ABSTRACT
Various Listeria monocytogenes strains may contaminate a single food product, potentially resulting in simultaneous exposure of consumers to multiple strains. However, due to bias in strain recovery, L. monocytogenes strains isolated from foods by selective enrichment (SE) might not always represent those that can better survive the immune system of a patient. We investigated the effect of cocultivation in tryptic soy broth with 0.6% yeast extract (TSB-Y) at 10°C for 8 days on (i) the detection of L. monocytogenes strains during SE with the ISO 11290-1:1996/Amd 1:2004 protocol and (ii) the in vitro virulence of strains toward the Caco-2 human colon epithelial cancer cell line following exposure to simulated gastric fluid (SGF; pH 2.0)-HCl (37°C). We determined whether the strains which were favored by SE would be effective competitors under the conditions of challenges related to gastrointestinal passage of the pathogen. Interstrain competition of L. monocytogenes in TSB-Y determined the relative population of each strain at the beginning of SE. This in turn impacted the outcome of SE (i.e., favoring survival of competitors with better fitness) and the levels exposed subsequently to SGF. However, strong growth competitors could be outcompeted after SGF exposure and infection of Caco-2 cells by strains outgrown in TSB-Y and underdetected (or even missed) during enrichment. Our data demonstrate a preferential selection of certain L. monocytogenes strains during enrichments, often not reflecting a selective advantage of strains during infection. These findings highlight a noteworthy scenario associated with the difficulty of matching the source of infection (food) with the L. monocytogenes isolate appearing to be the causative agent during listeriosis outbreak investigations.
IMPORTANCE This report is relevant to understanding the processes involved in selection and prevalence of certain L. monocytogenes strains in different environments (i.e., foods or sites of humans exposed to the pathogen). It highlights the occurrence of multiple strains in the same food as an important aspect contributing to mismatches between clinical isolates and infection sources during listeriosis outbreak investigations.
INTRODUCTION
Selective enrichment (SE) for detection of foodborne pathogens has been a fundamental tool in the food industry, critical for hygiene control and safety monitoring (1) while providing crucial information during trace-back investigations of foodborne outbreaks. However, selective culture-based enrichment procedures are associated with inherent bias since the use of selective agents and the presence of competing background microorganisms in food samples sometimes obstruct the isolation of a target pathogen and lead to false-negative results (2, 3).
Listeria monocytogenes stands out among the pathogens of major concern for food safety. This Gram-positive bacterium causes the rare but life-threatening disease listeriosis and manifests the interplay between saprophytic lifestyle and virulence (4, 5). Its ubiquity allows L. monocytogenes to easily enter the food chain, whereas the capacity to survive and grow in various habitats (e.g., cold, highly acidic, or osmotic environments) provides the microorganism with the potential to withstand extremely adverse conditions involved in food production or storage (6). After contaminated food is consumed, this remarkable adaptability also helps L. monocytogenes to remain viable during digestion, endure the passage to the intestine, and eventually infect susceptible hosts (7, 8).
The accurate detection of L. monocytogenes in foods is clearly of utmost importance. Nonetheless, the bias associated with enrichment protocols introduces recovery limitations and compromises the isolation of the pathogen. The interference of background food microflora (9, 10) or other Listeria spp. (particularly L. innocua) may mask the presence and diminish the detectability of L. monocytogenes (11–17).
Recent studies have addressed the issue of L. monocytogenes strain competition as a factor related to enrichment bias (18, 19). The efficiency of enrichment protocols in isolating all L. monocytogenes strains that might have contaminated the same food has reasonably become a subject of investigation; mixed populations of L. monocytogenes strains could be present in a single sample, and ingestion of more than one strain by the same individual is likely (20, 21). Apparently, the success of an enrichment protocol is dependent on detection of the infecting strain.
Among the 13 serotypes of L. monocytogenes, serotype 4b is considered the major outbreak-associated serotype, while 1/2a strains are more frequently food isolates (22). Such a food- or outbreak-strain correlation might be attributable not just to the particular genetic characteristics of strains that equip them with proper capabilities to survive or thrive under different conditions (e.g., in foods or during passage through the gastrointestinal tract [GIT]) but might also be the result of the potential failure of selective enrichment to detect all relevant strains in a food contaminated with multiple strains.
Considering the above, we investigated the effect of cocultivation on the recovery of L. monocytogenes strains after selective enrichment or after exposure to simulated gastric fluid (SGF) and subsequent infection of Caco-2 cells. We hypothesized that the selective enrichment would not always detect the strains that would survive better in gastric fluid and infect Caco-2 cells.
MATERIALS AND METHODS
Bacterial strains, culture, and growth conditions.
The L. monocytogenes strains used in this study are listed in Table 1. The selection of strains was performed according to two previous studies investigating the growth, virulence, and enrichment competition of L. monocytogenes strains (23, 24). Strains selected for resistance to rifampin (Rifambicin; AppliChem) or streptomycin (Streptomycin Sulfate Biochemica; AppliChem) according to the method described by Blackburn and Davies (25) were used for enabling selective enumeration of each strain in coculture.
TABLE 1.
Listeria monocytogenes strains used in the study
| Strain | Serotype(s) | MLST | Source | Yr of isolation | Country | Antibiotic(s) (MIC [μg/ml])a |
|---|---|---|---|---|---|---|
| C5 | 4b | ST2 | Cow feces | 2007 | Ireland | Streptomycin (2,000) |
| 6179 | 1/2a | ST121 | Cheese | 1999 | Ireland | Rifampin (>800) |
| ScottA | 4b | ST290 | Human isolate | 1983 | United States | Streptomycin (4,000), rifampin (>800) |
| PL25 | 1/2b (3b,7)b | ST59 | Ground pork | 2009 | Greece | Rifampin (800) |
The approximate MIC value was considered the minimum tested concentration (in micrograms per milliliter) of antibiotic at which no bacterial growth was observed after 24 h at 30°C. Bacterial growth was confirmed through measurements of the optical density at 600 nm (OD600). Streptomycin was evaluated at 0, 125, 250, 500, 1,000, 2,000, and 4,000 μg/ml. Rifampin was evaluated at 0, 200, 400, and 800 μg/ml.
The serovar-specific group was characterized by multiplex PCR according to Doumith et al. (59), and the serovars in parentheses were omitted on the basis of the multilocus sequence typing (MLST) classification results.
Strains were stored at −80°C in tryptic soy broth (Lab M) with 0.6% yeast extract (TSB-Y; pH 7.2) and 20% glycerol. During the experiments, all strains were maintained on tryptic soy agar (TSA; Lab M) supplemented with 0.6% yeast extract (Lab M) (TSA-Y) containing rifampin (50 μg/ml) or streptomycin (1,000 μg/ml).
For each strain, one single colony from a TSA-Y stock culture was transferred to 10 ml TSB-Y plus streptomycin (1,000 μg/ml) or rifampin (50 μg/ml) and incubated for 24 h at 30°C. Subsequently, 100 μl of the 24-h cultures was transferred to 10 ml of TSB-Y plus the corresponding antibiotic and incubated at 30°C for 18 h.
Inoculation of TSB-Y was performed as previously described for single or mixed listerial cultures (23). Briefly, the activated 18-h cultures (corresponding to approximately 109 CFU/ml) of L. monocytogenes strains were washed with Ringer solution (Lab M; LAB100Z), resuspended in 10 ml TSB-Y, and serially diluted in TSB-Y to obtain a final inoculum of approximately 103 CFU/ml. Strains were grown at 10°C for 8 days as single cultures or in combinations by mixing a rifampin-resistant strain with a streptomycin-resistant strain (ratio, 1:1; final volume, 10 ml). On days 2, 4, 6, and 8, cultures were sampled for determination of CFU and then used for enrichment experiments or exposure to simulated gastric fluid, as described below.
Enrichment of L. monocytogenes cocultures.
Enrichment of mixed listerial cultures was performed according to the ISO 11290-1:1996/Amd 1:2004 enrichment protocol (26) using the media recommended by the method description. There are also other standard protocols for the enrichment of L. monocytogenes available, such as the U.S. Food and Drug Administration (FDA) Bacterial Analytical Manual (BAM) method; this protocol has already been used to test competition of L. monocytogenes serotype 4b strains against strains of serotype 1/2a (19). We chose ISO protocols as reference methods widely used across laboratories in Europe and also regulated by the European Commission (27). We showed previously that coenrichment of L. monocytogenes strains—also used in this study—following the ISO method might favor the recovery of certain strains, resulting in a biased outcome (24). In addition, we have demonstrated previously that growth competition could occur between L. monocytogenes strains during their cocultivation in TSB-Y at 10°C (23). The 10°C temperature was initially chosen as one at which at we could observe equal levels of growth of all single cultures of L. monocytogenes strains, thus ensuring that the observed inhibition would not be the result of differences in the individual growth potentials of strains under the conditions tested. On the basis of these observations, we moved onward by investigating the effect of the duration of cocultivation on the detection of L. monocytogenes strains, simulating the conditions occurring during storage of a contaminated food. Therefore, on days 2, 4, 6, and 8 of incubation at 10°C, a 1-ml volume from each L. monocytogenes coculture (TSB-Y) was added to 9 ml of Half Fraser broth (HF; Lab M) and the reaction mixture was incubated at 30°C for 24 h. Subsequently, 100 μl of HF was transferred into 10 ml of Full Fraser broth (FF; Lab M) and the reaction mixture was incubated at 37°C for 48 h. After each enrichment step, the enrichment broths (HF and FF) were streaked (10 μl) onto Agar Listeria Ottavian Agosti (ALOA; Biolife 4016052) and the ALOA plates were incubated at 37°C for 2 days. Following incubation, all individual L. monocytogenes colonies were picked (1-μl inoculating loop) from plates and further streaked on TSA-Y containing rifampin or streptomycin in order to determine the percentage of colonies formed by each strain (streptomycin or rifampin resistant) among the total colonies appearing on the streaked plate. Furthermore, the CFU counts of each strain in the xenic cultures were determined after inoculation of HF and at the end of both enrichment steps. Each enrichment experiment was performed three independent times in triplicate, and each of the triplicate (HF or FF) cultures was streaked on two different ALOA plates. The number of isolated colonies ranged from 15 to 30 for each plate, thus resulting in a total of ca. 270 to 540 colonies per mixed culture for each enrichment step.
Exposure of L. monocytogenes cultures to simulated gastric fluid (SGF).
