ABSTRACT
Pseudomonas brassicacearum DF41 is a biocontrol agent that suppresses disease caused by the fungal pathogen Sclerotinia sclerotiorum. A number of exometabolites are produced by DF41, including the lipopeptide sclerosin, hydrogen cyanide (HCN), and degradative enzymes. The production of these compounds is controlled at both the transcriptional and posttranscriptional levels by quorum sensing (QS) and the Gac two-component regulatory system. In order to be successful, a biocontrol agent must persist in the environment at levels sufficient for pathogen control. Bacterivorous predators, including nematodes, represent a challenge to the establishment of introduced microorganisms. In the current study, DF41 was investigated for its ability to resist predation by Caenorhabditis elegans. We discovered that this bacterium is capable of killing C. elegans through two different mechanisms: the first involves exposure to toxic metabolites, and the second entails biofilm formation on the nematode head blocking the buccal cavity. Biofilm formation on nematodes, which has been reported only for Yersinia spp. and Xenorhabdus nematophila, is dependent upon the Gac system. Biofilms were not observed when bacteria were grown on NaCl-containing medium or on C. elegans biofilm-resistant mutants. Coculturing with nematodes led to the increased expression of the pdfRI-rfiA QS genes and hcnA, which is under QS control. HCN was the most nematicidal of the exometabolites, suggesting that this bacterium can respond to predator cues and upregulate expression of toxins accordingly. In summary, DF41 is able to respond to the presence of C. elegans, and through two distinct mechanisms, it can escape predation.
IMPORTANCE Pseudomonas brassicacearum DF41 can suppress fungal pathogens through a process known as biocontrol. To be successful, a biocontrol agent must be able to persist in the environment at levels sufficient for pathogen control. Predators, including the nematode Caenorhabditis elegans, represent a threat to persistence. The aim of the current study was to investigate the DF41-C. elegans interaction. We discovered that DF41 is able to escape predation through two distinct mechanisms. The first involves exposure to toxic bacterial metabolites, and the second entails the formation of a sticky coating on the nematode head, called a biofilm, which blocks feeding and causes starvation. We report here a pseudomonad forming biofilms on the C. elegans surface. When grown with C. elegans, DF41 exhibits altered gene expression and metabolite production, indicating that this bacterium can sense the presence of these predators and adjust its physiology accordingly.
INTRODUCTION
The production of extracellular metabolites by biocontrol bacteria is energetically costly; as such, these compounds are expected to impart a fitness advantage to the producer. It can be argued that reduced competition for resources is not sufficient to warrant the synthesis of inhibitory compounds. Rather, these products must provide additional advantages, such as reducing the threat of grazing predators through their repellent and/or cidal activities (1, 2). Nematodes are among the most abundant animals on the planet, and through their grazing pressure, they are believed to influence microbial community structure (3). Bacteria, in turn, have evolved defensive mechanisms to resist nematode grazing, including the production of exometabolites that act to deter and/or reduce predator populations. The model organism Caenorhabditis elegans has been widely employed in bacterium-nematode interaction studies because of its genetic tractability and the comprehensive array of mutants available (4). For pseudomonads that exhibit pathogenicity toward C. elegans, killing ensues through two distinct mechanisms. On rich media, which support the production of high levels of toxic metabolites, death occurs through intoxication, a mechanism known as fast killing (5–8). Conversely, low-nutrient media result in the production of sublethal levels of exometabolites, and slow killing occurs over the course of several days. In this instance, bacterial colonization of the intestinal tract ultimately leads to nematode death (8).
In nature, the bulk of bacterial biomass exists as an adherent community of cells encased in an extrapolymeric matrix, collectively known as a biofilm. The ability to form biofilms has many advantages, including protection from environmental assaults that would threaten planktonic cells (9). Biofilms exhibit elevated resistance to biocidal agents, immune components, desiccation, and UV radiation (9, 10). For a select group of bacteria, the ability to form biofilms allows them to escape predation by grazing nematodes. In 2002, Darby and coworkers reported that Yersinia pestis is able to form biofilms on the head and body of C. elegans, blocking feeding and ultimately causing starvation (11). Similarly, Xenorhabdus nematophila (12) and some but not all strains of Yersinia pseudotuberculosis (13) are capable of establishing nematode-associated biofilms. Screening of an array of 26 pathogenic bacteria revealed that C. elegans surface colonization is not a common trait (14, 15). Even the notorious infection-related biofilm formers Pseudomonas aeruginosa and uropathogenic Escherichia coli were found to lack this ability, leading to the conclusion that this defense strategy is employed by only a few bacterial species (15).
Pseudomonas brassicacearum strain DF41 is a biocontrol agent capable of suppressing disease caused by the plant-pathogenic fungus Sclerotinia sclerotiorum (Lib.) de Bary in both greenhouse and field studies (16, 17). This bacterium produces an arsenal of extracellular metabolites, including degradative enzymes, hydrogen cyanide (HCN), and a novel lipopeptide called sclerosin (17, 18). Sclerosin has been confirmed to be essential for biocontrol, as a sclerosin-deficient mutant, DF41-1278, exhibits dramatically reduced fungal suppression (17). A complex regulatory cascade oversees the production of DF41 exometabolites, including the GacS/GacA two-component system, the stationary-phase sigma factor RpoS, and the stringent response (17, 19). In addition, DF41 has a quorum-sensing (QS) system composed of a LuxR-type transcriptional activator (PdfR) and an acyl-homoserine lactone (AHL) synthase (PdfI), encoded by pdfR and pdfI, respectively (20). Immediately downstream of pdfI lies a third gene in the QS locus, called rfiA. RfiA belongs to a distinct group of LuxR regulators characterized as having a C-terminal helix-turn-helix DNA binding motif but lacking an N-terminal AHL binding domain (20, 21). AHL signaling molecules are not involved in DF41 biocontrol, because an AHL-deficient strain exhibits no discernible phenotype (20). RfiA, on the other hand, was found to be essential, as an rfiA mutation results in a complete loss of antifungal activity (20). RfiA is believed to control expression of the PdfABC efflux pump involved in exometabolite export. In the ΔrfiA mutant background, metabolites that are normally excreted accumulate to elevated levels within the mutant cells (20).
