ABSTRACT
A defining feature of mycobacterial redox metabolism is the use of an unusual deazaflavin cofactor, F420. This cofactor enhances the persistence of environmental and pathogenic mycobacteria, including after antimicrobial treatment, although the molecular basis for this remains to be understood. In this work, we explored our hypothesis that F420 enhances persistence by serving as a cofactor in antimicrobial-detoxifying enzymes. To test this, we performed a series of phenotypic, biochemical, and analytical chemistry studies in relation to the model soil bacterium Mycobacterium smegmatis. Mutant strains unable to synthesize or reduce F420 were found to be more susceptible to a wide range of antibiotic and xenobiotic compounds. Compounds from three classes of antimicrobial compounds traditionally resisted by mycobacteria inhibited the growth of F420 mutant strains at subnanomolar concentrations, namely, furanocoumarins (e.g., methoxsalen), arylmethanes (e.g., malachite green), and quinone analogues (e.g., menadione). We demonstrated that promiscuous F420H2-dependent reductases directly reduce these compounds by a mechanism consistent with hydride transfer. Moreover, M. smegmatis strains unable to make F420H2 lost the capacity to reduce and detoxify representatives of the furanocoumarin and arylmethane compound classes in whole-cell assays. In contrast, mutant strains were only slightly more susceptible to clinical antimycobacterials, and this appeared to be due to indirect effects of F420 loss of function (e.g., redox imbalance) rather than loss of a detoxification system. Together, these data show that F420 enhances antimicrobial resistance in mycobacteria and suggest that one function of the F420H2-dependent reductases is to broaden the range of natural products that mycobacteria and possibly other environmental actinobacteria can reductively detoxify.
IMPORTANCE This study reveals that a unique microbial cofactor, F420, is critical for antimicrobial resistance in the environmental actinobacterium Mycobacterium smegmatis. We show that a superfamily of redox enzymes, the F420H2-dependent reductases, can reduce diverse antimicrobials in vitro and in vivo. M. smegmatis strains unable to make or reduce F420 become sensitive to inhibition by these antimicrobial compounds. This suggests that mycobacteria have harnessed the unique properties of F420 to reduce structurally diverse antimicrobials as part of the antibiotic arms race. The F420H2-dependent reductases that facilitate this process represent a new class of antimicrobial-detoxifying enzymes with potential applications in bioremediation and biocatalysis.
INTRODUCTION
The redox cofactor F420 [7,8-didemethyl-8-hydroxy-5-deazariboflavin-5′-phosphoryllactyl(glutamyl)n glutamate] is unique to bacteria and archaea. This deazaflavin is structurally similar to flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) but behaves more similarly electrochemically to the nicotinamides NAD and NADP (1). An obligate two-electron carrier, F420 mediates hydride transfer to or from a wide range of organic carbon compounds. Its low standard redox potential of −340 mV enables it to catalyze otherwise challenging reduction reactions, for example, those of enone, imine, enamine, and nitro groups (2–5). Due to these unique properties, F420 has been harnessed by bacteria and archaea in a wide range of metabolic pathways. The cofactor is best known for its role in methanogens, where it serves as the main catabolic cofactor (6). However, we have shown it is also synthesized by aerobic bacteria from at least three phyla, the Actinobacteria, Chloroflexi, and Proteobacteria (7). While the cofactor is of secondary importance to nicotinamides in bacteria, an increasing number of redox reactions in central and secondary metabolic pathways have been shown to depend on it (1). These include the biodegradation of aflatoxins and nitrophenols (2, 4), the biosynthesis of tetracycline and lincosamide antibiotics (8, 9), and the reduction of biliverdin, quinones, and mycolic acids (5, 10, 11). In turn, it was recently recognized that F420 may have a significant role in shaping the biological and chemical compositions of soil ecosystems (7).
In recent years, F420 has inspired interest for its role in the redox metabolism of mycobacteria, an actinobacterial genus of environmental and medical significance. Most of these studies have focused on the soil saprophyte Mycobacterium smegmatis and the human pathogen Mycobacterium tuberculosis. These studies have shown that F420 is dispensable for growth but essential for the survival of antibiotic stress, oxidative stress, and hypoxia, for reasons still incompletely understood (10, 12, 13). In this genus, the cofactor is synthesized by four known enzymes (FbiC, CofC, CofD, and CofE) and is reduced by the F420-dependent glucose 6-phosphate dehydrogenase (Fgd) (14). The physiological roles of the cofactor are elicited by F420H2-dependent reductases that couple the reoxidation of F420H2 to the reduction of organic compounds (1, 5, 11, 15). Genomic and biochemical studies have shown that mycobacteria synthesize a multiplicity of F420H2-dependent reductases from two superfamilies, the flavin/deazaflavin oxidoreductases (FDORs) and the luciferase-like hydride transferases (LLHTs). The soil organism M. smegmatis encodes some 31 FDORs and 45 LLHTs that are predicted to bind F420 (1, 5, 15). A smaller number of these proteins are encoded in the reduced genomes of the human pathogens M. tuberculosis and Mycobacterium leprae (5, 15).
The recently discovered FDOR superfamily is of particular interest because of its physiological and pharmacological roles in mycobacteria (1, 5). This diverse family is subdivided into 16 subgroups (A1 to A4, AA1 to AA6, and B1 to B6), with genes encoding the A1 and B1 subgroups in multiple copies in individual mycobacterial genomes (2, 5, 16, 17). To date, the physiological role of just one mycobacterial FDOR has been fully defined, a biliverdin reductase from the B3 subfamily that reduces the heme oxygenation product biliverdin to the potent antioxidant bilirubin (5, 18). Other subgroups have been implicated in quinone and fatty acid metabolism (5, 10). Several FDORs of the manifold A1 and B1 subgroups are also known for their ability to reduce several medically important exogenous compounds, including furanocoumarin natural products (e.g., aflatoxins) (2, 16) and clinical nitroimidazole prodrugs (i.e., delamanid and pretomanid) (19–21). Our previous structural studies have shown that the F420H2 moiety binds at an amphiphilic conserved F420-binding domain, whereas the substrate binds in a more variable substrate-binding pocket (1, 2, 5). The promiscuous FDORs of the A1 subgroups are able to accommodate a range of substrates in wide pockets through a combination of hydrophobic and hydrophilic interactions involving conserved aromatic residues. Hydride transfer occurs directly between C-5 of F420H2 to the proximal electrophilic alkene or imine groups of the substrate (2, 5, 18, 22).