SGF was prepared according to the method described by Barmpalia-Davis et al. (28) and consisted of the following reagents (per liter): 0.4 g glucose (Riedel de Haën, Switzerland), 3.0 g yeast extract (Lab M Limited, United Kingdom), 1.0 g Bacto peptone (Lab M Limited, United Kingdom), 4.0 g porcine mucin (Sigma-Aldrich Co., USA), 0.5 g cysteine (Sigma-Aldrich Co., USA), 0.08 g NaCl (Merck KGaA, Germany), 0.4 g NaHCO3 (PanReac AppliChem, Spain), 0.04 g K2HPO4 (Merck KGaA, Germany), 0.008 g CaCl2-2H2O (Merck KGaA, Germany), 0.008 g MgSO4·7H2O (Mallinckrodt Pharmaceuticals, Ireland), 1.0 g xylan (Sigma-Aldrich, Co., USA), 3.0 g soluble starch (Merck KGaA, Germany), 2.0 g pectin (Sigma-Aldrich, Co., USA), and 1 ml Tween 80 (Scharlab S.L., Spain). The components were mixed, and the fluid was autoclaved. Prior to use, the solution was adjusted to 37°C, 3.0 g pepsin from porcine stomach mucosa (≥400 U/mg protein) (Sigma-Aldrich, Co., USA) was added to the solution, and the pH of SGF was adjusted to 2.0 using 6 N HCl under aseptic conditions.
The survival of L. monocytogenes strains in SGF was evaluated for single and mixed TSB-Y (10°C) cultures as follows: on days 2, 4, 6, and 8 of incubation, 2-ml volumes of the cultures were centrifuged (10,000 × g for 1 min), resuspended in 2 ml of SGF (37°C), and incubated in a water bath at 37°C for total exposure times of 18, 48, 60, and 90 min, respectively. During exposure of the strains to SGF, the cultures were sampled at specific time points (depending on the day) and the surviving populations were enumerated by plating appropriate serial dilutions on TSA-Y or TSA-Y containing rifampin or streptomycin. The experiment was performed three independent times in triplicate.
In vitro virulence potential of L. monocytogenes strains.
The tumor-derived Caco-2 human intestinal epithelial cell line (American Type Culture Collection [ATCC]) was used for the in vitro virulence assays; Caco-2 cells were grown in a mixture consisting of Eagle's minimum essential medium (MEM) supplemented with 15% (vol/vol) fetal bovine serum (FBS) inactivated at 56°C for 30 min, 2 mM l-glutamine, 100 U/ml penicillin-streptomycin, and 1% (vol/vol) nonessential amino acids (all from Biochrom) at 37°C in a humidified atmosphere (95% relative humidity) containing 5% CO2.
On the basis of the growth curves of L. monocytogenes strains at 10°C and their capacity to survive in SGF (to ensure a sufficient number of survivors), the in vitro virulence potential of the strains was evaluated after incubation for 6 and 8 days at 10°C in TSB-Y and subsequent exposure to SGF for 20 and 30 min, respectively. Also, due to high levels of the population differences at the selected time points, the combination of strain C5 and strain 6179 was not selected for in vitro virulence assays.
Invasion efficiency and intracellular proliferation were assessed for L. monocytogenes strains in Caco-2 cell monolayers, as previously described (23). Briefly, Caco-2 cells were seeded into 24-well tissue culture plates (Greiner Bio-One) in MEM supplemented with 15% (vol/vol) FBS until confluence was reached. At 24 h prior to the experiment, culture medium was aspirated and replaced by MEM without antibiotics and containing 0.1% (vol/vol) FBS.
L. monocytogenes strains were cultivated at 10°C as described above except for the use of a different culture volume, which was set at 30 ml TSB-Y in 50-ml plastic tubes. On day 6 or 8 of incubation, bacterial cells were exposed to SGF (20 ml of culture centrifuged and resuspended in 20 ml of SGF) for 20 or 30 min, respectively, at 37°C. Following exposure to SGF, bacterial cultures were centrifuged (5,000 × g for 5 min at 37°C) and resuspended in prewarmed MEM (37°C) to obtain a multiplicity of infection of ∼25. Caco-2 cell monolayers were infected with the cultures for 1 h at 37°C; at 60 min postinfection, Caco-2 cells were washed twice with Dulbecco's phosphate-buffered saline (DPBS) and incubated in MEM containing 0.1% FBS and 100 μg/ml gentamicin (Biochrom). After 45 min (invasion assay) or 4 h (intracellular proliferation assay), Caco-2 cells were washed twice with DPBS and lysed with 1 ml of cold 0.1% Triton X-100 (Applichem). The 45-min or 4-h suspension was used for enumeration of viable L. monocytogenes cells, the levels of which were determined by plating appropriate dilutions on TSA-Y or TSA-Y supplemented with rifampin or streptomycin. Invasion efficiency was reported as follows:
| (1) |
Intracellular replication of L. monocytogenes was expressed as intracellular growth coefficient (IGC) values; IGC was calculated using the following fraction:
| (2) |
In addition, the total in vitro virulence potential of L. monocytogenes strains was described as the percentage of the initial inoculum that was recovered and enumerated after the proliferation assay.
The in vitro virulence properties of L. monocytogenes strains were determined for (i) mixed cultures, (ii) single-strain cultures, (iii) single-strain cultures combined in mixtures before exposure to SGF, and (iv) single-strain cultures without prior exposure of cultures to SGF. The experiments were performed three independent times in triplicate.
Statistical analysis and curve fitting.
Data analysis was performed using Microsoft Excel 2011 and SPSS 22.0 for Mac (SPSS Inc., Chicago, IL, USA). The Tukey's honestly significant difference (HSD) test was used for multiple comparisons regarding cell concentration or in vitro virulence or to determine differences between the means of the Weibull model parameters for comparisons of conditions. For all pairwise comparisons, the Student t test was used. Differences were considered to be significant for P values of <0.05.
For the SGF assays, the mean log10 CFU counts for the strains were plotted against sampling times and the Weibull inactivation model was fitted to the experimental data, using Microsoft Excel GInaFIT add-in software (version 1.6). The software tool was used for the calculation of the estimates for delta (δ) and p values. The delta value is the time for the first log reduction expressed in minutes, and p is a shape factor indicating whether the curve is concave (p < 1), convex (p > 1), or linear (p = 1). On the basis of the δ and p values, the time for 4 log inactivation (t4D) was estimated to enable comparisons of curves with varying p values using the Weibull equation and, in particular, the following formula:
| (3) |
RESULTS
Growth of L. monocytogenes strains in TSB-Y.
The growth of single and mixed cultures of L. monocytogenes strains at 10°C was evaluated in our previous study (23). The population of each strain (grown individually or in coculture in TSB-Y) after 2, 4, 6, and 8 days of incubation is given in Table 2 as Log10(N0) [where Log10(N0) represents the measured population (log CFU/milliliter) of L. monocytogenes strains in TSB-Y and the initial inoculum used for enrichments or exposure to SGF on each day]. Cocultivation of C5 with 6179 or ScottA inhibited the growth of the two latter strains, resulting in their lower numbers compared to C5 on days 6 and 8. Similarly, PL25 suppressed the growth of ScottA, which did not manage to attain more than ca. 6 log CFU/ml compared to the 9 log CFU/ml of PL25. Cocultivation of C5 with PL25 resulted in equivalent levels of growth of the two strains.
