7. Squalene epoxidase (ERG-1) gene amplification showed one specific band (500 bp) in all the twenty Trichoderma isolates. The phylogenetic tree obtained by sequence analysis of ERG1 region of all the tested isolates is represented in Fig. 8. A neighbour-joining analysis of the alienable ERG1-sequences of all the tested isolates demonstrated two distinct phylogenetic clades. Clade A comprised mainly T. harzianum (UNT60, UNT64, UNT68, NAT69 and UNT70), T. viride (UNT09, DET02 and NAT03), T. koningii (UNS63) and T. virens (UNS28) and showed very high homology to the nearest ERG1 sequence of H. lixii, T. arundinaceum and T. reesei submitted in NCBI GenBank. Clade B represented four isolates of T. harzianum (ALT73, DET89, HAT96, and DET94), two isolates of T. koningii (UNT38), two isolates of T. asperellum (UNT13 and UNT70) and three isolates of T. virens (ALS47, UNS30 and NAS46) and showed heterogeneity with respect to the ERG1 sequence of H. lixii, T. arundinaceum and T. reesei.
Fig. 7.
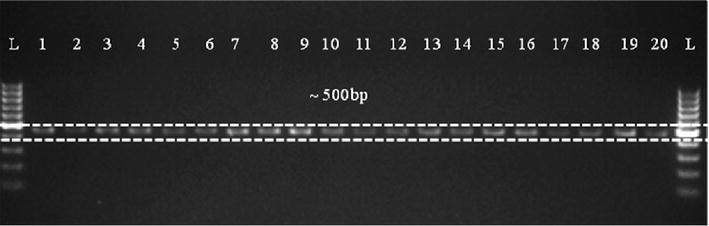
PCR amplification of squalene epoxidase (ERG1) gene, showing ~500 bp amplicon in Trichoderma isolates having distinct geographical lineages. Lanes 1–20 are different Trichoderma isolates as mentioned in Table 1. L is a 100-bp DNA marker
Fig. 8.
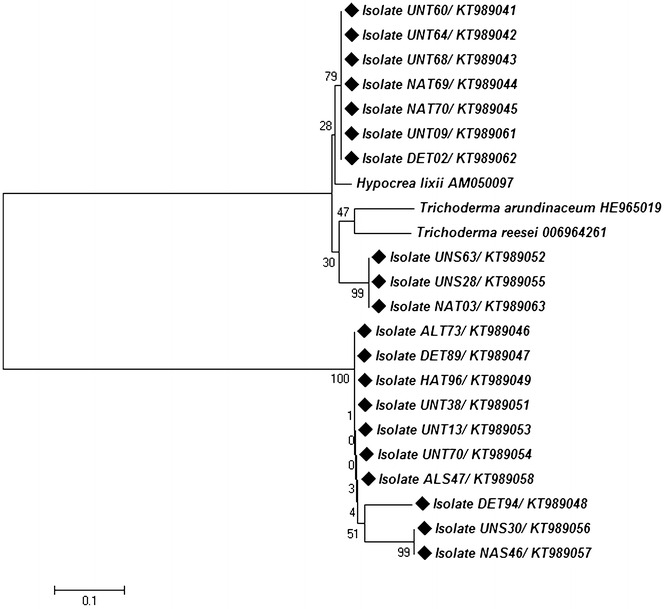
Phylogenetic analysis of the ERG1 sequences from different isolates of Trichoderma isolates from tomato rhizosphere. Tree was constructed by the neighbour-joining method. The numbers given over branches indicate bootstrap coefficient
Discussion
Microbial inoculants with antagonistic properties towards fungal plant pathogens have a potential to replace chemical pesticides since they are known for growth promotion and disease reduction in crops. Several species of Trichoderma have been used as biological control agents to manage diseases of vegetable and other crops (Solanki et al. 2011; Srivastava et al. 2012; Al-Sadi et al. 2015). In the present study, twenty isolates of Trichoderma collected from rhizosphere soil of tomato were phenotypically, biochemically and genetically characterized to identify and screen the most efficient antagonistic against four tomato fungal pathogens (FOL, AA, CG and RS). All the tested isolates grew considerably faster than the fungal pathogens and quickly controlled the pathogens. The ability to grow rapidly gives antagonists an important advantage in competition for space and nutrients with pathogen (Benítez et al. 2004; El_Komy et al. 2015). Nine isolates (UNT68, DET94, HAT96, UNT38, UNS30, DET02, ALS47, UNS28 and UNT09) showed significant per cent mycelium inhibition against the test pathogens. These isolates overgrew and sporulated on the pathogen colonies. In the interaction zone, the mycelia of all the fungal pathogens had abnormal morphology and lysed, which implies the occurrence of strong mycoparasitism. These results are in conformity with previous studies where Trichoderma isolates showed high capabilities as versatile biocontrol agents (Trillas et al. 2006; Tondje et al. 2007; de los Santos-Villalobos et al. 2013). Interestingly, the interaction of indigenous Trichoderma isolates with four different fungal pathogens resulted in significantly different