Despite the progress in understanding its biochemical basis, it remains unclear whether substrate promiscuity in the FDOR superfamily is physiologically significant and, if so, how this promiscuity has been shaped by natural selection. We recently proposed that mycobacteria evolved a multiplicity of promiscuous F420H2-dependent reductases in order to enhance the range of substrates which they may biodegrade (1, 5). These enzymes may use electrons derived from the cytosolic reservoir of glucose 6-phosphate (12) to reductively transform exogenous compounds, leading to their detoxification and/or biomineralization. To explore this hypothesis, this paper investigates the role of F420H2-dependent reductases in the metabolism of a range of antibiotics and xenobiotics. By combining cellular physiology, protein biochemistry, and analytical chemistry approaches, we resolved that F420 is crucial for the ability of mycobacteria to resist a range of antimicrobial compounds and demonstrated that F420H2-dependent FDORs can likely reductively detoxify some of these compounds.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
This study employed Mycobacterium smegmatis mc2155 (23) and two derivative strains containing disruptions in the genes fgd and fbiC with kanamycin resistance cassettes (2). The strains were grown on LBT (lysogeny broth containing 0.05% Tween 80) and maintained on LBT agar. Kanamycin at 20 μg · ml−1 was used to propagate the mutant strains. For all physiological experiments, liquid cultures (30 ml) were grown to mid-exponential phase (optical density at 600 nm [OD600], ≈0.5) in 125-ml aerated conical flasks in a rotary incubator (200 rpm, 37°C).
Phenotypic experiments.
Twenty antimicrobial compounds were sourced from Sigma-Aldrich and dissolved into 1 M working stocks in dimethyl sulfoxide (DMSO). The structures of the compounds tested are shown in Table S1 in the supplemental material. Phenotypic experiments studied the effect of these compounds on the growth and survival of M. smegmatis and derivative strains. MICs were determined by a standard protocol (24) by serially diluting the compounds in 96-well plates and measuring growth inhibition by OD600 following incubation (400 rpm, 37°C, 12 h). Strain viability following challenge with antimicrobial agents was measured by challenging mid-exponential-phase cultures with the compound of interest at a concentration of 5 times the MIC of the wild-type strain. At regular time intervals, 1-ml cultures were centrifuged (16,000 × g, 1 min) and resuspended in 1 ml of HBTP (Hartmans de Bont minimal medium supplemented with 0.05% tyloxapol) (25) two times to remove the compounds. CFU · ml−1 were subsequently counted by spot-plating serially diluted cultures, as previously described (26). No CFU or MIC differences were observed for DMSO-only controls. To test the capacity of strains to grow on compounds as a sole source of carbon, exponential-phase cultures were inoculated to a starting OD600 of 0.005 into HBTP medium (25) supplemented with either 1 mM or 10 mM the compounds of interest, and growth was determined by the OD600 following incubation (200 rpm, 37°C, 24 h). Eight compounds were tested: coumarin, methoxsalen, imperatorin, menadione, azure B, malachite green, crystal violet, and phenol red.
Protein expression and purification.
Six F420H2-dependent reductases of the flavin/deazaflavin oxidoreductase (FDOR) superfamily (MSMEG_5998, MSMEG_2027, MSMEG_6325, MSMEG_3380, MSMEG_0048, and MSMEG_5170 loci) and the F420-reducing glucose 6-phosphate dehydrogenase (Fgd) were recombinantly overexpressed in Escherichia coli BL21(DE3) and purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography, as previously described (2, 5). F420 was extracted, purified, and concentrated from a recombinant F420 overexpression strain of M. smegmatis mc24517 (27) by a combination of anion-exchange chromatography, hydrophobic interaction chromatography, and rotary evaporation, as previously described (28).
Enzymatic assays.
Two assays were optimized to monitor F420H2-coupled substrate reduction. A high-performance liquid chromatography (HPLC)-based method monitored the reduction of substrates by measuring changes in their absorbance at λmax (maximum absorbance wavelength) or their fluorescence at λem max (maximum emission wavelength) in aerobic vials containing the FDOR of interest, F420, Fgd, and glucose 6-phosphate. The second method monitored the reoxidation of F420H2 by the absorbance at 420 nm in anoxic mixtures containing prereduced F420H2 and one of the FDORs. The rates obtained for the two methods were similar (see Table S2 in the supplemental material), demonstrating that rates of F420H2 reoxidation are proportional to those with substrate reduction. However, the second method proved more reliable and systematic, in part because it better discriminated between enzyme-dependent and enzyme-independent F420H2-mediated substrate reduction. Using this method, we assayed the specific activities of the six purified FDORs for 20 compounds. All assays were prepared in a nitrogen glove box and used buffers that were degassed by sonication. F420 was enzymatically reduced overnight with 1 μM Fgd and purified by spin filtration using previously reported methods (5). All reaction mixtures contained degassed Tris buffer (200 mM Tris, 0.1% [wt/vol] Triton X-100 [pH 8.0]) sequentially supplemented with 50 μM substrate, 25 μM F420H2, and, depending on reactivity, between 100 nM and 1 μM the FDOR. Reaction rates were monitored by recording the initial linear increase in absorbance at 420 nm using an Epoch 2 microplate spectrophotometer (BioTek). Specific activities were calculated by subtracting rates of no-enzyme controls and expressed in nmol · min−1 · μmol enzyme−1, as previously described (2).
LC-MS analysis.
For liquid chromatography-mass spectrometry (LC-MS) analysis of the FDOR-mediated reactions, 100 μM standards of malachite green and methoxsalen were prepared in 20 mM Tris buffer (pH 8.0). The mass spectra of their FDOR reaction products were measured after the addition of 100 μM glucose 6-phosphate, 10 μM F420, 1 μM Fgd, and 1 μM MSMEG_2027, followed by incubation at 37°C for 1 h. An Agilent 1100 series binary LC equipped with a diode array detector and in-line time of flight mass spectrometer (LC-MSD TOF) were used, with mass data obtained by electrospray ionization (ESI) in positive ion mode. Samples were separated on an Agilent Poroshell 120 EC-C18 column (2.7 μm, 2.1 by 100 mm), utilizing a gradient protocol composed of buffer A, with ammonium formate plus 0.1% formic acid; and buffer B, with acetonitrile plus 0.1% formic acid. The applied gradient was as follows: 10% buffer B from 0.5 min to 60% buffer B at 6.50 min, and then increased to 90% buffer B at 7 min, at a flow rate of 0.4 ml · min−1. The Agilent MassHunter quantitative analysis software was used to analyze product formation.
