ABSTRACT
Fructophilic lactic acid bacteria (FLAB) are strongly associated with the gastrointestinal tracts (GITs) of Apis mellifera L. worker bees due to the consumption of fructose as a major carbohydrate. Seventy-seven presumptive lactic acid bacteria (LAB) were isolated from GITs of healthy A. mellifera L. adults, which were collected from 5 different geographical locations of the Apulia region of Italy. Almost all of the isolates showed fructophilic tendencies: these isolates were identified as Lactobacillus kunkeei (69%) or Fructobacillus fructosus (31%). A high-throughput phenotypic microarray targeting 190 carbon sources was used to determine that 83 compounds were differentially consumed. Phenotyping grouped the strains into two clusters, reflecting growth performance. The utilization of phenolic acids, such as p-coumaric, caffeic, syringic, or gallic acids, as electron acceptors was investigated in fructose-based medium. Almost all FLAB strains showed tolerance to high phenolic acid concentrations. p-Coumaric acid and caffeic acid were consumed by all FLAB strains through reductases or decarboxylases. Syringic and gallic acids were partially metabolized. The data collected suggest that FLAB require external electron acceptors to regenerate NADH. The use of phenolic acids as external electron acceptors by the 4 FLAB showing the highest phenolic acid reductase activity was investigated in glucose-based medium supplemented with p-coumaric acid. Metabolic responses observed through a phenotypic microarray suggested that FLAB may use p-coumaric acid as an external electron acceptor, enhancing glucose dissimilation but less efficiently than other external acceptors such as fructose or pyruvic acid.
IMPORTANCE Fructophilic lactic acid bacteria (FLAB) remain to be fully explored. This study intends to link unique biochemical features of FLAB with their habitat. The quite unique FLAB phenome within the group lactic acid bacteria (LAB) may have practical relevance in food fermentations. The FLAB phenome may have implications for the levels of hexose metabolism products in fermented foods, as well as food probiotication. Due to the harsh conditions of honeybees' GITs, these bacteria had to develop specific physiological and biochemical characteristics, such as tolerance to phenolic acids. The screening of FLAB strains based on metabolic pathways involving phenolic acids may allow the selection of starter cultures with both technological and functional beneficial attributes. Bioconversion of phenolic compounds may contribute to the aroma attributes and biofunctionality of fermented foods. Thus, the selection of FLAB strains as starter cultures with specific enzymatic activities involving phenolic acids may have a promising role in food fermentations.
INTRODUCTION
Fructophilic lactic acid bacteria (FLAB), described only recently (1–9), belong to a special group of lactic acid bacteria (LAB) that prefer fructose instead of glucose as a carbon source. These bacteria have been isolated from specific ecological fructose-rich niches such as flowers, fruits, and fermented food based-fruits and, only recently, from the gastrointestinal tracts (GITs) of several insects (e.g., bumblebees, honeybees, tropical fruit flies, and Camponotus ants), which have a fructose-based diet (4, 10, 11). Among these insects, social honeybees are those of greater interest because of their economic and ecological importance for honeybee products (e.g., propolis, royal jelly, honey, and pollen) and especially for crop pollination. Despite a global increase in the population of domesticated bees according to the FAO data (12), honeybees are facing growing adversity. Localized declines in bee populations have occurred in many European countries (13–15). To understand and to prevent the decrease, several studies have been addressed to investigate the symbiotic and pathogenic microbial interactions (16–19). The observations made as of to date indicate the honeybees' GITs harbor a core microbiota dissimilar to those of other animals, including humans (16–24). FLAB are strongly associated with honeybees' GIT compartments, where the enhancement of intestinal barrier function was found to be a consequence of the limited translocation of foreign antigens or pathogens in the gut due to commensal and probiotic bacteria (6, 19, 24–26). Consequently, it is not surprising that FLAB may have a positive role in honeybee health, representing one of the main mechanisms of defense from pathogens in bees. FLAB species are separated into obligately and facultative fructophilic species, based on biochemical features (1). The first group includes Lactobacillus kunkeei and Fructobacillus species, which grow on d-fructose and on d-glucose only when pyruvate or oxygen is available as an external electron acceptor. Obligately FLAB convert glucose into almost equimolar amounts of lactic acid and acetic acid and trace amounts of ethanol. The low synthesis of ethanol is due to the absence of acetaldehyde dehydrogenase and a weak activity of alcohol dehydrogenase (7). The second group includes Lactobacillus florum and a few biotypes of Lactobacillus brevis (2, 4, 5, 27). Facultative FLAB grow well on fructose and are able to grow on glucose at a delayed rate if electron acceptors are absent. Conversion of glucose to lactic acid and ethanol at a ratio of 1:1:0.2 has been reported for facultatively FLAB. The insufficient NAD+ regeneration and the requirement of an external electron acceptor for growth on glucose are the hallmark fructophilic characteristics (1, 7, 8).
The honeybees' GIT represent a rich source of FLAB. These bacteria had to develop specific physiological and biochemical features and show signs of niche-specific regressive evolution at the genomic level (7, 28–30). The homeostasis of the honeybees' GIT microbiota may be affected by several drivers, including diet—especially those that are chemically defined. Secondary metabolites, including tannins, alkaloids, and terpenes, and especially phenols as phenolic acids and flavonoids, have been found in pollen (31–36). p-Coumaric acid, a phenolic compound, is a structural component of sporopollenin, which makes up the principal matrices comprising the outer wall of the pollen grain (35). Pollen secondary metabolites may exert a beneficial effect on pollinators via antimicrobial activity (36). Some studies suggest that the pollen secondary metabolites may also be the cause for bacterial endosymbiosis (21). Similarly to plant-associated bacteria (37), FLAB colonizing honeybees' GITs might have modulated specific molecular defense mechanisms to ensure the tolerance against the broad range of plant secondary metabolites ingested by bees as part of their diet. To the best of our knowledge, the adaptation to and metabolism of phenolic acids have been investigated exclusively for Lactobacillus plantarum and lactic acid bacteria used for wine making (e.g., Oenococcus oeni) and rarely for other Lactobacillus spp., Weissella spp., and Leuconostoc mesenteroides (38, 39). To date, the metabolism of phenolic acids by FLAB has not yet been investigated, and more in general, FLAB remain an unexplored group. Confirming this, only 15 published items on FLAB were retrieved from the main literature databases in July 2016. Understanding of the ecological significance and the potential these bacteria may have in food fermentations should be deepened. L. kunkeei bacteria also play a key role in the lactic acid fermentation of pollen to bee bread due to their acid-, oxygen-, and osmo-tolerant features and are among the small subset of bacteria isolated from pure honey (25, 40). Although the mechanisms underlying the bee bread production are not completely elucidated, LAB have been supposed to affect the pollen maturation process lowering the pH and modifying the sugar and amino acid profiles (25, 41). This study aimed at investigating the metabolism of phenolic acids by FLAB as a potential alternative energy route using a high-throughput phenotypic microarray targeting 190 carbon sources. The understanding of FLAB metabolism under the conditions used in this study should help elucidate the effects of phenolic acids on bacterial growth, physiology, and fermentation end products.
MATERIALS AND METHODS
Honeybee intestinal tract sampling.