amounts of pathogen inhibition. For instance, DET94 had very strong inhibitory effect on the growth of FOL, RS and CG pathogens, whereas moderate inhibition effect was recorded in case of AA. These results are consistent with the findings of Markovich and Kononova (2003). They reported that the mycoparasitic capacity of various species and isolates of Trichoderma differs. There are several mechanisms involved in Trichoderma antagonism, namely, antibiosis whereby the antagonist fungus produces antibiotics, competes for nutrients and mycoparasitism, whereas Trichoderma directly attacks the plant pathogen by excreting lytic enzymes such as chitinases and β-1,3 glucanases (Kubicek et al. 2001; Radjacommare et al. 2010; Solanki et al. 2011). Such hydrolytic enzymes partially degrade the pathogen cell wall that leads to parasitization (Howell 2003). Also in the present study antagonistic isolates (UNT68, DET94, HAT96, UNT38, UNS30, DET02, ALS47, UNS28 and UNT09) with the highest levels of enzyme activities showed the strong inhibitory effect on the growth of fungal plant pathogens. Similar observations were made by Howell (2003), wherein the activity of lytic enzymes (chitinases and β-1,3 glucanses) was responsible for lysis of R. solani hyphae through digestion of major cell wall components. There was a positive relationship between the antagonistic capacity of the Trichoderma isolates and the production of chitinase and β-1,3-glucanases. Thus, efficient antagonistic isolates inhibited fungal growth through the production of lytic enzymes. On parallel lines, Lopes et al. (2012) reported a positive correlation between the lytic enzymes activities and the antagonism capacity of T. asperellum against Sclerotinia sclerotiorum. Moreover, Qualhato et al. (2013) and El_Komy et al. (2015) reported that there was a positive correlation between the amounts of secreted cell-wall degrading enzymes by Trichoderma strains and their ability to control plant pathogenic fungi.
Taxonomic knowledge on Trichoderma isolates is important for identification and characterization of potential biocontrol species and to avoid potential risk from introducing an unknown fungal species into the rhizosphere of a given ecosystem. A combination of morphological and molecular methods is desirable for the reliable and accurate identification of Trichoderma spp. The few morphological characteristics with limited variation in Trichoderma spp. may lead to an overlap and wrong identification of the species (Galarza et al. 2015). In present study, Trichoderma isolates were categorized on the basis of description and keys given by Gams and Bissett (1998). As a result, ellipsoidal and sub-globose to globose condial structures resembled with T. harzianum, while ellipsoidal and ovoid shaped conidia were matched with T. virens isolates; as previously mentioned by Choi et al. (2003). However, some isolates showing overlapping characters and resembling with T. koningii, T. viride and T. asperellum could not be separated using the morphology-based method. Thus, molecular identification of Trichoderma isolates at the species level was done on the basis of TEF-1a gene as it has been reported to be better for distinguishing Trichoderma spp. (Samuels 2006).
