Abstract
Filopodia are thin, fingerlike structures that contain bundled actin filaments and project from the cell periphery. These structures are dogmatically endowed with the ability to sense cues in the microenvironment, implying that filopodia foster local signal transduction, yet their small diameter hampers the imaging of dynamic processes therein. To overcome this challenge, we analyzed total internal reflection fluorescence images of migrating fibroblasts coexpressing either a plasma membrane marker or tagged AktPH domain, a translocation biosensor for signaling through the phosphoinositide 3-kinase pathway, along with a cytosolic volume marker. We devised a scheme to estimate the radii of filopodia using either the membrane marker or volume marker data, and we used that information to account for geometry effects in the biosensor data. With conservative estimates of relative target molecule abundance, it is revealed that filopodia typically harbor higher densities of 3′ phosphoinositides than adjacent regions at the cell periphery. In this context at least, the analysis supports the filopodial signaling hypothesis.
Main Text
In migrating cells, various cytoskeletal arrays assemble in concert with motor proteins to generate forces that drive cell locomotion. At the leading edge of a motile cell, two prominent F-actin architectures are typically observed: the dendritic actin network that is characteristic of broad protrusions (lamellipodia), and bundled actin filaments that are typically present in thin (∼100 nm diameter), fingerlike projections called filopodia (1). Both structures promote formation of adhesion complexes, which in turn interact with F-actin as a mechanical clutch (2, 3). Adhesion complexes also mediate signal transduction, which affects F-actin polymerization and myosin contractility (4).
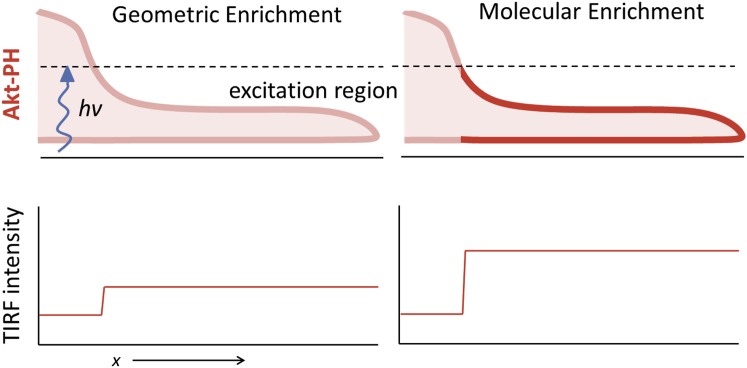
Recently, we studied the interplay among these dynamical systems, as it relates to the coupling of adhered filopodia and the emergence of nascent lamellipodia; we showed that adhesion-based signaling mediated the coupling (5). Enriched type I phosphoinositide 3-kinase (PI3K) signaling was detected by total internal reflection fluorescence (TIRF) microscopy in filopodia before the onset of lamellipodial protrusion, using fluorescent protein-tagged Akt PH domain (AktPH) as a translocation biosensor; however, we were unable to interpret the intensity data in terms of the density of AktPH recruited to filopodia. The analysis was confounded because the entirety of a thin filopod might be significantly excited by TIRF, exaggerating the membrane-bound contribution to the intensity (Fig. 1). Accounting for this effect of filopod geometry is the rationale for this study.
Figure 1.
Illustration of enrichment due to filopod geometry versus enrichment of bound biosensor. To see this figure in color, go online.
TIRF illumination selectively excites fluorophores in close proximity to a glass-aqueous interface. The local fluorescence intensity, f(x,y), is generally related to the fluorophore concentration, [F](x,y,z), as follows (6):
| (1) |
The penetration depth, d ∼100 nm, describes the decay of TIRF excitation with distance from the surface, z; d is proportional to the excitation wavelength and readily calculated (6). The coefficient α accounts for the properties of the fluorophore and of the image acquisition.