Healthy adult worker bees of the species Apis mellifera L. (10 bees for each apiary) were collected between June and November 2015 from 5 different apiaries of the Apulia region of Italy placed in 5 different geographical locations (Mottola, 40°44′23.71″N, 16°59′27.70″E; Noci, 40°44′52.82″N, 17°8′57.43″E; Ostuni, 40°44′21.35″N, 17°34′30.36″E; Bari, 41°6′42.25″N, 16°52′54.27″E; and Valenzano, 41°1′23.03″N, 16°54′15.94″E) and immediately transported to the Department of Soil, Plant and Food Science Laboratory at the University of Bari Aldo Moro. Aiming to avoid cross contamination with the external surface of the bee body, the surface was sterilized by being submerged in 5% sodium hypochlorite solution and washed with sterile 0.1% peptone saline solution supplemented with Tween 80 (0.9% [wt/vol] NaCl, 0.1% [wt/vol] Tween 80, and 0.1% [wt/vol] peptone). The whole intestinal tracts (esophagus to rectum) were obtained through aseptic excisions. The intestinal tracts were placed in sterile tubes containing 1 ml of sterile 0.1% peptone saline solution supplemented with Tween 80.
Isolation, typing, and identification of LAB.
One milliliter of intestinal tract suspension was diluted in 9 ml of sterile sodium chloride (0.9% [wt/vol]) solution and homogenized. Serial dilutions were made and plated on fructose-yeast extract-polypeptone (FYP) agar (10 g d-fructose, 10 g yeast extract, 5 g polypeptone, 2 g sodium acetate, 0.5 g Tween 80, 0.2 g MgSO4·7H2O, 0.01 g MnSO4·4H2O, 0.01 g FeSO4·7H2O, 0.01 g NaCl, 0.05 g cycloheximide, and 0.05 g sodium azide per liter of distilled water [pH 6.8]) supplemented with 0.5% CaCO3 (wt/vol) and incubated aerobically at 30°C for 24 to 48 h (1). At least 10 colonies were isolated from the highest dilutions of the FYP plates, based on the morphology and size of the clearance zone surrounding the colonies, and streaked on FYP agar. The clearance zone surrounding the colonies indirectly indicates the hydrolysis of CaCO3 reacting with organic acids synthesized by bacteria. Gram-positive and catalase-negative isolates were cultivated in FYP broth at 30°C for 24 h and restreaked on FYP agar. Genomic DNA was extracted from 2 ml of FYP culture broth of each isolate using the DNeasy blood and tissue kit (Qiagen, Valencia, CA), according to the manufacturer's instructions. Two primer pairs, LacbF/LacbR and LpCoF/LpCoR (Sigma Chemical Co., Milan, Italy), were used to amplify 16S rRNA gene fragment of lactic acid bacteria (42). The expected amplicons of ca. 1,400 and 1,000 bp were eluted from the gel and purified by the Nucleospin gel and PCR cleanup kit (Macherey-Nagel, Düren, Germany). PCR products were separated by electrophoresis, purified as described above, and subjected to Sanger sequencing (43). Taxonomic strain identification was performed by comparing the sequences of each isolate with those reported in the NCBI Reference Sequence (RefSeq) database (44). Strains showing homology of at least 97% were considered to belong to the same species (45). Randomly amplified polymorphic DNA-PCR (RAPD-PCR) was carried out to exclude clonal relatedness. RAPD-PCR analysis was performed as described by Di Cagno et al. (46), using primers M13, P7, and P4 (Invitrogen Life Technologies, Milan, Italy). Cultures were maintained as stocks in 15% (vol/vol) glycerol at −80°C and routinely propagated at 30°C for 24 h in FYP broth.
To investigate whether or not isolates were fructophilic lactic acid bacteria (FLAB), isolates were inoculated into FYP broth and glucose-yeast extract-polypeptone (GYP) broth (identical to FYP broth but with fructose replaced with 1% [wt/vol] glucose). The 24-h-old cells grown aerobically in FYP broth were harvested by centrifugation (10,000 × g for 10 min at 4°C), washed twice in 50 mmol liter−1 sterile potassium phosphate buffer (pH 7.0), resuspended in FYP or GYP to a final optical density at 620 nm (OD620) of 0.025 (corresponding to ca. 7.0 log CFU ml−1) and incubated anaerobically at 30°C for 24 h. Anaerobic conditions were achieved and maintained through anaerobic atmosphere-generating system with AnaeroGen bags (AnaeroGen system; Oxoid, Ltd., Basingstoke, Hampshire, England). Differences in growth among the strains were monitored by recording optical density readings at 620 nm. Isolates that grew well in FYP broth but poorly in GYP broth were confirmed as being FLAB (1).
The kinetics of growth in FYP broth incubated anaerobically at 30°C for 24 h and GYP broth (identical to FYP broth, but with fructose replaced with 1% [wt/vol] glucose) incubated aerobically under stirring conditions (200 rpm) at 30°C for 24 h were determined and modeled according to the Gompertz equation as modified by Zwietering et al. (47): y = k + A exp{−exp[(μmax e/A)(λ − t) + 1]}, where k is the initial level of the dependent variable to be modeled (OD620 units), A is the difference in cell density between inoculation and the stationary phase, μmax is the maximum growth rate (expressed as OD620 units per hour), λ is the length of the lag phase (expressed in hours), and t is the time.
Carbohydrate consumption and main fermentation end products.
The supernatants recovered from FYP cultures incubated anaerobically and GYP cultures incubated aerobically under stirring conditions (200 rpm) were filtered through a Millex-HA 0.22-μm-pore-size filter (Millipore Co.) and used to determine carbohydrates, lactic acid, acetic acid, acetaldehyde, and ethanol by high-performance liquid chromatography (HPLC). An Äkta purifier system (GE Healthcare) was equipped with an Aminex HPX-87H column (ion exclusion; Bio-Rad), a UV detector operating at 210 nm, and a PerkinElmer 200a refractive index detector (PerkinElmer, Waltham, MA) operating at 32°C. Elution was at 35°C with a flow rate of 0.6 ml min−1, and 10 mmol liter−1 H2SO4 was used as the mobile phase (48).
Phenotypic microarray analysis.
Differences in phenotype during growth in FYP broth incubated anaerobically and in GYP broth incubated aerobically under stirring conditions (200 rpm) were monitored using the OmniLog phenotype microarray (PM) technology (Biolog). PM plates (Biolog) containing 190 carbon sources (PM1 and PM2) were used. Phenotypic microarray analyses were performed with two biological replicates for each growth condition in accordance with the manufacturer's instructions. Cells were collected after 36 h at 30°C. Cells were washed in 50 mmol liter−1 sterile potassium phosphate buffer (pH 7.0), diluted (to achieve 65% transmittance) in inoculating fluid (Biolog), and used to inoculate the PM plates. One-hundred microliters of cell suspension was added to each well. Plates were incubated for 24 h at 30°C in an OmniLog automated incubator/reader (Biolog). During incubation, reduction of tetrazolium dye by respiring cells was measured in each well every 15 min by the OmniLog system. Cellular respiration activity was evaluated as the area of a region bounded by a color development time-series. The results were analyzed using the OmniLog PM software (Biolog) according to the manufacturer's instructions. Phenotypic assays were performed in duplicate with high reproducibility (R2 > 0.95 for each metabolite). We show only the most significant (P < 0.05) differences in metabolic activities under the experimental conditions of this study.