The present study also revealed the usefulness of DNA polymorphism techniques to detect genetic variation among antagonistic Trichoderma isolates. These techniques are important not only for understanding their ecological role in the rhizosphere, but also to characterize the biological control agents for registration and patenting biocontrol strains, recognizing the strains, quality checking during production and ecological characterization (Plimmer 1993). The study of DNA polymorphisms involves the selection of a target sequence, and several approaches have been used to achieve this task. One approach involves the exploitation of ubiquitously conserved known genes that display sequence variation. Identification of Trichoderma to the species level based on reference sequences from the National Center for Biotechnology Information correlated with phylogenetic analysis based on sequences of the ITS rRNA and the translation elongation factor gene (EF1a). However, the limited intraspecific variation within Trichoderma species based on sequences of the EF1a gene helped giving better resolution in separating Trichoderma species when compared to sequences of the ITS region (Al-Sadi et al. 2015). Thus, in present study, comparative nucleotide sequencing of EF1a gene was performed to distinguish and identify antagonistic Trichoderma isolates. Based on the sequence analysis of EF1 gene, the 20 antagonistic isolates were divided in five species: T. harzianum, T. koningii, T. asperellum, T. virens and T. viride. Another approach involves the screening of random parts of the genome to identify distinctive nucleotide sequences by techniques, such as RAPD, REP-, ERIC- and BOX-PCR. The results indicated that BOX elements and ERIC-PCR are suitable for the rapid genetic differentiation of Trichoderma isolates. Some of the Trichoderma isolates such as NAT70, UNT64 and ALS47 which were not differentiated by RAPD can be discriminated by BOX and ERIC-PCR banding patterns. In general, both techniques were found to produce reproducible results especially with purified genomic nucleic acid as a template, and when the primer concentration and composition of buffer were strictly controlled. It is also worth mentioning here that ERIC-, REP- and BOX-PCR marker systems revealed >60% intra-species variability among Trichoderma isolates, although clustering on the basis of antagonism, geographical origin and hydrolytic enzyme production was not detected. Additionally, the present study was unable to correlate biomarker variation with fungal growth inhibition activity of Trichoderma isolates. These findings are in agreement with earlier studies, where no defined correlations between genetic variability assessed by random markers (e.g. RAPD) and the ability of Trichoderma isolates to inhibit fungal mycelia growth were obtained (Sharma et al. 2009; El_Komy et al. 2015). This may be due to the ubiquitous nature and seemingly random chromosomal distribution of random repeats in Trichoderma genome, giving rise to simultaneous PCR amplification of multiple genomic regions (Rai et al. 2016). The high genotypic variability among Trichoderma isolates could be associated with mutations in priming sites, rearrangements of chromosomal segments or recombination process in fungal genomes (Kumar et al. 2012, 2013b). However, genetic variability among Trichoderma isolates in addition to their differences in fungal growth inhibition toward fungal plant pathogens suggest that combinations of isolates could further be applied in both greenhouse and field studies to manage tomato diseases.
Terpene compounds (e.g., ergokonins and viridins) are involved in the biocontrol process due to their antifungal properties (Malmierca et al. 2015). Similar to this, the present study also documented the possibility of squalene epoxidase driven triterpene biosynthesis mechanism in biocontrol of tomato wilt and foliar blight diseases. Furthermore, PCR based detection of ERG1 gene in antagonistic isolates confirmed the presence of gene at molecular level and Blastn and Blastp results showed the maximum homology with a squalene epoxidase gene. Phylogenetic analysis of squalene epoxidase gene (ERG1) sequences revealed close relatedness of ERG-1sequences with earlier reported sequences of H. lixii, T. arundinaceum and T. reesei. However, ERG1 gene also showed heterogeneity among some antagonistic isolates and it may be possible that squalene epoxidase driven triterpene biosynthesis have an important role in biocontrol mechanisms of tested isolates.