The aim of the current study was to elucidate the interaction between DF41 and the bacterivorous predator C. elegans. Specifically, we were interested to learn whether DF41 is able to resist nematode grazing and what role, if any, DF41 exoproducts play in the bacterium-nematode interaction. We discovered that DF41 is capable of killing C. elegans through two distinct modes; the first involves exposure to toxic metabolites, while the second entails biofilm formation on the nematode surface. It has been reported that bacteria are capable of responding to predator cues through altered expression of toxin-encoding genes (7, 22, 23). To see if the same holds true for DF41, we cocultured bacteria with C. elegans and monitored the expression of genes associated with exometabolite production. In the presence of C. elegans, several genes were upregulated, including hcnA (hydrogen cyanide) and the QS genes pdfRI and rfiA. Our findings indicate that through soluble chemical cues and/or direct contact, DF41 is able to perceive the presence of C. elegans and adjust its physiology accordingly.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli was cultured at 37°C on lysogeny broth (LB) agar (Difco Laboratories, Detroit, MI). Pseudomonas strains were routinely cultured on LB, King's B (KB) (24), or in M9 minimal salts medium supplemented with 0.2% glucose (M9-Glc) at 28°C, unless otherwise indicated. The antibiotics used were 100 μg/ml ampicillin (Amp), 15 μg/ml gentamicin (Gm), and 15 μg/ml tetracycline (Tc) for E. coli, and 20 μg/ml Gm, 40 μg/ml piperacillin (Pip), and 15 μg/ml Tc for DF41. All antibiotics were obtained from Research Products International Corp. (Mt. Prospect, IL).
TABLE 1.
Bacterial strains, plasmids, and primers used in the study
| Strain, plasmid, or primer | Relevant genotype, phenotype, or sequencea | Reference or source |
|---|---|---|
| Strains | ||
| C. elegans | ||
| N2 | Wild-type isolate | CGC, University of Minnesota, MN |
| AT6 | srf-2(yj262) | CGC, University of Minnesota, MN |
| DC9 | bah-3(br9) | CGC, University of Minnesota, MN |
| CB4856 | Wild-type Hawaiian isolate | CGC, University of Minnesota, MN |
| P. brassicacearum | ||
| DF41 | Rifr wild type (canola root tip isolate) | 16 |
| DF41-1278 | Rifr lp::Tn5-1063 genomic insertion | 17 |
| DF41hcn | DF41 with the pKNOCK-Tc vector inserted into the hcn gene | This study |
| AI-deficient mutant | DF41 carrying pME6863 | 20 |
| DF41rfiA | DF41 with Gmr cassette inserted into rfiA gene | 20 |
| DF41gacS | Rifr gacS::Tn5-1063 genomic insertion | 17 |
| DF41-rfp | DF41 containing mCherry expressed from pMCh-23 | 18 |
| DF41-1278-rfp | DF41-1278 containing mCherry expressed from pMCh-23 | 18 |
| DF41hcn-rfp | DF41hcn containing mCherry expressed from pMCh-23 | This study |
| AI-deficient-rfp mutant | DF41-6863 containing mCherry expressed from pMCh-23 | This study |
| DF41rfiA-rfp | DF41rfiA containing mCherry expressed from pMCh-23 | This study |
| DF41gacS-rfp | PA23gacS containing mCherry expressed from pMCh-23 | This study |
| E. coli | ||
| DH5α | supE44 ΔU169 (Φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Gibco |
| DH5α λpir | λpir lysogen of DH5α | 46 |
| OP50 | Laboratory strain for maintenance of C. elegans; uracil auxotroph | 25 |
| Chromobacterium violaceum CVO26 | Autoinducer synthase (cviI) mutant from C. violaceum ATCC 31532 autoinducer biosensor | 29 |
| Plasmids | ||
| pME6863 | pME6000 carrying the aiiA gene from Bacillus sp. A24 under the constitutive Plac promoter | 47 |
| pCR2.1 | TA cloning vector, Ampr | Invitrogen |
| pKNOCK-Tc | Suicide vector for insertional mutagenesis; R6K ori Rp4 oriT Tcr | 48 |
| pRK600 | Contains tra genes for mobilization, Chlr | 49 |
| pCRhcnABC | 1.6-kb hcnABC fragment in pCR2.1 | This study |
| pKNOCK-hcnABC | 1.6-kb fragment from hcnABC in pKNOCK-Tc | This study |
| pUCP23 | Broad-host-range vector, Ampr Gmr | 50 |
| pRSET-B mCherry | mCherry expression vector, f1 ori, Ampr | 51 |
| pMCh-23 | pUCP23 carrying the mCherry red fluorescent protein gene | 18 |
| Primers | ||
| hcnA-FOR | 5′-ATGGGCGTATGCCACTGC-3′ | This study |
| hcnC-REV | 5′-TAAGCACACGACGCGCCG-3′ | This study |
| sclerosin-FOR | 5′-CCACAAACGGCATTTGCTGG-3′ | This study |
| sclerosin-REV | 5′-AGTTTGCTAAGGACCGCTGC-3′ | This study |
| hcnC-FOR | 5′-TACGTGGCGCAGAAAGACAACG-3′ | This study |
| hcnC-REV | 5′-TTGCACCAACCCTTCGATCTCG-3′ | This study |
| pdfR-FOR | 5′-AGCATCATCGCCAACCAACACC-3′ | This study |
| pdfR-REV | 5′-GTTTTTTCCCAGTGCGAGCCAG-3′ | This study |
| pdfI-FOR | 5′-ACCGTTGACAGACGCAATATCG-3′ | This study |
| pdfI-REV | 5′-AGCGTTCTTGCTAAGGACCTCC-3′ | This study |
| rfiA-FOR | 5′-GCACCTGAACTTGCCGAACAAC-3′ | This study |
| rfiA-REV | 5′-GCATCCATCGGATAAGCGAACG-3′ | This study |
| gacS-FOR | 5′-TGGTGCAAACCCTGCTCGAAG-3′ | This study |
| gacS-REV | 5′-TCTGCACGTCCATCAACACCAG-3′ | This study |
Rifr, rifampin resistance; Tc, tetracycline; Gmr, gentamicin resistance; Ampr, ampicillin resistance; Chlr, chloramphenicol resistance; Carb, carbenicillin.
Nematode strains and culture conditions.
The C. elegans strains used in this work include wild-type Bristol N2, AT6 [srf-2(yj262)], DC9 [bah-3(br9)], and CB4856, which were maintained at 15°C on nematode growth medium (NGM) (25) supplemented with E. coli OP50. The protocols available in WormBook were used in order to obtain the synchronous cultures (26). L4-stage hermaphrodites were used in the studies described herein.
Nucleic acid manipulation.
Cloning, purification, electrophoresis, and other manipulations of DNA were performed using standard techniques (27).
Construction of a DF41 hcn mutant.