MIC of phenolic acids.
The MICs of phenolic acids (p-coumaric, caffeic, syringic, and gallic acids) during the growth of FLAB were determined by the critical dilution assay (39, 49). Phenolic compounds were dissolved in methanol and diluted in FYP broth to a final concentration of 50 mmol liter−1. The pH was adjusted to 7.0 with 2 N NaOH, in order to exclude the effect of pH on the activity of phenolic acids. Serial 2-fold dilutions were made with FYP broth (pH 7.0) in sterile 96-well microtiter plates (Greiner Labortechnik). Bacterial strains were subcultured in FYP broth under the above-described conditions. Logarithmic-phase cells (ca. 8 log CFU ml−1) were harvested by centrifugation (8,000 × g for 10 min), washed twice with 10 mmol liter−1 phosphate buffer (pH 7.0), and adjusted to ca. 5 log CFU ml−1. Each well was inoculated (10% [vol/vol]). The final concentration of phenolic acids in sterile 96-well microtiter plates ranged from 45.5 mmol liter−1 to 0.7 mmol liter−1. Microtiter plates were incubated for 24 h. After incubation, bacterial growth was determined by measuring the optical density at 620 nm (OD620). The MIC was defined as the lowest concentration of phenolic acids inhibiting bacterial growth. Control wells contained all of the components except phenolic acids, which were replaced with distilled water (positive control) or with chloramphenicol (100 μg ml−1; negative control).
Metabolism of phenolic acids during growth in FYP broth.
FYP broth was supplemented with p-coumaric, caffeic, syringic, or gallic acid at a concentration of 1 mmol liter−1 (39, 49, 50). Cells from an overnight culture were inoculated (5% [vol/vol]) into supplemented FYP medium and incubated for 24 h at 30°C. Sterile media containing the corresponding phenolic compounds, without a bacterial inoculum, were used as the control. Viable cells were enumerated by surface plating onto FYP agar. The pH was measured by a Foodtrode electrode (Hamilton, Bonaduz, Switzerland). After incubation, cells were removed by centrifugation (10,000 × g for 10 min), and the supernatant was acidified to pH 1.5 with hydrochloric acid. Ethyl acetate (3 ml) was used for liquid-liquid extraction. The extracts (20 μl) were analyzed by high-performance liquid chromatography (HPLC), using a Äkta purifier system (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) equipped with an XTerra MS C18 column (particle size, 5 μm; 4.6 by 250 mm [Waters, Brussels, Belgium]) and a diode array detector (DAD). DAD detection was carried out between 100 and 400 nm. Eluent A consisted of 0.1% (vol/vol) formic acid in HPLC-grade water, and eluent B consisted of 0.1% (vol/vol) formic acid in acetonitrile (30% [vol/vol]) and HPLC-grade water (10% [vol/vol]). Extracts were eluted with the following gradient: 0% eluent B (3 min), 0 to 40% eluent B (10 min), 40 to 60% eluent B (60 min), 60 to 100% eluent B (10 min), 100% eluent B (20 min), and 100 to 0% eluent B (15 min). The efficiency of conversion of hydroxycinnamic acids into their derivatives was calculated as follows: amt (mmol) of end products produced in a defined period/amt (mmol) of hydroxycinnamic acid consumed in the same period × 100. External standards analyzed under the same conditions were used for the identification and quantification of phenolic acids and phenolic acid derivatives and used for identification by comparison of retention time and UV absorbance.
Growth in GYP supplemented with p-coumaric acid.
FLAB strains showing phenolic acid reductase activity were grown (with the initial cell number corresponding to ca. log 7 CFU ml−1) anaerobically at 30°C for 24 h in GYP supplemented with p-coumaric acid at a concentration of 2 mmol liter−1. This concentration was well below the MIC. GYP not supplemented with phenolic acid was used as the control. Differences in phenotype during growth in GYP supplemented with p-coumaric acid at 2 mmol liter−1 were monitored using OmniLog PM technology (Biolog) as described above. The NAD+/NADH ratio was determined as described by Filannino et al. (39). Cells were recovered through centrifugation (10,000 × g for 10 min at 4°C) and washed in cold phosphate-buffered saline (Sigma, St. Louis, MO). Cells (ca. 9 log CFU ml−1) were suspended in 400 μl of NADH/NAD extraction buffer (Abcam, Cambridge, MA), transferred into Lysing matrix E tubes (MP Biomedicals, Santa Ana, CA), and broken with a FastPrep FP120 instrument (Bio 101, Savant, Inc., Holbrook, NY) for 40 s at a speed of 6.0. The levels of NAD+ and NADH were determined according to the guidelines provided by the manufacturer (Abcam). The results are expressed as the NAD+/NADH ratio.
Statistical analyses.
Analyses were performed in triplicate with three biological replicates for each condition. Data were subjected to one-way analysis of variance (ANOVA); paired comparison of treatment means was achieved by Tukey's procedure at P < 0.05, using the statistical software Statistica for Windows (Statistica 7.0 per Windows). The metabolic fingerprints of FLAB strains as determined by OmniLog PM Technology (Biolog) were subjected to permutation analysis using PermutMatrix.
Accession number(s).
Sequences have been deposited in GenBank under the accession numbers given in Table 1.
TABLE 1.
Representative FLAB strains of each cluster obtained by combined RAPD patterns isolated from Apis mellifera L. bee gut
| Strain | No. of clustersa | Accession no.b | Geographical location (GPS) |
|---|---|---|---|
| L. kunkeei | |||
| BIII60 | CL-6 | KX833121 | Mottola (40°44′23.71″N, 16°59′27.70″E) |
| BIII59 | CL-2 | KX833122 | |
| B17 | CL-5 | KX833123 | Valenzano (41°1′23.03″N, 16°54′15.94″E) |
| B7 | CL-13 | KX833124 | |
| B23I | CL-10 | KX833125 | Bari (41°6′42.25″N, 16°52′54.27″E) |
| B4I | CL-7 | KX833126 | |
| BV61 | CL-3 | KX833127 | Noci (40°44′52.82″N, 17°8′57.43″E) |
| BV20 | CL-4 | KX833128 | |
| BVI14 | CL-8 | KX833129 | Ostuni (40°44′21.35″N, 17°34′30.36″E) |
| BVI17 | CL-9 | KX833130 | |
| BVI52 | CL-12 | KX833131 | |
| F. fructosus | |||
| MBIII2 | CL-15 | KX833132 | Mottola (40°44′23.71″N, 16°59′27.70″E) |
| MBIII5 | CL-14 | KX833133 | |
| B5 | CL-16 | KX833134 | Valenzano (41°1′23.03″N, 16°54′15.94″E) |
| B4 | CL-11 | KX833135 | |
| B1 | CL-1 | KX833136 |
Numbers of randomly amplified polymorphic DNA-PCR (RAPD-PCR) clusters. Clusters (CL) are listed with numbers from 1 to 16. RAPD-PCR analysis was carried out to exclude clonal relatedness.
The 16S rRNA gene sequences generated in this study were deposited in GenBank under the accession numbers shown.
