Abstract
Membrane proteins act as a central interface between the extracellular environment and the intracellular response and as such represent one of the most important classes of drug targets. The characterization of the molecular properties of integral membrane proteins, such as topology and interdomain interaction, is key to a fundamental understanding of their function. Atomic force microscopy (AFM) and force spectroscopy have the intrinsic capabilities of investigating these properties in a near-native setting. However, atomic force spectroscopy of membrane proteins is traditionally carried out in a crystalline setup. Alternatively, model membrane systems, such as tethered bilayer membranes, have been developed for surface-dependent techniques. While these setups can provide a more native environment, data analysis may be complicated by the normally found statistical orientation of the reconstituted protein in the model membrane. We have developed a model membrane system that enables the study of membrane proteins in a defined orientation by single-molecule force spectroscopy. Our approach is demonstrated using cell-free expressed bacteriorhodopsin coupled to a quartz glass surface in a defined orientation through a protein anchor and reconstituted inside an artificial membrane system. This approach offers an effective way to study membrane proteins in a planar lipid bilayer. It can be easily transferred to all membrane proteins that possess a suitable tag and can be reconstituted into a lipid bilayer. In this respect, we anticipate that this technique may contribute important information on structure, topology, and intra- and intermolecular interactions of other seven-transmembrane helical receptors.
Introduction
The characterization of transmembrane proteins is key to a better understanding of essential processes in life. Transmembrane proteins account for ∼30% of all proteins (1) and act as sensors, catalysts, receptors, transporters, and channels. Thus, they play an important role in almost all cellular processes and are associated with a broad range of different diseases (2, 3, 4, 5). Among the membrane proteins, the class of seven-transmembrane helical (7TM) proteins, which includes G-protein-coupled receptors (GPCRs), have a very central part in a variety of sensing and signaling pathways, as well as physiological responses, making them a prominent target for drug development.
Transmembrane proteins easily lose their functionality and denature when removed from their natural membrane environment. One approach to study functional membrane proteins is the development of simple artificial membrane systems that reduce the complexity but still mimic the most important properties of biological membranes. A promising system is a tethered bilayer lipid membrane (tBLM). Lipids of the first layer of the membrane are anchored covalently to a solid substrate through a spacer. This spacer, e.g., a polymer, acts as a cushion that compensates surface roughness, mimicking a cytoskeleton, and can additionally create an ion reservoir beneath the membrane (6, 7, 8). The spacer can be bound to the substrate first, binding the lipid to the spacer in a subsequent step (9). tBLMs have proven to be stable for days and can even be used for weeks when covered by a hydrogel (9). Binding of the membrane to the spacer occurs through an anchoring molecule, a lipid or hydrophobic chain. Important while binding the anchoring molecule is the grafting density, i.e., the ratio of anchored to not-anchored lipids. Although a high grafting density leads to high electrical resistance, it can also hinder the incorporation of proteins (8) or the diffusion of incorporated proteins through the membrane (10). In addition to tBLMs, so-called protein tethered membranes can be formed as well. In this case, the anchoring lipid or hydrophobic chain is directly substituted with the membrane protein, which is coupled to the surface, e.g., through a complex between a surface bound nitrilotriacetic acid (NTA) and the protein histidine-tag (His-tag) (11).
Alternatively, membrane proteins can be studied in other artificial membrane systems, e.g., liposomes (12), nanodiscs (13), black or bilayer lipid membranes (BLMs) (8), and solid supported BLMs (sBLM) (8).
The formation of a tBLM can be done by self-assembly (14). One way of self-assembly is to adsorb and then spread out whole vesicles over the surface (15, 16). This way, proteins can be reconstituted already into the prepared vesicles before the final bilayer formation (17) or can be added to the finished bilayer on the surface (18).
Proteins embedded in a tBLM can be studied by atomic force microscopy (AFM). AFM-based single-molecule force spectroscopy can be used to obtain information on dissociation rates (19), energy barriers (20, 21), Gibbs free energy (19, 21), the form of a binding potential (21), and inter- and intramolecular interactions (20, 22, 23, 24), as well as the folding of proteins and their constitution inside a membrane (20).
In this study, we demonstrate a method of forming a tBLM on quartz glass with the incorporated 7TM model membrane protein bacteriorhodopsin (BR) in a defined orientation. BR has been studied extensively (25, 26, 27) and offers high stability. In addition, the covalently bound retinal allows direct insight into its folding state through absorption measurements (e.g., at 555 nm). We investigated the conformation of BR inside the presented tBLM by AFM-based force spectroscopy. Our data are in accordance with expected force-distance curves of a 7TM membrane protein (13, 28). The orientation of BR in purple membranes can be predetermined through AFM imaging (29, 30). Thus, the characteristics of force-distance curves of BR pulled from the extracellular side and the cytoplasmic side are already known, which in turn can be used to process the data of statistically oriented BR. In general, knowledge of the orientation will substantially facilitate data analysis and interpretation. In our model system, predefined orientation is achieved through first functionalizing the quartz glass by silanization. Afterwards, a polyethylene glycol (PEG) is coupled as a spacer. To reconstitute the protein on the surface in a defined orientation, a protein anchor is bound to the spacer in addition to the lipid anchors. We make use of cell-free protein expression, a method that is suitable to produce 1) a broad range of different membrane proteins (31), and 2) large amounts of protein in the presence or absence of cofactors such as ligands, detergents, or lipids. We could show before that cell-free expression of BR enables analysis of the pure protein that is not biased by copurification of, e.g., coordinated lipids, which are normally present when BR is extracted from the native purple membrane (32). To test our model system, we use cell-free expressed BR that contains a His-tag and a surface-coupled protein anchor consisting of Tris-NTA (trisNTA). Our results show the specific coupling of the protein to the surface and the successful formation of a tBLM containing the membrane protein in a defined orientation.
Materials and Methods
All water used was purified by the Milli-Q Integral Water Purification System (Merck Millipore, Darmstadt, Germany). Specific buffer compositions are described in the Supporting Material. All given cantilever values are nominal values.
Lipids, detergents, and stock solutions
1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC; >99% purity), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(succinyl) Sodium salt (SuccinylPE; >99% purity), and n-dodecylphosphocholine (DPC; >99% purity) were purchased as powders from Avanti Polar Lipids (Alabaster, AL). n-Dodecyl-β-D-maltopyranoside (DDM; Anagrade) was purchased from Affymetrix (Santa Clara, CA). Carboxy-functionalized tert-butyl ester (OtBu)-protected trisNTA (33) was provided by the group of Prof. Dr. Jacob Piehler of the University of Osnabrück. Stock solutions in chloroform (for spectroscopy, 99+% purity, stabilized with amylene; Acros Organics, Geel, Belgium) were created as follows: 0.026 g SuccinylPE, 0.022 g DPPC, and 0.005 g trisNTA were dissolved in 1 mL chloroform and stored at −80°C.
