Abstract
Neuronal transmitters are released from nerve terminals via the fusion of synaptic vesicles with the plasma membrane. Vesicles attach to membranes via a specialized protein machinery composed of membrane-attached (t-SNARE) and vesicle-attached (v-SNARE) proteins that zipper together to form a coiled-coil SNARE bundle that brings the two fusing membranes into close proximity. Neurotransmitter release may occur either in response to an action potential or through spontaneous fusion. A cytosolic protein, Complexin (Cpx), binds the SNARE complex and restricts spontaneous exocytosis by acting as a fusion clamp. We previously proposed a model in which the interaction between Cpx and the v-SNARE serves as a spring to prevent premature zippering of the SNARE complex, thereby reducing the likelihood of fusion. To test this model, we combined molecular-dynamics (MD) simulations and site-directed mutagenesis of Cpx and SNAREs in Drosophila. MD simulations of the Drosophila Cpx-SNARE complex demonstrated that Cpx’s interaction with the v-SNARE promotes unraveling of the v-SNARE off the core SNARE bundle. We investigated clamping properties in the syx3-69 paralytic mutant, which has a single-point mutation in the t-SNARE and displays enhanced spontaneous release. MD simulations demonstrated an altered interaction of Cpx with the SNARE bundle that hindered v-SNARE unraveling by Cpx, thus compromising clamping. We used our model to predict mutations that should enhance the ability of Cpx to prevent full assembly of the SNARE complex. MD simulations predicted that a weakened interaction between the Cpx accessory helix and the v-SNARE would enhance Cpx flexibility and thus promote separation of SNAREs, reducing spontaneous fusion. We generated transgenic Drosophila with mutations in Cpx and the v-SNARE that disrupted a salt bridge between these two proteins. As predicted, both lines demonstrated a selective inhibition in spontaneous release, suggesting that Cpx acts as a fusion clamp that restricts full SNARE zippering.
Introduction
Neurotransmitters are released from nerve terminals via the fusion of synaptic vesicles with the neuronal plasma membrane. Vesicles are attached to the membrane through the assembly of a coiled-coiled SNARE protein complex (1). The SNARE complex forms a parallel four-helix bundle comprising the vesicle transmembrane protein synaptobrevin (Syb), the plasma membrane protein syntaxin (Syx), and the membrane-anchored protein SNAP25 (2). Stimulus-evoked vesicle fusion occurs in response to Ca2+ influx triggered by an action potential. Release of transmitters can also occur spontaneously in the absence of stimulation, a process that is essential for normal neuronal development and homeostasis (3). Both evoked and spontaneous synaptic transmissions are regulated by the cytosolic protein Complexin (Cpx) through its interaction with the SNARE bundle (4). Cpx inhibits spontaneous release and promotes evoked transmission (5). Structurally, Cpx is comprised of several domains, including two helical regions (the central helix and the accessory helix (AH)), and it directly binds the SNARE complex and forms a SNARE-Cpx five-helix protein bundle (4). Cpx inhibits, or clamps, spontaneous fusion via the interaction of its AH with the SNARE bundle (6). However, the specific molecular mechanism by which Cpx prevents spontaneous vesicle fusion is still debated.
Extensive evidence suggests that before fusion occurs, a vesicle is docked to the plasma membrane in a prefusion state (1, 7). The prefusion state of the SNARE bundle is likely to represent a partially zippered SNARE complex comprised of Syx and SNAP25 (t-SNARE) with a partially unraveled Syb (v-SNARE) (8), although an alternative view suggests that SNARE zippering occurs rapidly and without stable intermediate states (9). It was suggested that approximately half of the helical region of Syb is separated from the bundle, and that the Cpx AH inserts between the t-SNARE and Syb, substituting for Syb in binding the t-SNARE complex (10, 11). In this model, the bridging of multiple SNAREs by Cpx would both promote evoked release and inhibit spontaneous fusion. However, both theoretical (12) and experimental (13, 14) studies suggested that the separation of the SNARE bundle is likely to be less radical, and that it probably involves only one or two helical turns. In addition, several studies demonstrated that different Cpx domains are responsible for regulating evoked and spontaneous fusion (6, 15, 16). Thus, the question of Cpx’s mechanistic action in clamping fusion is still debated (5, 17, 18).
Computational molecular modeling represents a powerful tool for understanding the dynamics of the SNARE complex before fusion (12, 19, 20, 21, 22, 23). Based on molecular-dynamics (MD) simulations of the SNARE-Cpx complex, we previously proposed a model whereby Cpx dynamically interacts with Syb, preventing it from fully zippering onto the SNARE bundle (23). Here, we used this model to generate testable predictions for site-directed mutagenesis in Drosophila. We combined MD simulations with site-directed mutagenesis, Drosophila genetics, and focal electrical recordings of synaptic activity to probe how Cpx interacts with the SNARE bundle to clamp spontaneous fusion.
Materials and Methods
Fly stocks
Drosophila melanogaster were cultured on standard medium at 25°C. The following fly stocks were used: Canton-S (wild-type (WT); Bloomington Drosophila Stock Center, Indiana University), cpx null mutant cpxSH1 (24), temperature-sensitive Syx mutant syx3-69 (25), and n-syb null mutant n-sybΔF33 (26). Since the homozygote n-sybΔF33 line is embryonic lethal (26), the n-sybΔF33 stock was maintained over the balancer chromosome TM6Tb.
To generate the transgenic lines UAS-CpxE34A and UAS-n-SybK83A, we used QuikChange (Stratagene, La Jolla, CA) for site-directed mutagenesis on the existing cloned Cpx (isoform 7A (24, 27)) and n-Syb (28). PCR products were subcloned into a pValum construct, allowing use of the Gal4/UAS expression system (29). Constructs were injected into yv; attP 3rd chromosome docking strains by BestGene (Chino Hills, CA).
The C155 elav-GAL4 driver was used for neuronal expression of transgenes. We generated lines with overexpressed (OE) mutated and WT Cpx (CpxE34A OE and CpxWT OE), as well as mutated and WT n-Syb (n-SybK83A OE and n-SybWT OE). The mutations were combined to generate lines with both mutated proteins and the respective controls (CpxE34A/n-SybK83A OE and CpxWT/n-SybWT OE).
The UAS lines used in the study were recombined into the cpxSH1 or n-sybΔF33 null mutant background to generate CpxE34A,cpxSH1 and n-SybK83A,n-sybΔF33 lines, as well as their respective controls (CpxWT,cpxSH1 and n-SybWT,n-sybΔF33).
Electrophysiology
Experiments were performed on type Ib boutons of muscles 6 and 7 at abdominal segments 2–4 of the 3rd instar larvae in HL3 solution containing (in mM) 70 NaCl, 5 KCl, 20 MgCl2, 10 NaHCO3, 5 trehalose, 115 sucrose, 5 HEPES, and 0.25, 1, or 2.5 CaCl2. Excitatory postsynaptic currents (EPSCs) were recorded focally (30) as previously described (31, 32) from boutons visualized with differential interference contrast optics using macropatch electrodes with a 5 μm tip diameter and 1–1.2 MΩ resistance. The electrode tips were fire polished and bent to enable recording under a 60× water immersion objective with a 2 mm working distance. The seal resistance was monitored continuously, and only experiments in which the seal resistance stayed constant (within the range of 0.89–0.91 MΩ) throughout the experiment were analyzed. Recordings were digitized with a Digidata A/D board and Axoscope software (Axon Instruments), and analyzed offline employing in-house-written Quantan software (33).