RESULTS
Identification and typing of LAB.
Seventy-seven Gram-positive, catalase-negative, nonmotile rods able to grow at 15°C and to acidify FYP broth were identified by partial sequencing of the 16S rRNA. The following species were identified: Lactobacillus kunkeei (n = 52 isolates), Fructobacillus fructosus (n = 23), Lactobacillus plantarum (n = 1), and Lactobacillus fermentum (n = 1). For all L. kunkeei strains, the top BLAST hit (99% identity) from BLAST analysis of the 16S rRNA sequence was L. kunkeei YH-15 (NR_026404.1) previously isolated from grape juice. For all F. fructosus strains, the top BLAST hit (99% identity) was F. fructosus NBRC 3516 (NR_113579.1). L. kunkeei was widespread throughout all of the sampling places, whereas F. fructosus was found only in two geographical locations (Mottola and Valenzano). All L. kunkeei and F. fructosus strains grew well in FYP broth (OD620s ranging from 0.74 to 1.77) but poorly in GYP broth under anaerobic conditions (OD620s ranging from 0.024 to 0.165) and were thus regarded as FLAB. Only L. plantarum and L. fermentum strains grew well in both FYP and GYP under anaerobic conditions. Strains recognized as FLAB were subjected to RAPD-PCR analysis in order to exclude clonal relatedness. Conventional analyses of community profiles based on partial sequencing of the 16S rRNA gene may fail to show the strains' differences. Although 16S rRNA sequences indicate a close phylogenetic relationship, even closely related strains may display massive differences in functional gene content, due to the dynamic nature of bacterial genomes (17, 21). The reproducibility of the RAPD fingerprints was assessed by comparing the PCR products obtained from three separate cultures of the same strain. Primers M13, P7, and P4 generated different patterns and were used for cluster analysis. At the similarity level of 90%, isolates were grouped into 16 clusters (data not shown). The representative strains for each cluster group were used for further analysis (Table 1). RAPD-PCR patterns showed a relative high diversity at the strain level among all of the sampling places, and at least two unique patterns were found for each geographical location.
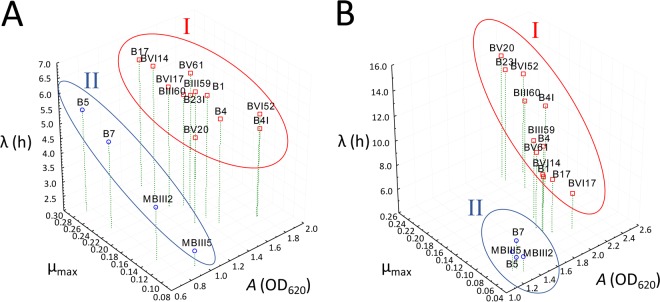
The kinetics of growth in FYP broth incubated anaerobically and GYP broth incubated aerobically under stirring conditions were determined. All FLAB strains grew under both the conditions, but the increases of cell density (A) depended on the strains and on the medium (Fig. 1; see Table S1 in the supplemental material). GYP broth under aerobic conditions induced the highest A (1.25 to 2.38 OD620 units; median, 1.92 OD620 units) and the longest lag phase (λ) (4.89 to 13.23 h; median, 8.91 h) compared to FYP (0.74 to 1.78 OD620 units and 2.52 to 6.59 h, respectively). Based on the A value, strains were grouped in two clusters (I and II). Ten L. kunkeei strains and two F. fructosus strains (cluster I) showed the highest values of A both in FYP (1.31 to 1.77 OD620 units) and in GYP (1.84 to 2.38 log OD620 units). The lowest values of A in the same media were found for L. kunkeei B7 and F. fructosus MBIII2, MBIII5, and B5 (cluster II) (0.74 to 0.94 and 1.25 to 1.36 OD620 units, respectively) isolated from apiaries placed in Mottola and Valenzano. Strains belonging to cluster I showed an average yield of lactic acid, acetic acid, and ethanol at a ratio of 1:1:0.1 or 0.8:1:0.1 from the fermentation of d-glucose and 0.7:1:0.1 from the fermentation of d-fructose (Tables 2 and 3). A ratio of 0.6:1:0.1 (lactic acid-acetic acid-ethanol) from the fermentation of d-glucose or d-fructose was found for strains belonging to cluster II. All strains produced d-mannitol from the fermentation of d-fructose (Table 2). The final pH values after 24 h of incubation ranged from 4.31 ± 0.02 to 4.42 ± 0.01 in FYP and from 3.99 ± 0.02 to 4.50 ± 0.02 in GYP.
FIG 1.
Three-dimensional (3D) scatter plot based on parameters of the growth kinetics of fructophilic lactic acid bacteria (FLAB) strains during growth in fructose-yeast extract-polypeptone (FYP) broth incubated anaerobically at 30°C for 24 h (A) and glucose-yeast extract-polypeptone (GYP) broth incubated aerobically under stirring conditions (200 rpm) at 30°C for 24 h (B). Growth kinetics were modeled according to the Gompertz equation as modified by Zwietering et al. (21): y = k + A exp{−exp[(μmax e/A)(λ − t) + 1]}, where k is the initial level of the dependent variable to be modeled (OD620 units), A is the difference in cell density between inoculation and the stationary phase of growth, μmax is the maximum growth rate (expressed as OD620 units per hour), λ is the length of the lag phase (expressed in hours), and t is the time. Strains were categorized into two clusters, I and II, according to the differences in cell density between inoculation and the stationary phase of growth (A).
TABLE 2.