AFM
AFM images were taken with an AFM Nanowizard III (JPK Instruments, Berlin, Germany). Imaging was done at room temperature (RT) using contact mode with the probe OMCL-TR400PSA (Cantilever 2, Asylum Research, Mannheim, Germany) with a spring constant (kc) of 0.02 N/m, a resonance frequency (f) in water of 11 kHz, and a tip radius (r) of 15 nm, or using tapping mode with the probe SNL-10 (Cantilever A, kc = 0.35 N/m, f = 65 kHz in air, r = 2 nm; Bruker, Billerica, MA) in buffer P1.
AFM-based force spectroscopy
Force-distance curves were measured using a Nanowizard III (JPK Instruments). The same probes as for imaging in contact mode (OMCL-TR400PSA, Cantilever 2) were used. All experiments were performed at RT in buffer P1. Force-distance curves were acquired as follows. Starting away from the surface, the cantilever is moved downward at a constant speed (200 nm/s). When the surface is reached, the cantilever is moved farther until it reaches a defined maximal deflection (0.4 mV). This position is held for 1 s before retraction of the cantilever (200 nm for force-distance curves unfolding BR, 100–400 nm for force-distance curves measuring adhesion). This cycle is repeated at different positions on the surface (acquiring ∼500 force-distance curves per position for experiments unfolding BR and ∼100 force-distance curves per position during adhesion experiments). The position on the surface was changed two to three times per experiment. Experimental conditions were repeated in two independent measurements. A fresh cantilever was used for each measurement. As force measurements require the precise knowledge of the cantilever’s spring constant, kc, the thermal noise method (34) is used for calibration. Additionally, the cantilever’s sensitivity (deflection → force) is calibrated by measuring the deflection of the cantilever approached against the surface. Calibration was performed at the beginning of each experiment and between positions on the surface.
X-ray photoelectron spectroscopy
Cleaned quartz glass surfaces and silanized quartz glass surfaces were characterized with x-ray photoelectron spectroscopy (XPS) using an ESCALAB MK II spectrometer (Thermo Fisher Scientific, Oberhausen, Germany).
Functionalization steps
Cleaning of quartz glass according to a previously described protocol (35) was followed by silanization with (3-aminopropyl)trimethoxysilane (APTES; 97%, Sigma Aldrich, St. Louis, MO) and subsequent binding of the spacer Mmt-NH-PEG12-COO-Tfp (PEG; Iris Biotech, Marktredwitz, Germany). To this spacer was coupled the protein anchor trisNTA alone, the lipid anchor SuccinylPE alone, and a mixture of both anchors 1:2 (mol/mol). The detailed coupling steps are described in the Supporting Material.
Lipid suspension
30 mM DPPC stock (230 μL) was transferred into a sample vial and the chloroform was evaporated under nitrogen. Buffer P1 (1 mL) with 0.1% DPC (w/v) was added to create the final lipid suspension.
Binding of BR and membrane formation on functionalized quartz glass
After rinsing the functionalized slides (1 cm2) 10 times with water, the surface was covered with 50 mM EDTA (disodium salt dehydrate, molecular biology grade; AppliChem, Darmstadt, Germany) solution for 5 min. After removal of the EDTA solution, a 10 mM nickel(II) chloride (98%, for analysis; Grüssing, Filsum, Germany) solution was added to the surface for 5 min. The solution was removed from the surface and replaced with buffer DDM-W1. The surface was rinsed once with protein buffer DDM-P1, and 20 μL of a 10 μM BR solution in buffer DDM-P1 was added to 180 μL lipid suspension. This solution was placed onto the surface and allowed to incubate for 2 h. The sample was rinsed 10 times with 250 μL buffer P1 and covered with 50 μL of the same buffer for AFM measurements.
Membrane formation on mica
Lipid suspension (50 μL) was incubated on a round mica surface (Ø 10 mm, Ted Pella, Redding, CA) for 2 h. The surface was rinsed five times with 100 μL P1 and covered with 50 μL of the same buffer for AFM measurements.
Membrane formation with reconstituted BR on mica
A 10 μM BR solution in buffer DDM-P1 (5 μL) was added to 45 μL lipid suspension. The mixture was incubated on a round mica surface (Ø 10 mm) for 2 h. The surface was rinsed five times with 100 μL P1 and covered with 50 μL of the same buffer for AFM measurements.
Dot blot
Specifically bound BR should only be rinsed from a functionalized surface with a buffer containing imidazole. After rinsing the surface once with buffer missing imidazole and once with buffer containing imidazole, a simplified Western blot (dot blot) was utilized to test the specific binding of BR to the functionalized surface. The procedure is described in detail in the Supporting Material.
Cloning and cell-free expression
Cell-free expression of BR was carried out as described before (32). An Escherichia coli-based system following previously published protocols (36) was used. Dialysis-mode reactions were carried out in the absence of retinal and detergents. The resulting protein pellet was washed with S30 buffer and directly refolded or stored at −20°C. The refolding procedure is described in more detail in the Supporting Material.
Results
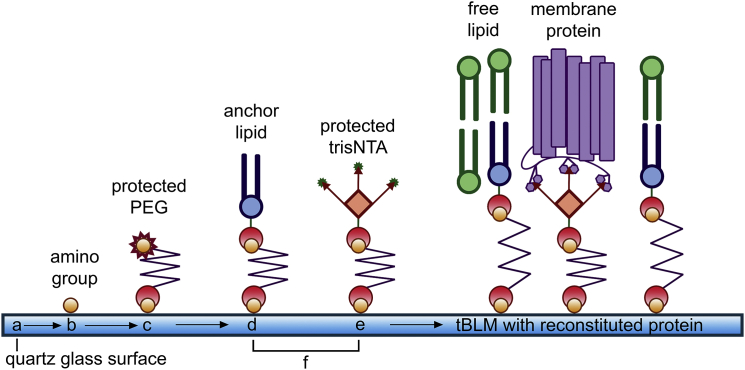
In our study, we aimed to reconstitute BR as a model membrane protein in a defined orientation in artificial tBLMs to perform single-molecule force spectroscopic studies. The process is depicted schematically in Fig. 1. The clean quartz glass surface (Fig. 1 a) is coated with APTES for amino functionalization (Fig. 1 b). After this, a PEG with an activated carboxyl group and a protected amino group is coupled to the surface (Fig. 1 c). Following the deprotection of the PEG’s amino group, the binding of an anchoring lipid (SuccinylPE) (Fig. 1 d) and a protein anchor (trisNTA) (Fig. 1 e) were investigated both separately and as a mixture of both anchors (Fig. 1 f). The protein is reconstituted on the surface together with the free lipid (DPPC) supported by detergent (DPC).