In conclusion, the present study provides preliminary information on the biological control of tomato diseases by correctly identifying the fungal antagonists. Correct identification will provide information on understanding the interparasitic relationship with target pathogens and the subsequent environmental fate of the antagonist needed for effective application. Further, combined studies including biological, biochemical and molecular technologies, are essential to select indigenous antagonistic Trichoderma isolates that can be used under different environmental conditions. Genetic variability of squalene epoxidase (ERG1) gene among these isolates in addition to their differences in aggressiveness toward multiple fungal pathogens suggest that combinations of isolates could further be applied in both greenhouse and field studies to obtain resistance against multiple fungal pathogens in tomato crop. However, further experiments are needed to validate the role of squalene epoxidase driven triterpene biosynthesis in biocontrol mechanisms of tested isolates.
Authors’ contributions
SR carried out the sampling and performed assays for the screening of antagonistic Trichoderma. PLK participated in the design of the study, performed statistical analysis and helped to draft the manuscript. SK participated in the identification of Trichoderma isolates, sequence alignment and drafted the manuscript. AKS carried out the molecular characterization of antagonists and sequence submission. PWR conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Shalini Rai, Email: shalinimicro09@gmail.com.
Prem Lal Kashyap, Phone: +911772621978, Email: plkashyap@gmail.com, Email: Prem.Kashyap@icar.gov.in.
Sudheer Kumar, Email: sudheer.icar@gmail.com.
Alok Kumar Srivastava, Email: aloksrivastva@gmail.com.
Pramod W. Ramteke, Email: pwramteke@yahoo.com
References
- Al-Sadi AM, Al-Oweisi FA, Edwards SG, Al-Nadabi H, Al-Fahdi AM. Genetic analysis reveals diversity and genetic relationship among Trichoderma isolates from potting media, cultivated soil and uncultivated soil. BMC Microbiol. 2015;15:147. doi: 10.1186/s12866-015-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anees M, Tronsmo A, Edel-Hermann V, Hjeljord LG, Héraud C, Steinberg C. Characterization of field isolates of Trichoderma antagonistic against Rhizoctonia solani. Fungal Biol. 2010;114:691–701. doi: 10.1016/j.funbio.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Benítez T, Rincón MA, Limón MC, Codón CA. Biocontrol mechanisms of Trichoderma strains. Int Microbiol. 2004;7:249–260. [PubMed] [Google Scholar]
- Błaszczyk L, Popiel D, Chełkowski J, Koczyk G, Samuels GJ, Sobieralski K, Siwulski M. Species diversity of Trichoderma in Poland. J Appl Genet. 2011;52:233–243. doi: 10.1007/s13353-011-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoza RE, Malmierca MG, Hermosa MR, Alexander NJ, et al. Identification of loci and functional characterization of trichothecene biosynthetic genes in the filamentous fungus Trichoderma. Appl Environ Microbiol. 2011;77:4867–4877. doi: 10.1128/AEM.00595-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoza RE, Malmierca MG, Gutiérrez S. Overexpression of erg1 gene in Trichoderma harzianum CECT 2413: effect on the induction of tomato defence-related genes. J Appl Microbiol. 2014;117:812–823. doi: 10.1111/jam.12574. [DOI] [PubMed] [Google Scholar]