Whole-cell consumption assays.
Whole-cell assays measured the consumption of the antimicrobial compounds malachite green and methoxsalen. Mid-exponential-phase cells grown in LBT were centrifuged (16,000 × g, 10 min) and resuspended in a buffer containing 50 mM MOPS (pH 7.4), 25 mM NaCl, and 25 mM KCl. The antimicrobial compound of interest was then added to a final concentration of 1/2 the MIC of the fbiC mutant strain. At regular intervals, samples of 200 μl were centrifuged to remove cells (16,000 × g, 5 min), and supernatants were analyzed via a high-performance liquid chromatography–diode array detection (HPLC-DAD) protocol. The analysis was carried out on an Agilent 1200 series system equipped with autosampler and a Poroshell 120 EC-C18 2.7-μm, 2.1 by 100-mm column. A gradient consisting of buffer A (ultrapure water plus 0.1% formic acid) and buffer B (acetonitrile plus 0.1% formic acid) was applied as follows: 0 to 1 min 10% buffer B, 1 to 5 min 90% buffer B, 5 to 10 min 10% buffer B, 10 to 12 min 10% buffer B. Degradation of malachite green (retention time, 7.2 min) and methoxsalen (retention time, 6.9 min) was measured by tracking the loss of absorbance at 615 nm and 310 nm, respectively. Concentration was calculated by integrating the area of the absorbance peaks and calibrating with standards of known concentration.
RESULTS
F420-deficient mutants are hypersusceptible to growth inhibition by antimicrobial compounds.
We compared the susceptibility of wild-type and F420-deficient mutant strains to growth inhibition by 20 antimicrobial agents. Two mutant strains were employed: one unable to synthesize F420 (fbiC::kan mutant) due to disruption of the gene encoding the first enzyme in the F420 biosynthesis pathway (FbiC), the other unable to reduce F420 (fgd::kan mutant) due to disruption of the gene encoding F420-dependent glucose 6-phosphate dehydrogenase (Fgd). Six of the compounds were first- and second-line clinical antimycobacterial agents. The remainders were aminocoumarins/furanocoumarins, quinolones/fluoroquinolones, quinone analogues, and arylmethane compounds containing enone, lactone, or imine functionalities known to be catalytically compatible with F420H2-dependent reductases (2, 5, 10, 16).
The MICs for the fgd and fbiC mutants were lower than those of the wild type for 18 of the 20 compounds (Table 1). The mutant strains showed greatly increased susceptibility to four compound classes usually resisted by mycobacteria: furanocoumarins, quinone analogues, arylmethane dyes, and quinolones. Whereas wild-type strains were only compromised by high concentrations of these compounds, the mutants were susceptible even at subnanomolar levels. In addition, the mutants exhibited small but reproducible increases in susceptibility to growth inhibition to the five first-line clinical antimycobacterials tested. Similar MICs were observed for the fgd and fbiC mutants, which is consistent with FbiC being essential for F420 biosynthesis and Fgd being the main route for F420 reduction in M. smegmatis (1, 14). The slight increase in susceptibility of the fbiC mutant over the fgd mutant for three of the compounds suggests that some residual functions of F420 can be maintained in the absence of Fgd.
TABLE 1.
Antibiotic susceptibility of F420-deficient mycobacteriaa
| Substrate | MIC (μg · ml−1) |
MIC ratio (WT/mutant)b | ||
|---|---|---|---|---|
| WT | fgd::kan | fbiC::kan | ||
| Quinone analogues | ||||
| Menadione | 32 | 2 | 2 | 16 |
| 1,4-Naphthoquinone | 32 | 1 | 1 | 32 |
| 1,2-Naphthoquinone | 32 | 2 | 2 | 16 |
| Furanocoumarins and aminocoumarins | ||||
| Methoxsalen | 64 | 1 | 1 | 64 |
| Angelicin | 128 | 1 | 1 | 128 |
| Imperatorin | 32 | 0.5 | 0.5 | 64 |
| Novobiocin | 0.5 | 0.5 | 0.5 | 1 |
| Arylmethane dyes | ||||
| Malachite green | 64 | 1 | 0.5 | 64, 128 |
| Crystal violet | 16 | 1 | 0.5 | 16, 32 |
| Phenol red | 8 | 0.5 | 0.5 | 16 |
| Azure B | 16 | 2 | 2 | 8 |
| Quinolones and fluoroquinolones | ||||
| Oxolinic acid | 16 | 0.125 | 0.125 | 128 |
| Nalidixic acid | 128 | 1 | 1 | 128 |
| Ciprofloxacin | 0.25 | 3.1 × 10−2 | 7.8 × 10−3 | 8 |
| Moxifloxacin | 6.3 × 10−2 | 6.3 × 10−2 | 3.1 × 10−2 | 1, 2 |
| Clinical antimycobacterials | ||||
| Rifampin | 4 | 2 | 2 | 2 |
| Ethambutol | 0.5 | 0.13 | 0.13 | 4 |
| Isoniazid | 32 | 16 | 16 | 2 |
| Pyrazinamide | 512 | 256 | 256 | 2 |
| Clofazimine | 8 | 4 | 4 | 2 |
MICs are shown for exponential-phase wild-type (WT), fgd mutant, and fbiC mutant cultures. MICs were determined by serially diluting antibiotic stocks in 96-well plates and measuring growth inhibition by the OD600. Five first-line antimycobacterials (rifampin, ethambutol, isoniazid, pyrazinamide, and clofazimine) and 1 second-line antimycobacterial (moxifloxacin) were tested.
Cells with two values are the ratios for the fgd::kan mutant and fbiC::kan mutant, respectively.