Figure 1.
Schematic representation of the functionalization and coupling steps. The cleaned quartz glass surface (a) is first functionalized using APTES (b) to create amino groups. To these amino groups a PEG linker (c) is coupled with its activated carboxyl group and protected amino group. After deprotection of the PEG linker amino group, the surface can be functionalized in three ways, with only the lipid anchor bound to PEG (d), with only the protein anchor (protected trisNTA) bound to PEG (e), or with both anchors bound simultaneously to PEG (f). Afterward, a mixture of the protein with free lipid is applied to the surface, forming the final tBLM with reconstituted protein. To see this figure in color, go online.
Surface composition before and after silanization
The atomic composition of a pure quartz glass surface was compared to a silanized surface using XPS. In the survey spectrum of cleaned quartz glass (Fig. S1 a), binding energies corresponding to fluorine (F), sodium (Na), and carbon (C) levels are detected apart from expected silicon (Si) and oxygen (O) levels. A survey spectrum after silanization with APTES shows binding energies corresponding to levels in O and Si (Fig. S1 b). Additionally, there is an increase of C from 12.8 atomic percent (at.%) to 41.6 at.% and an appearance of 7.2 at.% N.
Changes in surface topography
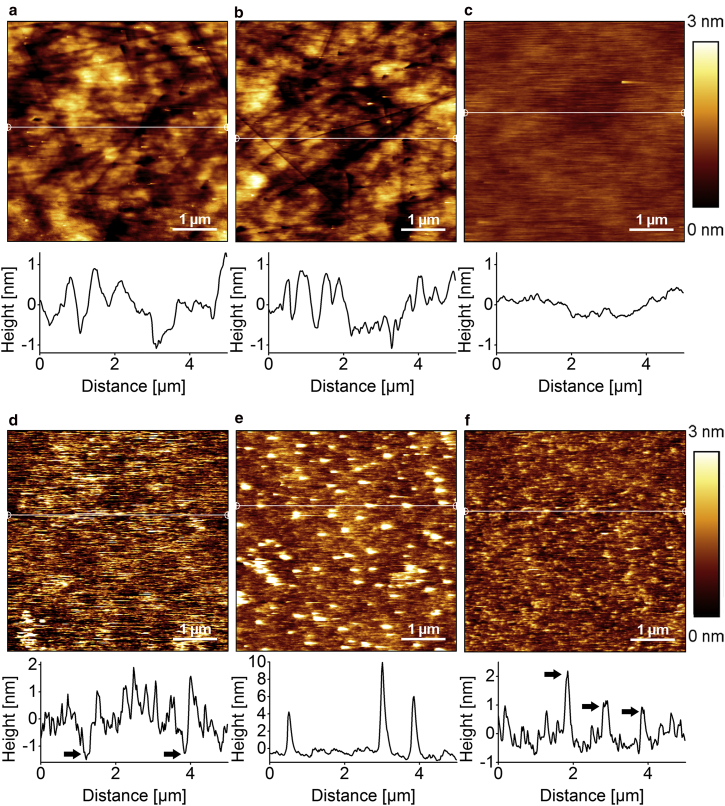
The functionalization of quartz glass surfaces was controlled by AFM to be able to observe the changes in surface topography after each functionalization step. The surface roughness is calculated as the arithmetic mean (i.e., the average roughness, Ra) of absolute height values for each surface. The AFM image of quartz glass (Fig. 2 a) after the cleaning procedure shows the standard surface topography of quartz glass. The average surface roughness is 511 picometers (pm). Holes and scratches, which are typical for quartz glass, are also visible. The surface topography of cleaned quartz glass is similar after silanization (Fig. 2 b), which is also represented by the average surface roughness. A surface functionalized with PEG is shown in Fig. 2 c. The average surface roughness has decreased to 154.1 pm and there are no longer holes or scratches visible.
Figure 2.
AFM scans (5 μm × 5 μm) in contact mode in buffer P1, with corresponding height profiles, represented as white horizontal lines in each image, below each scan. (a) The cleaned quartz glass surface shows a roughness of 511 PM. (b) Silanized quartz glass with APTES shows no change in surface roughness. (c) Binding PEG to silanized quartz reduces the roughness to 154.1 PM. (d) A surface functionalized only with the anchoring lipid SuccinylPE. (e) A surface functionalized only with the protein anchor trisNTA. (f) A surface functionalized with a mixture of 2:1 (mol/mol) SuccinylPE:trisNTA. To see this figure in color, go online.
The surface was functionalized further with the lipid anchor SuccinylPE only (Fig. 2 d), with trisNTA only (Fig. 2 e), and with a mixture of 2:1 (mol/mol) SuccinylPE/trisNTA (Fig. 2 f). The surface with the lipid anchor alone shows an increased average roughness of 585.5 pm compared to the previous step (Fig. 2 c). Some small regions indicate an additional depth of 1 nm (arrows). The surface functionalized solely with trisNTA also shows a generally higher average roughness and round structures of 4–10 nm height and 200 nm diameter distributed over the whole surface. A mixture of SuccinylPE and trisNTA yields an average roughness of 332.6 pm, but distributed protruding structures of 1–3 nm height can be seen (arrows).