To generate an hcn mutant, an internal region of the DF41 hcn gene cluster was PCR amplified using primers hcnA-FOR1 and hcnC-REV1 (Table 1). A TOPO kit (Thermo Fisher Scientific, Waltham, MA, USA) was employed to clone the 1.6-kb PCR product into the pCR2.1-TOPO vector, creating pCRhcnABC-41. To liberate the insert, pCRhcnABC-41 was digested with HindIII and XhoI and subcloned into the same sites of the suicide vector pKNOCK-Tc. Triparental mating between the donor [E. coli DH5α λpir(pKNOCKhcnABC)], helper [E. coli DH5α(pRK600)], and recipient (DF41) strains was performed to interrupt the wild-type hcnABC gene cluster as a single crossover insertion selected using Tc. PCR was used to confirm the hcn mutation, and the mutant was found to be devoid of HCN production when tested using Cyantesmo paper (Macherey-Nagel GmbH and Co., Germany).
Nematode slow- and fast-killing assays.
Slow-killing assays were performed by spotting 10 μl of a 1:10 dilution of an overnight bacterial culture grown in KB, LB, or NGM onto a 35 by 10-mm agar plate of the same medium. After 24 h of incubation at 28°C, the plates were cooled to room temperature and seeded with 30 L4-stage nematodes. The plates were incubated at 20°C, and the nematodes were scored for viability by examining them with a stereomicroscope over a 10-day period. Nematodes without detectable movement were considered dead after confirmatory prodding with a nematode pick. Three replicates were included for each trial, and the assays were repeated three times independently. Fast-killing assays were carried out in a similar manner, except that brain heart infusion (BHI) agar (Difco) with and without FeCl3 (100 μM) was used as the medium, and nematodes were monitored at 4-, 6-, 8-, 10-, 12-, and 24-h time points.
Microscopic imaging of Caenorhabditis elegans.
Bacterial strains harboring pMCh-23, which encodes constitutively expressed mCherry fluorescent protein, were spotted onto agar plates and incubated overnight at 28°C. Plates were cooled to room temperature prior to seeding with nematodes, followed by incubation at 20°C. Nematodes were mounted onto 2% agarose pads on glass microscope slides and anesthetized with 10 mmol/liter levamisole (Sigma) in M9 medium. The nematodes were examined with a Zeiss LSM 700 confocal laser scanning microscope (Carl Zeiss Microscopy GmbH, Göttingen, Germany).
Biofilm formation.
To analyze the ability of DF41 and derivative strains to form biofilms, a 96-well plate assay was employed, as detailed in a study by Berry et al. (17).
Analysis of metabolite and regulatory gene expression in DF41 in the presence and absence of Caenorhabditis elegans.
Gene expression was monitored in DF41 grown in the presence and absence of C. elegans. Approximately 200 nematodes were added to a 3-ml volume of DF41 culture (optical density at 600 nm [OD600], 0.1; M9-Glc), which was incubated at room temperature for 72 h with shaking. A 1-ml volume of culture was spun for 1 min at 1,000 × g to pellet the nematodes, after which 400 μl of the bacterial cell suspension was added to 800 μl of RNAprotect (Qiagen, Valencia, CA, USA). Total RNA was extracted using the RNeasy minikit (Qiagen), and residual genomic DNA was removed by treatment with Turbo RNase-free DNase I (Ambion, Carlsbad, CA, USA). cDNA was generated by reverse transcription using the Maxima first-strand cDNA synthesis kit (Thermo Scientific, Rockford, IL, USA) under the following conditions: initial heating at 25°C for 10 min, 50°C for 15 min for reverse transcription, and 85°C for 5 min for enzyme denaturation. The primer sequences for the genes of interest are listed in Table 1. Reverse transcription-quantitative PCR (qRT-PCR) was performed using a CFX Connect real-time system (Bio-Rad, Ontario, Canada) and SsoFast EvaGreen Supermix (Bio-Rad). The PCR conditions included an initial denaturation at 98°C for 2 min, followed by 40 cycles of 98°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Reactions were performed in triplicate, and experiments were repeated three times with four biological replicates. Relative gene expression was determined using the comparative threshold cycle (CT) method, as described by Livak and Schmittgen (28).
Analysis of HCN and AHL signal production.
DF41 was grown in the presence and absence of C. elegans for 72 h at room temperature on M9-Glc agar plates sealed with Parafilm. HCN production was monitored using Cyantesmo paper, which turns blue in the presence of this volatile compound. The production of AHL molecules was assessed qualitatively by spotting 5 μl of culture grown for 72 h in the presence and absence of C. elegans onto plates seeded with Chromobacterium violaceum CVO26. This bacterial strain is able to produce the purple pigment violacein only in the presence of exogenous autoinducer (29). Samples were analyzed in triplicate, and the experiments were repeated twice.
Statistical analysis.
An unpaired Student t test was used for statistical analysis of biofilm formation, qRT-PCR gene expression, and AHL production. The log rank (Mantel-Cox) test was applied for statistical analysis of pairwise comparisons in the fast and slow nematode killing assays. Nematodes that died from crawling off the plate were excluded from statistical analysis.
RESULTS
Pseudomonas brassicacearum DF41 kills Caenorhabditis elegans through production of toxic metabolites.
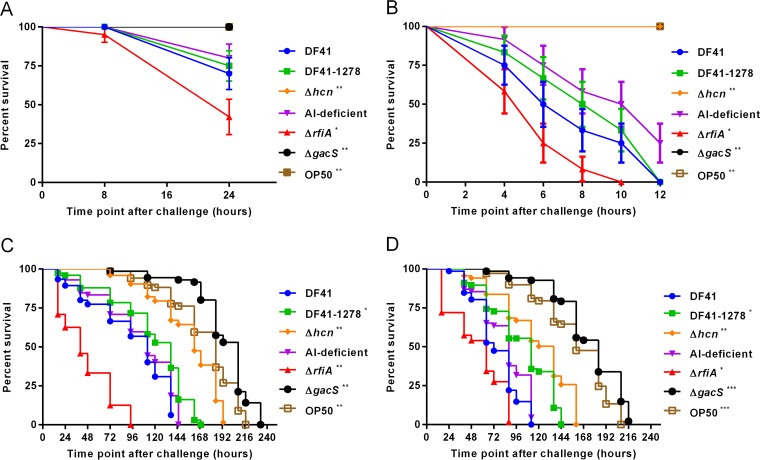
When DF41 and derivative strains, including DF41-1278, DF41hcn, autoinducer (AI)-deficient DF41, DF41rfiA, and DF41gacS, were grown on BHI, which supports fast killing by pseudomonads (7, 8), differences in lethality were observed; however, none elicited 100% nematode killing at 24 h (Fig. 1A). The lowest level of toxicity was exhibited by the gacS and hcn mutants. The sclerosin-deficient strain, DF41-1278, was similar to wild type, indicating that this lipopeptide (LP) does not contribute to fast killing. Similarly, the autoinducer (AI)-deficient DF41 strain (pME6863) exhibited no change in lethality. The most toxic strain was the ΔrfiA mutant. HCN production was increased in this background compared to the DF41 wild type (data not shown), likely accounting for the high nematicidal activity.