Molecular modeling
We obtained the structure of the Drosophila SNARE-Cpx complex by homology modeling of the mammalian complex simulated in our earlier study (23). Single-point mutations were performed with the use of Visual Molecular Dynamics software (VMD, University of Illinois, Urbana-Champaign, IL). MD simulations were performed as described previously (23). Briefly, the water box (150 × 70 × 70 A) contained 150 mM KCl with all the negative charges neutralized. MD simulations were performed using the NAMD package (34) and CHARMM22 force field (35) with periodic boundary conditions and Ewald electrostatics in the NTP ensemble (Berendsen barostat and Langevin thermostat) at 300 K temperature, with a 2 fs time step for the simulations at equilibrium and a 1 fs time step for the simulations under external forces. Computations were performed using the Extreme Science and Engineering Discovery Environment (XSEDE). PyMol software (Schrödinger) was used for molecular graphics, and VegaZZ software (Drug Design Laboratory) was used for trajectory analysis.
Results
Cpx may act as a spring to unravel the SNARE bundle
Our MD simulations of the mammalian SNARE-Cpx complex (23) suggested that the Cpx AH interacts tightly with Syb in solution, in contrast to the structure observed after crystallization (4). Notably, only 70% of residues are conserved between mammalian and Drosophila Cpx-SNARE complexes (Fig. S1 in the Supporting Material), and thus it is unclear whether these structural and dynamic properties are conserved. To address this issue, we created a molecular model of the Drosophila SNARE-Cpx complex by employing homology modeling coupled with MCM optimization, and performed a 230 ns MD simulation of the equilibrated complex in a water/ion environment (Fig. S2, A and B). We found that the Drosophila SNARE-Cpx complex formed a tight bundle, similar to what was observed for the mammalian complex, and was stabilized by a salt bridge (Fig. S2 A, magenta line, and B) between K83 of Syb (numeration per mammalian homologs) and E34 of Cpx. In contrast to the mammalian complex, this salt bridge was not stable, and the hydrogen bond between residues K83 of Syb and E34 of Cpx continuously formed and broke over the timeframe of the simulation (Fig. S2 A, magenta line). Interestingly, the distance between the membrane proximal residues of Syx and Syb (Fig. S2 A, blue line) demonstrated considerable fluctuations, with the maximal openings of the SNARE bundle coinciding with disruptions in the Syb-Cpx salt bridge (Fig. S2 A, arrows). Notably, the presence of Cpx significantly enhanced these fluctuations (Fig. S2 C). This result suggests that Cpx may destabilize the C-terminus of the SNARE bundle, and that the bundle would be unraveled more rapidly in the presence of Cpx.
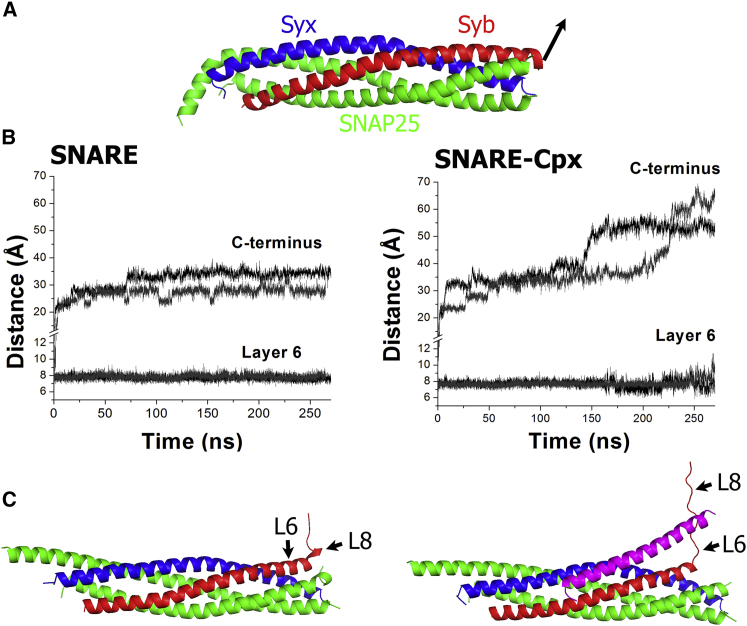
To test whether this is the case, we performed MD simulations of the Drosophila SNARE and SNARE-Cpx complexes under a force of 2 kcal/Mol/Å (Fig. 1 A), corresponding to the force produced by membrane electrostatic repulsion (23). In the absence of Cpx, a rapid unraveling of the C-terminal helical turn of Syb was observed; however, all the remaining layers of Syb remained intact over the entire 270 ns period of the simulation (Fig. 1 B, left). At the end of the trajectory (Fig. 1 C, left), the complex had an unstructured C-terminal helical turn, and layer 8 was slightly separated from the core bundle. However, all the other layers of the SNARE bundle remained intact.
Figure 1.
Cpx interaction with Syb on the SNARE bundle in Drosophila promotes Syb unraveling from the bundle. (A) An external force of 2 kcal/Mol/Å was applied to Syb. (B) MD trajectories of the SNARE and SNARE-Cpx complexes under the external force. The graphs show the distances between the Cα atoms of the C-terminus residues of Syb and Syx, as well as between the Cα atoms of the Syb and Syx residues forming layer 6. Each graph shows two replicas started from subsequent trajectory points. Note that the SNARE-Cpx complex demonstrates a faster unraveling of Syb. Layer 6 of the SNARE complex remains intact over the length of the simulation, whereas in the SNARE-Cpx complex, layer 6 is destabilized at the end of the trajectory (170–270 ns). (C) The final states of the SNARE and SNARE-Cpx complexes (Cpx, magenta) reveal a more radical unraveling in SNARE-Cpx. Note that the SNARE complex is zippered up to layer 8 (L8), whereas in the SNARE-Cpx complex, layers L6–L8 of Syb are unraveled.
In contrast, in the SNARE-Cpx complex, the same force produced unraveling of layers 8 and 7 at 150–200 ns of the trajectory (Fig. 1 B, right), producing a separation of ∼5.5 nm between the terminal residues of Syb and Syx. In the final state of the SNARE-Cpx complex (Fig. 1 C, right), layers 7 and 8 of Syb were unraveled and separated from the bundle. Importantly, the Cpx AH interacted with Syb, stabilizing the partially unraveled state of the bundle.
Notably, in a complex with the fully assembled SNARE bundle, the Cpx helical structure was slightly bent between its central and accessory helixes (Figs. S2 B and S3). In contrast, a partially unraveled complex Cpx structure (Figs. 1 B and S3) represented a continuous helix, thus residing in a more energetically favorable state. These simulations suggest a mechanical model whereby Cpx in a complex with the SNARE bundle may act as a spring that shifts the equilibrium toward a partially unraveled SNARE complex state.
If our model is correct, mutations that disrupt the interactions of the Cpx AH with the SNARE bundle would be likely to affect Cpx clamping properties. This would be manifested as a selective increase or decrease in spontaneous synaptic activity. We tested this prediction in two ways. First, we took advantage of the temperature-sensitive paralytic mutant syx3-69 (25), which has increased spontaneous activity (36) and possibly an altered Cpx conformation (23). Second, we generated new mutations that affect the interaction of the Cpx AH with the SNARE bundle, and investigated evoked and spontaneous transmissions in the mutant lines.