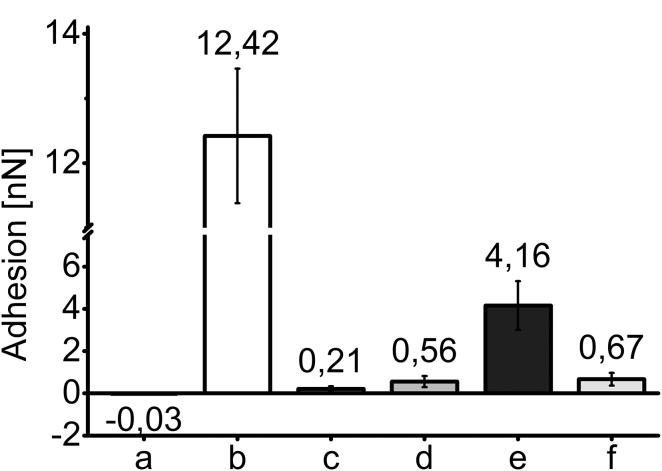
Changes in adhesion
Force-distance curves were performed between surface functionalization steps to observe the changes in cantilever-surface interaction (adhesion). This was done to observe whether adhesion on the functionalized surfaces remained constant over the majority of the surface, which would be an indication of complete and homogeneous functionalization. In Fig. 3, the mean adhesion of the cantilever on the surface is depicted for all functionalization steps, and for surfaces functionalized with SuccinylPE only, trisNTA only, or a mixture of the two. For quartz glass (Fig. 3 a), no adhesion can be observed. APTES-functionalized surfaces (Fig. 3 a) show a strong increase in adhesion force, from −25 pN ± 8 pN to 12,420 pN ± 1039 pN. After the second functionalization step, the strong adhesion forces diminish to 212 pN ± 125 pN for PEG (Fig. 3 c). From this step on, three functionalization ways were chosen. Surfaces functionalized with the lipid anchor SuccinylPE (Fig. 3 d) show an increase of adhesion forces to 556 pN ± 262 pN. Surfaces functionalized only with the protein anchor trisNTA (Fig. 3 e) show a higher increase in adhesion to 4160 pN ± 1152 pN, whereas surfaces with both anchors in a 2:1 (mol:mol) SuccinylPE:trisNTA mixture (Fig. 3 f) show an adhesion force of 669 pN ± 300 pN. Detailed distribution of adhesion forces as well as exemplary force-distance curves for each functionalization step can be seen in Fig. S2.
Figure 3.
Mean adhesion measured through force-distance curves with AFM. An average of 410 ± 38 force-distance curves were analyzed per experimental condition. (a) Mean adhesion for cleaned quartz glass (−0.025 nN ± 0.008 nN). (b) Mean adhesion for silanized quartz glass (12.42 nN ± 1.04 nN). (c) Mean adhesion for silanized quartz glass functionalized with PEG (0.21 nN ± 0.13 nN). (d) Mean adhesion for SuccinylPE bound to PEG (0.56 nN ± 0.26 nN). (e) Mean adhesion of trisNTA bound to PEG (4.16 nN ± 1.15 nN). (f) Mean adhesion of a 2:1 (mol/mol) mixture of SuccinylPE:trisNTA bound to PEG (0.67 nN ± 0.30 nN).
Protein complex formation
To test the successful immobilization of BR through a complex between the protein’s N-terminal deca-histidine-tag (His10-tag) and the trisNTA on the surface, specific elution of the protein by imidazole-containing buffer was performed and visualized by dot blot. To accomplish this, BR was first immobilized on the surface functionalized with the protein anchor and the lipid anchor. Then the surface was rinsed first with a buffer without imidazole. The volume of this rinsing step was collected as the rinsing fraction. Afterward, the surface was rinsed again with the buffer containing imidazole. The volume of this elution step was collected as the elution fraction. Together with a buffer control of the buffer containing imidazole, the fractions were distributed on a dot blot. As can be seen in Fig. S3, only the elution fraction (C) shows luminescence.
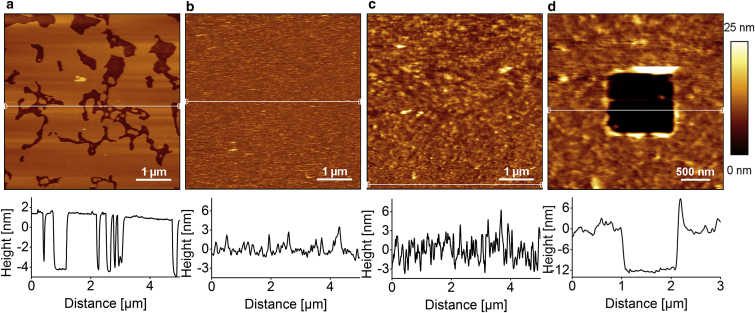
BR in membranes
Vesicle spreading on mica is an often-used method of creating solid sBLMs (15, 16, 18). A trial of the lipid mixture with BR was performed on mica to investigate possible effects of BR on bilayer formation. Fig. 4 a shows a mica surface after incubation of the lipid alone. The surface shows a homogeneous coverage with a lipid bilayer membrane (light gray area) with a few defects (dark gray areas). The corresponding height profile shows a membrane height of 5 nm and a low roughness of the membrane surface. Compared to an sBLM without BR, the bilayer containing BR (Fig. 4 b) has a much higher average roughness of 709.4 pm. All in all, the surface shows a complete coverage with no membrane defects. When spreading the lipid mixture with BR onto a functionalized quartz glass surface (Fig. 4 c), this high average roughness not only persists but increases to 1094 pm. However, the protein-lipid layer is spread over the whole surface. It has to be noted that during the scan process (bottom to top), the structures on the surface of the protein-containing membrane appear to increase in size, which is likely due to lipid adsorption to the cantilever tip.
Figure 4.
AFM scans (5 μm × 5 μm for a–c; 3 μm × 3 μm for d) in tapping mode in buffer P1, with corresponding height profiles, represented as white horizontal lines in each image, below each scan. (a) Membrane formation on a mica surface. (b) A mica surface covered with membrane containing BR. (c) A functionalized glass surface covered with the membrane containing BR. (d) Scratching was performed with the cantilever on a surface functionalized with lipid and protein anchor and incubated with the protein lipid mixture. To see this figure in color, go online.
After scratching with the cantilever tip on a surface covered with the protein-lipid mixture (Fig. 4 d), a depth of 10 nm can be measured.