TABLE 2.
Inactivation kinetics of L. monocytogenes strains in SGF as described by the Weibull modela
| Day and strain | Combination | Log10(N0)b | Deltac | pd | t4De |
|---|---|---|---|---|---|
| 2 | |||||
| C5 | Single | 5.33 ± 0.00 | 0.40 ± 0.00ab | 0.41 ± 0.00 | 12.06 ± 0.00abc |
| +6179 | 5.07 ± 0.17 | 0.29 ± 0.02a | 0.37 ± 0.01 | 13.14 ± 1.78abc | |
| +ScottA | 5.17 ± 0.00 | 0.14 ± 0.00a | 0.30 ± 0.00 | 14.22 ± 0.00bc | |
| +PL25 | 5.13 ± 0.00 | 0.17 ± 0.00a | 0.30 ± 0.00 | 16.38 ± 0.00c | |
| 6179 | Single | 5.13 ± 0.10 | 0.47 ± 0.06ab | 0.45 ± 0.01 | 10.26 ± 0.59ab |
| +C5 | 5.10 ± 0.01 | 0.31 ± 0.18ab | 0.36 ± 0.05 | 14.58 ± 1.27bc | |
| ScottA | Single | 5.44 ± 0.03 | 0.34 ± 0.04ab | 0.42 ± 0.01 | 8.94 ± 0.08a |
| +C5 | 5.03 ± 0.06 | 0.65 ± 0.06b | 0.43 ± 0.01 | 16.20 ± 0.00c | |
| +PL25 | 5.00 ± 0.16 | 0.27 ± 0.15a | 0.35 ± 0.07 | 13.86 ± 2.80bc | |
| PL25 | Single | 5.41 ± 0.00 | 0.26 ± 0.00a | 0.37 ± 0.00 | 11.52 ± 0.00ab |
| +C5 | 5.36 ± 0.00 | 0.35 ± 0.00ab | 0.43 ± 0.00 | 9.12 ± 0.00a | |
| +ScottA | 5.15 ± 0.00 | 0.19 ± 0.17a | 0.34 ± 0.09 | 11.04 ± 1.19ab | |
| 4 | |||||
| C5 | Single | 7.84 ± 0.28 | 0.56 ± 0.01a | 0.55 ± 0.00 | 7.11 ± 0.13a |
| +6179 | 7.54 ± 0.00 | 1.55 ± 0.00abc | 0.73 ± 0.00 | 10.44 ± 0.00abc | |
| +ScottA | 7.45 ± 0.37 | 1.33 ± 0.28ab | 0.71 ± 0.04 | 9.54 ± 1.02ab | |
| +PL25 | 7.08 ± 0.34 | 1.76 ± 0.59abcd | 0.75 ± 0.08 | 11.25 ± 1.40abc | |
| 6179 | Single | 7.33 ± 0.00 | 1.66 ± 0.00abcd | 0.76 ± 0.00 | 10.44 ± 0.00abc |
| +C5 | 7.08 ± 0.01 | 2.60 ± 0.13bcd | 0.90 ± 0.01 | 12.24 ± 0.25abc | |
| ScottA | Single | 7.00 ± 0.90 | 3.41 ± 0.25d | 0.96 ± 0.15 | 14.91 ± 2.25c |
| +C5 | 6.67 ± 0.09 | 1.56 ± 1.36abc | 0.60 ± 0.18 | 14.25 ± 3.61bc | |
| +PL25 | 6.47 ± 0.38 | 3.31 ± 0.11cd | 1.00 ± 0.00 | 13.32 ± 0.51bc | |
| PL25 | Single | 7.82 ± 0.00 | 1.77 ± 0.00abcd | 0.80 ± 0.00 | 10.26 ± 0.00abc |
| +C5 | 7.74 ± 0.00 | 1.80 ± 0.00abcd | 0.81 ± 0.00 | 10.08 ± 0.00abc | |
| +ScottA | 7.60 ± 0.00 | 0.96 ± 0.00ab | 0.63 ± 0.00 | 9.00 ± 0.00ab | |
| 6 | |||||
| C5 | Single | 9.50 ± 0.08 | 5.19 ± 3.29ab | 0.87 ± 0.21 | 24.90 ± 7.21ab |
| +6179 | 8.93 ± 0.28 | 4.62 ± 0.86ab | 0.82 ± 0.04 | 25.50 ± 2.97ab | |
| +ScottA | 9.26 ± 0.27 | 6.28 ± 4.22ab | 0.92 ± 0.26 | 27.60 ± 7.64ab | |
| +PL25 | 8.30 ± 0.67 | 10.07 ± 0.82ab | 1.06 ± 0.13 | 38.10 ± 2.97ab | |
| 6179 | Single | 9.18 ± 0.23 | 1.73 ± 0.17a | 0.61 ± 0.01 | 17.10 ± 1.27a |
| +C5 | 7.95 ± 0.86 | 2.87 ± 1.59a | 0.66 ± 0.08 | 23.04 ± 7.47a | |
| ScottA | Single | 8.85 ± 0.58 | 5.48 ± 2.62ab | 0.86 ± 0.15 | 27.30 ± 5.52ab |
| +C5 | 7.60 ± 0.29 | 8.24 ± 2.52ab | 0.98 ± 0.17 | 33.90 ± 2.12ab | |
| +PL25 | 6.65 ± 0.44 | 22.01 ± 20.39b | 2.12 ± 2.01 | 45.90 ± 10.61b | |
| PL25 | Single | 9.26 ± 0.16 | 5.28 ± 1.72ab | 0.86 ± 0.15 | 27.00 ± 0.85ab |
| +C5 | 9.22 ± 0.04 | 5.69 ± 2.55ab | 0.84 ± 0.21 | 29.70 ± 1.27ab | |
| +ScottA | 8.93 ± 0.21 | 8.73 ± 4.82ab | 1.04 ± 0.28 | 32.70 ± 6.36ab | |
| 8 | |||||
| C5 | Single | 9.33 ± 0.04 | 4.81 ± 4.94a | 0.67 ± 0.31 | 30.30 ± 13.18ab |
| +6179 | 9.33 ± 0.11 | 6.49 ± 4.48ab | 0.81 ± 0.19 | 33.90 ± 9.62ab | |
| +ScottA | 9.25 ± 0.49 | 3.63 ± 1.67a | 0.68 ± 0.11 | 27.00 ± 5.47a | |
| +PL25 | 9.16 ± 0.34 | 4.25 ± 1.89a | 0.69 ± 0.09 | 28.17 ± 4.46ab | |
| 6179 | Single | 9.44 ± 0.11 | 4.68 ± 4.57a | 0.70 ± 0.24 | 29.10 ± 12.61ab |
| +C5 | 7.72 ± 0.38 | 4.30 ± 3.52 | 0.68 ± 0.21 | 29.10 ± 9.62ab | |
| ScottA | Single | 9.40 ± 0.09 | 7.03 ± 2.75ab | 0.79 ± 0.12 | 40.50 ± 5.62abc |
| +C5 | 7.64 ± 0.17 | 17.25 ± 5.58bc | 1.21 ± 0.26 | 54.84 ± 9.63bc | |
| +PL25 | 6.59 ± 0.00 | 19.90 ± 3.06c | 1.20 ± 0.25 | 65.70 ± 7.14c | |
| PL25 | Single | 9.41 ± 0.05 | 7.76 ± 4.20abc | 0.84 ± 0.20 | 40.20 ± 5.79abc |
| +C5 | 9.17 ± 0.28 | 5.20 ± 1.70ab | 0.74 ± 0.09 | 33.60 ± 4.16ab | |
| +ScottA | 9.28 ± 0.25 | 9.19 ± 7.27abc | 0.91 ± 0.31 | 38.40 ± 13.95ab |
Data represent mean values ± standard deviations (SD) of the results of three independent experiments performed in triplicate. Single or mixed cultures of L. monocytogenes strains were grown in TSB-Y for 8 days at 10°C before exposure to SGF. Different lowercase superscript letters indicate significant differences among values within the same column.
Log10(N0), measured population (log CFU/milliliter) of L. monocytogenes strains in TSB-Y and the initial inoculum used for enrichments or exposure to SGF on each day.
Delta, time (in minutes) for the first decimal reduction.
p, shape parameter.
t4D, time (min) for 4-decimal reduction.
Growth of L. monocytogenes strains in enrichment broths.
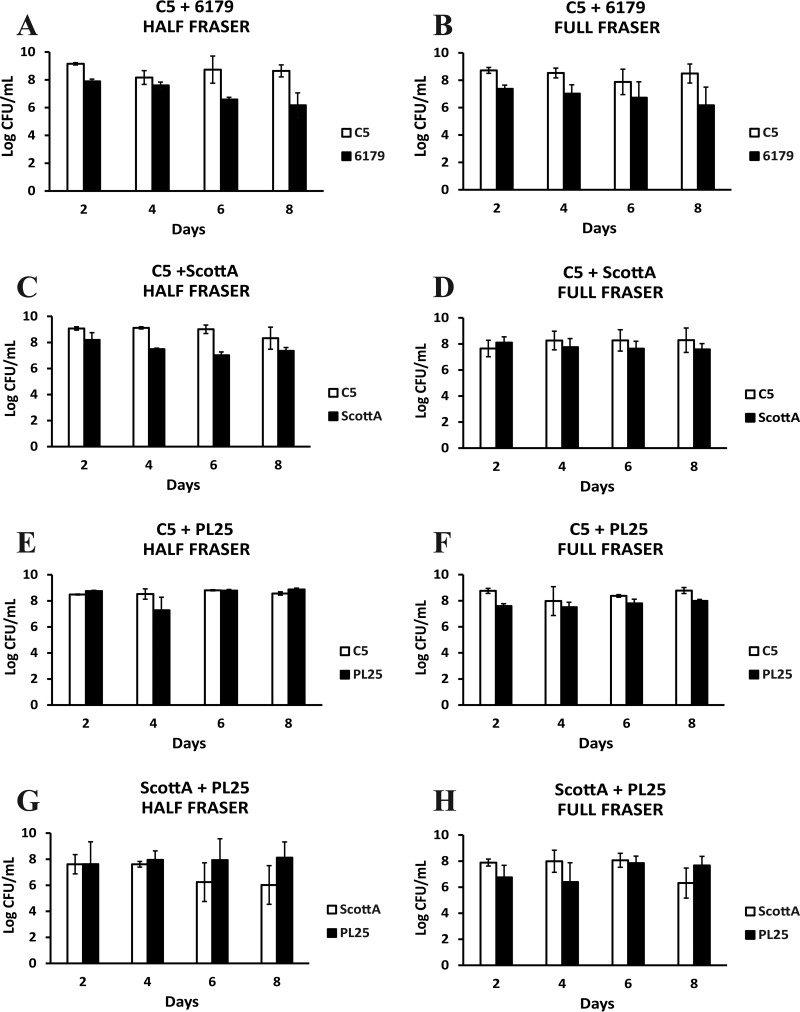
Cocultivation of L. monocytogenes strains for 2, 4, 6, and 8 days in TSB-Y was followed by their enrichment according to the ISO method. All strains in mixed cultures reached 6 to 9 log CFU/ml after incubation in HF and FF enrichment broths, and any observed differences between the final cell densities of two strains in a mixed culture did not exceed 3 log CFU/ml at the end of the two enrichment steps (Fig. 1). After enrichment of a C5 plus 6179 coculture previously grown in TSB-Y for 2 and 4 days, the population of 6179 in enrichment broths increased up to ca. 7.5 log CFU/ml, while the population of C5 was constantly ca. 9 log CFU/ml (Fig. 1A and B). The levels of the 6th- and 8th-day cocultures of 6179 did not increase in HF, and the cell density after enrichment was similar to the initial level added to HF. In the C5 plus ScottA combination, ScottA had a CFU count that was ca. 1.5 log CFU/ml lower than that of C5 in HF, but no significant population differences were observed for the two strains in FF (Fig. 1C and D). Regarding C5 and PL25, both strains reached ca. 8 to 9 log CFU/ml in HF and FF regardless of the day on which TSB-Y composites were subjected to enrichment (Fig. 1E and F). In the combination of ScottA plus PL25, the 6th- and 8th-day cells of PL25 reached a higher final population (ca. 8.0 log CFU/ml) than the 6th- and 8th-day cells of ScottA (ca. 6.0 log CFU/ml) after incubation in HF (Fig. 1G and H). However, there were no significant population differences for the two strains in FF.
FIG 1.
Numbers (log CFU per milliliter) of L. monocytogenes strains C5, 6179, ScottA, and PL25 following incubation for 24 h at 30°C in Half Fraser (A, C, E, and G) or 48 h at 37°C in Full Fraser (B, D, F, and H) enrichment broth. Selective enrichment was performed for cocultures of L. monocytogenes strains C5 plus 6179 (A and B), C5 plus ScottA (C and D), C5 plus PL25 (E and F), and ScottA plus PL25 (G and H) after incubation for 2, 4, 6, or 8 days at 10°C in TSB-Y. Bars represent mean values ± standard deviations (SD) of results of three independent experiments performed in triplicate.
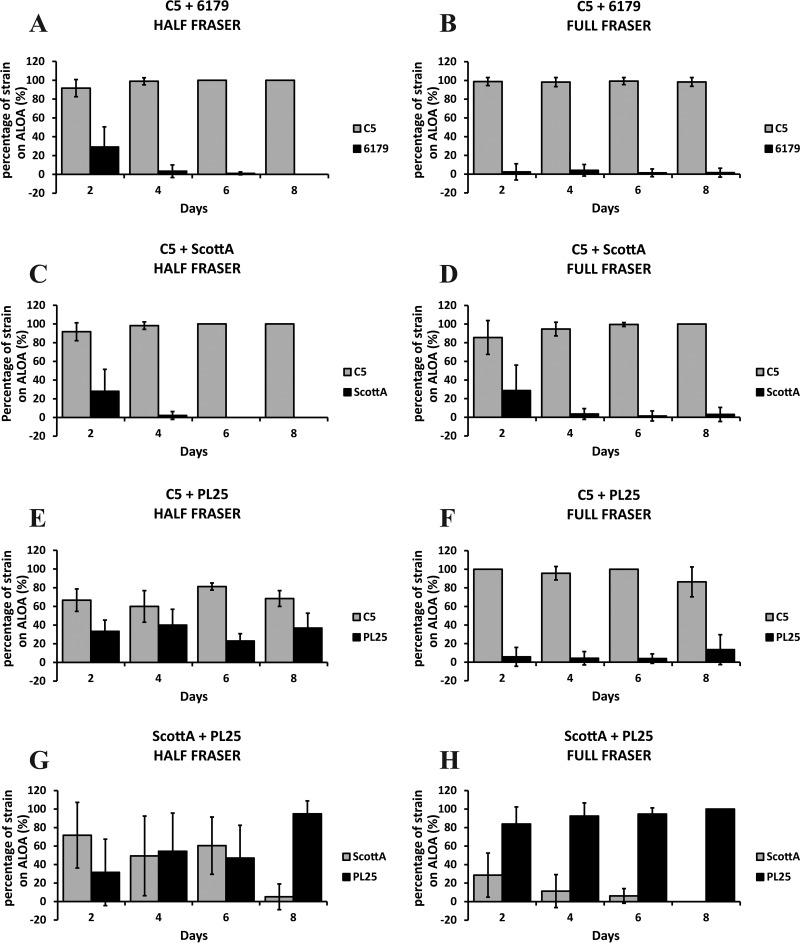
Recovery of the L. monocytogenes strains on selective agar.