Fructose consumption and synthesis of main products by FLAB strains during growth in FYP broth incubated anaerobically at 30°C for 24 h
| Strain | Concn of fructose consumed (mmol liter−1) | Concn of end product (mmol liter−1)a |
Molar ratio of lactic acid-acetic acid-ethanol-mannitol | |||
|---|---|---|---|---|---|---|
| Lactic acid | Acetic acid | Ethanol | Mannitol | |||
| L. kunkeei | ||||||
| BIII60 | 68.5 ± 1.2 AB | 32.3 ± 0.5 AB | 43.2 ± 0.9 D | 5.7 ± 0.5 AB | 25.9 ± 0.5 BCD | 0.7:1:0.1:0.6 |
| BIII59 | 66.3 ± 1.4 BC | 31.2 ± 0.9 BC | 42.9 ± 0.8 DE | 5.4 ± 0.4 AB | 25.7 ± 0.4 BCD | 0.7:1:0.1:0.6 |
| B17 | 67.4 ± 0.9 ABC | 30.3 ± 0.4 D | 42.3 ± 0.7 DE | 5.3 ± 0.2 B | 25.4 ± 0.5 CDE | 0.7:1:0.1:0.6 |
| B7 | 61.4 ± 1.1 D | 26.5 ± 0.7 E | 41.7 ± 0.5 EF | 4.1 ± 0.2 C | 25.0 ± 0.6 DEF | 0.6:1:0.1:0.6 |
| B23I | 66.4 ± 1.3 BC | 31.4 ± 0.6 BC | 42.8 ± 0.6 DE | 5.2 ± 0.3 B | 25.7 ± 0.4 BCD | 0.7:1:0.1:0.6 |
| B4I | 69.2 ± 1.2 A | 33.5 ± 0.9 A | 46.2 ± 0.5 A | 6.4 ± 0.8 A | 27.7 ± 0.6 A | 0.7:1:0.1:0.6 |
| BV61 | 67.8 ± 1.6 ABC | 31.6 ± 0.3 C | 43.0 ± 0.4 D | 5.9 ± 0.6 AB | 25.8 ± 0.4 BCD | 0.7:1:0.1:0.6 |
| BV20 | 66.3 ± 1.0 BC | 32.0 ± 0.3 BC | 43.4 ± 0.7 CD | 5.7 ± 0.5 AB | 26.0 ± 0.3 BC | 0.7:1:0.1:0.6 |
| BVI14 | 66.9 ± 1.2 ABC | 31.8 ± 0.6 BC | 42.8 ± 0.6 DE | 6.0 ± 0.4 A | 25.7 ± 0.4 BCD | 0.7:1:0.1:0.6 |
| BVI17 | 68.9 ± 1.4 AB | 30.9 ± 0.5 CD | 44.3 ± 0.4 BC | 5.2 ± 0.5 AB | 26.6 ± 0.5 AB | 0.7:1:0.1:0.6 |
| BVI52 | 68.1 ± 1.3 AB | 32.9 ± 0.7 AB | 45.3 ± 0.6 AB | 6.2 ± 0.5 A | 27.2 ± 0.6 A | 0.7:1:0.1:0.6 |
| F. fructosus | ||||||
| MBIII2 | 63.2 ± 1.1 D | 25.4 ± 0.6 EF | 40.9 ± 0.7 F | 3.6 ± 0.2 D | 24.5 ± 0.4 F | 0.6:1:0.1:0.6 |
| MBIII5 | 64.8 ± 1.2 CD | 26.1 ± 0.5 E | 41.3 ± 0.5 F | 3.8 ± 0.2 CD | 24.8 ± 0.4 EF | 0.6:1:0.1:0.6 |
| B5 | 62.9 ± 1.1 D | 24.5 ± 0.8 F | 40.1 ± 0.4 G | 3.0 ± 0.3 E | 24.1 ± 0.5 F | 0.6:1:0.1:0.6 |
| B4 | 68.7 ± 1.8 A | 31.4 ± 0.8 BC | 43.9 ± 0.3 C | 6.0 ± 0.5 AB | 26.3 ± 0.3 B | 0.7:1:0.1:0.6 |
| B1 | 68.4 ± 1.3 A | 30.2 ± 0.9 CD | 42.9 ± 0.6 D | 5.8 ± 0.2 A | 25.7 ± 0.9 B | 0.7:1:0.1:0.6 |
Means within a column with different letters are significantly different (P < 0.05).
TABLE 3.
Glucose consumption and synthesis of main products by FLAB strains during growth in GYP broth incubated aerobically under stirring conditions (200 rpm) at 30°C for 24 h
| Strain | Concn of glucose consumed (mM) | Concn of end product (mM)a |
Molar ratio of lactic acid-acetic acid-ethanol | ||
|---|---|---|---|---|---|
| Lactic acid | Acetic acid | Ethanol | |||
| L. kunkeei | |||||
| BIII60 | 60.3 ± 1.6 ABCD | 46.7 ± 0.9 BC | 58.3 ± 0.9 ABC | 6.2 ± 0.2 AB | 0.8:1:0.1 |
| BIII59 | 57.9 ± 1.8 CD | 45.4 ± 0.9 CDE | 56.4 ± 0.8 D | 5.8 ± 0.4 ABC | 0.8:1:0.1 |
| B17 | 56.4 ± 2.3 D | 44.3 ± 0.7 E | 57.4 ± 0.8 CD | 6.3 ± 0.3 AB | 0.8:1:0.1 |
| B7 | 51.9 ± 1.9 EF | 28.7 ± 0.3 F | 48.1 ± 0.7 F | 4.4 ± 0.4 D | 0.6:1:0.1 |
| B23I | 62.3 ± 1.1 A | 47.2 ± 0.8 AB | 57.6 ± 0.6 C | 6.4 ± 0.3 AB | 0.8:1:0.1 |
| B4I | 61.1 ± 1.8 ABC | 46.9 ± 0.9 BC | 57.6 ± 0.7 BC | 5.6 ± 0.2 C | 0.8:1:0.1 |
| BV61 | 59.4 ± 1.4 B | 45.7 ± 0.5 CD | 58.2 ± 0.8 ABC | 5.7 ± 0.4 BC | 0.8:1:0.1 |
| BV20 | 64.3 ± 2.5 A | 46.4 ± 0.7 BC | 59.3 ± 0.8 A | 5.4 ± 0.2 C | 0.8:1:0.1 |
| BVI14 | 61.3 ± 1.5 ABC | 43.9 ± 0.8 E | 57.6 ± 0.9 ABCD | 5.6 ± 0.3 C | 0.8:1:0.1 |
| BVI17 | 62.4 ± 1.6 AB | 44.7 ± 0.8 DE | 59.4 ± 1.1 AB | 5.8 ± 0.4 ABC | 0.8:1:0.1 |
| BVI52 | 62.2 ± 1.4 AB | 47.8 ± 0.7 AB | 60.8 ± 1.8 A | 6.7 ± 0.5 A | 0.8:1:0.1 |
| F. fructosus | |||||
| MBIII2 | 52.9 ± 1.2 EF | 28.4 ± 0.5 F | 48.8 ± 0.8 F | 4.2 ± 0.2 D | 0.6:1:0.1 |
| MBIII5 | 54.2 ± 1.4 E | 27.8 ± 0.5 F | 50.4 ± 0.5 E | 4.6 ± 0.2 D | 0.6:1:0.1 |
| B5 | 52.7 ± 0.8 EF | 28.1 ± 0.4 F | 50.5 ± 0.8 E | 4.3 ± 0.3 D | 0.6:1:0.1 |
| B4 | 50.3 ± 1.8 F | 46.6 ± 0.7 BC | 46.3 ± 0.7 G | 6.6 ± 0.5 AB | 1:1:0.1 |
| B1 | 61.4 ± 2.3 AB | 48.9 ± 0.7 A | 58.3 ± 1.1 AB | 5.3 ± 0.2 C | 0.8:1:0.1 |
Means within a column with different letters are significantly different (P < 0.05).
Phenotypic microarray analysis.