FIG 1.
Fast-killing (A and B) and slow-killing (C and D) of Caenorhabditis elegans by Pseudomonas brassicacearum DF41. Kaplan-Meier survival plots of C. elegans N2 (n = 25) fed E. coli OP50, DF41, or derivative strains propagated on BHI agar (A), BHI agar supplemented with 100 μM FeCl3 (B), NGM agar (C), and KB agar (D). Each data point represents the average of three biological replicates; error bars indicate the standard deviation (SD). Experiments were performed three times; one representative data set is shown. Asterisks indicate significant difference from the wild type as determined by the log rank test (*, P < 0.01; **, P < 0.001; ***, P < 0.0001). Note that in panels A and B, symbols representing the OP50, Δhcn mutant, and ΔgacS mutant strains overlap.
Although DF41 produces HCN, it does so at levels lower than what we have observed for other biocontrol strains, including Pseudomonas chlororaphis PA23 and Pseudomonas protegens Pf5 (data not shown). Supplementing media with iron has been reported to elevate HCN production (30); therefore, BHI containing 100 μm FeCl3 was employed in the aforementioned assays to see if more rapid killing would ensue. While the same relative toxicity pattern was observed, the presence of iron did enhance overall lethality (Fig. 1B). When nematodes were placed on BHI (100 μM FeCl3), the DF41 wild-type and sclerosin mutant strains caused 100% killing after 12 h. As before, the ΔrfiA mutant strain was the most toxic, killing 100% of the nematodes by 10 h, while the HCN-deficient hcn and gacS mutants were completely benign (Fig. 1B). Collectively, these findings indicate that on BHI (100 μM FeCl3), HCN is the primary compound responsible for fast killing of C. elegans. Due to reduced production of this volatile compound on media containing low FeCl3, however, rapid intoxication does not occur.
Slow killing of Caenorhabditis elegans by biofilm-dependent and biofilm-independent mechanisms.
For the slow-killing assays, DF41 and mutant strains were grown on NGM and KB agar prior to seeding with nematodes. On NGM agar, the survival of the nematodes decreased over the course of days (Fig. 1C); survival rates were strain dependent but nevertheless consistent with lethality mediated by bacterial colonization rather than intoxication. Under these conditions, the gacS mutant was found to be less toxic than the hcn mutant. In addition to HCN, the gacS mutant fails to produce degradative enzymes and antibiotic compounds, including sclerosin (17), some of which may be contributing to toxicity. The sclerosin-deficient mutant (DF41-1278) showed reduced nematicidal effects compared to the wild type, indicating that this LP is involved in slow killing, albeit modestly. As before, the greatest toxicity was observed for the ΔrfiA mutant, which killed 100% of the nematodes within 96 h (Fig. 1C).
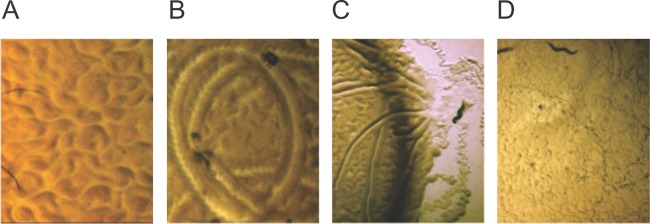
Slow killing of C. elegans was also observed when nematodes were fed bacteria growing on KB medium (Fig. 1D). However, there was one dramatic difference compared to nematodes fed DF41 and derivative strains grown on NGM. After 24 h, we noticed that biofilms began to form on the C. elegans head and body, which dramatically impacted nematode physiology and behavior (Fig. 2). Microscopic visualization at low magnification revealed that when nematodes were placed on NGM supplemented with the standard lab strain E. coli OP50, the nematodes moved freely through the bacterial lawns, generating sinusoidal track marks known as “skd” marks (Fig. 2A). Conversely, on DF41 lawns grown on KB medium, aberrant skd marks were formed (Fig. 2B and C), and the nematodes fishtailed through the viscous bacterial lawn like a car moving through deep snow. Moreover, the animals were frequently observed to migrate away from the bacteria, possibly in an attempt to remove their matrix entrapment (Fig. 2C). Over time, the biofilm-coated nematodes became progressively thinner, with some disintegrating completely, presumably due to starvation or exposure to toxic bacterial metabolites (data not shown).
FIG 2.
Track marks of nematodes fed on E. coli OP50 (A), DF41 (B and C), and a DF41 gacS mutant (D).
DF41 biofilm formation on Caenorhabditis elegans is dependent on GacS.
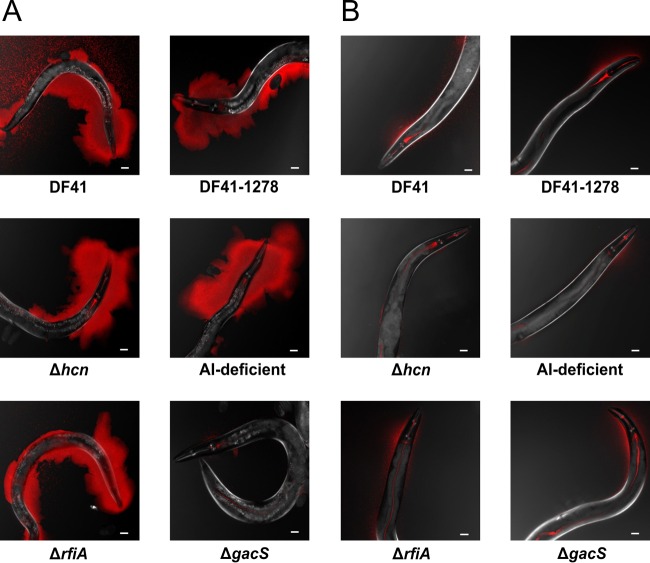
Next, we examined whether all of the DF41 derivative strains were capable of forming biofilms on the C. elegans head and body. On KB medium, the Δhcn, ΔrfiA, sclerosin-lacking (DF41-1278), and AI-deficient mutant strains all coated the surface of C. elegans and resulted in aberrant skd marks (data not shown); however, C. elegans growth on and movement through the gacS bacterial lawn resembled those observed for E. coli OP50 (Fig. 2D). Confocal microscopic analysis of red fluorescent protein (RFP)-expressing bacteria revealed that, with the exception of the gacS mutant, all of the strains formed a thick biofilm on the C. elegans surface after 24 h (Fig. 3A). In all cases, bacteria are visible in the mouth, grinder, and intestinal track of the nematodes, indicating that bacterial ingestion had occurred prior to biofilm formation blocking the buccal cavity.