Following incubation in enrichment broths, the cocultures of L. monocytogenes strains were streaked on selective ALOA plates. Strain C5 systematically accounted for at least 80% of the total ALOA colonies in testing against 6179 (Fig. 2A and B) or ScottA (Fig. 2C and D). In fact, after the 4th day of their coincubation in TSB-Y, all the enrichments resulted in the dominance of C5 and minor or zero recovery of ScottA and 6179 on ALOA, streaked from either Half Fraser broth or Full Fraser broth. In the presence of strain PL25, the dominance of C5 on ALOA was marginal after the first enrichment step, with 60% of the total colony count belonging to this strain (Fig. 2E). However, following the second enrichment, the rate of recovery of PL25 was dramatically reduced and C5 was almost exclusively isolated from the surface of ALOA regardless of the day on which enrichment was performed (Fig. 2F). When PL25 and ScottA were grown together in TSB-Y, the effect of cocultivation time on the recovery of strains on ALOA was considerable (Fig. 2G and H). After 2 days of coincubation with PL25 and subsequent enrichment in HF, ScottA dominated on ALOA, accounting for ca. 70% of the total isolated colonies (Fig. 2G). Following coincubation of the strains for 4 or 6 days in TSB-Y and enrichment in HF, the colony percentage of both strains was ca. 50%. After 8 days in TSB-Y and subsequent coenrichment of PL25 plus ScottA in HF, the relative proportions of the two strains on ALOA were reversed compared to those seen at the beginning of incubation, and PL25 prevailed, with over 95% of the total colonies belonging to this strain. Notably, after the second enrichment step in FF, PL25 was always the dominant strain on ALOA (Fig. 2H). Only 30% of the isolated ALOA colonies were confirmed to represent ScottA, following 2 days of coincubation with PL25 in TSB-Y and two subsequent enrichment steps. In addition, we observed again a declining trend regarding the recovery of ScottA on ALOA over the course of incubation in TSB-Y. After 8 days of coincubation with PL25 in TSB-Y and double enrichment, ScottA could not be detected on ALOA plates streaked from FF (i.e., 0% of the colony count).
FIG 2.
Percentages of L. monocytogenes strains C5, 6179, ScottA, and PL25 on ALOA after enrichment in Half Fraser (A, C, E, and G) and Full Fraser (B, D, F, and H) enrichment broth. Selective enrichment followed by streaking on ALOA was performed for cocultures of L. monocytogenes strains C5 plus 6179 (A and B), C5 plus ScottA (C and D), C5 plus PL25 (E and F), and ScottA plus PL25 (G and H) after incubation for 2, 4, 6, or 8 days at 10°C in TSB-Y. Bars represent mean values ± SD of results of three independent experiments performed in triplicate.
Overall, the recovery of strains on ALOA was dependent on their population at the end of the enrichment and this was associated with the strain-specific levels attained from the preceding growth in TSB-Y, with the latter determining the fitness of competing strains.
Survival of L. monocytogenes strains in SGF.
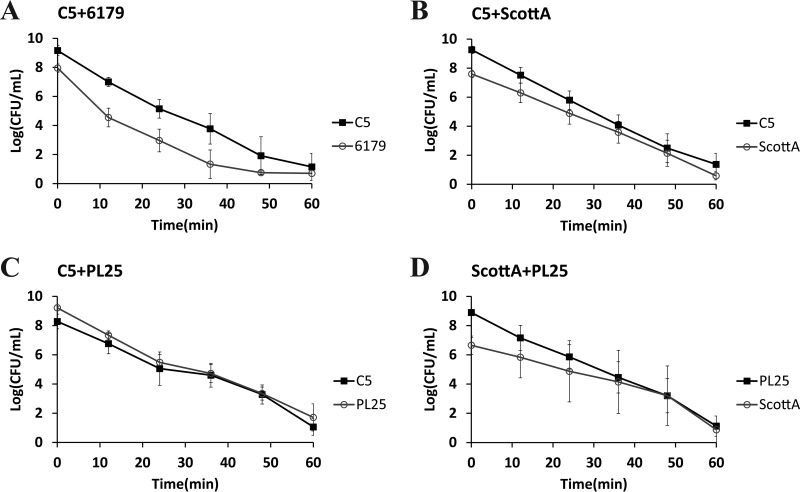
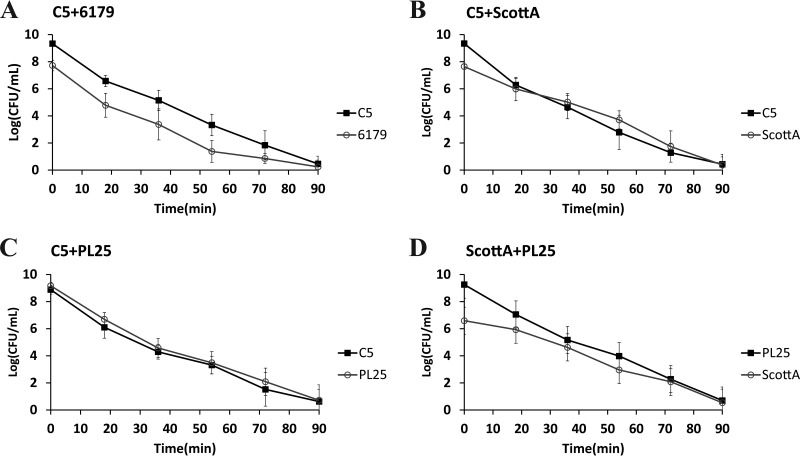
The survival of L. monocytogenes strains in SGF (pH 2.0, 37°C) was tested after growth in TSB-Y as single or mixed cultures for 2, 4, 6, or 8 days at 10°C. Following 2 or 4 days in TSB-Y, exposure of single cultures or composites to SGF resulted in the rapid inactivation of all strains (data not shown in graphs). A 4-log reduction of the initial populations or even a reduction below the enumeration limit (1 log CFU/ml) was noticed after a very short time (ca. 9 to 16 min) (Table 2); thus, any observed differences regarding the resistance of strains to acid stress, albeit statistically significant, were not considered relevant in the context of the study. When L. monocytogenes strains were grown in cocultures for 6 or 8 days, although their survival in SGF increased compared to that observed after 2 and 4 days of incubation prior to gastric challenge, their inactivation kinetics did not significantly differ from the kinetics seen with their respective monocultures (Table 2). In addition, significant differences were not observed in the SGF survival rates of 6179 and C5 (Fig. 3A and 4A and Table 2), but due to its lower initial cell density, 6179 was inactivated sooner than C5. ScottA displayed increased acid resistance compared to C5 and PL25 after 6 or 8 days of coincubation with the latter strains in TSB-Y. Despite having lower initial populations than C5 and PL25, ScottA showed an overall higher survival rate in SGF as indicated by the smoother slope of its inactivation curve (Fig. 3B and D and 4B and D). When C5 and PL25 were paired, the two strains produced almost identical inactivation curves (Fig. 3C and 4C).
FIG 3.
Survival of L. monocytogenes strains C5, 6179, ScottA, and PL25 in SGF (pH 2.0, 37°C), after coculture of C5 plus 6179 (A), C5 plus ScottA (B), C5 plus PL25 (C), and ScottA plus PL25 (D) for 6 days at 10°C in TSB-Y. Data points represent mean values ± SD of results of three independent replicates performed in triplicate.
FIG 4.
Survival of L. monocytogenes strains C5, 6179, ScottA, and PL25 in SGF (pH 2.0, 37°C), after coculture of C5 plus 6179 (A), C5 plus ScottA (B), C5 plus PL25 (C), and ScottA plus PL25 (D) for 8 days at 10°C in TSB-Y. Data points represent mean values ± SD of results of three independent replicates performed in triplicate.
Taking the data together, the cocultivation of L. monocytogenes strains did not have a profound role in the sensitization or resistance of cells to gastric acid stress, but overall, it contributed to strain-specific reductions by impacting the level of each strain in the composite at the beginning of exposure to SGF.
In vitro virulence of L. monocytogenes strains after exposure to SGF.
The efficiency of L. monocytogenes strains with respect to invasion and proliferation in Caco-2 cells after cocultivation and exposure to SGF was studied. We wanted to investigate whether the strains that were grown in mixed culture and that tended to be more easily recovered by enrichment and streaking were also capable of outcompeting the others during infection of intestinal epithelial cells.
The infection of Caco-2 cells was performed after incubation of L. monocytogenes cultures for 6 and 8 days in TSB-Y at 10°C and subsequent exposure to SGF (pH 2.0, 37°C) for 20 and 30 min, respectively; at these time points, the population of all L. monocytogenes strains in the different cocultures was ca. 106 CFU/ml, except for 6179 in coculture with C5, where 6179 had significantly lower cell density, and for this reason that combination was not used for in vitro virulence tests.
After incubation for 6 days in TSB-Y and subsequent exposure to SGF, the efficiency of L. monocytogenes strains mainly with respect to penetration but also with respect to proliferation into Caco-2 cells was poor (data not shown). In many cases, their numbers were below the detection limit (1 CFU/ml of Triton X-100 cell suspension). When possible, their total in vitro virulence was estimated (see Table S1 in the supplemental material).