Force spectroscopy
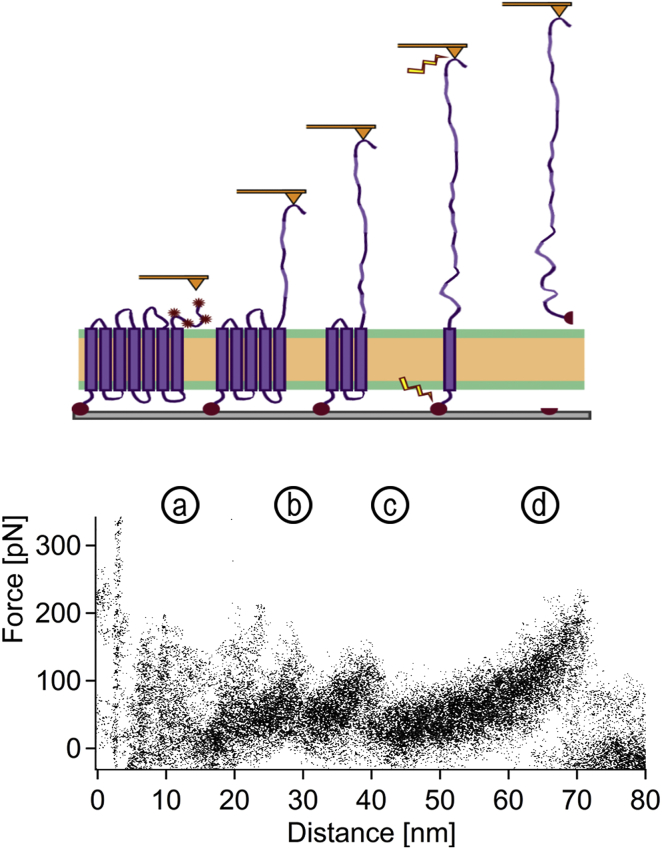
Force spectroscopy measurements were performed as described in Materials and Methods on surfaces with BR bound to the protein anchor and reconstituted into the lipid/detergent membrane. Only force-distance curves showing more than two force peaks and the last force peak at a peak position of >60 nm with a measured force of >100 pN were selected, to assure that only force-distance curves on specifically oriented and complex-coupled BR were used for analysis. Fig. S4 shows three representative force-distance curves that were selected according to these criteria. Force spectroscopic data was smoothed using moving-average filtering. In Fig. 5, selected force curves were overlaid to show the four characteristic peaks for the unfolding of BR as a seven-α-helical protein (13, 25, 26, 28). Although the first peak (Fig. 5 a) is inhomogeneous, the following peaks (Fig. 5 b–d) can be distinguished. The second and third unfolding peaks remain at forces <150 pN, whereas the last peak shows a force of ∼200 pN.
Figure 5.
BR unfolding curves from the model tBLM. As a 7TM membrane protein, the unfolding of BR will lead to four characteristic peaks in a force-distance curve. (a) The first peak shows the stretching of the terminus and unfolding of the first two transmembrane helices (TMs). As the cantilever can adsorb to the terminus at different positions, the first peak is not resolved clearly. This is followed by the pairwise unfolding of the next four TMs during the next two peaks (b and c). The last peak (d) shows the pulling of the seventh TM until BR dissociates from the tip or from the membrane. In our case, this means the dissociation between the His10-tag and the trisNTA. To see this figure in color, go online.
Discussion
Substrate characterization by XPS
To analyze the quartz glass surface and verify the first functionalization step, the surface was measured before and after silanization using XPS. A pure quartz glass surface shows the presence of a few undesired elements, namely fluorine, carbon, and sodium. As even quartz glass has some impurities, this is not surprising. However, any sign of these elements disappears after silanization with APTES. Given the structure and composition of APTES, and considering the theoretical distribution on the surface (Fig. S5 a), a theoretical composition of the silanized surface is shown in Fig. S5 b. It is also taken into account that XPS measures not only the topmost layer of a surface but can penetrate further. Thus our XPS results of the silanized surface (Fig. S5 b) are in accordance with a successful coverage of the surface with APTES.
Surface characterization by topography and adhesion
Surface topography
The surface functionalization steps were further characterized by AFM. AFM imaging offers the possibility of observing the changes in surface topography. A comparison of AFM imaging data of a clean quartz glass surface (Fig. 2 a) with a silanized quartz glass surface (Fig. 2 b) shows no change in average surface roughness and characteristics of the surface, in line with the small molecule size and flexibility of APTES. The short PEG linker that is coupled subsequently to the surface can have a length of 4 nm in its stretched conformation. This linker is flexible and will either coil on the surface or stretch to accommodate for surface roughness and differences between the bilayer and protein. AFM data of this stage (Fig. 2 c) shows that the holes and scratches found on quartz glass disappear, suggesting that the linker can compensate for surface roughness. The homogeneity and absence of defects also indicates a complete coverage with PEG. Coupling of only the anchoring lipid SuccinylPE to PEG yields a surface with an increased average roughness (Fig. 2 d). Additionally, buffer repelling properties, observed during the experiments, suggest strong hydrophobic characteristics of the surface. This can be explained by the hydrophobic chains of the anchoring lipid orientating up and away from the substrate, thereby creating a hydrophobic surface. The roughness of the surface is due to the length of the anchoring lipid (2 nm). The surface is again homogeneous and shows no defects. Coupling only the protein anchor, trisNTA, to the surface leads to the appearance of round structures ∼4–10 nm in height and 200 nm in diameter. This can indicate the aggregation of an excess amount of protein anchor. When the protein anchor is diluted with the lipid anchor 1:2 (mol/mol), the aggregates are no longer present. This and the uniform surface structure indicate a homogeneous mixture of lipid and protein anchor.
Adhesion
The differences in adhesion after each step are indicative of a successful functionalization process. Furthermore, a homogeneous adhesion force throughout the investigated surface implies a homogeneous surface functionalization. The strong adhesion forces for a surface functionalized only with protein anchor can relate to the presence of aggregates on the surface. Thus, even though the surface functionalized with the mixture shows only slightly higher mean adhesion forces than the surface only functionalized with the lipid anchor, a homogeneous and well distributed protein anchor is assumed due to the previously described changes in surface topography.
Specificity of protein binding
To test the specificity of protein binding to the surface and exclude unspecific adsorption, BR was coupled to a functionalized surface. The surface was then rinsed with a buffer without and with imidazole. As only imidazole-containing buffer can elute proteins with a poly-histidine-tag from NTA groups, BR will only be detected (using a dot blot system with anti-His antibodies) for the elution fraction containing imidazole. To exclude a possible binding of the antibody to the imidazole, imidazole-containing buffer was applied to the membrane as a buffer control. The results of the dot blot clearly show that the protein could only be eluted from the surface with imidazole. These data verify not only the specific binding of the protein to the surface anchor, but also the integrity of the previous coupling step leading to accessible trisNTA.