FIG 3.
Microscopic analysis of N2 (A) and Δsrf-2 mutant (strain AT6) (B) worms fed rfp-tagged DF41 and derivative strains on KB. Confocal images were taken with Zeiss LSM 700 confocal laser scanning microscope under ×10 magnification. Scale bar represents 20 μm.
C. elegans mutants resistant to biofilm formation by Y. pestis, Y. pseudotuberculosis, and X. nematophila have been identified (13, 31, 32). To determine whether similar mechanisms of DF41 nematode attachment were at play, we examined several biofilm-resistant C. elegans strains, namely, the srf-2(yj262) and bah-3(br9) mutants and a naturally biofilm-resistant Hawaiian isolate (CB4856), growing on DF41. In all cases, biofilm formation was markedly reduced on the nematodes (Fig. 3B).
NaCl impacts DF41 biofilm development on the surface of Caenorhabditis elegans.
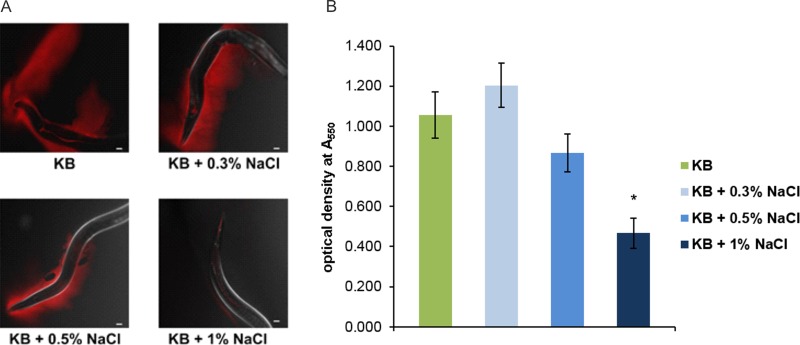
The fact that biofilms developed on the nematode surface when bacteria were grown on KB, but not BHI, NGM, or LB, suggested that medium composition has an impact on this trait. One obvious difference in the composition of these media is salt content. The three media that did not promote biofilm formation contained NaCl (NGM [0.3%], BHI [0.5%], and LB [1%]), whereas KB medium does not. This finding prompted us to investigate whether NaCl affects the ability of DF41 to form biofilms on the surface of C. elegans. Nematodes fed DF41 grown on KB medium containing 0.5% NaCl resulted in a noticeable decrease in biofilm formation on the head and body (Fig. 4A). The effects were even more dramatic when the concentration of NaCl increased to 1%, in which case virtually no biofilms were observed on the nematodes (Fig. 4A).
FIG 4.
(A) Microscopic analysis of N2 worms fed rfp-tagged DF41 propagated on KB supplemented with NaCl (0 to 1%). Confocal images were taken with Zeiss LSM 700 confocal laser scanning microscope under ×10 magnification. Scale bar represents 20 μm. (B) In vitro biofilm formation by DF41 in KB broth supplemented with NaCl (0 to 1%). Each value represents the mean from three biological replicates ± standard error. The data point marked with an asterisk indicates statistical significance (P < 0.01).
Biofilm formation on abiotic surfaces is affected by NaCl but not GacS.
Factors affecting attachment to abiotic surfaces are expected to be dramatically different from those involved in biofilm formation on biotic surfaces. As such, we employed a 96-well microtiter plate assay to assess the ability of DF41 and derivative strains to form adherent biomass under different medium conditions. While there was no significant difference in the ability of the strains to form biofilms on plastic, we did notice that in KB, biofilm formation was nearly double that observed for bacteria grown in NGM and LB (data not shown). These findings suggest that NaCl may be impacting biofilm formation not only on biotic surfaces but abiotic substrates as well. To test this hypothesis, we examined the adherent DF41 biomass after 24 h of static growth in KB supplemented with NaCl (0% to 1%). As illustrated in Fig. 4B, a significant decrease in biofilm formation was observed in KB containing 1% NaCl. No differences in growth rate were observed when bacteria were grown in the presence of NaCl (data not shown); as such, reduced biofilm formation is not a consequence of altered growth rate.
DF41 gene expression is affected by growth in the presence of Caenorhabditis elegans.
To investigate whether DF41 is able to sense the presence of C. elegans, either through direct contact or soluble chemical cues, we monitored bacterial gene expression upon growth in the presence and absence of these predators. Both regulatory genes (gacS, pdfR, pdfI, and rfiA) and the biosynthetic loci encoding HCN and sclerosin were analyzed. The most dramatic increase in gene expression was observed for hcnA, which was elevated over 5-fold in the presence of the nematodes; conversely, sclerosin biosynthetic gene activity was not affected (Fig. 5). Expression levels of the QS regulatory genes pdfR, pdfI, and rfiA were all significantly elevated upon C. elegans coculture (Fig. 5). Transcription of gacS, on the other hand, remained unaffected. We observed increased HCN and AHL signal production in DF41 cells cocultured with C. elegans (Fig. 6), consistent with the increase in hcnA and pdfI transcription, respectively.
FIG 5.
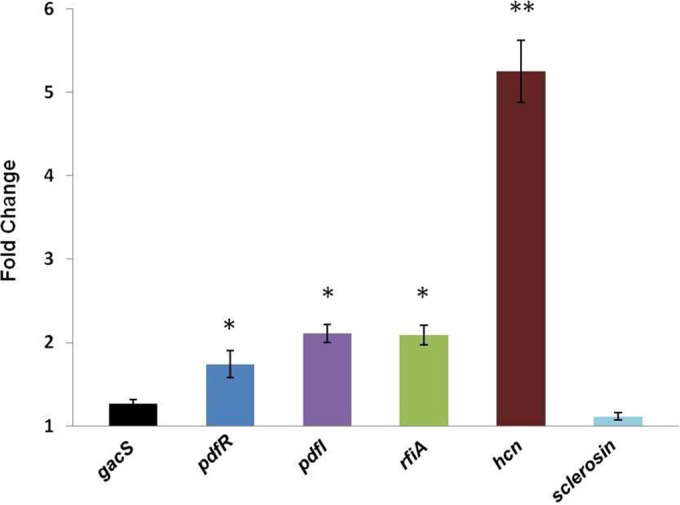
The impact of C. elegans coculture on DF41 biocontrol gene expression. qRT-PCR analysis was used to quantitatively assess gene expression in the presence and absence of nematodes. Expression levels in the absence of C. elegans were normalized to 1; differentially expressed genes are indicated with asterisks (*, P < 0.05; **, P < 0.001). Data shown are the means; error bars indicate SD.