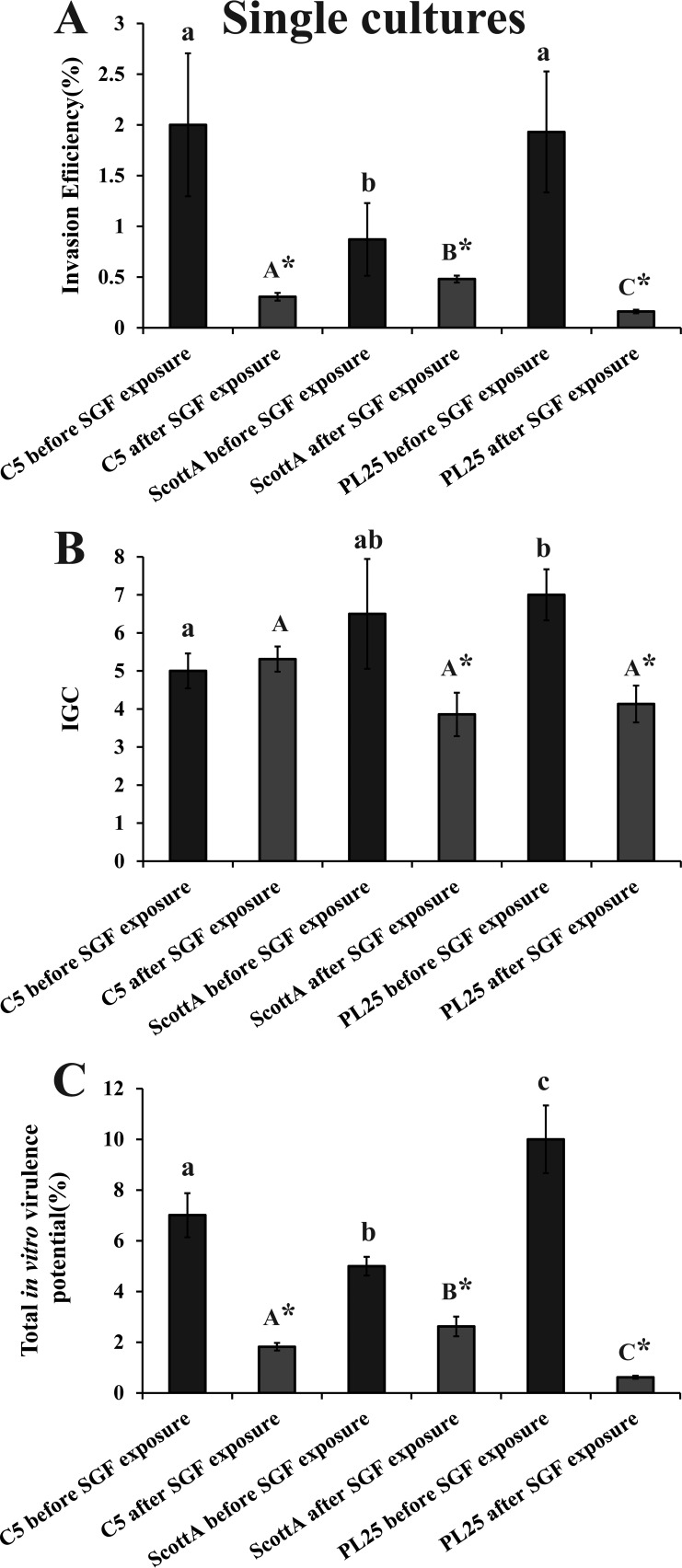
Before exposure to SGF, strains C5 and PL25 were more invasive than ScottA (Fig. 5A), while PL25 had slightly higher IGC values than ScottA and significantly higher IGC values than C5 (Fig. 5B). The exposure to SGF reduced the invasion of all three strains into epithelial cells but to different degrees depending on the strain, with ScottA being identified as the most invasive strain after SGF passage followed by C5 and PL25 (Fig. 5A). Due to the decrease also in the intracellular growth of ScottA and PL25, the three L. monocytogenes strains had similar IGC values after exposure to SGF (Fig. 5B). In total, the virulence potential of ScottA, following SGF exposure, was slightly but significantly (P < 0.05) higher than that of C5, which was more virulent than PL25 (Fig. 5C).
FIG 5.
Percent invasion (A), intracellular growth (IGC) (B), and percent total in vitro virulence (C) of L. monocytogenes strains C5, ScottA, and PL25 as determined using Caco-2 cells, after growth in TSB-Y at 10°C for 8 days without exposure to SGF (dark bars) or after growth in TSB-Y at 10°C for 8 days and subsequent exposure to SGF (pH 2.0, 37°C) for 30 min (light bars). Caco-2 cells were infected for 1 h with bacteria and incubated for 45 min (invasion) or 4 h (intracellular growth) with gentamicin. Total in vitro virulence was calculated as the percentage of initial bacteria recovered at the end of the assay. Data represent means ± standard errors of the means (SEM) of results of three biological replicates performed in triplicate. Asterisks (*) indicate significant differences between the results determined for a strain prior to and after exposure to SGF. Small letters indicate significant differences between strains prior to exposure to SGF. Capital letters indicate significant differences between strains after exposure to SGF (P < 0.05).
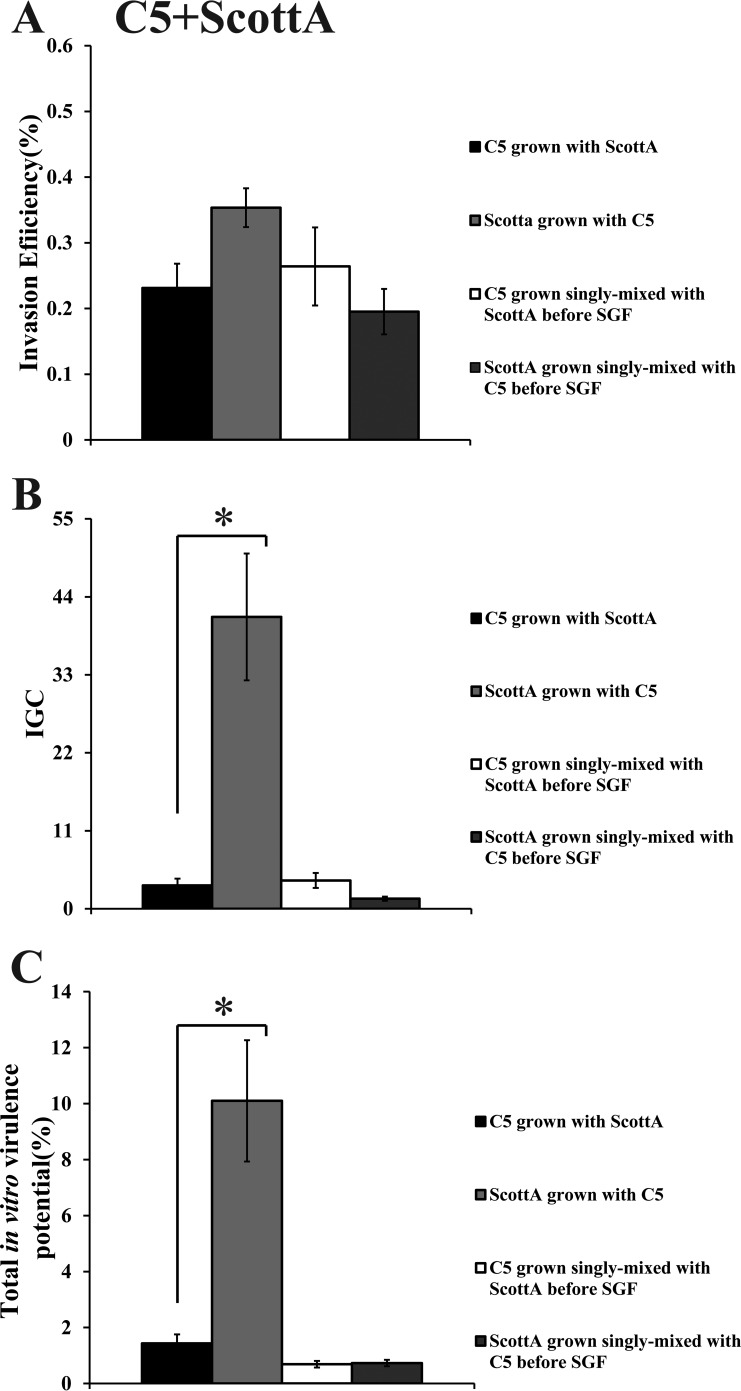
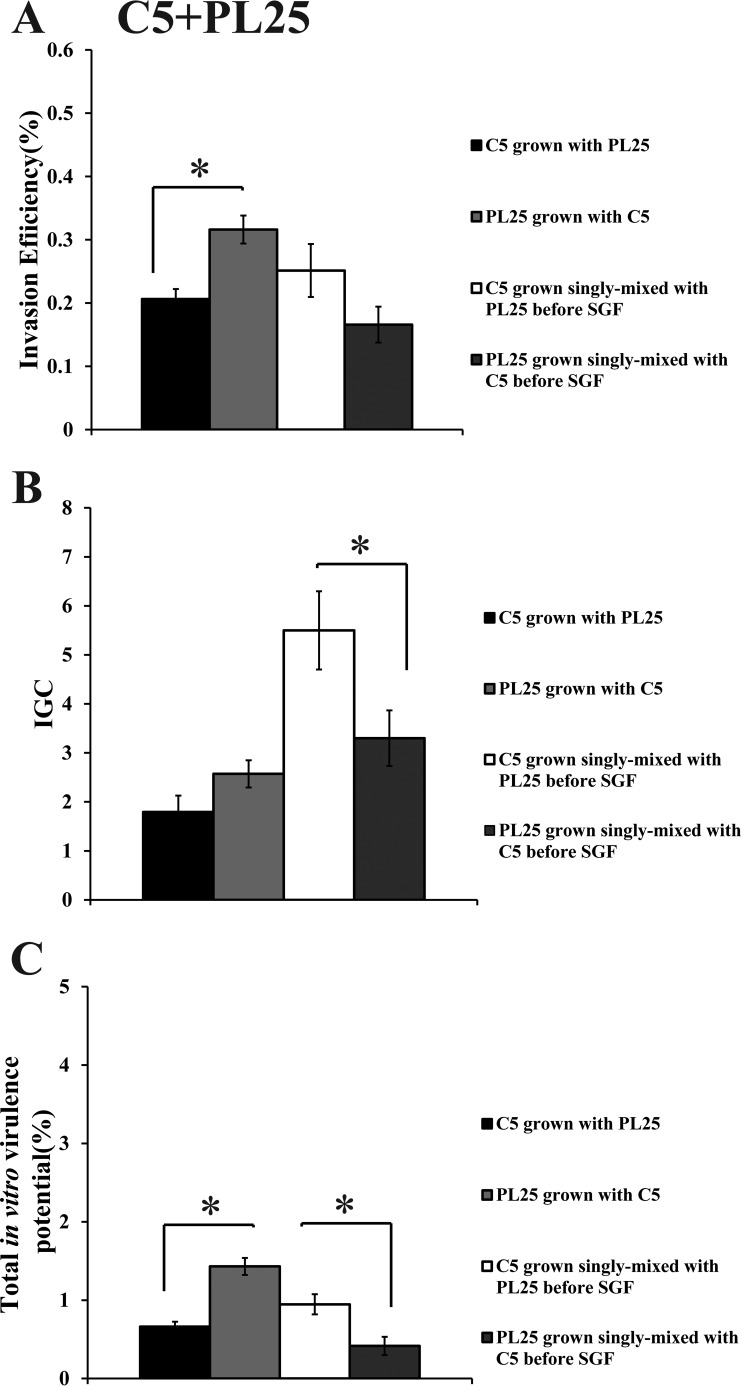
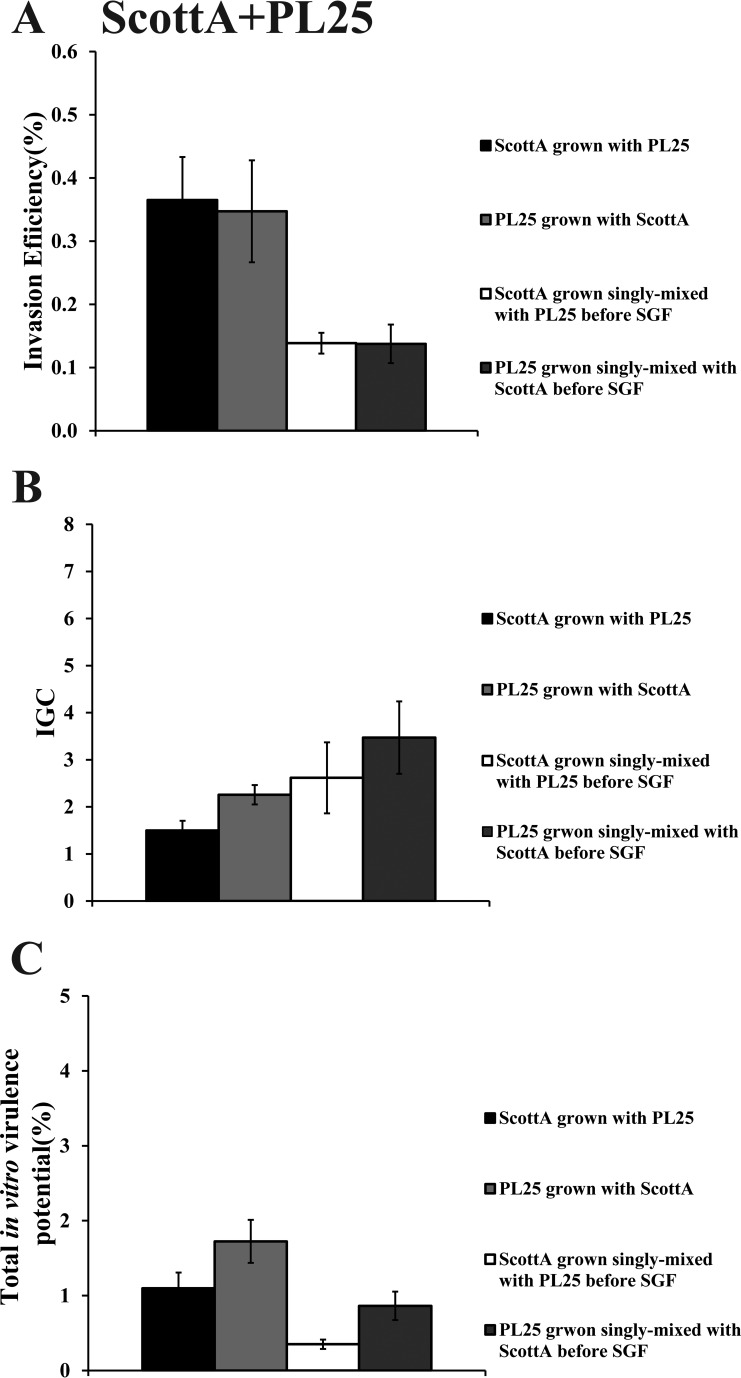
When C5 was cocultivated with ScottA, the two strains displayed comparable levels of invasion efficiency (Fig. 6A). However, the intracellular growth of ScottA was markedly increased in the presence of C5, resulting in a higher number of intracellular bacteria for ScottA after 4 h (Fig. 6B). The CFU of C5 at the end of the virulence assay corresponded only to 2% of the initial infecting population count compared to 10% of ScottA (Fig. 6C). Interestingly, when the two strains were combined before SGF exposure, they showed no differences in their in vitro virulence properties (Fig. 6). With regard to C5 and PL25, the invasion efficiency of PL25 (Fig. 7A) and the total number of CFU recovered from Caco-2 cells at the end of the assay (Fig. 7C) were higher than the levels seen with C5. In contrast, C5 was more efficient in multiplying in epithelial cells in the absence of previous coincubation with PL25 (Fig. 7B). As for ScottA and PL25, they managed to invade and proliferate in Caco-2 cells at similar levels (Fig. 8).
FIG 6.
Percent invasion (A), intracellular growth (IGC) (B), and percent total in vitro virulence (C) of L. monocytogenes strains C5 and ScottA as determined using Caco-2 cells after (i) cocultivation in TSB-Y at 10°C for 8 days and subsequent exposure to SGF (pH 2.0, 37°C) for 30 min or (ii) growth as a single culture in TSB-Y at 10°C for 8 days, mixing at equal volumes, and subsequent exposure to SGF (pH 2.0, 37°C) for 30 min. Caco-2 cells were infected for 1 h with bacteria and incubated for 45 min (invasion) or 4 h (intracellular growth) with gentamicin. Total in vitro virulence was calculated as the percentage of initial bacteria recovered at the end of the assay. Data represent means ± SEM of results of three biological replicates performed in triplicate. Asterisks (*) indicate significant differences between two strains in the same combination (P < 0.05).
FIG 7.
Percent invasion (A), intracellular growth (IGC) (B), and percent total in vitro virulence (C) of L. monocytogenes strains C5 and PL25 as determined using Caco-2 cells after (i) cocultivation in TSB-Y at 10°C for 8 days and subsequent exposure to SGF (pH 2.0, 37°C) for 30 min or (ii) growth as a single culture in TSB-Y at 10°C for 8 days, mixing at equal volumes, and subsequent exposure to SGF (pH 2.0, 37°C) for 30 min. Caco-2 cells were infected for 1 h with bacteria and incubated for 45 min (invasion) or 4 h (intracellular growth) with gentamicin. Total in vitro virulence was calculated as the percentage of initial bacteria recovered at the end of the assay. Data represent means ± SEM of results of three biological replicates performed in triplicate. Asterisks (*) indicate significant differences between two strains in the same combination (P < 0.05).
FIG 8.