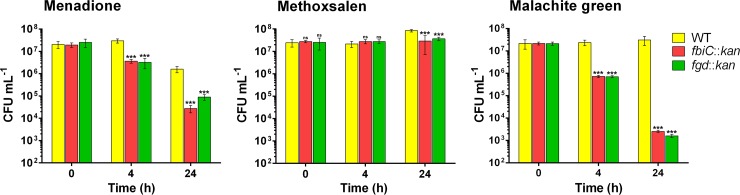
CFU measurements confirmed that loss of function of F420 reduces the capacity of M. smegmatis to survive treatment with representative antimicrobial compounds (Fig. 1). Whereas malachite green proved bacteriostatic to the wild-type strain at high concentrations, the dye rapidly killed F420 mutant strains. Likewise, the bactericidal agent menadione killed the fbiC mutant and, to a lesser extent, the fgd mutant strains at higher rates than for the wild-type strain. At 24 h following administration, we observed reductions in the survival of the fbiC mutant strain versus the wild type of 10,000-fold for malachite green and 100-fold for menadione (P < 0.001). Despite inhibiting growth of the fbiC mutant at subnanomolar concentrations (Table 1), the furanocoumarin methoxsalen proved bacteriostatic to all three strains even at a high concentration (640 μg · ml−1). CFU and OD measurements confirmed that wild-type cells resumed growth on LBT medium following initial growth inhibition (Fig. 1). This suggests that the strain detoxified the furanocoumarin to sub-MIC levels. The mutant strains also exhibited a modest acceleration in killing following administration of the clinical antimycobacterial agent rifampin (see Fig. S1 in the supplemental material).
FIG 1.
F420-dependent survival of M. smegmatis following antimicrobial challenge. The wild-type (WT), fgd mutant, and fbiC mutant strains were challenged with three antimicrobials (menadione, methoxsalen, and malachite green) during mid-exponential growth. The survival of the strains, as determined by CFU, is shown following administration of antimicrobials at 5 times the MIC of the wild-type strain. Error bars represent standard deviations from the results from three biological replicates. P values were calculated by comparing CFU counts between wild-type and mutant strains. ***, P < 0.001; ns, not significant.
F420H2-dependent reductases directly reduce antimicrobial compounds.
We hypothesized that the increased susceptibility of the mutant strains to many of the compounds tested was due to their inability to detoxify the compounds using F420H2-dependent reductases. To test this, we recombinantly expressed and purified six F420H2-dependent FDORs (see Fig. S2 in the supplemental material) from M. smegmatis. Enzymes were selected to cover a range of FDOR subgroups while focusing on A1 and B1 enzymes that have previously been shown to display the greatest substrate promiscuity (2, 5, 16). We spectroscopically measured the rates of F420H2-dependent reduction of the compounds, employing anaerobic conditions to prevent low-level spontaneous F420H2 reoxidation. These enzymes catalyzed the F420H2-dependent reduction of 10 of the 20 compounds tested (Fig. 2). Consistent with previous observations on the reactivity of F420H2 (11), several compounds were additionally reduced by F420H2 in an enzyme-independent manner (see Fig. S3 in the supplemental material).
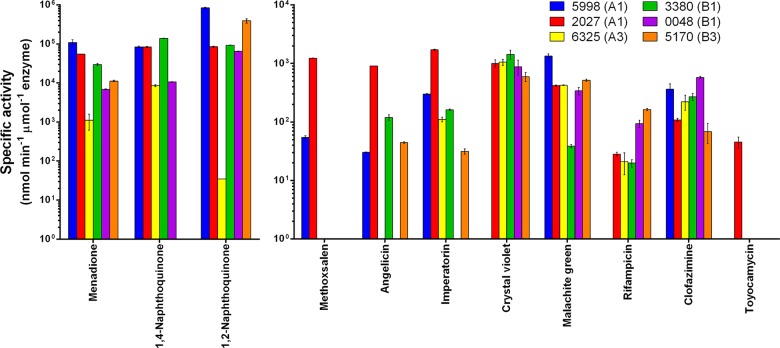
FIG 2.
Reduction of antimicrobials by F420H2-dependent reductases from Mycobacterium smegmatis. Six characterized F420H2-dependent reductases (MSMEG_5998, MSMEG_2027, MSMEG_6325, MSMEG_3380, MSMEG_0048, and MSMEG_5170) were recombinantly expressed and purified from E. coli. The specific activity for each substrate was determined by recording the rate of F420H2 reoxidation by absorbance at 420 nm. Error bars show standard deviations from the results from three independent replicates. No F420H2-dependent reduction of ethambutol, isoniazid, pyrazinamide, oxolinic acid, novobiocin, nalidixic acid, ciprofloxacin, or moxifloxacin was observed. Significant rates of enzyme-independent F420H2 reoxidation were observed for phenol red and azure B (see Fig. S3 in the supplemental material).
FDORs transformed quinone analogues at high rates (>100,000 nmol · min−1 · μmol enzyme−1) and furanocoumarins at low rates (>1,000 nmol · min−1 · μmol enzyme−1). In addition, all four arylmethane compounds were also reduced by F420H2 through both FDOR-facilitated (malachite green and crystal violet) (Fig. 2) and nonenzymatic (azure B, phenol red, and malachite green) activities (see Fig. S3 in the supplemental material). While it is well established that mycobacteria reductively decolorize malachite green in an F420-dependent manner (29), the enzymatic determinants of this process have remained elusive until now. Two of the six clinical antimycobacterials, namely, rifampin and clofazimine, were also reduced, although at very low rates of below 100 and 600 nmol · min−1 · μmol enzyme−1, respectively. All six FDORs tested had some degree of promiscuity, although they varied significantly in their substrate ranges and reaction rates. The A1 enzymes (MSMEG_5998 and MSMEG_2027) exhibited the widest substrate range and generally the highest rates of the enzymes, with MSMEG_2027 having detectable activity with all 10 compounds for which FDOR activity was observed. Members of the B1 subgroup (MSMEG_0048 and MSMEG_3380) had high activities against quinones and arylmethanes but not furanocoumarins, whereas representatives of the A3 (MSMEG_6325) and B3 (MSMEG_5170) subgroups generally catalyzed reactions at fractional rates compared to the other FDORs. This is consistent with our previous studies showing most FDOR-A and FDOR-B enzymes can reduce aflatoxins, but at rates varying by four orders of magnitude (2, 16).