The ability of Cpx to unravel the SNARE bundle is compromised in the syx3-69 mutant and spontaneous synaptic activity is selectively increased
We employed the paralytic mutant syx3-69 (25) to investigate how the mutation in Syx affects the attachment of Cpx to the SNARE bundle and SNARE unraveling, as well as spontaneous and evoked synaptic transmissions. The syx3-69 mutant has a single-point mutation T254I in Syx (T251I per mammalian numeration), which is positioned at layer 7 of the SNARE bundle (36) and is in proximity to the Cpx AH. MD simulations of the mammalian SNARE-Cpx complex suggested that the T251I mutation in Syx may alter the attachment of Cpx AH to the SNARE bundle (23). Notably, spontaneous activity is increased in the syx3-69 mutant (23, 36), in line with the hypothesis that this mutation may compromise the interaction of the Cpx AH with the SNARE bundle and thus mimic Cpx deficiency in the regulation of spontaneous activity. To test this hypothesis, we combined modeling and experimentation to test whether Cpx’s clamping properties are disrupted in the syx3-69 mutant. To do this, we 1) investigated spontaneous and evoked release components in the syx3-69 mutant; 2) employed MD simulations in equilibrium to test whether the Cpx AH conformation is altered in the mutated Drosophila SNARE-Cpx complex, as was the case for the mammalian complex (23); and 3) employed MD simulations under external forces to test whether Syb unraveling is altered in the mutant.
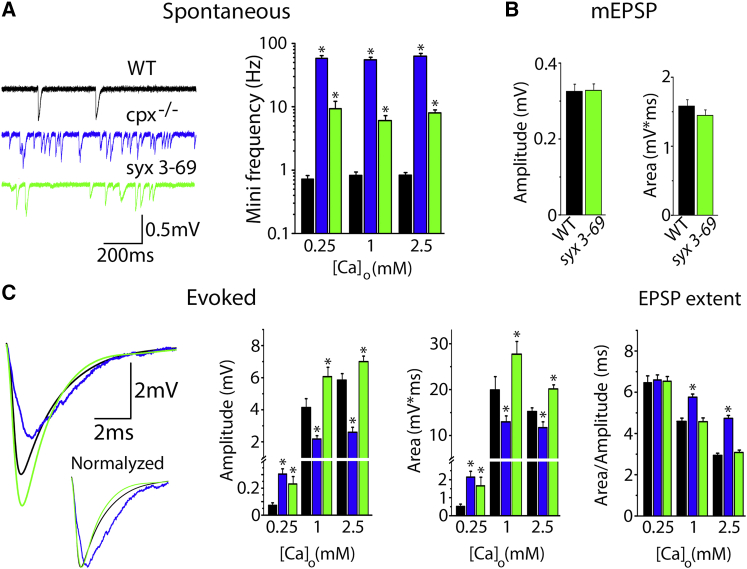
To accurately quantify spontaneous and evoked transmissions in the syx3-69 mutant and compare them with those observed in the WT and cpx−/− lines, we employed focal recordings of synaptic responses from visualized boutons (31, 32). Since it was previously demonstrated (37) that Cpx affects evoked transmission in a Ca2+-dependent way, we monitored spontaneous and evoked activities at different Ca2+ levels. Spontaneous activity was increased ∼70-fold in the cpx−/− line and ∼15-fold in the syx3-69 line (Fig. 2 A), and this effect did not depend on the Ca2+ concentration. The miniature EPSC (mEPSC) amplitude and area were not affected in syx3-69 (Fig. 2 B; we were not able to accurately quantify mini sizes in the cpx−/− line, since the mEPSCs overlapped due to their high frequency).
Figure 2.
The syx3-69 mutation partially mimics the effect of Cpx deletion on spontaneous, but not evoked, release. (A) Cpx deletion and syx3-69 mutation increase spontaneous activity in a Ca2+-independent manner. Black, WT; blue, cpx−/−; green, syx3-69. (B) The syx3-69 mutation does not affect quantal size. (C) The syx3-69 mutation does not mimic the effect of Cpx deletion on evoked release. Cpx deletion inhibits release and produces EPSC broadening in a Ca2+-dependent manner, whereas the syx3-69 mutation increases evoked release in a Ca2+-independent manner and does not affect the release time course. EPSC traces represent an average over the 300 recorded sweeps in a single experiment at 2.5 mM Ca2+. Data were collected from at least 20 recording sites in at least seven larvae for each line under each condition.
The amplitude and area of evoked EPSCs showed a dependence on Cpx that was influenced by Ca2+ levels (Fig. 2 C), in agreement with prior studies (37). At 1 and 2.5 mM Ca2+, Cpx deletion produced a significant decrease in evoked release, as well as EPSC broadening. At the lowest Ca2+ level (0.25 mM), evoked release in cpx−/− was significantly increased (note the increase in EPSC amplitude and area in Fig. 2 C), and the EPSC time course was not affected (note the unchanged EPSC extent in Fig. 2 C). This Ca2+-dependent effect of Cpx deletion was not mimicked by the syx3-69 mutation. In contrast, the syx3-69 mutant showed a modest but significant increase in evoked release at all Ca2+ levels, and this increase was not accompanied by any change in the time course of release (note the unchanged EPSC extent in the syx3-69 mutant). This increase in evoked release observed in the syx3-69 mutant may be associated with an increased number of synapses (38).
Thus, both the syx3-69 mutation and the Cpx deletion showed a very prominent and Ca2+-independent increase in spontaneous release, although the effect of the syx3-69 mutation was significantly weaker. The Cpx deletion also produced a Ca2+-dependent effect on the evoked-release component, and this effect was not mimicked by the syx3-69 mutation. Notably, the increase in evoked release produced by the syx3-69 mutation was modest (∼20%) and contrasts with the dramatic effect of this mutation on spontaneous release (∼700%). Thus, syx3-69 shows a very prominent and selective increase in the spontaneous release component. We hypothesized that this increase may be associated with a conformational change in Cpx and the displacement of the Cpx AH in the syx3-69 mutant (23). To test this hypothesis, we employed MD simulations of the mutated Drosophila SNARE-Cpx complex.