Percent invasion (A), intracellular growth (IGC) (B), and percent total in vitro virulence (C) of L. monocytogenes strains ScottA and PL25 as determined using Caco-2 cells after (i) cocultivation in TSB-Y at 10°C for 8 days and subsequent exposure to SGF (pH 2.0, 37°C) for 30 min or (ii) growth as a single culture in TSB-Y at 10°C for 8 days, mixing at equal volumes, and subsequent exposure to SGF (pH 2.0, 37°C) for 30 min. Caco-2 cells were infected for 1 h with bacteria and incubated for 45 min (invasion) or 4 h (intracellular growth) with gentamicin. Total in vitro virulence was calculated as the percentage of initial bacteria recovered at the end of the assay. Data represent means ± SEM of results of three biological replicates performed in triplicate. Asterisks (*) indicate significant differences between two strains in the same combination (P < 0.05).
DISCUSSION
In two previous publications, we confirmed our hypothesis that growth, virulence, and enrichment competition may take place between L. monocytogenes strains (23, 24). In the present study, we used the knowledge obtained by our previous findings to investigate a particularly relevant topic: the potential failure of enrichment protocols to detect the L. monocytogenes strains responsible for listeriosis cases or outbreaks. We demonstrated that L. monocytogenes strains which were well-suited to coping with barriers relevant to GIT were sometimes underrepresented during selective enrichment.
The process of selective enrichment is considered biased since it relies upon the ability of a pathogen to counteract the adverse conditions induced by growth-inhibiting selective agents, food-related compounds, and competing food microflora (29–31). In a previous work (24) which also included the strains of this study, we observed preferential selection of certain L. monocytogenes strains after their coenrichment with the ISO protocol. At the beginning of enrichment, the initial populations of the strains were adjusted to the same level. In the present study, the populations of strains were developed naturally as a result of their coincubation in TSB-Y. This determined their initial levels before enrichment. Keys et al. (32) reported that high initial population differences between L. monocytogenes and L. innocua in enrichment broth restrict the presence of L. monocytogenes in the confluent layer of the streaked selective plate while enabling L. innocua to develop individual isolated colonies. Likewise, we observed that the strains which were outcompeted during growth (see also reference 23) were also underrecovered after enrichment. This suggests that if a product is contaminated with two different strains of L. monocytogenes, then strain competition during storage might result in the strain with the growth disadvantage being missed during enrichment. In fact, if population differences increase with storage time, then the likelihood of the outgrown strain being underdetected during enrichment also increases. In line with our previous findings (24), we showed that the ratios of two strains after the first enrichment step can change substantially following the second enrichment step. For instance, the probability that a strain would become totally undetectable might be higher after the second enrichment step. According to Gnanou Besse et al. (33), who proposed a 24-h reduction in the duration specified the ISO protocol, the latter scenario might be related to the production of inhibitory factors (e.g., phages or phage tails, namely, monocins) by competing strains over the last 24 h of the second enrichment cycle. LiCl, a principal selective agent present in Fraser broth and ALOA, has been reported to induce the production of such inhibitory compounds (34). Furthermore, poor recovery after the second enrichment step could be the result of the inability of the strain to remain viable throughout the whole duration of the procedure (33, 35).
The viability and competitive fitness of different L. monocytogenes strains contaminating the same sample are also crucial for food ingestion and the evolution of a possible infection. Investigating the effect of cocultivation on the survival of L. monocytogenes strains in SGF, we illustrated the fact that cocultivation has an indirect effect on the survival of strains in SGF through the following succession: strain competition determines the associations of strains during growth in TSB-Y (see also reference 23) and defines the population of each strain upon entry in the gastric fluid. As a result, despite the similar inactivation rates, the populations of two competing strains in SGF could be different at each time point due to differences in their initial cell density (e.g., see the case of C5 plus 6179). Previous studies have suggested that the inoculum size can affect bacterial inactivation kinetics with lower inocula, resulting in faster inactivation (28, 36). On the other hand, we showed that after 6 or 8 days of cocultivation in TSB-Y, the lower population levels of ScottA compared to C5 or PL25 populations did not lead to faster elimination of ScottA in SGF. Thus, despite being a weak competitor during growth in TSB-Y, this strain was an efficient survivor in SGF, which points out that some L. monocytogenes strains might be outgrown on foods due to competition but could be nonetheless be adept at passing the gastric barrier and reaching the small intestine.