If we consider a translocation biosensor such as AktPH, a fraction of it is in complex with its target(s) at the plasma membrane, and a fraction resides in the adjacent cytoplasm. Using the notation of Haugh (7), we take the local concentration of unbound biosensor as [B] and the local density of membrane-bound complex as C. For a semiinfinite region, such as the center of the cell, and a gap height h between the glass surface and the cell, Eq. 1 is solved as follows (6):
| (2) |
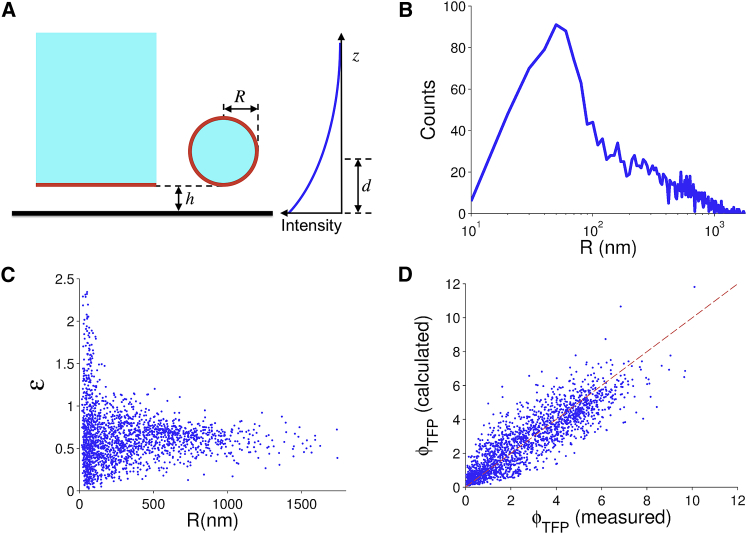
The solution of Eq. 1 for a different geometry is not as straightforward. To compare the TIRF intensities of individual filopodia to that of a semiinfinite region, their geometries must be taken into account (Fig. 2 A). In the analyses presented here, we consider each filopod as a cylinder with length, L, and radius, R, with the latter typically below the limit of optical resolution.
Figure 2.
Properties of adhered filopodia inferred from TIRF images. (A) Illustration of TIRF illumination for a semiinfinite region of a cell versus the circular cross section of a filopod. (B) Histogram of filopod radii determined from images of the membrane marker, GPI-tdTomato. (C) With R determined from b, the volume marker (TFP) images were used to estimate the void fraction εf for each filopod. (D) With R determined from b and assuming εf = 0.62 for each filopod, the predicted value of Φ for the volume marker (Eq. 5) is compared to the measured value. To see this figure in color, go online.
A cohort of nine mouse fibroblasts (NIH3T3; American Type Culture Collection, Manassas, VA) that coexpressed GPI-tdTomato, a membrane marker, and cytosolic teal fluorescent protein (TFP) were imaged by TIRF microscopy as reported in Johnson et al. (5). For each filopod (n = 1,916) and marker, we determined the dimensionless quantity, Φ, defined here as follows:
| (3) |
Here, ff and Af are the mean intensity and area of the filopod region, and fc is the mean intensity of the cell center (see Supporting Materials and Methods and Fig. S1 in the Supporting Material). Each filopod was tracked over sequential frames (5), and the measured value of Φ is an average over the filopod’s lifetime. Assuming a uniform membrane marker and our geometric model, and neglecting the possible difference in h between the two regions, we derived an expression for Φ as a function of ρ = R/d. We refer to this function as Φmem (all derivations are provided in Supporting Materials and Methods):
| (4) |
We applied this model to the GPI-tdTomato data (with d for 561 nm excitation) to estimate R for each filopod (Fig. 2 B). The distribution peaks at ≈50 nm, consistent with ultrastructural measurements (1). For the cytosolic marker, TFP, we similarly derived an equation for Φ, Φcyt (with d for 442 nm excitation):
| (5) |
The coefficient εf was added to account for the void fraction inside filopodia (relative to the cytosol in the center region). Using the values of R estimated from the GPI-tdTomato data, the values of εf were estimated from the TFP data. The results reveal a reasonable peak εf value of 0.62, with large variability only among filopodia with the lower estimates of R (Fig. 2 C). Supposing that the mean value of εf were assumed for all filopodia, and using the same estimates of R from before, the values of Φ for TFP were predicted (Eq. 5) and compared to the measured values. A reasonable agreement was found (Fig. 2 D; coefficient of determination = 0.72). We conclude that our geometric model is suitable for characterizing filopodia.