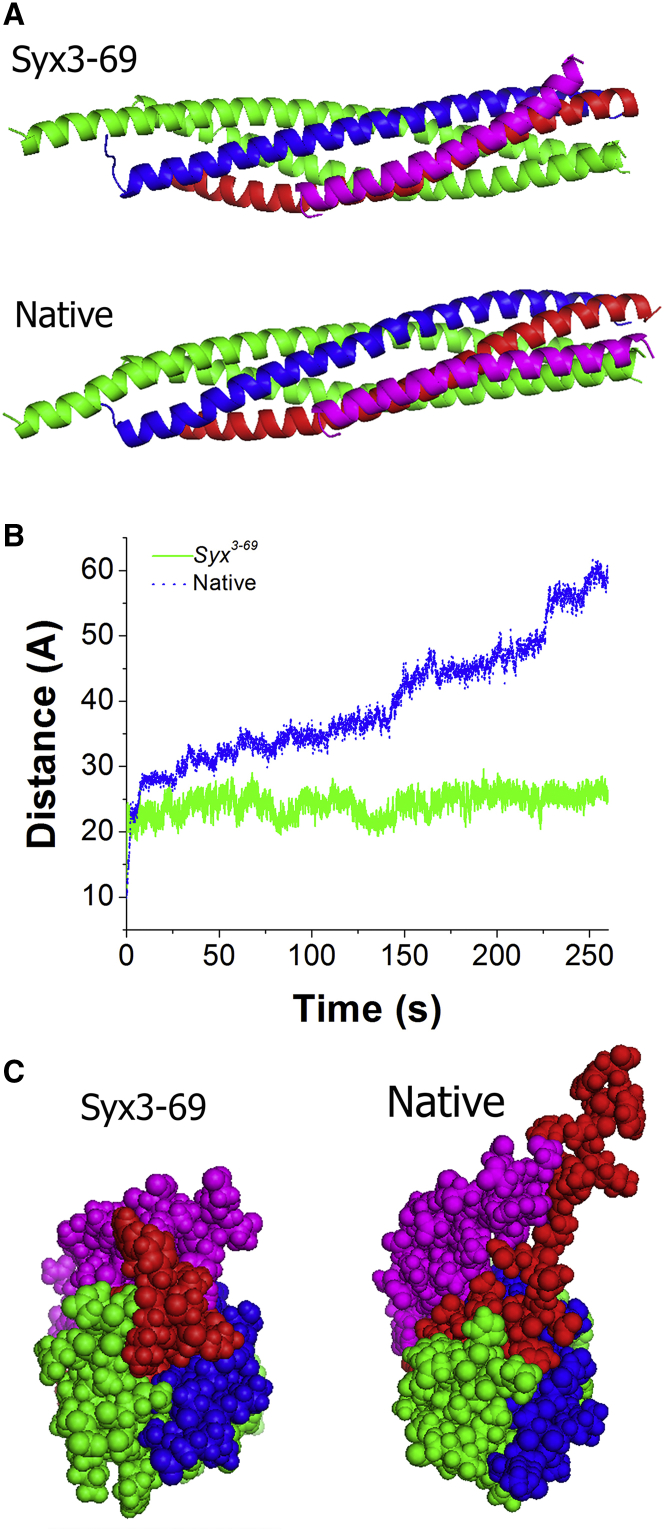
First, we investigated whether the Syx T251I mutation (T254 in Drosophila) alters the interaction of Drosophila Cpx with the SNARE bundle at equilibrium. We performed a 260 ns MD simulation of the mutated SNARE-Cpx complex, starting from a conformation identical to the SNARE-Cpx structure at the end of the MD trajectory at equilibrium (Fig. S2 B). After 95 ns of simulation, the Cpx AH separated from the SNARE bundle, and after a further 30 ns it stabilized in a conformation whereby the Cpx AH did not interact with SNAP25 and deviated from the orientation parallel to the SNARE bundle (Fig. S4, A and B). Thus, we found that the syx3-69 mutation produced a deviation of the Cpx AH from the Drosophila SNARE bundle (see Fig. 4 A). We next tested whether the mutation affects the stability of the SNARE C-terminus. We found that the distance between the Syb and Syx C-termini fluctuated more broadly in the native SNARE-Cpx complex than in the mutant (Fig. S4 C), suggesting that the mutation may stabilize the SNARE C-terminus and makes it less prone to unraveling.
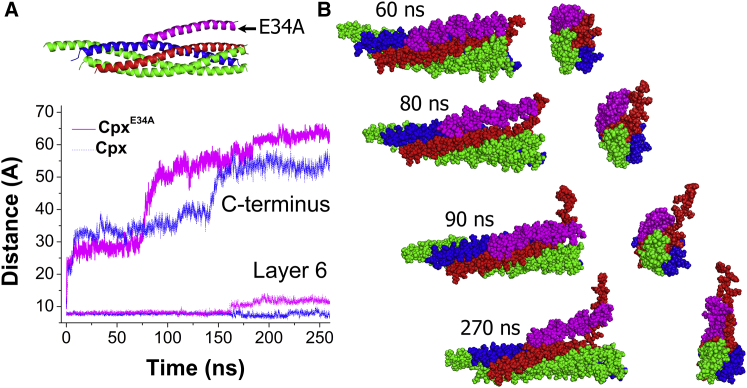
Figure 4.
The E34A mutation in Cpx promotes Syb unraveling. (A) The mutation accelerates unraveling of the bundle under external forces (the blue dotted line represents the average for the MD replicas of the native SNARE-Cpx complex; Fig. 1B). (B) Subsequent time points along the trajectory of the mutant complex under external forces suggest the following scenario for Syb unraveling: 1) the partially unraveled Syb C-terminus contacts Cpx (60 ns); 2) Cpx transitions from a closed to an open state, separating Syb from the bundle (80 ns); Cpx transitions back to the closed state, with Cpx AH moving along the partially unraveled Syb C-terminus (90 ns); and 4) Cpx transitions into the open state, further unraveling Syb (270 ns). Two perpendicular views are shown at each time point.
To test whether this is the case, we performed MD simulations of the mutant complex under external forces as described above. Notably, the mutant SNARE-Cpx complex showed a very modest unraveling under external forces (Fig. 3, B and C), unlike the native complex. An examination of the Syb and Cpx conformations in both native and mutated complexes reveals that Cpx is likely to hinder Syb unraveling in the mutated complex (Fig. 3 C, left). In contrast, in the native complex, the Cpx AH is positioned so that it can serve as a climbing step for Syb unraveling (Fig. 3 B, right). These results argue that fusion clamping might be selectively manipulated by altering the interaction of the Cpx AH and Syb. To test whether this is the case, we combined modeling and experimentation to predict and test the effect of the mutations that alter this interaction.
Figure 3.
The syx3-69 mutation produces a deviation of Cpx from the bundle, which hinders unraveling of Syb. (A) SNARE-Cpx complex with and without the mutation. Both states are captured at the end of the respective MD trajectories. In the absence of the mutation, Cpx lies roughly parallel to the bundle, but in the mutant complex it deviates from the bundle. (B) The mutation slows down the unraveling of Syb from the SNARE bundle under external forces (the graph shows the distance between the C-terminal residues of Syb and Syx; the blue line corresponds to the average of the two SNARE-Cpx replica; Fig. 2B). (C) Final states of the mutated and native complexes. In the mutant complex, Cpx (magenta) is positioned over Syb (red) and hinders unraveling, whereas in the native complex, Cpx is positioned to promote unraveling.
Disrupting a salt bridge between Cpx and Syb promotes the ability of Cpx to unravel the SNARE bundle and produces a superclamping phenotype
Since the interaction between the Cpx AH and Syb is stabilized by a salt bridge between K83 of Syb and E34 of Cpx (Fig. S2 B), we questioned how the disruption of this salt bridge would affect the protein dynamics and the spontaneous transmitter release. This manipulation could partially mimic the Cpx null phenotype, eliminating the interaction of the Cpx AH with the core SNARE bundle. Conversely, disrupting this salt bridge could destabilize the SNARE C-terminus and enhance the Cpx clamping function. To discriminate between these two possibilities, we performed MD simulations to investigate how disruption of the Cpx E34-Syb K83 salt bridge affects the dynamics and stability of the SNARE C-terminus.
We generated a model of the mutated SNARE-Cpx complex with the E34A substitution in the Cpx AH, employing the final structure of the SNARE-Cpx trajectory (Fig. S2 B). First, we performed a 300 ns MD simulation of the mutated complex and examined the interaction of the Cpx AH with the SNARE bundle. We found that the distance between the Cpx AH and Syb increased in the mutated complex (Fig. S5). In the predominant state of the E34A complex, the Cpx AH was positioned in parallel to the bundle (Fig. S5, B and C, state 1), although transient states were observed in which the Cpx AH was radically separated from the bundle (Fig. S4, B and C, state 3) or interacted with SNAP25 (Fig. S5, B and C, state 2). These simulations indicate that the salt bridge between the conserved residues E34 of Cpx and K83 of Syb is a critical determinant of the interaction between Cpx and the SNARE bundle.