After the exposure of L. monocytogenes to the primary physical stress of high acidity, crossing the barrier of intestinal epithelium signifies the entry of the pathogen in the host and triggers the early events of infection (37, 38). The intermediate passage of L. monocytogenes through the highly acidic (pH 2.0) SGF after incubation in TSB-Y and before infection of Caco-2 cells, as performed in our study, had a major influence on the virulence of L. monocytogenes by significantly reducing the virulence characteristics of L. monocytogenes strains. The encounter of L. monocytogenes with acidic environments is known to induce the transcription of virulence-associated genes (e.g., inlA, which mediates the entry of L. monocytogenes in epithelial cells, and prfA, a key regulator of L. monocytogenes virulence) regulated by the stress-responsive alternative sigma factor σΒ (39, 40). However, consistently with our results, there is also evidence for attenuated invasion of L. monocytogenes in Caco-2 cells or decreases in the levels of virulence-related genes after exposure to low pH (41, 42). In addition, despite the higher invasion efficiency of L. monocytogenes after adaptation to sublethal acid conditions, Garner et al. (43) have demonstrated that this elevated invasiveness was reduced to the levels seen prior to adaptation following exposure of L. monocytogenes to simulated gastric fluid. The cocultivation of strains followed by their passage through SGF seemed to affect the selection of efficient competitors during invasion and multiplication in Caco-2 cells. The probability of a strain dominating throughout the infection process was dependent on the individual virulence potential of each strain and was also associated with the combination of the strains. Previously (23), we suggested that cocultivation of strains might influence the transcription of virulence genes as demonstrated by Tan et al. (44), who investigated virulence gene expression of L. monocytogenes in the presence of Bifidobacterium longum. Furthermore, we hypothesized that competition between L. monocytogenes strains might take place upon the approach to host cells. This hypothesis seems to be supported by our present results, which showed that culturing of strains individually, but combining them prior to SGF exposure, could impact their competition in Caco-2 cells. As previously discussed, for probiotic bacteria capable of reducing the in vitro virulence of L. monocytogenes, physical blocking of adhesion and invasion sites on the surface of epithelial cells could explain the competitive advantage of a strain regarding invasion (45). Likewise, competition in the host cytoplasm might influence intracellular processes and contribute to the dominance of certain strains during infection.
Our findings do not suggest a link of the L. monocytogenes competitive advantage to strain serotype, sequence type, or strain origin. Strain C5, a serotype 4b dairy farm environmental isolate (ST2), was a strong growth competitor which managed to dominate on ALOA during mixed enrichments and displayed the highest recovery rate regardless of the competing strain. In contrast, C5 was outcompeted when confronted with gastrointestinal challenges. The second serotype 4b strain, the clinical isolate ScottA (ST290), which was a poor competitor during growth in TSB-Y and enrichment, performed remarkably well in gastric fluid and epithelial cells. PL25, a serotype 1/2b minced pork isolate (ST59), diminished the growth and detection of ScottA but could not efficiently compete against the latter strain in Caco-2 cells. This was reversed when PL25 was combined with C5. Finally, strain 6179 (ST121) was always outcompeted during growth and enrichment despite being a serotype 1/2a cheese isolate that persisted for over 8 years. This strain was not included in the assays performed with Caco-2 cells, but it harbors a truncated inlA, which would most likely result in attenuated virulence compared to that seen with competing strains, similarly to previous studies (23, 46). The limited number of tested strains and strain combinations in our study did not allow us to establish a generic pattern. In line with this statement, Gorski et al. (19) could not confirm that serotype 1/2a L. monocytogenes strains would be fitter than serotype 4b strains during enrichment performed with the FDA BAM protocol. Furthermore, there have been controversial results regarding the serotype- or origin-dependent survival of L. monocytogenes under acidic conditions (47–49). Also, there is no solid evidence available supporting a distinct link between virulence and origin or serotype of L. monocytogenes (50–52). Thus, in the absence of consistent trends, existing reports acknowledge the role of strain-to-strain variations and specificity regarding responses to stressful challenges (e.g., selective enrichment) and infectivity of L. monocytogenes (53, 54). Such interstrain variations might be the result of differences in the genome content of different L. monocytogenes strains. Previous studies have identified the presence of strain-specific virulence-associated genes in different L. monocytogenes strains (55) or of proteins potentially related to L. monocytogenes contact-dependent growth inhibition (46). Gene nucleotide polymorphisms, such as premature stop codons in inlA or prfA, which result in virulence attenuation, might also justify the hypothesis of a disadvantage of L. monocytogenes strains during virulence competition (54). As aforementioned, the production of monocins might confer a competitive advantage to the producing L. monocytogenes strains during selective enrichment. The monocin locus, a highly conserved cryptic prophage region that includes the lma operon, has been shown to play a role also in the intracellular growth of L. monocytogenes inside macrophages (56, 57), and the presence of a complete lma operon in a L. monocytogenes strain has been suggested to be involved in the finding that its virulence was higher than that seen with a strain harboring a truncated lma operon (58). Nevertheless, besides the interstrain genomic differences which might explain strain advantages or disadvantages under certain environments, the stimuli and conditions that trigger the expression of factors related to enrichment or virulence competition are also unknown and may well be subject to strain variations. In this study, the responses of ScottA might be an indication of reduced detectability of human isolates during selective enrichment but enhanced effectiveness with respect to outcompeting other strains during exposure to host barriers. The reported findings could serve as a basis for validation of our implications via further, more in-depth research involving a larger set of strains and focusing on the underlying mechanisms.
Conclusions.
Our results demonstrate that the occurrence of multiple L. monocytogenes strains in a single food sample can complicate downstream investigations and effective source attribution not only due to genetic and phenotypic diversity between strains but also due to their interactions. The succession of steps included in this study did not entirely simulate the passage of contaminated food through the GIT in vivo. L. monocytogenes faces various stresses before it reaches enterocytes, and such stresses affect the behavior of the pathogen. Future studies incorporating the simulation of additional compartments of the GIT and challenges encountered by L. monocytogenes strains in the protocol until infection of Caco-2 cells would strengthen our implications. Finally, potential in vivo experiments could allow us to accurately assess strain competition during infection.
Supplementary Material
ACKNOWLEDGMENTS
Strains 6179 and C5 were kindly provided by K. Jordan, Teagasc Food Research Centre, Moorepark, Fermoy, Cork, Ireland. Human colorectal epithelial adenocarcinoma Caco-2 cells were generously provided by A. Pintzas and G. Panayotou. Multilocus sequence typing of the strains used in the study was performed in collaboration with Kathrin Rychli according to the Institute Pasteur website (http://bigsdb.pasteur.fr/listeria/primers_used.html) in the Institute for Milk Hygiene, University of Veterinary Medicine, Vienna, Austria.
This study was supported by the 7th Framework Programme project PROMISE (project number 265877).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02120-16.
REFERENCES
- 1.Gracias KS, McKillip JL. 2004. A review of conventional detection and enumeration methods for pathogenic bacteria in food. Can J Microbiol 50:883–890. doi: 10.1139/w04-080. [DOI] [PubMed] [Google Scholar]
- 2.Pettengill JB, McAvoy E, White JR, Allard M, Brown E, Ottesen A. 2012. Using metagenomic analyses to estimate the consequences of enrichment bias for pathogen detection. BMC Res Notes 5:378. doi: 10.1186/1756-0500-5-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorski L. 2012. Selective enrichment media bias the types of Salmonella enterica strains isolated from mixed strain cultures and complex enrichment broths. PLoS One 7:e34722. doi: 10.1371/journal.pone.0034722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray MJ, Freitag NE, Boor KJ. 2006. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect Immun 74:2506–2512. doi: 10.1128/IAI.74.5.2505-2512.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes- from saprophyte to intracellular pathogen. Nat Rev Microbiol 7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandhi M, Chikindas ML. 2007. Listeria: a foodborne pathogen that knows how to survive. Int J Food Microbiol 113:1–15. doi: 10.1016/j.ijfoodmicro.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Gahan CGM, Hill C. 2005. Gastrointestinal phase of Listeria monocytogenes infection. J Appl Microbiol 98:1345–1353. doi: 10.1111/j.1365-2672.2005.02559.x. [DOI] [PubMed] [Google Scholar]
- 8.Gahan CGM, Hill C. 2014. Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front Cell Infect Microbiol 4:9. doi: 10.3389/fcimb.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dailey RC, Martin KG, Smiley RD. 2014. The effects of competition from non-pathogenic foodborne bacteria during the selective enrichment of Listeria monocytogenes using buffered Listeria enrichment broth. Food Microbiol 44:173–179. doi: 10.1016/j.fm.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Zeyara SA, Jarvis B, Mackey BM. 2011. The inhibitory effect of natural microflora of food on growth of Listeria monocytogenes in enrichment broths. Int J Food Microbiol 145:98–105. doi: 10.1016/j.ijfoodmicro.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 11.Dailey RC, Welch LJ, Hitchins AD, Smiley RD. 2015. Effect of Listeria seeligeri or Listeria welshimeri on Listeria monocytogenes detection in and recovery from buffered Listeria enrichment broth. Food Microbiol 46:528–534. doi: 10.1016/j.fm.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Besse NG, Barre L, Buhariwalla C, Vignaud ML, Khamissi E, Decourseulles E, Nirsimloo M, Chelly M, Kalmokoff M. 2010. The overgrowth of Listeria monocytogenes by other Listeria spp. in food samples undergoing enrichment cultivation has a nutritional basis. Int J Food Microbiol 136:345–351. doi: 10.1016/j.ijfoodmicro.2009.10.025. [DOI] [PubMed] [Google Scholar]
- 13.Curiale MS, Lewus C. 1994. Detection of Listeria monocytogenes in samples containing Listeria innocua. J Food Prot 57:1048–1051. [DOI] [PubMed] [Google Scholar]
- 14.Carvalheira A, Eusébio C, Silva J, Gibbs P, Teixeira P. 2010. Influence of Listeria innocua on the growth of Listeria monocytogenes. Food Control 21:1492–1496. doi: 10.1016/j.foodcont.2010.04.021. [DOI] [Google Scholar]