Membrane and protein reconstitution
Artificial lipid membranes have been observed to undergo phase transitions from more liquid unordered phases to ordered gel/crystalline phases at specific temperatures. The phase transition temperature depends on the membrane composition. As the integration of proteins into lipid membranes and their functionality is highly dependent on the membrane fluidity, we aimed to provide a lipid membrane system that is stable but still offers enough fluidity for the integration of membrane proteins. Future studies may benefit from the possibility of a careful selection of lipids that are compatible with the setup. We chose DPPC as the tBLM main component due to its good properties to form stable membranes on mica. DPPC has been used previously to create stable sBLMs (37, 38). However, as the phase transition occurs at a temperature of 42°C, those sBLMs are in the ordered gel-to-crystal-like phase at RT (39, 40). Adding a detergent like DPC weakens lipid-lipid interactions and increases the permeability of the bilayer to proteins (41). A membrane mimetic system consisting of a lipid and detergent mixture has been described previously (42). The formation of an sBLM on mica by a mixture of DPPC and DPC was tested, and a homogeneous bilayer formation with a few defects could be detected by AFM. The bilayer height of 5 nm is in accordance with bilayer heights in previous studies (43, 44). Reconstitution of BR into the lipid/detergent mixture and applying this mixture to a mica surface leads to coverage of the surface with a homogeneous layer (Fig. 4 b). Defects could not be observed. However, compared to an sBLM without BR (Fig. 4 a), the roughness of the membrane surface has increased. It is known that membrane-integrated proteins can protrude out of the membrane (45). The roughness of the membrane could thus indicate protrusions of BR out of the membrane. The protrusions persist on membranes formed on functionalized quartz glass (Fig. 4 c). The roughness is even more pronounced, as functionalized quartz glass in itself provides a rough substrate (Fig. 2 f). The increase in size of the structures observed on the surface with scan direction (bottom to top) is a broadening effect of the cantilever tip. Especially soft materials, like membranes and proteins, in our case, most likely the free lipid DPPC, can adsorb to the tip during scanning and increase the effective tip diameter. Every structure will then be broadened by the tip diameter. This is why for the first 500 nm the surface structure appears comparable to a membrane with protein on mica and becomes increasingly dissimilar during the scan process. For this reason, the height profile was measured during the first 500 nm of the scan. As a control, the membrane was also formed on a functionalized surface without BR (Fig. S6 a). Here, the surface was again homogeneously covered with the tBLM with only a few small defects. The surface roughness of the membrane was only influenced by the roughness of the quartz glass and the underlying functionalization, indicating that the high roughness of Fig. 4 c is caused by protrusions of BR. In contrast, a membrane on a surface functionalized only with trisNTA and no lipid anchor (Fig. S6 b) could not be formed on quartz glass under the same conditions.
Scratching of the membrane with BR on quartz glass (Fig. 4 d) revealed a height of 10 nm and protein clusters protruding 1–2 nm out of the membrane. When taking into account the length of the PEG linker and the height of the sBLM on mica (4 nm + 5 nm), the measured height of 10 nm is in accordance with expectations.
Force spectroscopy
Force-distance curves of force spectroscopy measurements performed on BR reconstituted into tBLMs showed the typical pattern of four peaks, consistent with a functional reconstitution of BR. In Fig. S7, a representative BR force-distance curve from BR purple membranes is compared to the measured force-distance curves of Fig. 5. The first peak of the measured force curves of BR in the tBLM was inhomogeneous, which can be attributed to the unspecific adhesion positions of the terminus to the tip (28). The second peak seems to favor lower forces and follows the shape of the intermediate peaks found in force-distance curves of purple membranes (30). Considering the average noise of the recorded force-distance curves (±30 pN) the third peak is in accordance with unfolding forces of BR from purple membranes and nano-discs (13). During the last unfolding event, BR can either dissociate from the tip or be pulled out of the membrane completely. In the case of being pulled out completely, BR would have to also dissociate from the protein anchor. It has been shown previously on soluble proteins that the dissociation forces between triNTA and His6 can be between 100 and 400 pN (46). Our results indicate that the last peak of our force-distance curves represents these dissociation forces. Although this influence of the protein anchor on the last unfolding event should be considered during analysis, it also shows that BR reconstituted into the here-presented tBLM is coupled to the protein anchor. In addition, in the case of BR not being inserted in a defined orientation, two sets of force-distance curves are expected (one pulled from the N- and one from the C-terminus), which can be the case for reconstitution of BR into nanodiscs (13) and force spectroscopic measurements performed on BR crystals (30). As BR force-distance curves from both termini are indeed very similar, the high forces detected for the last peak indicate a complex formation between the His-tag and trisNTA, thus supporting a defined orientation of BR. Should force-distance curves be measured that do not show the dissociation forces of the complex, they can be sorted out before analysis of the data.
Apart from the influence of the trisNTA, there could also be an influence of the PEG linker on the last unfolding event. As we expect a typical stretching behavior of the PEG linker (47, 48), we do not analyze the PEG-linker elasticity directly. However, it can be assumed that the influence of the PEG-linker elasticity is minimal due to its short length.
Even though higher forces for the last peak can be seen in our experimental setup, we also have to note the stronger noise. As this high noise can indicate misfolding or denaturation of BR, we cannot clearly determine whether BR is still in its functional state after reconstitution. The functional form of the protein has been checked before reconstitution into the membrane through ultraviolet-visible spectroscopy (described in the Supporting Material). An example absorbance spectrum of a 1:10 diluted solution of cell-free expressed and purified BR is shown in Fig. S8. Due to the very low protein concentration and the experimental setup, we can, however, only assume the functional form of the protein via the measured force curves. However, we anticipate that further optimization of tBLM composition, which has to be adjusted to every new protein of interest, will allow for further stabilization of the protein conformation inside the tBLM.
Conclusions
Gaining knowledge about the structure of membrane proteins is an important endeavor to reliably study the cause of diseases and facilitate drug development. In this respect, single-molecule AFM force spectroscopy offers unique possibilities in particular for the investigation of multipass membrane proteins. We here showed that AFM force spectroscopy can be carried out in a tBLM system, and our method allows for filtering out force-distance curves specific to a defined orientation. Although the tBLM system offers a near-native environment that can be adjusted to accommodate different membrane proteins, the predefined orientation can simplify data analysis by removing a second contribution arising from different orientations and by better defining the starting conditions for each protein. In our work, we show a five-step functionalization and reconstitution process that we successfully apply to the cell-free expressed 7TM protein BR that has been frequently used as a model for GPCRs. We anticipate that the presented approach can be transferred to a broad range of target membrane proteins and surface-based techniques and may help to provide new insights into structure, function, and interactions of these important biological systems.
Author Contributions
D.H., F.O., and A.M.B. designed research; S.E. and A.M.B. performed research; F.O. and A.B contributed analytic tools; A.B. and A.M.B. analyzed data; and M.E., A.B., and A.M.B. wrote the manuscript.