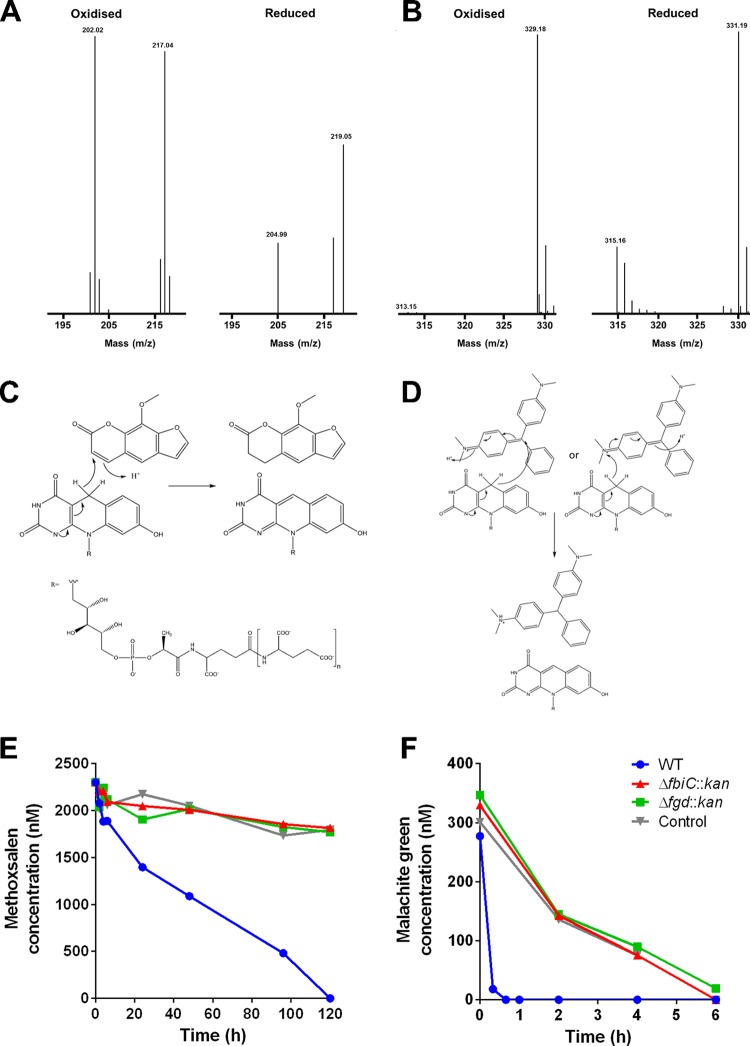
HPLC experiments with selected compounds validated that substrates were reduced by F420H2-dependent reductases. This occurred at rates similar to that of F420H2 reoxidation, suggesting an equimolar reaction (see Table S2 in the supplemental material). LC-MS analysis of two of the compounds, methoxsalen and malachite green, demonstrated the emergence of reduced forms of the compounds, with [M+H] ions corresponding to a 2-Da increase in the reaction product spectrum (Fig. 3A and B). This is consistent with direct hydride transfer occurring from the C-5 atom of F420H2 to the electrophilic double bonds of the substrates (Fig. 3C and D), coupled with addition of a proton from either solvent or a proximate residue, as observed for other FDOR-dependent reactions (2, 5, 17, 18, 30). Our previous structural studies provide a basis for understanding how these substrates can be accommodated into the substrate-binding sites of FDORs through both hydrophilic and hydrophobic interactions (1, 2, 5, 18). All other compounds reduced by F420H2 also contain functional groups known to be reduced by FDORs (1), namely, enones (quinone analogues), lactones (furanocoumarins), and imines (arylmethane dyes, clofazimine, and rifampin). On this basis, we propose probable reaction mechanisms for their reductions in Fig. S4 in the supplemental material. Two interesting exceptions are the quinolones (oxolinic acid and nalidixic acid), which were not reduced by FDORs despite containing enone moieties and inhibiting the growth of F420 mutant strains. It is possible that these compounds are instead substrates for other F420H2-dependent enzymes, such as the luciferase-like hydride transferases (LLHTs).
FIG 3.
F420-dependent antimicrobial detoxification in vitro and in vivo. (A and B) Mass spectra of methoxsalen (A) and malachite green (B) before and after reduction with the FDOR MSMEG_2027. Major peaks corresponding to oxidized (m/z 217) and reduced (m/z 219) methoxsalen and oxidized (m/z 329) and reduced (m/z 331) malachite green were observed. For both compounds, there are also ionization fragments corresponding to the demethylated compounds. (C and D) Plausible mechanisms for FDOR-mediated hydride transfer from F420H2 to methoxsalen (C) and malachite green (D) are shown based on the mass spectra. (E and F) F420-dependent reduction of methoxsalen (E) and malachite green (F) in resting cells of Mycobacterium smegmatis. The concentration of the antimicrobial following administration at 1/2 the MIC of the fbiC mutant strain is shown over time for the wild-type strain, fbiC mutant, fgd mutant, and a no-culture control.
F420 is required for detoxification of antimicrobial compounds.
With the exception of the quinolones, all compounds that strongly inhibited growth of F420 mutant strains were reduced by FDORs (see Fig. S5 in the supplemental material). The correlation between the phenotypic and biochemical data suggests that F420H2-dependent reductases directly detoxify many of these compounds, and the loss of these activities results in enhanced growth inhibition. It is impossible to test this hypothesis with targeted FDOR knockouts, as M. smegmatis encodes multiple F420H2-dependent reductases with overlapping substrate specificities, including five enzymes each from the promiscuous fast-acting A1 and B1 subgroups. Instead, we substantiated this hypothesis by comparing the abilities of the wild-type and mutant strains to detoxify model antimicrobial compounds from two distinct chemical classes that were reduced in vitro by FDORs. The furanocoumarin methoxsalen and the arylmethane malachite green were selected on the basis that their reduction can be sensitively monitored via diode array detection when supplemented in cultures at sub-MIC levels.
The wild-type strain transformed the two antimicrobial compounds tested. All the malachite green added was consumed to below-detectable limits within 40 min, while methoxsalen was more slowly degraded over 120 h (Fig. 3E and F). In contrast, the concentrations of the two compounds in the mutant cultures remained similar to those in the no-culture controls (although, in accordance with previous reports [31], significant culture-independent decolorization of malachite green was also observed). The rates of biotransformation observed suggest that the wild-type strain may survive malachite green treatment and recover from methoxsalen-induced growth inhibition (Fig. 1) by reductively detoxifying these compounds. Malachite green may have been transformed more rapidly than methoxsalen in vivo because FDORs able to reduce the compound are more numerous and faster acting (Fig. 2). In combination, these results show that phenotypic resistance (Fig. 1), in vitro reactivity (Fig. 2), and in vivo reactivity (Fig. 3) to methoxsalen and malachite green are strongly correlated and F420 dependent.
We previously predicted that F420H2-dependent reductases might also support growth of mycobacteria by reductively activating exogenous compounds for use as sources of carbon and energy (1, 2, 16). However, we confirmed in vivo that neither wild-type nor mutant strains could grow when inoculated with minimal salts medium supplemented with any one of eight known FDOR substrates as the sole carbon or energy source. This is again consistent with our hypothesis that mycobacteria employ F420H2-dependent reductases to support detoxification rather than growth.