Next, we questioned whether this mutation would affect unraveling of the SNARE complex. To address this question, we performed MD simulations of the mutated complex under external forces, as described above, using conformation 1 (Figs. S5 C and 4 A) as the initial state. We found that the mutation drastically promoted unraveling of the SNARE bundle under external forces (Fig. 4 A). The separation of the layers proceeded more rapidly than in the native Cpx/SNARE complex, and at the end of the 270 ns trajectory, layer 6 of Syb was separated from the bundle in the mutant complex, in contrast to the native complex. An examination of the intermediate conformations of the mutant complex reveals that in the initial stages of unraveling, Syb forms contacts with Cpx (Fig. 4 B; 60 ns time point). Subsequently, as Cpx transitions from the closed state (1) to the open state (3) (Fig. S5 C), it may promote the unraveling of Syb. Since Cpx has formed fairly tight contacts with a partially unraveled Syb at this stage, Cpx may act as a lever to further unravel Syb and separate it from the bundle (Fig. 4 B, 80 ns time point). This process may continue to promote further Syb unraveling (as illustrated by the 90 ns and 270 ns time points of the trajectory; Fig. 4 B).
These simulations demonstrate that weakening the interaction of Syb and Cpx within the bundle by disrupting the Syb K83-Cpx E34 salt bridge increases the flexibility of the Cpx AH, promoting unraveling of the SNARE bundle under external forces. Thus, our simulations predict that disrupting the Syb K83-Cpx E34 salt bridge would enhance the clamping function of Cpx and create a phenotype in which spontaneous release is reduced.
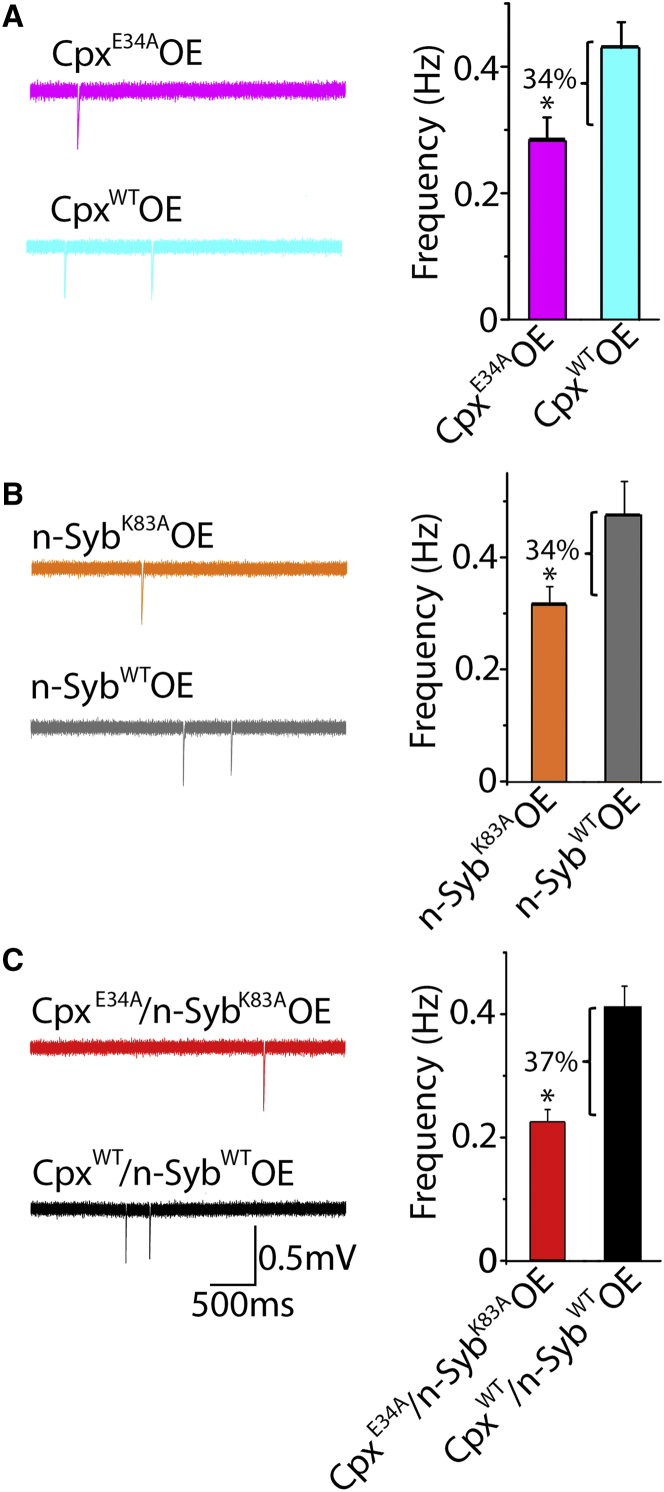
To test this prediction, we generated Drosophila transgenic strains with residue E34 (E40 in Drosophila) mutated to Ala. We monitored spontaneous activity at boutons of the CpxE34A OE larvae and found that the frequency of minis was reduced compared with the CpxWT OE control (Fig. 5 A), as predicted by our model. To further assay whether the effect of the E34A mutation in Cpx is due to the disruption of the Cpx E34-Syb K83 salt bridge, we generated a line with the K83A mutation in n-Syb (K100A in Drosophila). We found that spontaneous activity was reduced in the n-SybK83A OE line compared with the n-SybWT OE control line (Fig. 5 B), demonstrating a superclamped phenotype in the SybK83A OE line. Finally, we combined the two mutations and recorded spontaneous activity in the CpxE34A/n-SybK83A OE line versus the control CpxWT/n-SybWT OE (Fig. 5 C). We reasoned that if the effect of each mutation is due to disrupting the interaction between Cpx and n-Syb proteins, then the effect of the two mutations combined should be similar to the effect of each single mutation. Indeed, the effect of the two mutations combined (37%) was similar to the effect of a single mutation in either Cpx or n-Syb (34%).
Figure 5.
(A–C) CpxE34A (A) and n-SybK83A (B) mutations similarly reduce spontaneous activity, and combining the two mutations (C) does not produce a further decrease in the mEPSC frequency. Data were collected from at least 20 recordings sites in at least five larvae for each genotype. To see this figure in color, go online.
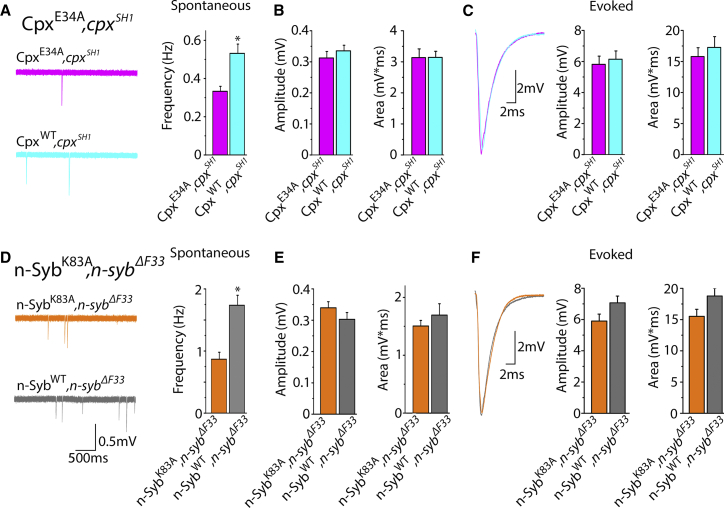
We next tested whether the CpxE34A and n-SybK83A mutations selectively affect the rate of spontaneous release or they also alter evoked fusion. The CpxE34A mutation was introduced into the cpx−/− background (CpxE34A,cpxSH1 line), and the Cpx rescue line expressing WT Cpx in the cpx−/− background (CpxWT,cpxSH1) was used as a control. We ascertained that Cpx expression levels were similar in both lines (Fig. S6). We monitored spontaneous activity at boutons of the CpxE34A,cpxSH1 larvae and found that the frequency of minis was significantly reduced (Fig. 6 A). Notably, the effect of the E34A mutation was similar to the effect of Cpx overexpression (37). Since the loss of Cpx effects evoked release most prominently at 2.5 mM Ca2+ (Fig. 3), we monitored evoked release under that condition. The mutation did not affect either EPSC amplitude or area (Fig. 6 C). Similarly, the amplitude and area of mEPSCs were not affected (Fig. 6 B), indicating a selective effect on spontaneous release.