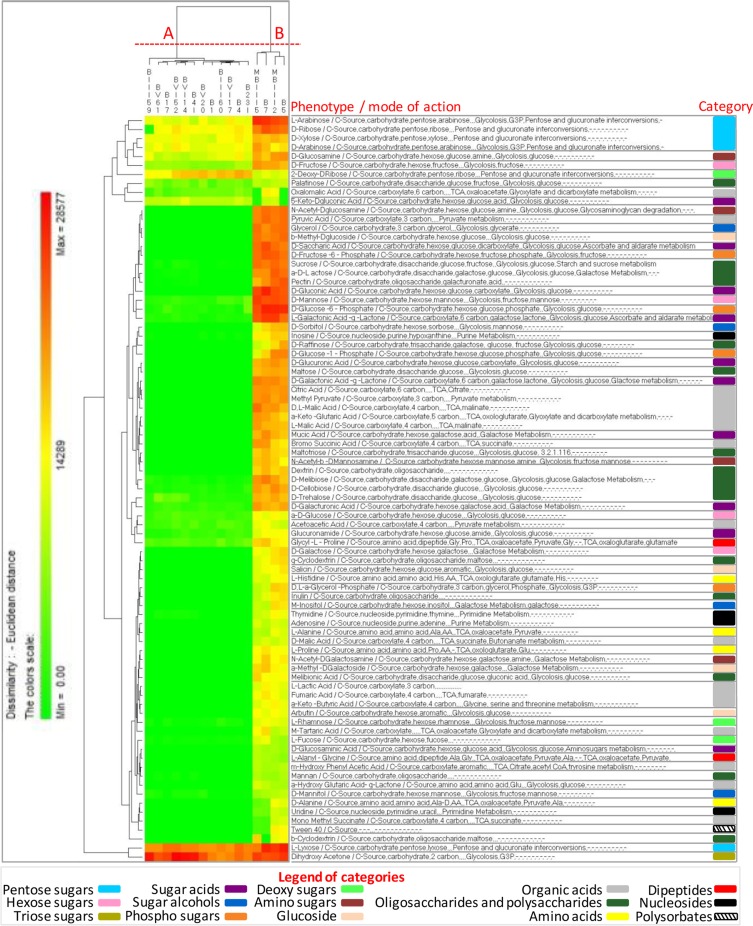
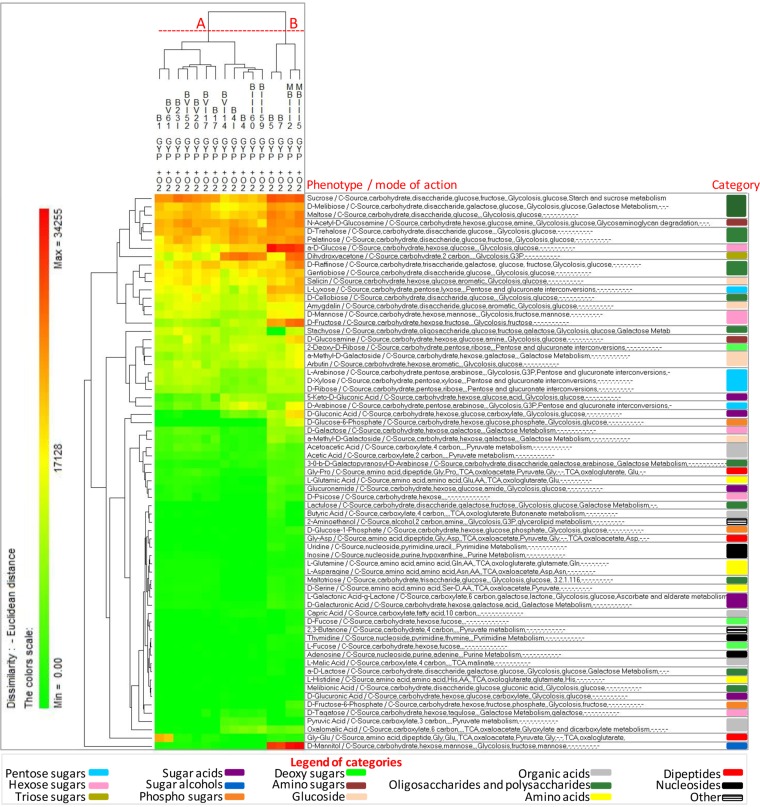
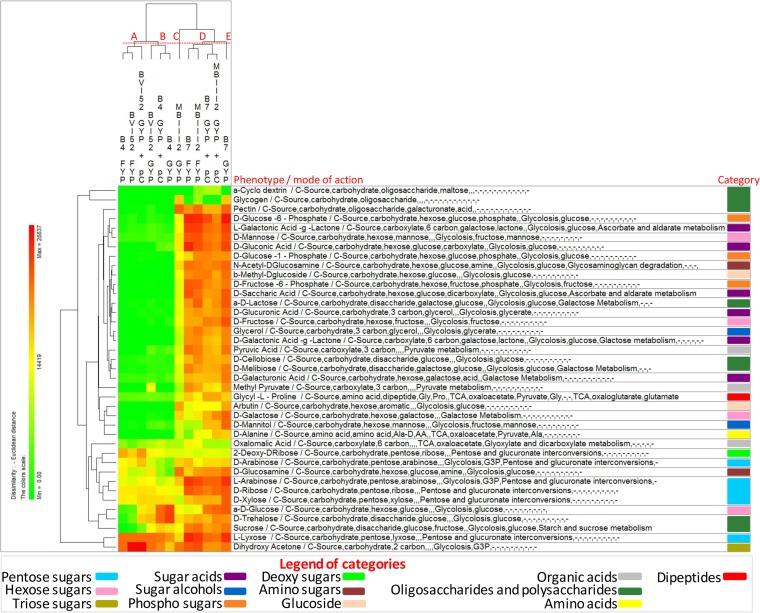
All FLAB strains were also phenotypically characterized using the OmniLog PM technology (Biolog) (Fig. 2 and 3; see Data Set S1 in the supplemental material). The range of phenotypes analyzed included the transport, uptake, and catabolism of 190 carbon sources. After incubation in FYP broth under anaerobic conditions (Fig. 2), 83 carbon sources were differentially consumed, which were mainly involved in pentose and glucuronate interconversions, the tricarboxylic acid (TCA) cycle, glycolysis, pyruvate metabolism, and galactose metabolism (see Data Set S1). Strains were grouped into two clusters, A and B, based on phenotype profiles. Strain clustering reflected growth performance, where cluster A grouped strains with the highest value of A and cluster B those with the lowest value. Certain substrates, such as pentose sugars (e.g., arabinose, ribose, xylose, and lyxose), fructose, palatinose, dihydroxyacetone, oxalomalic acid, or glycyl-l-proline, were commonly consumed by both clusters. Compared to cluster A, cluster B grouped strains consuming preferentially several additional substrates, such as hexose sugars (e.g., mannose and glucose), disaccharides (e.g., sucrose, maltose, and cellobiose), heteropolysaccharide (e.g., pectin), and carboxylic acids (e.g., malic and fumaric acids). After incubation in GYP broth under aerobic conditions (Fig. 3), 67 carbon sources were differentially consumed, which were mainly involved in glycolysis, the TCA cycle, pyruvate metabolism, galactose metabolism, and pentose and glucuronate interconversions (see Data Set S1). Similarly to FYP broth, strains were also grouped into two clusters, A and B, based on phenotype profiles, reflecting growth performance.
FIG 2.
Pseudo-heat map showing the phenotypes of fructophilic lactic acid bacteria (FLAB) strains during growth in fructose-yeast extract-polypeptone (FYP) broth incubated anaerobically at 30°C for 36 h. Each phenotype profile was assayed for growth in the presence of 190 carbon sources using the OmniLog phenotypic microarrays. The color scale shows the cellular respiration activity, evaluated through the OmniLog automated incubator/reader (Biolog) as the area of a region bounded by a color development time series. Sources not listed have not been consumed by any of the strains. Phenotype categories are differentiated with different colors.