DISCUSSION
To summarize, this study makes three interrelated findings. First, the cofactor F420 is critical for resistance of mycobacteria to antimicrobial compounds. Strains unable to synthesize or reduce this cofactor were hypersusceptible to most antimicrobial compounds tested, resulting in logarithmic increases in susceptibility for three compound classes (quinone analogues, furanocoumarins, and arylmethanes). Second, at least six F420H2-dependent reductases are capable of reducing antimicrobial compounds, including representatives of the three above-mentioned compound classes. Third, strains unable to utilize F420H2 were unable to reduce representatives of these compounds and exhibited accelerated death. On this basis, we propose that mycobacteria employ some F420H2-dependent reductases to reductively detoxify various antimicrobial compounds.
Environmental and pathogenic mycobacteria alike are renowned for their intrinsic resistance to antimicrobial compounds. Contributing to this capacity are their impermeable cell walls, abundance of efflux pumps, and diversity of chemical detoxification mechanisms (32). Mycobacteria produce a wide range of characterized enzymes capable of degrading antimicrobial compounds, including beta-lactamases (33), cytochrome P450 oxidases (34), arylamine N-acetyltransferases (35), and nitroreductases (36). We propose that F420H2-dependent reductases of the FDOR superfamily enhance this arsenal by reductively activating organic compounds otherwise recalcitrant to biodegradation. Such enzymes can reduce a wide range of functional groups (e.g., lactones, enones, and imines) in structurally diverse monocyclic and polycyclic organic compounds by hydride transfer. It is possible to rationalize why some FDOR-reduced substrates are less toxic to the cell based on the mode-of-action of the compounds: reduction of quinones will prevent spontaneous formation of semiquinone species that induce the production of reactive oxygen species (ROS) (10, 37); reduction of the central carbon of arylmethanes will produce nonplanar molecules unable to intercalate DNA (38); and reduction of furanocoumarins stimulates hydrolytic ring opening that prevents binding to topoisomerase I (16, 39). As with cytochrome P450 oxidases, however, the cellular consequences of F420H2-dependent transformations will depend on the compound reduced. While these enzymes will detoxify antimicrobial compounds if the reduced compounds are less toxic to the cell, as appears to be case for the furanocoumarins, quinones, and arylmethanes tested, their effects will be null if oxidized and reduced forms of the antimicrobial compound have similar efficacy, as may be the case for clofazimine and rifampin. Furthermore, some F420H2-dependent reductases activate certain prodrugs, as reflected by the transformation of the clinical nitroimidazoles delamanid and pretomanid by the FDOR-A1 enzyme Rv3547 (20, 30).
Natural selection for enzymatic strategies to reductively detoxify a wide range of antimicrobial natural products may explain the number, diversity, and promiscuity of F420H2-dependent reductases found in mycobacteria. While the genus Mycobacterium is renowned for its pathogens, the majority of its species are saprophytic soil organisms, including its deepest-branching members (40). We have demonstrated that FDORs can degrade fungal aflatoxins (2) and plant furanocoumarins, as well as quinone groups, which serve as building blocks for various streptomycete antibiotics. Based on their chemical structures, other substrates potentially compatible with FDORs include numerous natural products of plant (e.g., flavonoids), bacterial (e.g., germicidins and mitomycins), and fungal (e.g., ochratoxins and patulin) origin. Our recent evolutionary and structural analyses of the FDOR superfamily support the contention that the most numerous subclasses of this family are also its most promiscuous. Whereas some phylogenetically clustered subgroups of this family appear to be functionally constrained (e.g., B2 and B4 subgroups), the A1 and B1 enzymes have multiplied and diversified in function over the course of mycobacterial evolution (5). Our structural analyses have shown that enzymes from these subgroups have evolved wide substrate-binding pockets, particularly the recently solved MSMEG_2027 structure, which allow them to accommodate substrates as structurally diverse as menadione, malachite green, and methoxsalen (1, 2, 5). Docking studies have suggested that these enzymes bind quinone and coumarin compounds through multiple hydrophobic interactions with their phenyl rings and stabilizing Tyr-mediated hydrogen-bonding interactions with their ketone groups (2, 5). Thus, by encoding multiple reductases with broad specificities, mycobacteria increase the range of compounds they can reductively detoxify.
In addition to facilitating direct detoxification mechanisms, it is possible that F420 also enhances the capacity of mycobacteria to resist antimicrobial agents through mechanisms independent of detoxification. We found that the mutant strains unable to make F420H2 were significantly more susceptible to quinolones and first-line clinical antimycobacterials, even though they were not substrates for the FDORs we tested. Other groups have similarly shown that M. tuberculosis mutants unable to synthesize F420 are hypersusceptible to isoniazid and moxifloxacin (10). It cannot be ruled out that the numerous other F420H2-dependent reductases of the FDOR and LLHT superfamilies metabolize such compounds; this is possible in the cases of oxolinic acid and nalidixic acid, which harbor compatible enone functionalities and cause major growth inhibition. For the antimycobacterials, for which more mild phenotypes were observed, it is more likely that more general loss of functionality of F420-dependent oxidoreductases results in pleiotropic effects on mycobacterial physiology that hypersensitize cells to antimicrobial stress. While the roles of this cofactor in mycobacterial physiology remain incompletely defined, F420 has been linked to maintaining redox homeostasis (10), maintaining cell wall structure (11), and generating antioxidants (5). Disruption of any of these processes could result in broad increases in antimicrobial susceptibility.
The findings from this work have wider environmental, medical, and industrial significance. F420H2-dependent reductases appear to contribute to the antibiotic arms race through dual roles in actinobacteria: whereas streptomycetes use such enzymes to synthesize antibiotics (1, 8), mycobacteria harness them for detoxification. This supports our recent hypothesis that F420 may significantly shape the biological and chemical composition of soil ecosystems (7). The results also have obvious clinical significance given that FDORs have been retained in pathogenic mycobacteria (1), and previous studies have shown that F420 is critical to antimicrobial resistance of M. tuberculosis (1, 10, 13). However, there is currently no evidence that F420H2-dependent reductases directly contribute to the detoxification of clinical antimycobacterial compounds. As recently reviewed (1), the substrate promiscuity of the F420H2-dependent reductase superfamily also opens up potential applications for these enzymes in bioremediation and industrial biocatalysis; for example, FDORs may be used to decolorize environmental toxins, such as malachite green (31), or catalyze stereospecific imine reductions in asymmetric synthesis (41). There is now need for further biochemical studies to fully understand the basis and range of substrate promiscuity of F420H2-dependent reductases, as well as targeted phenotypic studies of individual FDORs to better define their physiological and pharmacological roles in mycobacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chris Coppin (CSIRO) for assistance with figure preparation, Ghader Bashiri (University of Auckland) for providing the fbiABC overexpression plasmid, and William Jacobs, Jr. (Albert Einstein College of Medicine) for supplying Mycobacterium smegmatis mc24517.