Figure 6.
Disrupting the interaction between Cpx and Syb selectively reduces spontaneous release. (A–C) The CpxE34A,cpxSH1 line has reduced spontaneous activity compared with the control rescue line CpxWT,cpxSH1. The mEPSC frequency is decreased by the mutation (A), but the mEPSC amplitude and area (B), and EPSC and area (C) are not affected. (D–F) The n-SybK83A,n-sybΔF33 line has reduced spontaneous activity compared with the control rescues n-SybWT,n-sybΔF33 (D), but the mini amplitude and area (E), as well as evoked release (F), are not affected. Data were collected from at least 20 recordings sites in at least six larvae for each genotype.
We then introduced the n-SybK83A mutation into the n-syb null background. Expression of the n-SybK83A UAS construct rescued the embryonic lethality of the n-syb null mutant n-sybΔF33, indicating that the K83A mutation is functional for restoring synaptic transmission compared with the null. However, we found that spontaneous activity was selectively reduced in the n-SybK83A,n-sybΔF33 mutant compared with its control, n-SybWT,n-sybΔF33, demonstrating a superclamped phenotype in the n-SybWT,n-sybΔF33 line. Importantly, evoked release was not affected by the mutation (Fig. 6, D–F), demonstrating that the SybK83A mutation selectively affects the rate of spontaneous release.
These results demonstrate that disrupting the salt bridge between the Cpx AH and Syb produces a selective inhibition of spontaneous release, and suggest a new mechanism whereby Cpx prevents spontaneous release as a vesicle fusion clamp.
Discussion
We combined molecular modeling with targeted mutagenesis to investigate how Cpx clamps spontaneous fusion. Our modeling suggests that the Cpx AH interacts with Syb and may act as a spring in promoting partial unraveling of the C-terminus of Syb from the SNARE bundle. This unraveling would likely serve to move the synaptic vesicle farther from the plasma membrane, reducing the chance that spontaneous release would occur in the absence of other fusion-promoting factors such as Ca2+-bound synaptotagmin.
Our MD simulations of both mammalian (23) and Drosophila (this study) SNARE-Cpx complexes suggest that the Cpx AH forms tight contacts with Syb. It should be noted that the structure of the SNARE-Cpx complex observed by crystallography (4) shows a slight separation between the Cpx AH and Syb. However, several lines of evidence suggest that the interaction between the Cpx AH and Syb is likely to take place in solution. The initial evidence comes NMR experiments (4) showing that the presence of Cpx affects the SNARE C-terminus, and in particular Syb residue F77 at layer 6, suggesting that Cpx is likely to interact with the SNARE C-terminus in solution. Furthermore, the MD simulations started from this x-ray structure show a fairly stable salt bridge between the Cpx AH and Syb in both mammalian (23) and Drosophila (this study) isoforms. This is not a trivial result, since the homology between mammalian and Drosophila SNAREs and Cpx is fairly low (Fig. S1). Importantly, our MD structure was not trapped in a tight bundle conformation representing a local energy minimum, since the Drosophila SNARE-Cpx complex shows several points along the MD trajectory where the Cpx AH separates from the bundle (Fig. S2, purple line). Altogether, these data strongly argue the Cpx AH comes into contact with Syb in solution. Our model suggests that this interaction contributes to Cpx’s clamping function by providing a spring from the AH of Cpx to allow binding and unraveling of Syb off the SNARE bundle, thus increasing the distance between the two membranes.
To initially test this model, we performed MD simulations of the SNARE complex under external forces. Our simulations demonstrated that Syb separates from the core SNARE bundle more radically when Cpx is attached to the bundle. This result suggests that in the absence of Cpx, full assembly of the SNARE complex is more likely, providing an explanation for Cpx’s function in clamping spontaneous fusion.
Next, we took advantage of the syx3-69 mutant (25, 36), which has a single-point mutation in Syx, with the mutated residue lying in proximity to the Cpx AH. We demonstrated a prominent and selective increase in spontaneous release in the syx3-69 mutant. Our MD simulations of the mutated SNARE-Cpx complex suggested that the Cpx conformation is altered in this mutant both in mammalian (23) and Drosophila (this study) forms. The MD simulations under external forces suggested that in the mutated complex, Cpx hinders Syb unraveling, thus tightening the SNARE complex. These results demonstrated that our model provides a parsimonious explanation for the poor-clamp phenotype in the syx3-69 mutant.
We then employed the model to design new mutations in the SNARE-Cpx complex that would alter Cpx clamping. Since our MD simulations suggested that a salt bridge between Syb and Cpx may facilitate the clamping function by enhancing the Cpx-Syb interaction, we questioned how disrupting this salt bridge would affect the separation of Syb from the core SNARE bundle. To address this question, we performed MD simulations of the mutated complex with the disrupted salt bridge, and then generated two Drosophila lines with mutated SNARE-Cpx complexes to test the predictions derived from molecular modeling. First, we investigated the E34A mutation in the Cpx AH. The MD simulations at equilibrium demonstrated that the Cpx AH becomes transiently separated from the SNARE bundle in the mutated complex. We then questioned how this mutation would affect the separation of Syb from the core SNARE bundle. One possibility is that the mutation would partially mimic the Cpx null phenotype, since the interaction of the Cpx AH with the SNARE bundle would be weakened. In this case, Syb interaction with the core SNARE bundle would become tighter, as in the absence of Cpx. Another possibility is that the mutated Cpx AH would promote Syb unraveling even more strongly than in the native SNARE-Cpx complex, since the mutated Cpx AH remains in proximity to Syb and is involved in van der Waals interactions with Syb, being at the same time more mobile than in the native complex. To discriminate between these opposite scenarios, we performed MD simulations of the mutated complex under external forces. The results of the simulations supported the second scenario, suggesting that the E34A mutation would promote Syb unraveling from the core SNARE bundle, and predicted a superclamping phenotype in the CpxE34A mutant. To test this prediction, we generated this mutation in Drosophila and found that indeed the mutation selectively reduced spontaneous activity. To further test the model, we generated a line with a complementary mutation in n-Syb and found that this mutation also produced a selective decrease in spontaneous activity.
These results help define the mechanisms by which Cpx clamps spontaneous release. It is established that Cpx promotes evoked release in both invertebrates and vertebrates (39, 40). In contrast, suppression of spontaneous activity by Cpx is most pronounced in invertebrate preparations, including Drosophila (24) and Caenorhabditis elegans (41, 42), although the inhibitory Cpx action was also observed at mammalian synapses (43, 44). Importantly, different Cpx domains are responsible for the regulation of evoked and spontaneous release, and these two Cpx functions can be manipulated independently (6, 15, 37, 45, 46). Since the Cpx AH was shown to contribute to the regulation of spontaneous activity (6, 47), we focused on the molecular mechanics and dynamics that would enable the Cpx AH to regulate spontaneous fusion.