- Choi I-Y, Hong S-B, Yadav MC. Molecular and morphological characterization of green mold, Trichoderma spp. isolated from oyster mushrooms. Mycobiology. 2003;31:74–80. doi: 10.4489/MYCO.2003.31.2.074. [DOI] [Google Scholar]
- de los Santos-Villalobos S, Guzmàn-Ortiz DA, Gomez-Lim MA, Délano-Frier JP, et al. Potential use of Trichoderma asperellum (Samuels, Liechfeldt et Nirenberg) T8a as a biological control agent against anthracnose in mango (Mangifera indica L.) Biol Control. 2013;64:37–44. doi: 10.1016/j.biocontrol.2012.10.006. [DOI] [Google Scholar]
- El_Komy MH, Saleh AA, Eranthodi A, Molan YY. Characterization of novel Trichoderma asperellum isolates to select effective biocontrol agents against tomato Fusarium wilt. Plant Pathol J. 2015;31:50–60. doi: 10.5423/PPJ.OA.09.2014.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarza L, Akagi Y, Takao K, Kim CS, Maekawa N, et al. Characterization of Trichoderma species isolated in Ecuador and their antagonistic activities against phytopathogenic fungi from Ecuador and Japan. J Gen Plant Pathol. 2015;81:201–210. doi: 10.1007/s10327-015-0587-x. [DOI] [Google Scholar]
- Gams W, Bissett J. Morphology and Identification of Trichoderma. In: Harman GE, Kubicek CP, editors. Trichoderma and Gliocladium, basic biology, taxonomy and genetics. London: Taylor and Francis; 1998. pp. 3–34. [Google Scholar]
- Harman G, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- Howell CR. Mechanisms employed by Trichoderma species in the biological control of plant diseases, the history and evolution of current concepts. Plant Dis. 2003;87:4–10. doi: 10.1094/PDIS.2003.87.1.4. [DOI] [PubMed] [Google Scholar]
- Jain A, Singh A, Singh S, Singh HB. Biological management of Sclerotinia sclerotiorum in pea using plant growth promoting microbial consortium. J Basic Microbiol. 2015;55:961–972. doi: 10.1002/jobm.201400628. [DOI] [PubMed] [Google Scholar]
- Kashyap PL, Rai S, Kumar S, Srivastava AK, Anandaraj M, Sharma AK. Mating type genes and genetic markers to decipher intraspecific variability among Fusarium udum isolates from pigeonpea. J Basic Microbiol. 2015;55:846–856. doi: 10.1002/jobm.201400483. [DOI] [PubMed] [Google Scholar]
- Kashyap PL, Rai S, Kumar S, Srivastava AK. Genetic diversity, mating types and phylogenetic analysis of Indian races of Fusarium oxysporum f. sp. ciceris from chickpea. Arch Phytopathol Plant Prot. 2016 [Google Scholar]
- Körmöczi P, Danilović G, Manczinger L, Jovanović L, Panković D, et al. Species composition of Trichoderma isolates from the rhizosphere of vegetables grown in Hungarian soils. Fresenius Environ Bull. 2013;22:1736–1741. [Google Scholar]
- Kubicek CP, Mach RL, Peterbauer CK, Lorito M. Trichoderma: from genes to biocontrol. J Plant Pathol. 2001;83:11–24. [Google Scholar]
- Kullnig CM, Krupica T, Woo SL, Mach RL, Rey M, Benítez T, Lorito M, Kubicek CP. Confusion abounds over identities of Trichoderma biocontrol isolates. Mycol Res. 2001;105:769–772. doi: 10.1017/S0953756201229967. [DOI] [Google Scholar]
- Kumar S, Maurya D, Kashyap PL, Srivastava AK. Computational mining and genome wide distribution of microsatellites in Fusarium oxysporum f. sp. lycopersici. Not Sci Biol. 2012;4:127–131. [Google Scholar]