This work was supported by a CSIRO Office of the Chief Executive Postdoctoral Fellowship awarded to C.G., a CSIRO Office of the Chief Executive Ph.D. Scholarship awarded to S.A., and Australian Research Council grants (DE120102673 and DP130102144) awarded to C.J.J.
C.G., G.P., J.G.O., M.C.T., A.C.W., B.N., S.A., T.J., R.J.R., C.J.J., and B.M.L. designed the experiments. T.J., B.N., C.G., S.A., G.P., and M.C.T. performed the experiments. C.G., C.J.J., M.C.T., J.G.O., G.P., A.C.W., B.M.L., and R.J.R. supervised students. C.G., J.G.O., M.C.T., T.J., B.N., A.C.W., C.J.J., G.P., S.A., and B.M.L. analyzed the data. C.G., J.G.O., B.N., and T.J. wrote the paper.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02500-16.
REFERENCES
- 1.Greening C, Ahmed FH, Mohamed AE, Lee BM, Pandey G, Warden AC, Scott C, Oakeshott JG, Taylor MC, Jackson CJ. 2016. Physiology, biochemistry, and applications of F420- and Fo-dependent redox reactions. Microbiol Mol Biol Rev 80:451–493. doi: 10.1128/MMBR.00070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor MC, Jackson CJ, Tattersall DB, French N, Peat TS, Newman J, Briggs LJ, Lapalikar GV, Campbell PM, Scott C, Russell RJ, Oakeshott JG. 2010. Identification and characterization of two families of F420H2-dependent reductases from mycobacteria that catalyse aflatoxin degradation. Mol Microbiol 78:561–575. doi: 10.1111/j.1365-2958.2010.07356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Khullar A, Chou S, Sacramo A, Gerratana B. 2009. Biosynthesis of sibiromycin, a potent antitumor antibiotic. Appl Environ Microbiol 75:2869–2878. doi: 10.1128/AEM.02326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebert S, Rieger P-G, Knackmuss H-J. 1999. Function of coenzyme F420 in aerobic catabolism of 2,4,6-trinitrophenol and 2,4-dinitrophenol by Nocardioides simplex FJ2-1A. J Bacteriol 181:2669–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed FH, Carr PD, Lee BM, Afriat-Jurnou L, Mohamed AE, Hong N-S, Flanagan J, Taylor MC, Greening C, Jackson CJ. 2015. Sequence-structure-function classification of a catalytically diverse oxidoreductase superfamily in mycobacteria. J Mol Biol 427:3554–3571. doi: 10.1016/j.jmb.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 7.Ney B, Ahmed FH, Carere CR, Biswas A, Warden AC, Morales SE, Pandey G, Watt SJ, Oakeshott JG, Taylor MC, Stott MB, Jackson CJ, Greening C. 9 August 2016. The methanogenic redox cofactor F420 is widely synthesized by aerobic soil bacteria. ISME J doi: 10.1038/ismej.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, Bashiri G, Gao X, Sawaya MR, Tang Y. 2013. Uncovering the enzymes that catalyze the final steps in oxytetracycline biosynthesis. J Am Chem Soc 135:7138–7141. doi: 10.1021/ja403516u. [DOI] [PubMed] [Google Scholar]
- 9.Peschke U, Schmidt H, Zhang H-Z, Piepersberg W. 1995. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol Microbiol 16:1137–1156. doi: 10.1111/j.1365-2958.1995.tb02338.x. [DOI] [PubMed] [Google Scholar]
- 10.Gurumurthy M, Rao M, Mukherjee T, Rao SPS, Boshoff HI, Dick T, Barry CE, Manjunatha UH. 2013. A novel F420-dependent anti-oxidant mechanism protects Mycobacterium tuberculosis against oxidative stress and bactericidal agents. Mol Microbiol 87:744–755. doi: 10.1111/mmi.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purwantini E, Mukhopadhyay B. 2013. Rv0132c of Mycobacterium tuberculosis encodes a coenzyme F420-dependent hydroxymycolic acid dehydrogenase. PLoS One 8:e81985. doi: 10.1371/journal.pone.0081985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasan MR, Rahman M, Jaques S, Purwantini E, Daniels L. 2010. Glucose 6-phosphate accumulation in mycobacteria: implications for a novel F420-dependent anti-oxidant defense system. J Biol Chem 285:19135–19144. doi: 10.1074/jbc.M109.074310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purwantini E, Mukhopadhyay B. 2009. Conversion of NO2 to NO by reduced coenzyme F420 protects mycobacteria from nitrosative damage. Proc Natl Acad Sci U S A 106:6333–6338. doi: 10.1073/pnas.0812883106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bashiri G, Squire CJ, Moreland NJ, Baker EN. 2008. Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding. J Biol Chem 283:17531–17541. doi: 10.1074/jbc.M801854200. [DOI] [PubMed] [Google Scholar]
- 15.Selengut JD, Haft DH. 2010. Unexpected abundance of coenzyme F420-dependent enzymes in Mycobacterium tuberculosis and other actinobacteria. J Bacteriol 192:5788–5798. doi: 10.1128/JB.00425-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lapalikar GV, Taylor MC, Warden AC, Scott C, Russell RJ, Oakeshott JG. 2012. F420H2-dependent degradation of aflatoxin and other furanocoumarins is widespread throughout the Actinomycetales. PLoS One 7:e30114. doi: 10.1371/journal.pone.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lapalikar GV, Taylor MC, Warden AC, Onagi H, Hennessy JE, Mulder RJ, Scott C, Brown SE, Russell RJ, Easton CJ, Oakeshott JG. 2012. Cofactor promiscuity among F420-dependent reductases enables them to catalyse both oxidation and reduction of the same substrate. Catal Sci Technol 2:1560–1567. doi: 10.1039/c2cy20129a. [DOI] [Google Scholar]