FIG 6.
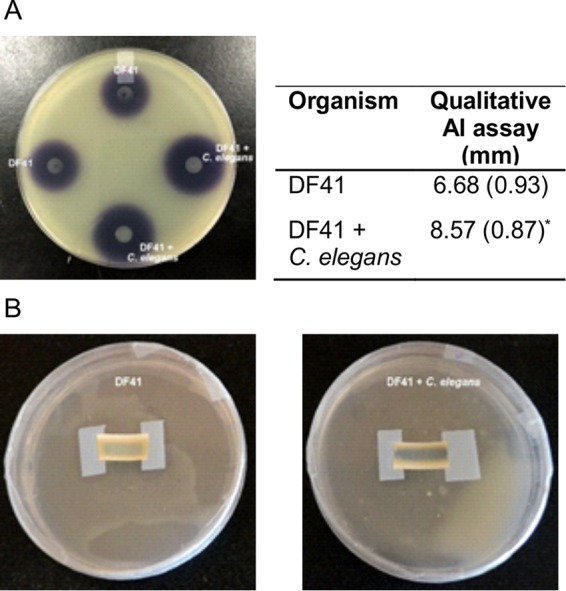
(A) Autoinducer production by DF41 grown in the presence and absence of nematodes detected by the AHL biosensor Chromobacterium violaceum CVO26. The mean (SD) of zones of activity (in millimeters) obtained from 10 replicates are shown in the table. The asterisk indicates a significant difference from the wild type (P < 0.001). (B) HCN production by DF41 grown in the presence and absence of C. elegans.
DISCUSSION
In the current study, we explored the interaction between the biocontrol agent P. brassicacearum DF41 and the bacterivorous predator C. elegans. We discovered that DF41 is among a select group of bacteria capable of establishing biofilms on the C. elegans surface, blocking feeding and ultimately starving the nematodes. To the best of our knowledge, biofilm formation on C. elegans has never been reported for a pseudomonad, pathogenic or otherwise.
It is well established that medium composition has a profound impact not only on secondary metabolites produced by pseudomonads (33) but also the nematicidal activity associated with these organisms (5–8, 34). Consequently, several different media were employed as part of our DF41-C. elegans interaction studies. On nutrient-rich BHI, which has been previously reported to support high exometabolite production and fast killing of C. elegans (6, 7), none of the strains tested led to 100% killing at 24 h (Fig. 1A). Because HCN is a nematicidal agent causing rapid paralysis of C. elegans (6, 8), we hypothesized that HCN levels were below those required for fast killing. When FeCl3 was added to the media to boost HCN production, the rate of killing by DF41 was enhanced such that all of the nematodes were dead by 12 h (Fig. 1B). With respect to relative toxicity, both in the presence and absence of FeCl3, the strain exhibiting the highest level of nematicidal activity was the rfiA mutant (Fig. 1A and B). The rfiA mutant exhibits increased accumulation of AI molecules and elevated expression of QS-controlled genes, including pdfI and hcnA (20). HCN analysis revealed elevated production of this volatile by the rfiA mutant (data not shown), which likely accounts for the increased nematicidal activity. We cannot rule out the possibility that yet-to-be identified metabolites trapped within these cells contribute to toxicity. HCN is clearly an important toxin, since the Δhcn and ΔgacS mutant cells, which are both devoid of HCN production (17), show significantly reduced toxicity, consistent with previous findings (6, 8). Sclerosin, on the other hand, did not contribute to killing under these conditions.
In slow-killing assays, which depend upon infection of the C. elegans intestine, killing occurred over the course of days (Fig. 1C and D). On both NGM and KB media, HCN, and to a lesser degree, sclerosin, contributed to the nematicidal effects; however, one dramatic difference was observed between the two media. After 24 h on KB, biofilms began to accumulate on the surface of the nematodes as they translocated through the bacterial lawns. The deleterious effect of biofilm formation is reflected by the fact that with the exception of the gacS mutant and OP50, which do not form biofilms, C. elegans survival decreased on bacteria propagated on KB (Fig. 1D).
In Y. pestis, Y. pseudotuberculosis, and X. nematophila, biofilm development on C. elegans is dependent upon the hmsHFRS gene cluster (11, 32). Analysis of the DF41 genome revealed the presence of a homologous hmsHFRS operon (hmsH, CD58_RS00620; hmsF, CD58_RS00625; hmsR, CD58_RS00630; and hmsS, CD58_RS00635) (35). This cluster showed the highest degree of similarity at the amino acid level with PgaABCD of Pseudomonas fluorescens. The DF41 HmsH protein is 91% identical to PgaA, a predicted poly-beta-1,6-N-acetyl-d-glucosamine (PGA) export porin (accession no. ALI07281.1). HmsF exhibits 94% sequence identity with PgaB, a PGA N-deacetylase (accession no. WP_024618876.1). HmsR is 99% identical to the PGA synthase PgaC (accession no. ALI07279.1), and HmsS exhibits 93% identity with the PGA biosynthesis protein PgaD (WP_003186997.1). In E. coli, PGA biosynthesis mediated by pgaABCD is essential for biofilm formation (36, 37). BLAST analysis revealed that homologs of the hmsHFRS operon are found in only a limited number of Pseudomonas species (Table 2). It is noteworthy that these genes are absent in P. aeruginosa, which does not form biofilms on C. elegans (15). Conversely, biocontrol strain P. chlororaphis PA23, which harbors this locus, was unable to form biofilms on the nematode surface under the conditions tested (data not shown). Additional work is required to establish a role for these genes in DF41 biofilm development; however, the fact that C. elegans mutants that are resistant to Yersinia and Xenorhabdus biofilms are resistant to those of DF41 suggests that a common ligand-receptor interaction is involved (15, 32).
TABLE 2.