Acknowledgments
We thank Mario Schneider for his help in programming analysis software, as well as the groups of P.D. Dr. M. Lohrengel, Prof. Dr. Seidel, and Prof. Dr. Weinkauf of the Heinrich-Heine-Universität Düsseldorf for providing access to, and assistance with measuring X-ray photoelectron spectroscopy. We thank the group of Prof. Dr. Jacob Piehler of the University of Osnabrück for providing the trisNTA and all derivatives.
We gratefully acknowledge support (and training) from the International NRW Research School BioStruct, granted by the Ministry of Innovation, Science and Research of the State North Rhine-Westphalia, the Heinrich-Heine-University of Düsseldorf, and the Entrepreneur Foundation at the Heinrich-Heine-University of Düsseldorf, as well as from the Deutsche Forschungsgemeinschaft through Sonderforschungsbereich 974 “Communication and System Relevance in Liver Damage and Regeneration,” through an Emmy Noether grant (ET 103/2-1) and through the Forschungskommission Düsseldorf.
Editor: Simon Scheuring.
Footnotes
Supporting Materials and Methods and eight figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30835-9.
Supporting Citations
Reference (49) appears in the Supporting Material.
Supporting Material
References
- 1.Liu J., Rost B. Comparing function and structure between entire proteomes. Protein Sci. 2001;10:1970–1979. doi: 10.1110/ps.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aridor M., Hannan L.A. Traffic jam: a compendium of human diseases that affect intracellular transport processes. Traffic. 2000;1:836–851. doi: 10.1034/j.1600-0854.2000.011104.x. [DOI] [PubMed] [Google Scholar]
- 3.Aridor M., Hannan L.A. Traffic jams II: an update of diseases of intracellular transport. Traffic. 2002;3:781–790. doi: 10.1034/j.1600-0854.2002.31103.x. [DOI] [PubMed] [Google Scholar]
- 4.Cotton R.G.H., Horaitis O., Human Genome Organization The HUGO Mutation Database Initiative. Pharmacogenomics J. 2002;2:16–19. doi: 10.1038/sj.tpj.6500070. [DOI] [PubMed] [Google Scholar]
- 5.Stenson P.D., Ball E.V., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 6.Heyse S., Stora T., Vogel H. Emerging techniques for investigating molecular interactions at lipid membranes. Biochim. Biophys. Acta. 1998;1376:319–338. doi: 10.1016/s0304-4157(98)00020-3. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka M., Sackmann E. Polymer-supported membranes as models of the cell surface. Nature. 2005;437:656–663. doi: 10.1038/nature04164. [DOI] [PubMed] [Google Scholar]
- 8.Köper I. Insulating tethered bilayer lipid membranes to study membrane proteins. Mol. Biosyst. 2007;3:651–657. doi: 10.1039/b707168j. [DOI] [PubMed] [Google Scholar]
- 9.Vockenroth I.K., Ohm C., Köper I. Stable insulating tethered bilayer lipid membranes. Biointerphases. 2008;3:FA68. doi: 10.1116/1.2912097. [DOI] [PubMed] [Google Scholar]
- 10.Deverall M.A., Gindl E., Naumann C.A. Membrane lateral mobility obstructed by polymer-tethered lipids studied at the single molecule level. Biophys. J. 2005;88:1875–1886. doi: 10.1529/biophysj.104.050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giess F., Friedrich M.G., Knoll W. The protein-tethered lipid bilayer: a novel mimic of the biological membrane. Biophys. J. 2004;87:3213–3220. doi: 10.1529/biophysj.104.046169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigaud J.-L., Lévy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 13.Zocher M., Roos C., Müller D.J. Single-molecule force spectroscopy from nanodiscs: an assay to quantify folding, stability, and interactions of native membrane proteins. ACS Nano. 2012;6:961–971. doi: 10.1021/nn204624p. [DOI] [PubMed] [Google Scholar]
- 14.Atanasova P.P., Atanasov V., Köper I. Anchor-lipid monolayers at the air-water interface; prearranging of model membrane systems. Langmuir. 2007;23:7672–7678. doi: 10.1021/la7002854. [DOI] [PubMed] [Google Scholar]
- 15.Sinner E.K., Knoll W. Functional tethered membranes. Curr. Opin. Chem. Biol. 2001;5:705–711. doi: 10.1016/s1367-5931(01)00269-1. [DOI] [PubMed] [Google Scholar]
- 16.Pfeiffer I., Petronis S., Zäch M. Vesicle adsorption and phospholipid bilayer formation on topographically and chemically nanostructured surfaces. J. Phys. Chem. B. 2010;114:4623–4631. doi: 10.1021/jp908283g. [DOI] [PubMed] [Google Scholar]
- 17.Roder F., Waichman S., Piehler J. Reconstitution of membrane proteins into polymer-supported membranes for probing diffusion and interactions by single molecule techniques. Anal. Chem. 2011;83:6792–6799. doi: 10.1021/ac201294v. [DOI] [PubMed] [Google Scholar]
- 18.Coutable A., Thibault C., Trévisiol E. Preparation of tethered-lipid bilayers on gold surfaces for the incorporation of integral membrane proteins synthesized by cell-free expression. Langmuir. 2014;30:3132–3141. doi: 10.1021/la5004758. [DOI] [PubMed] [Google Scholar]
- 19.Oberbarnscheidt L., Janissen R., Oesterhelt F. Direct and model free calculation of force-dependent dissociation rates from force spectroscopic data. Biophys. J. 2009;97:L19–L21. doi: 10.1016/j.bpj.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engel A., Gaub H.E. Structure and mechanics of membrane proteins. Annu. Rev. Biochem. 2008;77:127–148. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- 21.Harris N.C., Song Y., Kiang C.-H. Experimental free energy surface reconstruction from single-molecule force spectroscopy using Jarzynski’s equality. Phys. Rev. Lett. 2007;99:068101. doi: 10.1103/PhysRevLett.99.068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee G.U., Kidwell D.A., Colton R.J. Sensing discrete streptavidin-biotin interactions with atomic force microscopy. Langmuir. 1994;10:354–357. [Google Scholar]
- 23.Oberbarnscheidt L., Janissen R., Oesterhelt F. Single-molecule force spectroscopy measures structural changes induced by light activation and transducer binding in sensory rhodopsin II. J. Mol. Biol. 2009;394:383–390. doi: 10.1016/j.jmb.2009.07.083. [DOI] [PubMed] [Google Scholar]