- Kumar S, Rai S, Maurya DK, Kashyap PL, Srivastava AK, Anandaraj M. Cross-species transferability of microsatellite markers from Fusarium oxysporum for the assessment of genetic diversity in Fusarium udum. Phytoparasitica. 2013;41:615–622. doi: 10.1007/s12600-013-0324-y. [DOI] [Google Scholar]
- Kumar S, Singh R, Kashyap PL, Srivastava AK. Rapid detection and quantification of Alternaria solani in tomato. Sci Hortic. 2013;151:184–189. doi: 10.1016/j.scienta.2012.12.026. [DOI] [Google Scholar]
- Lopes FAC, Steindorff AS, Geraldine AM, Brandao RS, et al. Biochemical and metabolic profiles of Trichoderma strains isolated from common bean crops in the Brazilian Cerrado, and potential antagonism against Sclerotinia sclerotiorum. Fungal Biol. 2012;116:815–824. doi: 10.1016/j.funbio.2012.04.015. [DOI] [PubMed] [Google Scholar]
- Malmierca MG, Cardoza RE, Alexander NJ, McCormick SP, Collado IG, Hermosa R, et al. Relevance of trichothecenes in fungal physiology: disruption of tri5 in Trichoderma arundinaceum. Fungal Genet Biol. 2013;53:22–33. doi: 10.1016/j.fgb.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Malmierca MG, McCormick S, Cardoza RE, Monte E, et al. Trichodiene production in a Trichoderma harzianum erg1-silenced strain provides evidence of the importance of the sterol biosynthetic pathway in inducing plant defense-related gene expression. Mol Plant Microbe Interact. 2015;28:1181–1197. doi: 10.1094/MPMI-06-15-0127-R. [DOI] [PubMed] [Google Scholar]
- Markovich NA, Kononova GL. Lytic enzymes of Trichoderma and their role in plant defense from fungal diseases: a review. Appl Biochem Microbiol. 2003;39:341–351. doi: 10.1023/A:1024502431592. [DOI] [PubMed] [Google Scholar]
- Martínez-Medina A, Alguacil MDM, Pascual JA, Van Wees SCM. Phytohormone profiles induced by Trichoderma isolates correspond with their biocontrol and plant growth-promoting activity on melon plants. J Chem Ecol. 2014;40:804–815. doi: 10.1007/s10886-014-0478-1. [DOI] [PubMed] [Google Scholar]
- Mohapatra BR, Broersma K, Mazumder A. Comparison of five rep-PCR genomic fingerprinting methods for differentiation of fecal Escherichia coli from humans, poultry and wild birds. FEMS Microbiol Lett. 2007;277:98–106. doi: 10.1111/j.1574-6968.2007.00948.x. [DOI] [PubMed] [Google Scholar]
- Ospina-Giraldo MD, Royse DJ, Thon MR, Chen X, Romaine CP. Phylogenetic relationships of Trichoderma harzianum causing mushroom green mold in Europe and North America to other species of Trichoderma from world-wide sources. Mycologia. 1998;90:76–81. doi: 10.2307/3761014. [DOI] [Google Scholar]
- Plimmer JR. Regulatory problems associated with natural products and biopesticides. Pestic Sci. 1993;39:103–108. doi: 10.1002/ps.2780390203. [DOI] [Google Scholar]
- Qualhato FT, Lopes FAC, Steindorff AS, Brandão RS, Jesuino RSA, Ulhoa CJ. Mycoparasitism studies of Trichoderma species against three phytopathogenic fungi: evaluation of antagonism and hydrolytic enzyme production. Biotechnol Lett. 2013;35:1461–1468. doi: 10.1007/s10529-013-1225-3. [DOI] [PubMed] [Google Scholar]
- Radjacommare R, Venkatesan S, Samiyappan R. Biological control of phytopathogenic fungi of vanilla through lytic action of Trichoderma species and Pseudomonas fluorescens. Arch Phytopathol Plant Prot. 2010;43:1–17. doi: 10.1080/03235400701650494. [DOI] [Google Scholar]