- 18.Ahmed FH, Mohamed AE, Carr PD, Lee BM, Condic-Jurkic K, O'Mara ML, Jackson CJ. 2016. Rv2074 is a novel F420H2-dependent biliverdin reductase in Mycobacterium tuberculosis. Protein Sci 25:1692–1709. doi: 10.1002/pro.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurumurthy M, Mukherjee T, Dowd CS, Singh R, Niyomrattanakit P, Tay JA, Nayyar A, Lee YS, Cherian J, Boshoff HI, Dick T, Barry CE III, Manjunatha UH. 2012. Substrate specificity of the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis responsible for the bioreductive activation of bicyclic nitroimidazoles. FEBS J 279:113–125. doi: 10.1111/j.1742-4658.2011.08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh R, Manjunatha U, Boshoff HIM, Ha YH, Niyomrattanakit P, Ledwidge R, Dowd CS, Lee IY, Kim P, Zhang L, Kang S, Keller TH, Jiricek J, Barry CE III. 2008. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohamed AE, Ahmed FH, Arulmozhiraja S, Lin CY, Taylor MC, Krausz ER, Jackson CJ, Coote ML. 2016. Protonation state of F420H2 in the prodrug-activating deazaflavin dependent nitroreductase (Ddn) from Mycobacterium tuberculosis. Mol Biosyst 12:1110–1113. doi: 10.1039/C6MB00033A. [DOI] [PubMed] [Google Scholar]
- 22.Cellitti SE, Shaffer J, Jones DH, Mukherjee T, Gurumurthy M, Bursulaya B, Boshoff HI, Choi I, Nayyar A, Lee YS, Cherian J, Niyomrattanakit P, Dick T, Manjunatha UH, Barry CE III, Spraggon G, Geierstanger BH. 2012. Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824. Structure 20:101–112. doi: 10.1016/j.str.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 24.Blanco-Ruano D, Roberts DM, Gonzalez-Del-Rio R, Alvarez D, Rebollo MJ, Perez-Herran E, Mendoza A. 2015. Antimicrobial susceptibility testing for Mycobacterium sp. Methods Mol Biol 1285:257–268. doi: 10.1007/978-1-4939-2450-9_15. [DOI] [PubMed] [Google Scholar]
- 25.Greening C, Berney M, Hards K, Cook GM, Conrad R. 2014. A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci U S A 111:4257–4261. doi: 10.1073/pnas.1320586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berney M, Greening C, Conrad R, Jacobs WR, Cook GM. 2014. An obligately aerobic soil bacterium activates fermentative hydrogen production to survive reductive stress during hypoxia. Proc Natl Acad Sci U S A 111:11479–11484. doi: 10.1073/pnas.1407034111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bashiri G, Rehan AM, Greenwood DR, Dickson JMJ, Baker EN. 2010. Metabolic engineering of cofactor F420 production in Mycobacterium smegmatis. PLoS One 5:e15803. doi: 10.1371/journal.pone.0015803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isabelle D, Simpson DR, Daniels L. 2002. Large-scale production of coenzyme F420-5,6 by using Mycobacterium smegmatis. Appl Environ Microbiol 68:5750–5755. doi: 10.1128/AEM.68.11.5750-5755.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerra-Lopez D, Daniels L, Rawat M. 2007. Mycobacterium smegmatis mc2 155 fbiC and MSMEG_2392 are involved in triphenylmethane dye decolorization and coenzyme F420 biosynthesis. Microbiology 153:2724–2732. doi: 10.1099/mic.0.2006/009241-0. [DOI] [PubMed] [Google Scholar]
- 30.Manjunatha UH, Boshoff H, Dowd CS, Zhang L, Albert TJ, Norton JE, Daniels L, Dick T, Pang SS, Barry CE III. 2006. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 103:431–436. doi: 10.1073/pnas.0508392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones JJ, Falkinham JO III. 2003. Decolorization of malachite green and crystal violet by waterborne pathogenic mycobacteria. Antimicrob Agents Chemother 47:2323–2326. doi: 10.1128/AAC.47.7.2323-2326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almeida Da Silva PE, Palomino JC. 2011. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother 66:1417–1430. doi: 10.1093/jac/dkr173. [DOI] [PubMed] [Google Scholar]
- 33.Flores AR, Parsons LM, Pavelka MS Jr. 2005. Genetic analysis of the beta-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to beta-lactam antibiotics. Microbiology 151:521–532. doi: 10.1099/mic.0.27629-0. [DOI] [PubMed] [Google Scholar]
- 34.Ouellet H, Johnston JB, Ortiz de Montellano PR. 2010. The Mycobacterium tuberculosis cytochrome P450 system. Arch Biochem Biophys 493:82–95. doi: 10.1016/j.abb.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sim E, Sandy J, Evangelopoulos D, Fullam E, Bhakta S, Westwood I, Krylova A, Lack N, Noble M. 2008. Arylamine N-acetyltransferases in mycobacteria. Curr Drug Metab 9:510–519. doi: 10.2174/138920008784892100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manina G, Bellinzoni M, Pasca MR, Neres J, Milano A, Ribeiro ALDJL, Buroni S, Skovierova H, Dianiskova P, Mikusova K, Marak J, Makarov V, Giganti D, Haouz A, Lucarelli AP, Degiacomi G, Piazza A, Chiarelli LR, De Rossi E, Salina E, Cole ST, Alzari PM, Riccardi G. 2010. Biological and structural characterization of the Mycobacterium smegmatis nitroreductase NfnB, and its role in benzothiazinone resistance. Mol Microbiol 77:1172–1185. doi: 10.1111/j.1365-2958.2010.07277.x. [DOI] [PubMed] [Google Scholar]
- 37.Winterbourn CC. 2008. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol 4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava S, Sinha R, Roy D. 2004. Toxicological effects of malachite green. Aquat Toxicol 66:319–329. doi: 10.1016/j.aquatox.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Diwan R, Malpathak N. 2009. Furanocoumarins: novel topoisomerase I inhibitors from Ruta graveolens L. Bioorg Med Chem 17:7052–7055. doi: 10.1016/j.bmc.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 40.Hartmans S, de Bont JM, Stackebrandt E. 2006. The genus Mycobacterium–nonmedical, p 889–918. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 41.Schrittwieser JH, Velikogne S, Kroutil W. 2015. Biocatalytic imine reduction and reductive amination of ketones. Adv Synth Catal 357:1655–1685. doi: 10.1002/adsc.201500213. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.