- 15.Zitz U, Zunabovic M, Domig KJ, Wilrich P-T, Kneifel W. 2011. Reduced detectability of Listeria monocytogenes in the presence of Listeria innocua. J Food Prot 74:1282–1287. doi: 10.4315/0362-028X.JFP-11-045. [DOI] [PubMed] [Google Scholar]
- 16.Petran RL, Swanson KMJ. 1993. Simultaneous growth of Listeria monocytogenes and Listeria innocua. J Food Prot 56:616–618. [DOI] [PubMed] [Google Scholar]
- 17.Engelhardt T, Ágoston R, Belák Á, Mohácsi-Farkas C, Kiskó G. 2016. The suitability of the ISO 11290-1 method for the detection of Listeria monocytogenes. LWT Food Sci Technol 71:213–220. doi: 10.1016/j.lwt.2016.03.038. [DOI] [Google Scholar]
- 18.Bruhn JB, Vogel BF, Gram L. 2005. Bias in the Listeria monocytogenes enrichment procedure: lineage 2 strains outcompete lineage 1 strains in University of Vermont selective enrichments. Appl Environ Microbiol 71:961–967. doi: 10.1128/AEM.71.2.961-967.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorski L, Flaherty D, Mandrell RE. 2006. Competitive fitness of Listeria monocytogenes serotype 1/2a and 4b strains in mixed cultures with and without food in the U.S. Food and Drug Administration enrichment protocol. Appl Environ Microbiol 72:776–783. doi: 10.1128/AEM.72.1.776-783.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danielsson-Tham M-L, Bille J, Brosch R, Buchrieser C, Persson K, Rocourt J, Schwarzkopf A, Tham W, Ursing J. 1993. Characterization of Listeria strains isolated from soft cheese. Int J Food Microbiol 18:161–166. doi: 10.1016/0168-1605(93)90220-B. [DOI] [PubMed] [Google Scholar]
- 21.Tham W, Aldén J, Ericsson H, Helmersson S, Malmodin B, Nyberg O, Pettersson A, Unnerstad H, Danielsson-Tham ML. 2002. A listeriosis patient infected with two different Listeria monocytogenes strains. Epidemiol Infect 128:105–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kathariou S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J Food Prot 65:1811–1829. [DOI] [PubMed] [Google Scholar]
- 23.Zilelidou EA, Rychli K, Manthou E, Ciolacu L, Wagner M, Skandamis PN. 2015. Highly invasive Listeria monocytogenes strains have growth and invasion advantages in strain competition. PLoS One 10:e0141617. doi: 10.1371/journal.pone.0141617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zilelidou E, Manthou E, Skandamis P. 2016. Growth differences and competition between Listeria monocytogenes strains determine their predominance on ham slices and lead to bias during selective enrichment with the ISO protocol. Int J Food Microbiol 235:60–70. doi: 10.1016/j.ijfoodmicro.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Blackburn WC, Davies AR. 1994. Development of antibiotic-resistant strains for the enumeration of foodborne pathogenic bacteria in stored foods. Int J Food Microbiol 24:125–136. doi: 10.1016/0168-1605(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 26.International Organization for Standardization (ISO). 2004. Microbiology of food and animal feeding stuffs—horizontal method for the detection and enumeration of Listeria monocytogenes. Part 1: detection method. ISO standard 11290-1:1996 and Amd.1:2004. ISO, Geneva, Switzerland. [Google Scholar]
- 27.Commission Regulation. 2005. Commission regulation no. 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. Off J Eur Union 338:1–26. [Google Scholar]
- 28.Barmpalia-Davis IM, Geornaras I, Kendall PA, Sofos JN. 2008. Survival of Listeria monocytogenes in a simulated dynamic gastrointestinal model during storage of inoculated bologna and salami slices in vacuum packages. J Food Prot 71:2014–2023. [DOI] [PubMed] [Google Scholar]
- 29.Donnelly CW. 2002. Detection and isolation of Listeria monocytogenes from food samples: implications of sublethal injury. J AOAC Int 85:495–500. [PubMed] [Google Scholar]
- 30.in't Veld PH, Notermans SHW, van de Berg M. 1995. Potential use of microbiological reference materials for the evaluation of detection methods for Listeria monocytogenes and the effect of competitors: a collaborative study. Food Microbiol 12:125–134. doi: 10.1016/S0740-0020(95)80088-3. [DOI] [Google Scholar]
- 31.Gasanov U, Hughes D, Hansbro PM. 2005. Methods for the isolation and identification of Listeria spp. and Listeria monocytogenes: a review. FEMS Microbiol Rev 29:851–875. doi: 10.1016/j.femsre.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Keys AL, Dailey RC, Hitchins AD, Smiley RD. 2013. Postenrichment population differentials using buffered Listeria enrichment broth: implications of the presence of Listeria innocua on Listeria monocytogenes in food test samples. J Food Prot 76:1854–1862. doi: 10.4315/0362-028X.JFP-13-089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gnanou Besse N, Favret S, Desreumaux J, Decourseulles Brasseur E, Kalmokoff M. 2016. Evaluation of reduction of Fraser incubation by 24h in the EN ISO 11290-1 standard on detection and diversity of Listeria species. Int J Food Microbiol 224:16–21. doi: 10.1016/j.ijfoodmicro.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 34.Lemaître JP, Duroux A, Pimpie R, Duez JM, Milat ML. 2015. Listeria phage and phage tail induction triggered by components of bacterial growth media (phosphate, LiCl, nalidixic acid, and acriflavine). Appl Environ Microbiol 81:2117–2124. doi: 10.1128/AEM.03235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gnanou Besse N, Audinet N, Kérouanton A, Colin P, Kalmokoff M. 2005. Evolution of Listeria populations in food samples undergoing enrichment culturing. Int J Food Microbiol 104:123–134. doi: 10.1016/j.ijfoodmicro.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 36.Johnston MD, Simons EA, Lambert RJW. 2000. One explanation for the variability of the bacterial suspension test. J Appl Microbiol 88:237–242. doi: 10.1046/j.1365-2672.2000.00951.x. [DOI] [PubMed] [Google Scholar]
- 37.Vázquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Domínguez-Bernal G, Goebel W, González-Zorn B, Wehland J, Kreft J. 2001. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ireton K. 2007. Entry of the bacterial pathogen Listeria monocytogenes into mammalian cells. Cell Microbiol 9:1365–1375. doi: 10.1111/j.1462-5822.2007.00933.x. [DOI] [PubMed] [Google Scholar]
- 39.Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J Bacteriol 185:5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nadon CA, Bowen BM, Wiedmann M, Boor KJ. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect Immun 70:3948–3952. doi: 10.1128/IAI.70.7.3948-3952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang L, Olesen I, Andersen T, Fang W, Jespersen L. 2010. Survival of Listeria monocytogenes in simulated gastrointestinal system and transcriptional profiling of stress- and adhesion-related genes. Foodborne Pathog Dis 7:267–274. doi: 10.1089/fpd.2009.0361. [DOI] [PubMed] [Google Scholar]
- 42.Olesen I, Vogensen FK, Jespersen L. 2009. Gene transcription and virulence potential of Listeria monocytogenes strains after exposure to acidic and NaCl stress. Foodborne Pathog Dis 6:669–680. doi: 10.1089/fpd.2008.0243. [DOI] [PubMed] [Google Scholar]
- 43.Garner MR, James KE, Callahan MC, Wiedmann M, Boor KJ. 2006. Exposure to salt and organic acids increases the ability of Listeria monocytogenes to invade Caco-2 cells but decreases its ability to survive gastric stress. Appl Environ Microbiol 72:5384–5395. doi: 10.1128/AEM.00764-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan Q, Xu H, Chen T, Li P, Aguilar ZP, Xu D, Ming X, Xu F, Wei H. 2012. Differential expression of virulence and stress fitness genes during interaction between Listeria monocytogenes and Bifidobacterium longum. Biosci Biotechnol Biochem 76:699–704. doi: 10.1271/bbb.110832. [DOI] [PubMed] [Google Scholar]
- 45.Moroni O, Kheadr E, Boutin Y, Lacroix C, Fliss I. 2006. Inactivation of adhesion and invasion of food-borne Listeria monocytogenes by bacteriocin-producing Bifidobacterium strains of human origin. Appl Environ Microbiol 72:6894–6901. doi: 10.1128/AEM.00928-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitz-Esser S, Müller A, Stessl B, Wagner M. 2015. Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front Microbiol 6:380. doi: 10.3389/fmicb.2015.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramalheira R, Almeida M, Azeredo J, Brandão TRS, Almeida G, Silva J, Teixeira P. 2010. Survival of clinical and food isolates of Listeria monocytogenes through simulated gastrointestinal tract conditions. Foodborne Pathog Dis 7:121–128. doi: 10.1089/fpd.2009.0319. [DOI] [PubMed] [Google Scholar]
- 48.Barmpalia-Davis IM, Geornaras I, Kendall PA, Sofos JN. 2008. Differences in survival among 13 Listeria monocytogenes strains in a dynamic model of the stomach and small intestine. Appl Environ Microbiol 74:5563–5567. doi: 10.1128/AEM.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werbrouck H, Botteldoorn N, Ceelen L, Decostere A, Uyttendaele M, Herman L, Van Coillie E. 2008. Characterization of virulence properties of Listeria monocytogenes serotype 4b strains of different origins. Zoonoses Public Health 55:242–248. doi: 10.1111/j.1863-2378.2008.01127.x. [DOI] [PubMed] [Google Scholar]
- 50.Gray MJ, Zadoks RN, Fortes ED, Dogan B, Cai S, Chen Y, Scott VN, Gombas DE, Boor KJ, Wiedmann M. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl Environ Microbiol 70:5833–5841. doi: 10.1128/AEM.70.10.5833-5841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barbour AH, Rampling A, Hormaeche CE. 2001. Variation in the infectivity of Listeria monocytogenes isolates following intragastric inoculation of mice. Infect Immun 69:4657–4660. doi: 10.1128/IAI.69.7.4657-4660.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werbrouck H, Grijspeerdt K, Botteldoorn N, Van Pamel E, Rijpens N, Van Damme J, Uyttendaele M, Herman L, Van Coillie E. 2006. Differential inlA and inlB expression and interaction with human intestinal and liver cells by Listeria monocytogenes strains of different origins. Appl Environ Microbiol 72:3862–3871. doi: 10.1128/AEM.02164-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lianou A, Koutsoumanis KP. 2013. Strain variability of the behavior of foodborne bacterial pathogens: a review. Int J Food Microbiol 167:310–321. doi: 10.1016/j.ijfoodmicro.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 54.Orsi RH, den Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 55.Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, Rasko DA, Angiuoli SV, Gill SR, Paulsen IT, Peterson J, White O, Nelson WC, Nierman W, Beanan MJ, Brinkac LM, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Haft DH, Selengut J, Van Aken S, Khouri H, Fedorova N, Forberger H, Tran B, Kathariou S, Wonderling LD, Uhlich GA, Bayles DO, Luchansky JB, Fraser CM. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res 32:2386–2395. doi: 10.1093/nar/gkh562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klumpp J, Loessner MJ. 2013. Listeria phages: genomes, evolution, and application. Bacteriophage 3:e26861. doi: 10.4161/bact.26861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hain T, Ghai R, Billion A, Kuenne C, Steinweg C, Izar B, Mohamed W, Mraheil M, Domann E, Schaffrath S, Kärst U, Goesmann A, Oehm S, Pühler A, Merkl R, Vorwerk S, Glaser P, Garrido P, Rusniok C, Buchrieser C, Goebel W, Chakraborty T. 2012. Comparative genomics and transcriptomics of lineages I, II, and III strains of Listeria monocytogenes. BMC Genomics 13:144. doi: 10.1186/1471-2164-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rychli K, Müller A, Zaiser A, Schoder D, Allerberger F, Wagner M, Schmitz-Esser S. 2014. Genome sequencing of Listeria monocytogenes “Quargel” listeriosis outbreak strains reveals two different strains with distinct in vitro virulence potential. PLoS One 9:e89964. doi: 10.1371/journal.pone.0089964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.