Sequence identity of DF41 HmsHFRS with homologs found in other Pseudomonas species
| Bacterial strain | Amino acid identity (%) of homolog to DF41 HmsHFRS |
|---|---|
| P. chlororaphis strain PA23 | 78 |
| P. koreensis strain D26 | 80 |
| P. poae strain RE*1-1-14 | 74 |
| P. antarctica strain PAMC 27494 | 74 |
| P. azotoformans strain S4 | 75 |
| P. simiae strain WCS417 | 74 |
| P. trivialis strain IHBB745 | 74 |
| P. citronellolis strain P3B5 | 59 |
In E. coli, a csrA (rsmA) mutant was found to overproduce PGA and showed increased biofilm development (38, 39). CsrA belongs to the RsmA family of repressor proteins that bind mRNA at the ribosome-binding site, thereby blocking translation (40). The Gac two-component system is a positive activator of small RNAs that titrate out the RsmA proteins allowing target gene expression. Consequently, in a gac mutant, targets remain permanently repressed. The findings presented herein suggest that genes underlying DF41 biofilm formation on the surface of C. elegans are subject to Gac regulation, consistent with the CsrA control observed in E. coli. In another study, the LysR-type regulator NhaR was found to activate pgaABCD expression in response to 100 mM NaCl and high pH in E. coli (41). We, on the other hand, observed that the addition of sodium ions (0.5% [86 mM] and 1% [171 mM]) diminished the establishment of DF41 biofilms on both biotic and abiotic surfaces (Fig. 4). A negative correlation between NaCl and biofilm formation has been reported for Clostridium difficile and Streptococcus suis (42, 43). Collectively, these findings suggest that the impact of sodium on biofilm development is species dependent.
Prokaryotes and eukaryotes have cohabited the Earth for millions of years; it is not surprising, therefore, that chemical communication between the two facilitates mutual perception (44). The production of toxic secondary metabolites is energetically costly, and so limiting production to situations where these compounds provide a fitness advantage, when confronted by predators, for example, would be beneficial to the producer. We cocultured DF41 in the presence of C. elegans to see whether chemical cues and/or direct contact would lead to changes in the expression of select bacterial genes. Upon coculture, the most highly upregulated DF41 gene was hcnA (Fig. 5). Intriguingly, HCN also exhibited the greatest nematicidal activity (Fig. 1). Sclerosin, which exhibited almost no toxicity toward C. elegans, was unchanged with respect to gene expression in DF41 grown with the nematodes. These findings are similar to those reported by Jousset and coworkers, wherein cell-free supernatants of the amoeba Acanthamoeba castellanii were found to increase the expression of phlA (diacetylphloroglucinol [DAPG]), prnA (pyrrolnitrin), and hcnA in P. fluorescens CHA0 (22). Notably, there was a correlation between the level of expression and the toxicity of the encoded product, with the greatest increase in gene expression observed for DAPG, the most toxic metabolite tested (22). More recently, global transcriptomic analysis of P. fluorescens SS101 cocultured with the protist Naegleria americana revealed altered expression of 2.3% of the genome (2). Moreover, protozoan predation led to upregulated lipopeptide and putrescine biosynthesis, with the putrescine biosynthesis inducing trophozoite encystment and decreasing cyst viability (2). When we examined the impact of coculturing on DF41 regulatory gene expression, we discovered that pdfR, pdfI, and the cotranscribed rfiA were upregulated in the presence of C. elegans (Fig. 5). We have previously shown that hcn expression is under the control of the PdfRI QS system, whereas the sclerosin biosynthetic genes are not (20). At present, it is unclear whether upregulation of the hcn genes is mediated directly or indirectly through increased QS gene expression. Analysis of HCN and AHL signal production by DF41 revealed that both end products were upregulated by the presence of the nematodes, consistent with our gene expression analysis.
Being encased in an extrapolymeric matrix as a biofilm promotes bacterial survival and colonization of root surfaces (45). In addition to facilitating exchange of metabolic by-products and genetic information, this mode of growth affords inhabitants protection from toxic compounds and desiccation. We have made the exciting discovery that DF41 is capable of biofilm formation on the C. elegans anterior, blocking feeding and ultimately starving the animals. Thus, for DF41, biofilm formation offers protection from grazing predators. It is possible that biocontrol agents that show increased environmental persistence through biofilm formation do so through similar antipredator strategies. As we unravel the biotic and abiotic factors underlying this phenomenon, transkingdom signaling between plants, bacteria, and the predators that feed upon them is expected to play a central role.
ACKNOWLEDGMENTS
We are grateful to D. Haas for plasmid pME6863 and E. Pesci for P. aeruginosa QSC (pEAL01). C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (grant P40 OD010440).
Funding Statement
This work, including the efforts of Teresa R. de Kievit, was funded by Government of Canada | Natural Sciences and Engineering Research Council of Canada (NSERC) (RGPIN/249559-2012).
REFERENCES
- 1.Jousset A, Rochat L, Péchy-Tarr M, Keel C, Scheu S, Bonkowski M. 2009. Predators promote defence of rhizosphere bacterial populations by selective feeding on non-toxic cheaters. ISME J 3:666–674. doi: 10.1038/ismej.2009.26. [DOI] [PubMed] [Google Scholar]
- 2.Song C, Mazzola M, Cheng X, Oetjen J, Alexandrov T, Dorrestein P, Watrous J, van der Voort M, Raaijmakers JM. 2015. Molecular and chemical dialogues in bacteria-protozoa interactions. Sci Rep 5:12837. doi: 10.1038/srep12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerry BR, Hominick WM. 2002. Biological control, p 483–510. In Lee DL. (ed), The biology of nematodes. CRC Press, New York, NY. [Google Scholar]
- 4.Hope IA. 1999. C. elegans: a practical approach. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 5.Darby C, Cosma Cl Thomas JH, Manoil C. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96:15202–15207. doi: 10.1073/pnas.96.26.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallagher LA, Manoil C. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J Bacteriol 183:6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nandi M, Selin C, Brassinga AKC, Belmonte MF, Fernando WGD, Loewen PC, de Kievit TR. 2015. Pyrrolnitrin and hydrogen cyanide production by Pseudomonas chlororaphis strain PA23 exhibits nematicidal and repellent activity against Caenorhabditis elegans. PLoS One 10:e0123184. doi: 10.1371/J.poine.0123184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan MW, Mahajan-Miklos S, Ausubel FM. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu Rev Microbiol 49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 10.Davey ME, O'Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867. doi: 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darby C, Hsu JW, Ghori N, Falkow S. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243–244. doi: 10.1038/417243a. [DOI] [PubMed] [Google Scholar]
- 12.Couillault C, Ewbank JJ. 2012. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect Immun 70:4705–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshua GW, Karlyshev AV, Smith MP, Isherwood KE, Titball RW, Wren BW. 2003. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology 149:3221–3229. doi: 10.1099/mic.0.26475-0. [DOI] [PubMed] [Google Scholar]