- Rai P, Sharma A, Saxena P, Soni AP, et al. Comparison of molecular and phenetic typing methods to assess diversity of selected members of the genus Bacillus. Microbiology. 2015;84:236–246. doi: 10.1134/S0026261715020113. [DOI] [Google Scholar]
- Rai S, Kashyap PL, Kumar S, Srivastava AK, Ramteke PW. Comparative analysis of microsatellites in five different antagonistic Trichoderma species for diversity assessment. World J Microbiol Biotechnol. 2016;32:8. doi: 10.1007/s11274-015-1964-5. [DOI] [PubMed] [Google Scholar]
- Samuels GJ. Trichoderma: systematics, the sexual state, and ecology. Phytopathology. 2006;96:195–206. doi: 10.1094/PHYTO-96-0195. [DOI] [PubMed] [Google Scholar]
- Sharma K, Mishra AK, Misra RS. Morphological, biochemical and molecular characterization of Trichoderma harzianum isolates for their efficacy as biocontrol agents. J Phytopathol. 2009;157:51–56. doi: 10.1111/j.1439-0434.2008.01451.x. [DOI] [Google Scholar]
- Singh RK, Kumar DP, Singh P, Solanki MK, et al. Multifarious plant growth promoting characteristics of chickpea rhizosphere associated Bacilli help to suppress soil-borne pathogens. Plant Growth Regul. 2014;73:91–101. doi: 10.1007/s10725-013-9870-z. [DOI] [Google Scholar]
- Solanki MK, Singh N, Singh RK, Singh P, Srivastava AK, Kumar S, Kashyap PL, Arora DK. Plant defense activation and management of tomato root rot by a chitin-fortified Trichoderma/Hypocrea formulation. Phytoparasitica. 2011;39:471–481. doi: 10.1007/s12600-011-0188-y. [DOI] [Google Scholar]
- Solanki MK, Kumar S, Pandey AK, Srivastava S, Singh RK, et al. Diversity and antagonistic potential of Bacillus spp. associated to the rhizosphere of tomato for the management of Rhizoctonia solani. Biocontrol Sci Technol. 2013;22:203–217. doi: 10.1080/09583157.2011.649713. [DOI] [Google Scholar]
- Srivastava AK, Singh RN, Kumar S, Kashyap PL, Arora DK. Growth promotion and management of Alternaria leaf spot in chilli by Trichoderma harzianum. IJIH. 2012;2:158–163. [Google Scholar]
- Srivastava AK, Singh P, Singh RK, Kashyap PL, et al. Identification and characterization of ethanol utilizing fungal flora of oil refinery contaminated soil. World J Microbiol Biotechnol. 2014;30:705–714. doi: 10.1007/s11274-013-1497-8. [DOI] [PubMed] [Google Scholar]
- Tondje PR, Roberts DP, Bon MC, Widmer T, et al. Isolation and identification of mycoparasitic isolates of Trichoderma asperellum with potential for suppression of black pod disease of cacao in Cameroon. Biol Control. 2007;43:202–212. doi: 10.1016/j.biocontrol.2007.08.004. [DOI] [Google Scholar]
- Trillas MI, Casanova E, Cotxarrera L, Ordovas J, Borrero C, Aviles M. Composts from agricultural waste and the Trichoderma asperellum strain T-34 suppress Rhizoctonia solani in cucumber seedlings. Biol Control. 2006;39:32–38. doi: 10.1016/j.biocontrol.2006.05.007. [DOI] [Google Scholar]
- Woo SL, Ruocco M, Vinale F, Nigro M, Marra R, Nadia L, Pascale A, Lanzuise S, Manganiello G, Lorito M. Trichoderma based products and their widespread use in agriculture. Open Mycol J. 2014;8:71–126. doi: 10.2174/1874437001408010071. [DOI] [Google Scholar]
- Zachow C, Berg C, Müller H, Meincke R, Komon-Zelazowska M, et al. Fungal diversity in the rhizosphere of endemic plant species of Tenerife (Canary Islands): relationship to vegetation zones and environmental factors. ISME J. 2009;3:79–92. doi: 10.1038/ismej.2008.87. [DOI] [PubMed] [Google Scholar]