In another group of 21 cells coexpressing mCherry-AktPH and TFP, a total of 1336 filopodia were identified. This data set extends one described previously in Johnson et al. (5). To relate the measured TIRF intensity to the abundance of AktPH in filopodia, we combine the results given above to derive Φ for a translocation biosensor:
| (6) |
Equation 6 is simplified by assuming: biosensor-target binding at the center of the cell, Cc, is negligible; free biosensor is fast diffusing and therefore close to uniform ([B]f ≈ [B]c); and biosensor-target binding is close to equilibrium. Therefore,
| (7) |
where Tf is the density of (unbound) target in the filopod membrane, and KD is the equilibrium dissociation constant of biosensor-target binding. With prior estimates of ρ and εf, Eq. 7 allows estimation of Tf/dKD, a dimensionless measure of target abundance. The underlying assumptions are conservative. If there were substantial target density in filopodia, then local depletion of free biosensor is expected ([B]f < [B]c) (7); accounting for this, and allowing Cc to be significant, would only increase the estimate of Tf/dKD.
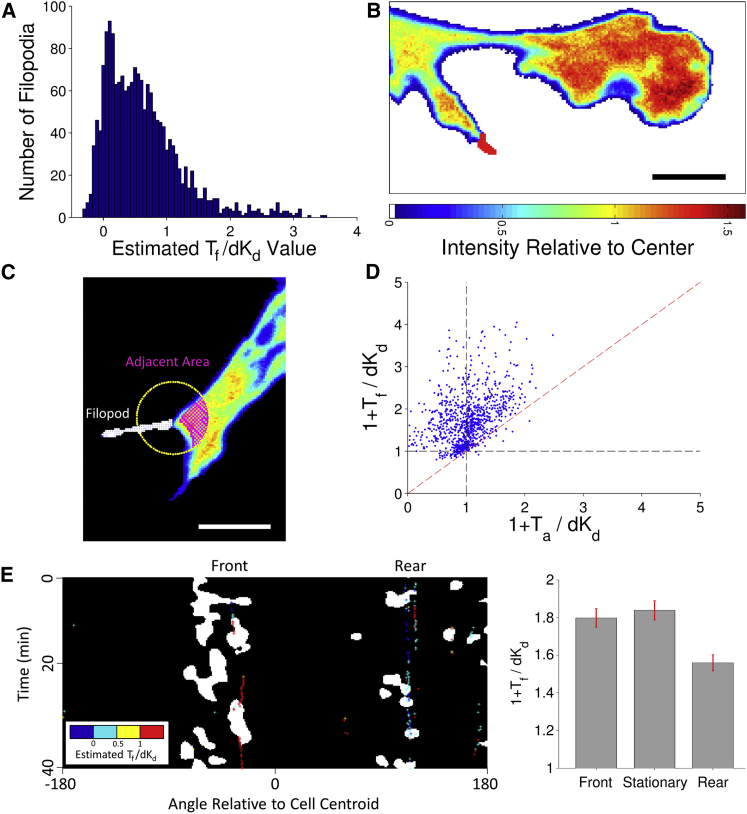
Assuming a value of εf = 0.62 based on the analysis shown in Fig. 1, C and D, the TFP data for this cohort yielded estimates of R for all filopodia. Turning to the mCherry-AktPH data, the assumed value of εf and the estimated value of R were used to estimate Tf/dKD for each filopod (Eq. 7, with d for 561 nm excitation). As expected, the vast majority of the Tf/dKD values are substantially positive (Fig. 3 A). Each Tf/dKD value can, in turn, be used to conservatively estimate an adjusted fluorescence, as if the filopod were flat and semiinfinite, comparable to intensities elsewhere in the contact area (Fig. 3 B):
| (8) |
With this approach, we compared the adjusted intensity of each filopod, 1+Tf/dKD, to that of the adjacent region, 1+Ta/dKD (Fig. 3 C; see Supporting Materials and Methods). According to this analysis, most filopodia (94%) showed enrichment of AktPH binding, even when the adjacent regions did not, and with few exceptions the enrichment in the filopod exceeded that of the adjacent region (95%; Fig. 3 D).
Figure 3.