- 24.Pelaseyed T., Zäch M., Hansson G.C. Unfolding dynamics of the mucin SEA domain probed by force spectroscopy suggest that it acts as a cell-protective device. FEBS J. 2013;280:1491–1501. doi: 10.1111/febs.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roychoudhury A., Haussinger D., Oesterhelt F. Effect of the compatible solute ectoine on the stability of the membrane proteins. Protein Pept. Lett. 2012;19:791–794. doi: 10.2174/092986612801619570. [DOI] [PubMed] [Google Scholar]
- 26.Roychoudhury A., Bieker A., Oesterhelt F. Membrane protein stability depends on the concentration of compatible solutes—a single molecule force spectroscopic study. Biol. Chem. 2013;394:1465–1474. doi: 10.1515/hsz-2013-0173. [DOI] [PubMed] [Google Scholar]
- 27.Wickstrand C., Dods R., Neutze R. Bacteriorhodopsin: would the real structural intermediates please stand up? Biochim. Biophys. Acta. 2015;1850:536–553. doi: 10.1016/j.bbagen.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 28.Baumann R.-P., Schranz M., Hampp N. Bending of purple membranes in dependence on the pH analyzed by AFM and single molecule force spectroscopy. Phys. Chem. Chem. Phys. 2010;12:4329–4335. doi: 10.1039/b919729j. [DOI] [PubMed] [Google Scholar]
- 29.Müller D.J., Schabert F.A., Engel A. Imaging purple membranes in aqueous solutions at sub-nanometer resolution by atomic force microscopy. Biophys. J. 1995;68:1681–1686. doi: 10.1016/S0006-3495(95)80345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessler M., Gaub H.E. Unfolding barriers in bacteriorhodopsin probed from the cytoplasmic and the extracellular side by AFM. Structure. 2006;14:521–527. doi: 10.1016/j.str.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 31.Klammt C., Schwarz D., Bernhard F. Cell-free production of G protein-coupled receptors for functional and structural studies. J. Struct. Biol. 2007;158:482–493. doi: 10.1016/S1047-8477(07)00164-5. [DOI] [PubMed] [Google Scholar]
- 32.Etzkorn M., Raschle T., Wagner G. Cell-free expressed bacteriorhodopsin in different soluble membrane mimetics: biophysical properties and NMR accessibility. Structure. 2013;21:394–401. doi: 10.1016/j.str.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lata S., Piehler J. Stable and functional immobilization of histidine-tagged proteins via multivalent chelator headgroups on a molecular poly(ethylene glycol) brush. Anal. Chem. 2005;77:1096–1105. doi: 10.1021/ac048813j. [DOI] [PubMed] [Google Scholar]
- 34.Butt H.-J., Jaschke M. Calculation of thermal noise in atomic I Calculation of thermal noise in atomic force microscopy. Nanotechnology. 1995;6:1–7. [Google Scholar]
- 35.Janissen R., Oberbarnscheidt L., Oesterhelt F. Optimized straight forward procedure for covalent surface immobilization of different biomolecules for single molecule applications. Colloids Surf. B Biointerfaces. 2009;71:200–207. doi: 10.1016/j.colsurfb.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Schwarz D., Junge F., Bernhard F. Preparative scale expression of membrane proteins in Escherichia coli-based continuous exchange cell-free systems. Nat. Protoc. 2007;2:2945–2957. doi: 10.1038/nprot.2007.426. [DOI] [PubMed] [Google Scholar]
- 37.Sullan R.M.A., Li J.K., Zou S. Direct correlation of structures and nanomechanical properties of multicomponent lipid bilayers. Langmuir. 2009;25:7471–7477. doi: 10.1021/la900395w. [DOI] [PubMed] [Google Scholar]
- 38.Giocondi M.-C., Yamamoto D., Le Grimellec C. Surface topography of membrane domains. Biochim. Biophys. Acta. 2010;1798:703–718. doi: 10.1016/j.bbamem.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 39.Giocondi M.C., Vié V., Le Grimellec C. In situ imaging of detergent-resistant membranes by atomic force microscopy. J. Struct. Biol. 2000;131:38–43. doi: 10.1006/jsbi.2000.4266. [DOI] [PubMed] [Google Scholar]
- 40.Choucair A., Chakrapani M., Johnston L.J. Preferential accumulation of Aβ1–42 on gel phase domains of lipid bilayers: an AFM and fluorescence study. Biochim. Biophys. Acta. 2007;1768:146–154. doi: 10.1016/j.bbamem.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Seddon A.M., Curnow P., Booth P.J. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 42.Vasudevan S.V., Schulz J., Cocco M.J. Protein folding at the membrane interface, the structure of Nogo-66 requires interactions with a phosphocholine surface. Proc. Natl. Acad. Sci. USA. 2010;107:6847–6851. doi: 10.1073/pnas.0911817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin H., Bhatia R., Lal R. Amyloid β protein forms ion channels: implications for Alzheimer’s disease pathophysiology. FASEB J. 2001;15:2433–2444. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- 44.Redondo-Morata L., Giannotti M.I., Sanz F. Influence of cholesterol on the phase transition of lipid bilayers: a temperature-controlled force spectroscopy study. Langmuir. 2012;28:12851–12860. doi: 10.1021/la302620t. [DOI] [PubMed] [Google Scholar]
- 45.Whited A.M., Park P.S.-H. Atomic force microscopy: a multifaceted tool to study membrane proteins and their interactions with ligands. Biochim. Biophys. Acta. 2014;1838(1 Pt A):56–68. doi: 10.1016/j.bbamem.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang J., Ebner A., Hinterdorfer P. Detection of metal binding sites on functional S-layer nanoarrays using single molecule force spectroscopy. J. Struct. Biol. 2009;168:217–222. doi: 10.1016/j.jsb.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Heymann B., Grubmüller H. Elastic properties of poly(ethylene-glycol) studied by molecular dynamics stretching simulations. Chem. Phys. Lett. 1999;307:425–432. [Google Scholar]
- 48.Oesterhelt F., Rief M., Gaub H.E. Single molecule force spectroscopy by AFM indicates helical structure of poly(ethylene-glycol) in water. New J. Phys. 1999;1:6. [Google Scholar]
- 49.Gruber, H. J. 2016. Amino-functionalization of AFM tips (and supports). Institute of Biophysics, Johannes Kepler University, Linz, Austria. May 6, 2016. http://www.jku.at/biophysics/content/e257042/e257048/03_AFM_tip_aminofunctionalization_2016_05_06_eng.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.