FIG 3.
Pseudo-heat map showing the phenotypes of fructophilic lactic acid bacteria (FLAB) strains during growth in glucose-yeast extract-polypeptone (GYP) broth incubated aerobically at 30°C for 36 h. Each phenotype profile was assayed for growth in the presence of 190 carbon sources using the OmniLog phenotypic microarrays. The color scale shows the cellular respiration activity, evaluated through the OmniLog automated incubator/reader (Biolog) as the area of a region bounded by a color development time series. Sources not listed have not been consumed by any of the strains. Phenotype categories are differentiated with different colors.
Antimicrobial activity of phenolic acids.
MICs were determined after 24 h of cultivation in FYP broth containing p-coumaric, caffeic, syringic, or gallic acids (45.5 to 0.7 mmol liter−1) (Fig. 4). The antimicrobial activity of p-coumaric and caffeic acids was not strain or species dependent. A MIC value of 22.7 mmol liter−1 was found for all strains, with the only exception of L. kunkeei BIII60, which showed a higher sensitivity to p-coumaric acid (MIC, 11.4 mmol liter−1). On the contrary, tolerance toward syringic and gallic acids strongly changed, depending on the strain. The highest MIC values for syringic acid (45.5 mmol liter−1) were found with L. kunkeei BV20, BVI14, BVI52, B17, and B7 and F. fructosus B1, whereas F. fructosus B5 was the most sensitive strain (11.4 mmol liter−1). L. kunkeei B23I, BVI14, and BVI17 and F. fructosus B1 showed the highest tolerance toward gallic acid (45.5 mmol liter−1). L. kunkeei BIII60, MBIII2, MBIII5, BVI52, and F. fructosus B5 were the most sensitive strains toward gallic acids (11.4 mmol liter−1). The capacity to tolerate phenolic acids was widespread throughout strains isolated from all five sampling places.
FIG 4.
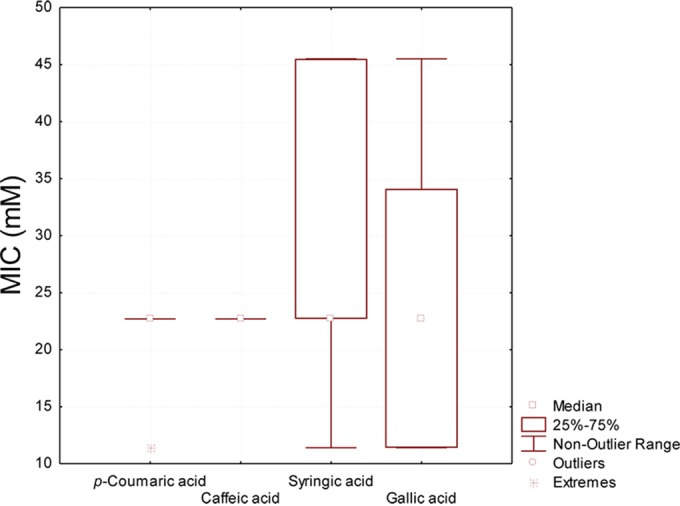
Box plots showing the MICs of p-coumaric, caffeic, syringic, and gallic acids toward fructophilic lactic acid bacteria (FLAB) strains. The final concentrations of the phenolic acids in the sterile 96-well microtiter plates ranged from 45.5 to 0.7 mmol liter−1. Further details are included in Materials and Methods. Analyses were performed in triplicate with three biological replicates for each condition.
Metabolism of phenolic acids during growth in FYP broth.
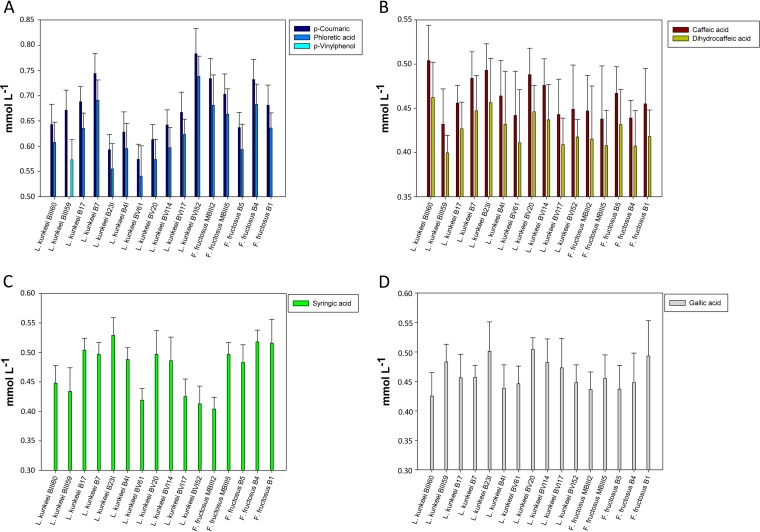
The metabolism of phenolic acids was further investigated in FYP broth supplemented with p-coumaric, caffeic, syringic, or gallic acid at a concentration of 1 mmol liter−1. Compared to the control, the supplementation with phenolic acids did not significantly (P > 0.05) affect the growth (data not shown). Substrate consumption and synthesis of metabolites were strain and phenolic acid dependent (Fig. 5). All FLAB strains consumed p-coumaric acid, and the percentages of degradation ranged from 78.3% (L. kunkeei BVI52) to 57.4% (L. kunkeei BV61). Metabolites were identified by comparison of retention time and UV absorbance with those of the external standards. Almost all of the strains reduced p-coumaric acid to phloretic acid (94.5 to 92.8% conversion efficiency), which was previously identified as a metabolite of p-coumaric acid (51). The only exception was found for L. kunkeei BIII59, which decarboxylated p-coumaric acid to p-vinylphenol (85.4% conversion efficiency). Similarly, all FLAB strains metabolized caffeic acid (50.4 to 43.2%) to dihydrocaffeic acid (93.6 to 91.4% conversion efficiency) through reductase activity. Syringic and gallic acids were also partially metabolized, with percentages of degradation ranging from 40.4 to 52.9% and from 42.5 to 50.4%, respectively. Related metabolites were not identified.
FIG 5.
Consumption (millimoles per liter) of p-coumaric (A), caffeic (B), syringic (C), and gallic (D) acids by fructophilic lactic acid bacteria (FLAB) strains during growth in fructose-yeast extract-polypeptone (FYP) broth supplemented with phenolic acids (1 mmol liter−1). The synthesis (millimoles per liter) of p-coumaric and caffeic acid derivatives (phloretic acid, p-vinylphenol, and dihydrocaffeic acid) is also reported. Analyses were performed in triplicate with three biological replicates for each condition.
Growth in GYP supplemented with p-coumaric acid.