Several molecular models of the Cpx-mediated fusion clamp have been proposed in previous studies. Since extensive evidence suggests that the intermediate state of the SNARE assembly is likely to be represented by partially unraveled Syb (13, 48, 49), it was initially proposed that the Cpx AH may insert between the Syb and the SNARE bundle, thus promoting a clamped state (50). Subsequently, this model was developed to include cooperation between several SNARE complexes bridged by Cpx (10, 11). This model proposes a clamped state of the SNARE complex with a radically unraveled C-terminus, whereby approximately a half of the helical region of Syb is separated from the bundle. However, theoretical studies suggest that such a radical separation of Syb from the bundle represents a high-energy state, and the electrical repulsion between the vesicle and the membrane would not be sufficient to maintain it (12, 23). Furthermore, recent studies suggest such a partially unraveled state of the SNARE complex is stabilized by Munc18 (51, 52, 53). Finally, a recent single-molecule study (14) convincingly demonstrated that the presence of Cpx stabilizes the partially unzippered SNARE complex at a distance of ∼1 nm between the Syx and Syb C-termini. Such a mild separation is in excellent agreement with the results of our MD simulations (Fig. 1), which suggest that the interaction of Cpx with Syb would promote unraveling of one or two helical turns of Syb (Fig. 1 C), increasing the separation of Syb and Syx by 1–1.5 nm (Fig. 1 B). Thus, it is likely that Cpx is required to stabilize the clamped state of the SNARE complex in the latest stages of SNARE bundle assembly.
Alternative models proposed that the Cpx AH inhibits release by producing electrostatic and steric repulsion between the vesicle and the membrane (17) or by stabilizing the overall helical structure of Cpx (18). Although these mechanisms could contribute to Cpx’s function, we show here that substitution of a single amino acid in Cpx (E34) significantly enhances its clamping function. Notably, a substitution of the same residue (E34F) was a part of a superclamping mutation that was previously examined in vitro (10) and in vivo (15). One could argue that due to the nature of the mutation (the replacement of Glu by Ala), it changes the net charge of the Cpx AH and alters its electrostatic properties, possibly preventing Cpx from cross-linking SNARE complexes (10). However, we show that mutating a matching residue in Syb has a similar effect. This result underscores the importance of chemical specificity in the interaction between Cpx and Syb, suggesting that the fusion clamp is mediated by the interaction between these two proteins. These results argue that the Cpx clamping function is defined to a large extent by its specific interaction with the SNARE bundle and, more specifically, with Syb.
Our results show that combining MD simulations with mutagenesis and genetics may facilitate the development of theoretical tools for targeted manipulation of the SNARE machinery and synaptic vesicle fusion. We show that MD simulations under external forces enable specific predictions of how single-point mutations in the SNARE proteins or Cpx would affect the pathways of SNARE assembly. Notably, recent advances in single-molecule experiments employing optical tweezers (54, 55) have provided opportunities to test such predictions directly, thus creating a potential bridge between molecular modeling and in vivo studies. The mutations examined in this study provide clear examples of the utility of MD simulations for predicting the outcome of mutagenesis. For example, it would be difficult to predict the effect of disrupting the interaction between Cpx and Syb based on intuitive considerations, since multiple scenarios are possible, and therefore the forces that affect the stability of the SNARE complex should be carefully quantified.
Together, the MD simulations combined with targeted mutagenesis suggest that Cpx is likely to clamp fusion by interacting with Syb and stabilizing the clamped state of the SNARE complex in which one or two C-terminal helical turns of Syb are unraveled.
Author Contributions
A.V. performed research and analyzed data. D.V. performed research. J.T.L. designed research and wrote the manuscript. M.B. designed and performed research, analyzed data, and wrote the manuscript.
Acknowledgments
This study was supported by a grant from the National Institutes of Health (R01 MH0999557 to M.B. and J.T.L.) and used XSEDE resources supported by National Science Foundation grant ACI-1053575.
Editor: Richard Bertram.
Footnotes
Six figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30817-7.
Supporting Material
References
- 1.Südhof T.C. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron. 2013;80:675–690. doi: 10.1016/j.neuron.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutton R.B., Fasshauer D., Brunger A.T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 3.Leitz J., Kavalali E.T. Ca2+ dependence of synaptic vesicle endocytosis. Neuroscientist. 2015;22:464–476. doi: 10.1177/1073858415588265. [DOI] [PubMed] [Google Scholar]
- 4.Chen X., Tomchick D.R., Rizo J. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 5.Trimbuch T., Rosenmund C. Should I stop or should I go? The role of complexin in neurotransmitter release. Nat. Rev. Neurosci. 2016;17:118–125. doi: 10.1038/nrn.2015.16. [DOI] [PubMed] [Google Scholar]
- 6.Xue M., Reim K., Rosenmund C. Distinct domains of complexin I differentially regulate neurotransmitter release. Nat. Struct. Mol. Biol. 2007;14:949–958. doi: 10.1038/nsmb1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Südhof T.C., Rothman J.E. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rizo J., Xu J. The synaptic vesicle release machinery. Annu. Rev. Biophys. 2015;44:339–367. doi: 10.1146/annurev-biophys-060414-034057. [DOI] [PubMed] [Google Scholar]
- 9.Jahn R., Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kümmel D., Krishnakumar S.S., Reinisch K.M. Complexin cross-links prefusion SNAREs into a zigzag array. Nat. Struct. Mol. Biol. 2011;18:927–933. doi: 10.1038/nsmb.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F., Pincet F., Rothman J.E. Complexin activates and clamps SNAREpins by a common mechanism involving an intermediate energetic state. Nat. Struct. Mol. Biol. 2011;18:941–946. doi: 10.1038/nsmb.2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortoul N., Singh P., Jagota A. Coarse-grained model of the snare complex determines the number of snares required for docking. Biophys. J. 2015;108:154a. doi: 10.1016/j.bpj.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter A.M., Wiederhold K., Sørensen J.B. Synaptobrevin N-terminally bound to syntaxin-SNAP-25 defines the primed vesicle state in regulated exocytosis. J. Cell Biol. 2010;188:401–413. doi: 10.1083/jcb.200907018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin L., Kim J., Shin Y.K. Complexin splits the membrane-proximal region of a single SNAREpin. Biochem. J. 2016;473:2219–2224. doi: 10.1042/BCJ20160339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho R.W., Kümmel D., Littleton J.T. Genetic analysis of the Complexin trans-clamping model for cross-linking SNARE complexes in vivo. Proc. Natl. Acad. Sci. USA. 2014;111:10317–10322. doi: 10.1073/pnas.1409311111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue M., Craig T.K., Rosenmund C. Binding of the complexin N terminus to the SNARE complex potentiates synaptic-vesicle fusogenicity. Nat. Struct. Mol. Biol. 2010;17:568–575. doi: 10.1038/nsmb.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trimbuch T., Xu J., Rosenmund C. Re-examining how complexin inhibits neurotransmitter release. eLife. 2014;3:e02391. doi: 10.7554/eLife.02391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radoff D.T., Dong Y., Dittman J.S. The accessory helix of complexin functions by stabilizing central helix secondary structure. eLife. 2014;3:e04553. doi: 10.7554/eLife.04553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindau M., Hall B.A., Sansom M.S. Coarse-grain simulations reveal movement of the synaptobrevin C-terminus in response to piconewton forces. Biophys. J. 2012;103:959–969. doi: 10.1016/j.bpj.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durrieu M.P., Lavery R., Baaden M. Interactions between neuronal fusion proteins explored by molecular dynamics. Biophys. J. 2008;94:3436–3446. doi: 10.1529/biophysj.107.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghahremanpour M.M., Mehrnejad F., Moghaddam M.E. Structural studies of SNARE complex and its interaction with complexin by molecular dynamics simulation. Biopolymers. 2010;93:560–570. doi: 10.1002/bip.21397. [DOI] [PubMed] [Google Scholar]