- 14.Alegado RA, Campbell MC, Chen WC, Slutz SS, Tan MW. 2003. Characterization of mediators of microbial virulence and innate immunity using Caenorhabditis elegans host-pathogen model. Cell Microbiol 5:435–444. doi: 10.1046/j.1462-5822.2003.00287.x. [DOI] [PubMed] [Google Scholar]
- 15.Tan L, Darby C. 2004. A moveable surface: formation of Yersina sp. biofilms on motile Caenorhabditis elegans. J Bacteriol 186:5087–5092. doi: 10.1128/JB.186.15.5087-5092.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savchuk SC, Dilantha Fernando WG. 2004. Effect of timing of application and population dynamics on the degree of biological control of Sclerotinia sclerotiorum by bacterial antagonists. FEMS Microbiol Ecol 49:379–388. doi: 10.1016/j.femsec.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 17.Berry C, Fernando WGD, Loewen PC, de Kievit TR. 2010. Lipopeptides are essential for Pseudomonas sp. DF41 biocontrol of Sclerotinia sclerotiorum. Biol Control 55:211–218. [Google Scholar]
- 18.Berry C, Brassinga AKC, Donald LJ, Fernando WGD, Loewen PC, de Kievit TR. 2012. Chemical and biological characterization of sclerosin, an antifungal lipopeptide. Can J Microbiol 58:1027–1034. doi: 10.1139/w2012-079. [DOI] [PubMed] [Google Scholar]
- 19.Manuel J, Berry C, Selin C, Fernando WGD, de Kievit TR. 2011. Repression of the antifungal activity of Pseudomonas sp. strain DF41 by the stringent response. Appl Environ Microbiol 77:5635–5642. doi: 10.1128/AEM.02875-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berry C, Nandi M, Manuel J, Brassinga AKC, Fernando WGD, Loewen PC, de Kievit TR. 2014. Characterization of the Pseudomonas sp. DF41 quorum sensing locus and its role in fungal antagonism. Biol Control 69:82–89. [Google Scholar]
- 21.de Bruijn I, Raaijmakers JM. 2009. Diversity and functional analysis of LuxR-type transcriptional regulators of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens. Appl Environ Microbiol 75:4753–4761. doi: 10.1128/AEM.00575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jousset A, Rochat L, Scheu S, Bonkowski M, Keel C. 2010. Predator-prey chemical warfare determines expression of biocontrol genes by rhizosphere-associated Pseudomonas fluorescens. Appl Environ Microbiol 76:5263–5268. doi: 10.1128/AEM.02941-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzola M, de Bruijn I, Cohen MF, Raaijmakers JM. 2009. Protozoan-induced regulation of cyclic lipopeptide biosynthesis is an effective predation defense mechanism for Pseudomonas fluorescens. Appl Environ Microbiol 75:6804–6811. doi: 10.1128/AEM.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med 44:301–307. [PubMed] [Google Scholar]
- 25.Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girard LR, Fiedler TJ, Harris TW, Carvalho F, Antoshechkin I, Han M, Sternberg PW, Stein LD, Chalfie M. 2007. WormBook: the online review of Caenorhabditis elegans biology. Nucleic Acids Res 35:D472–D475. doi: 10.1093/nar/gkl894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 28.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Latifi A, Winson MK, Foglino M, Bycroft BW, Stewart GSAB, Lazdunski A, Williams P. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol 17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 30.Michelsen CF, Stougaard P. 2012. Hydrogen cyanide synthesis and antifungal activity of the biocontrol strain Pseudomonas fluorescens In5 from Greenland is highly dependent on growth medium. Can J Microbiol 58:381–390. doi: 10.1139/w2012-004. [DOI] [PubMed] [Google Scholar]
- 31.Darby C, Chakrabarti A, Politz SM, Daniels CC, Tan L, Drace K. 2007. Caenorhabditis elegans mutants resistant to Yersinia biofilms. Genetics 176:221–230. doi: 10.1534/genetics.106.067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drace K, Darby C. 2008. The hmsHFRS operon of Xenorhabdus nematophila is required for biofilm attachment to Caenorhabditis elegans. Appl Environ Microbiol 74:4509–4515. doi: 10.1128/AEM.00336-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haas D, Keel C. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol 41:117–153. [DOI] [PubMed] [Google Scholar]
- 34.Cezairliyan B, Vinayavekhin N, Grenfell-Lee D, Yuen GJ, Saghatelian A, Ausubel FM. 2013. Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog 9:e1003101. doi: 10.1371/journal.ppat.1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loewen PC, Villenueva J, Fernando WG, de Kievit T. 2014. Genome sequence of Pseudomonas chlororaphis strain PA23. Genome Announc 2(4):e00689-14. doi: 10.1128/genomeA.00689-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF III, Romeo T. 2008. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesion poly-β-1,6-N-acetyl-d-glucosamine. J Bacteriol 190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Preston JR III, Romeo T. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesion required for biofilm formation. J Bacteriol 186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J Bacteriol 184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesion of Escherichia coli. Mol Microbiol 56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 40.Lapouge K, Schubert M, Allain FHT, Haas D. 2008. Gac/Rsm signal transduction pathway of g-proteobacteria: from RNA recognition to regulation of social behavior. Mol Microbiol 67:241–253. [DOI] [PubMed] [Google Scholar]
- 41.Goller C, Wang Itoh Y, Romeo T. 2006. The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcription, required for production of the biofilm adhesion poly-β-1,6-N-acetyl-d-glucosamine. J Bacteriol 188:8022–8032. doi: 10.1128/JB.01106-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ðapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, Scarselli M, Minton NP, Serruto D, Unnikrishnan M. 2013. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol 195:545–555. doi: 10.1128/JB.01980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawei G, Liping W, Chengping L. 2012. In vitro biofilm forming potential of Streptococcus suis isolated from human and swine in China. Braz J Microbiol 43:993–1004. doi: 10.1590/S1517-83822012000300021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes DT, Sperandio V. 2008. Inter-kingdom signaling: communication between bacteria and their hosts. Nat Rev Microbiol 6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris CE, Monier JM. 2003. The ecological significance of biofilm formation by plant-associated bacteria. Annu Rev Phytopathol 41:429–453. doi: 10.1146/annurev.phyto.41.022103.134521. [DOI] [PubMed] [Google Scholar]
- 46.House BL, Mortimer MW, Kahn ML. 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl Environ Microbiol 70:2806–2815. doi: 10.1128/AEM.70.5.2806-2815.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reimmann C, Ginet N, Michel L, Keel C, Michaux P, Krisnapillai V, Zala M, Heurlier K, Triandafillu K, Harms H, Défago G, Haas D. 2002. Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiology 148:923–932. doi: 10.1099/00221287-148-4-923. [DOI] [PubMed] [Google Scholar]
- 48.Alexeyev MF. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of Gram-negative bacteria. Biotechniques 26:824–826. [DOI] [PubMed] [Google Scholar]
- 49.Finan TM, Kunkel B, De Vos GF, Signer ER. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol 167:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 51.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]