Quantifying PI3K signaling in filopodia. (A) Histogram of Tf/dKD estimated from Eq. 7. (B) Adjusted fluorescence intensity of a filopod (arrow). Scale bar = 10 μm. (C) Pseudocolor TIRF image showing the specification of a region adjacent to a filopod. Scale bar = 10 μm. (D) The adjusted fluorescence of each filopod, 1+Tf/dKD, is compared to that of the adjacent region, 1+Ta/dKD. (E) Spatiotemporal map showing structures identified as lamellipodia (white) and filopodia (colored according to Tf/dKD). (Right) The bar graph shows the mean adjusted intensities of filopodia associated with front (n = 310), stationary (n = 547), and rear (n = 479) regions (see also Fig. S2). The error bars are 95% confidence intervals. To see this figure in color, go online.
To put this analysis in a dynamic context, a spatiotemporal map constructed from a time series of a representative cell displays the timing and locations of structures identified as lamellipodia and filopodia (Fig. 3 E) (5). In this sequence, a prominent filopod at the front of the cell adhered before the emergence of a lamellipod, consistent with our previous findings (5); this filopod showed strong enrichment of AktPH binding. At the same time, structures identified as filopodia (but exhibiting hallmarks of retraction fibers) were present at the trailing end of the cell and exhibited low enrichment of AktPH binding. The latter observation prevailed overall for filopodia emanating from regions identified as retracting (Figs. 3 E and S2).
Filopodia are broadly described as environmental sensors (1, 8), implying that signals are transduced from within. Here we have outlined a method for estimating the enrichment of fluorescent biosensors in adhered filopodia. We offer evidence that adhered filopodia of fibroblasts are indeed privileged loci for PI3K signaling, as inferred for neurons (9). Whether filopodia foster activation of PI3K or frustrate dephosphorylation of its lipid products remains to be explored. An alternate explanation is that filopodia offer a longer domain for lipid accumulation; the added length would ameliorate dilution by diffusion of the lipid away from the leading edge. This is plausible because we previously estimated a spatial range of ∼10 μm for this second messenger (10), substantially larger than the thickness of the dendritic F-actin network at the cell’s leading edge(s).
Author Contributions
H.E.J. performed research, and both authors designed research, analyzed data, and wrote the article.
Acknowledgments
This work was supported by grant No. R01-GM110155 from the National Institutes of Health.
Editor: Philip LeDuc.
Footnotes
Supporting Materials and Methods and two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30822-0.
Supporting Material
References
- 1.Mattila P.K., Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 2.Galbraith C.G., Yamada K.M., Galbraith J.A. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science. 2007;315:992–995. doi: 10.1126/science.1137904. [DOI] [PubMed] [Google Scholar]
- 3.Chan C.E., Odde D.J. Traction dynamics of filopodia on compliant substrates. Science. 2008;322:1687–1691. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- 4.Vicente-Manzanares M., Choi C.K., Horwitz A.R. Integrins in cell migration—the actin connection. J. Cell Sci. 2009;122:199–206. doi: 10.1242/jcs.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson H.E., King S.J., Haugh J.M. F-actin bundles direct the initiation and orientation of lamellipodia through adhesion-based signaling. J. Cell Biol. 2015;208:443–455. doi: 10.1083/jcb.201406102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reichert W.M., Truskey G.A. Total internal reflection fluorescence (TIRF) microscopy. I. Modelling cell contact region fluorescence. J. Cell Sci. 1990;96:219–230. doi: 10.1242/jcs.96.2.219. [DOI] [PubMed] [Google Scholar]
- 7.Haugh J.M. Live-cell fluorescence microscopy with molecular biosensors: what are we really measuring? Biophys. J. 2012;102:2003–2011. doi: 10.1016/j.bpj.2012.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davenport R.W., Dou P., Kater S.B. A sensory role for neuronal growth cone filopodia. Nature. 1993;361:721–724. doi: 10.1038/361721a0. [DOI] [PubMed] [Google Scholar]
- 9.Luikart B.W., Zhang W., Parada L.F. Neurotrophin-dependent dendritic filopodial motility: a convergence on PI3K signaling. J. Neurosci. 2008;28:7006–7012. doi: 10.1523/JNEUROSCI.0195-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider I.C., Parrish E.M., Haugh J.M. Spatial analysis of 3′ phosphoinositide signaling in living fibroblasts, III: influence of cell morphology and morphological polarity. Biophys. J. 2005;89:1420–1430. doi: 10.1529/biophysj.105.061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.