- 22.Bock L.V., Hutchings B., Woodbury D.J. Chemomechanical regulation of SNARE proteins studied with molecular dynamics simulations. Biophys. J. 2010;99:1221–1230. doi: 10.1016/j.bpj.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bykhovskaia M., Jagota A., Littleton J.T. Interaction of the complexin accessory helix with the C-terminus of the SNARE complex: molecular-dynamics model of the fusion clamp. Biophys. J. 2013;105:679–690. doi: 10.1016/j.bpj.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huntwork S., Littleton J.T. A complexin fusion clamp regulates spontaneous neurotransmitter release and synaptic growth. Nat. Neurosci. 2007;10:1235–1237. doi: 10.1038/nn1980. [DOI] [PubMed] [Google Scholar]
- 25.Littleton J.T., Chapman E.R., Ganetzky B. Temperature-sensitive paralytic mutations demonstrate that synaptic exocytosis requires SNARE complex assembly and disassembly. Neuron. 1998;21:401–413. doi: 10.1016/s0896-6273(00)80549-8. [DOI] [PubMed] [Google Scholar]
- 26.Deitcher D.L., Ueda A., Schwarz T.L. Distinct requirements for evoked and spontaneous release of neurotransmitter are revealed by mutations in the Drosophila gene neuronal-synaptobrevin. J. Neurosci. 1998;18:2028–2039. doi: 10.1523/JNEUROSCI.18-06-02028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buhl L.K., Jorquera R.A., Littleton J.T. Differential regulation of evoked and spontaneous neurotransmitter release by C-terminal modifications of complexin. Mol. Cell. Neurosci. 2013;52:161–172. doi: 10.1016/j.mcn.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiAntonio A., Burgess R.W., Schwarz T.L. Identification and characterization of Drosophila genes for synaptic vesicle proteins. J. Neurosci. 1993;13:4924–4935. doi: 10.1523/JNEUROSCI.13-11-04924.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 30.Dudel J. The effect of reduced calcium on quantal unit current and release at the crayfish neuromuscular junction. Pflugers Arch. 1981;391:35–40. doi: 10.1007/BF00580691. [DOI] [PubMed] [Google Scholar]
- 31.Akbergenova Y., Bykhovskaia M. Synapsin maintains the reserve vesicle pool and spatial segregation of the recycling pool in Drosophila presynaptic boutons. Brain Res. 2007;1178:52–64. doi: 10.1016/j.brainres.2007.08.042. [DOI] [PubMed] [Google Scholar]
- 32.Akbergenova Y., Bykhovskaia M. Stimulation-induced formation of the reserve pool of vesicles in Drosophila motor boutons. J. Neurophysiol. 2009;101:2423–2433. doi: 10.1152/jn.91122.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bykhovskaia M. Making quantal analysis more convenient, fast, and accurate: user-friendly software QUANTAN. J. Neurosci. Methods. 2008;168:500–513. doi: 10.1016/j.jneumeth.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackerell A.D., Jr. Empirical force fields for biological macromolecules: overview and issues. J. Comput. Chem. 2004;25:1584–1604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 36.Lagow R.D., Bao H., Zhang B. Modification of a hydrophobic layer by a point mutation in syntaxin 1A regulates the rate of synaptic vesicle fusion. PLoS Biol. 2007;5:e72. doi: 10.1371/journal.pbio.0050072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jorquera R.A., Huntwork-Rodriguez S., Littleton J.T. Complexin controls spontaneous and evoked neurotransmitter release by regulating the timing and properties of synaptotagmin activity. J. Neurosci. 2012;32:18234–18245. doi: 10.1523/JNEUROSCI.3212-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho R.W., Buhl L.K., Littleton J.T. Phosphorylation of complexin by PKA regulates activity-dependent spontaneous neurotransmitter release and structural synaptic plasticity. Neuron. 2015;88:749–761. doi: 10.1016/j.neuron.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reim K., Mansour M., Rosenmund C. Complexins regulate a late step in Ca2+-dependent neurotransmitter release. Cell. 2001;104:71–81. doi: 10.1016/s0092-8674(01)00192-1. [DOI] [PubMed] [Google Scholar]
- 40.Xue M., Stradomska A., Reim K. Complexins facilitate neurotransmitter release at excitatory and inhibitory synapses in mammalian central nervous system. Proc. Natl. Acad. Sci. USA. 2008;105:7875–7880. doi: 10.1073/pnas.0803012105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hobson R.J., Liu Q., Jorgensen E.M. Complexin maintains vesicles in the primed state in C. elegans. Curr. Biol. 2011;21:106–113. doi: 10.1016/j.cub.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin J.A., Hu Z., Dittman J.S. Complexin has opposite effects on two modes of synaptic vesicle fusion. Curr. Biol. 2011;21:97–105. doi: 10.1016/j.cub.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang S., Reim K., Taschenberger H. Complexin stabilizes newly primed synaptic vesicles and prevents their premature fusion at the mouse calyx of held synapse. J. Neurosci. 2015;35:8272–8290. doi: 10.1523/JNEUROSCI.4841-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maximov A., Tang J., Südhof T.C. Complexin controls the force transfer from SNARE complexes to membranes in fusion. Science. 2009;323:516–521. doi: 10.1126/science.1166505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaeser-Woo Y.J., Yang X., Südhof T.C. C-terminal complexin sequence is selectively required for clamping and priming but not for Ca2+ triggering of synaptic exocytosis. J. Neurosci. 2012;32:2877–2885. doi: 10.1523/JNEUROSCI.3360-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue M., Lin Y.Q., Rosenmund C. Tilting the balance between facilitatory and inhibitory functions of mammalian and Drosophila complexins orchestrates synaptic vesicle exocytosis. Neuron. 2009;64:367–380. doi: 10.1016/j.neuron.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X., Kaeser-Woo Y.J., Südhof T.C. Complexin clamps asynchronous release by blocking a secondary Ca(2+) sensor via its accessory α helix. Neuron. 2010;68:907–920. doi: 10.1016/j.neuron.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li F., Kümmel D., Pincet F. A half-zippered SNARE complex represents a functional intermediate in membrane fusion. J. Am. Chem. Soc. 2014;136:3456–3464. doi: 10.1021/ja410690m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pobbati A.V., Stein A., Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 50.Giraudo C.G., Garcia-Diaz A., Rothman J.E. Alternative zippering as an on-off switch for SNARE-mediated fusion. Science. 2009;323:512–516. doi: 10.1126/science.1166500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker R.W., Jeffrey P.D., Hughson F.M. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 2015;349:1111–1114. doi: 10.1126/science.aac7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen C., Rathore S.S., Shen J. The trans-SNARE-regulating function of Munc18-1 is essential to synaptic exocytosis. Nat. Commun. 2015;6:8852. doi: 10.1038/ncomms9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma L., Rebane A.A., Zhang Y. Munc18-1-regulated stage-wise SNARE assembly underlying synaptic exocytosis. eLife. 2015;4 doi: 10.7554/eLife.09580. pii:e09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao Y., Zorman S., Zhang Y. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zorman S., Rebane A.A., Zhang Y. Common intermediates and kinetics, but different energetics, in the assembly of SNARE proteins. eLife. 2014;3:e03348. doi: 10.7554/eLife.03348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.