In order to study the use of phenolic acids as external acceptors of electrons, the L. kunkeei BVI52 and B7 and F. fructosus B4 and MBIII2 strains showing the highest phenolic acid reductase activity were grown (initial cell number corresponding to ca. 7 log CFU ml−1) anaerobically at 30°C for 24 h in GYP broth supplemented with p-coumaric acid at a concentration of 2 mmol liter−1. p-Coumaric acid was chosen as the substrate since it is the main phenolic acid present in pollens and induced the highest percentages of degradation by FLAB strains (33, 35). The concentration of 2 mmol liter−1 matched conditions employed in previous investigations (39) and was well below the MIC of these phenolic acids against FLAB. Differences in phenotype during growth in GYP supplemented with p-coumaric acid 2 mmol liter−1 were monitored using the OmniLog phenotype microarray (PM) technology (Biolog) (Fig. 6; see Data Set S1 in the supplemental material). Only the most significant (P < 0.05) differences in metabolic activities under the experimental conditions of this study are shown. Thirty-nine carbon sources were differentially (P < 0.05) consumed, which were mainly involved in glycolysis and pyruvate metabolism, pentose and glucuronate interconversions, and the TCA cycle. Strains were grouped into five clusters based on phenotype profiles. Strain clustering reflected the growth conditions. Phenotype profiles of FLAB strains incubated in GYP supplemented with p-coumaric acid almost matched or approached those in FYP broth, more than profiles in GYP broth. The consumption of certain substrates was strain dependent, with a high metabolic variability in each biotype. The NAD+/NADH ratio was also determined. No significant (P > 0.05) differences were found, with the exception of L. kunkeei BVI52, which showed a higher (P < 0.05) ratio in GYP supplemented with p-coumaric acid (95 ± 2) compared to GYP broth (78 ± 3).
FIG 6.
Pseudo-heat map showing the phenotypes of Lactobacillus kunkeei BVI52 and B7 and Fructobacillus fructosus B4 and MBIII2 during growth in fructose-yeast extract-polypeptone broth (FYP), glucose-yeast extract-polypeptone broth (GYP), and GYP broth supplemented with p-coumaric acid at a concentration of 2 mmol liter−1 (GYP + pC) incubated anaerobically at 30°C for 36 h. Each phenotype profile was assayed for growth in the presence of 190 carbon sources using the OmniLog phenotypic microarrays. The color scale shows the cellular respiration activity, evaluated through the OmniLog automated incubator/reader (Biolog) as the area of a region bounded by a color development time series. Only the most significant (P < 0.05) differences in metabolic activities under the experimental conditions used in this study are shown. Phenotype categories are differentiated with different colors.
DISCUSSION
The microbiota associated with the honeybee Apis mellifera L. is complex and far from being fully understood. Social honeybees are economically and ecologically important for honeybee products and especially for crop pollination, which in turn is related to human food production (52). Lactobacilli have been found to be one of the dominating bacterial groups harbored within the gastrointestinal tract (GIT) of Apis mellifera L. (21, 22). Within this group, fructophilic lactic acid bacteria (FLAB) have been frequently identified, suggesting coevolution and mutualistic interactions with their hosts (6). Environmental selective pressure forced FLAB to develop specific physiological and biochemical features that allow them to grow and survive under harsh conditions, because of the constant nectar flow, secondary plant metabolites, high osmotic pressure, enzymes that discourage biofilm formation, and presence of microorganisms introduced by foraging. A positive effect on the intestinal barrier mechanism and the health status in honeybees by FLAB has been found (19, 24–26). Furthermore, the quite unique biochemical features of FLAB within the lactic acid bacteria (LAB) group may have practical relevance in food fermentation, affecting the levels of products of hexose metabolism in fermented foods, as well in food probiotication.
Seventy-seven presumptive LAB were isolated from GITs of healthy A. mellifera L. worker bees, which were collected from 5 different geographical locations of the Apulia region (Italy). With only two exceptions, all of the isolates showed fructophilic traits. Of the 75 FLAB isolates, 52 were identified as Lactobacillus kunkeei (69%) and 23 as Fructobacillus fructosus (31%). This result agrees with several previous findings whereby L. kunkeei has been found to be one of the most predominant LAB in honeybees (6). Anyway, a variety of culture-independent studies showed that the LAB community in the honeybees' GIT is dominated by Lactobacillus Firm-4 and Firm-5. We hypothesized that the prevalence of fructophilic species may be due in part to culturing bias (40). Apiaries located in Mottola and Valenzano showed the highest diversity at both the species and strain levels, although a relative high diversity at the strain level was found among all the five geographical locations, as shown by RAPD-PCR and phenotypic microarray analyses. Consistent with this finding, a polymorphism at strain level has been previously described in L. kunkeei and F. fructosus (53–55). Representative FLAB strains of each cluster group obtained by combined random amplification of polymorphic DNA patterns were phenotypically characterized using the OmniLog phenotype microarray (PM) Technology (Biolog). A high-throughput phenomic technique was applied, aiming to explore the metabolism of FLAB. Surprisingly, 83 carbon sources were differentially consumed, mainly involved in pentose and glucuronate interconversions, the tricarboxylic acid (TCA) cycle, glycolysis, pyruvate metabolism, and galactose metabolism. Phenotypes identified in FLAB may facilitate several metabolic processes in honeybees, such as sugar metabolism, pectin digestion, and nectar detoxification. Indeed, not all of the nectar sugars are equally used by honeybees, and mannose has been found to be toxic (22). To date, the literature reports that just a few carbohydrates are fermented by FLAB (4) and that Fructobacillus spp. lack pentose and glucuronate interconversions and do not metabolize mannose, galactose, starch, sucrose, amino sugars, or nucleotide sugars (8). Nevertheless, L. kunkeei and F. fructosus isolated from the honeybee's gut represent a relatively heterogeneous group, as they have been shown to comprise many plasmids with high diversity among different strains (53, 55). Plasmids may encode important metabolic traits, such as sugar catabolism. As mobile elements, plasmids may be lost or acquired and contribute to an important polymorphism among fructophilic strains. Phenome typing grouped the strains into two clusters (A and B) reflecting their growth performance. The lowest increment of cell density (cluster B) was associated with the preferential consumption of several additional substrates. This finding may be a sign of niche-specific regressive evolution of FLAB, particularly a metabolic simplification based on sugar availability (7, 28–30). We speculate that strains belonging to cluster B are characterized by a lower level of niche specialization and higher nutritional requirements, which in turn lead to lower growth performances.
Secondary plant metabolites, including tannins, phenolics, alkaloids, and terpenes, can also be found in pollen. These metabolites are usually interpreted as attractive and not deterrent to pollinators. Nevertheless, it is well known that phenolic acids exert a bactericidal activity (38, 49). The effect of phenolic acids on LAB was previously documented (38, 39, 49), but no studies have considered FLAB species. The capacity to tolerate phenolic acids is species and strain dependent. Depending on the chemical structure and concentration of phenolic acids of the food matrices, bacterial growth and viability are diversely affected. Low concentrations of gallic acid exert stimulatory effects on the growth and malolactic activity of LAB (38). Hydroxycinnamic acids may act as external electron acceptors for strictly heterofermentative LAB (39). Conversely, high concentrations of phenolic acids (>3 mmol liter−1) negatively affect the integrity of the cell wall and membrane and dissipate the pH gradient. Phenolic acids also delay the metabolism of carbohydrates by LAB (38, 51). Under the experimental conditions of this study, the antimicrobial activity of p-coumaric and caffeic acids was not strain or species dependent. On the contrary, tolerance toward syringic and gallic acids strongly changed as a function of the strain. Basically, FLAB strains showed higher MIC values than other LAB species or values comparable to those of the most tolerant species, as previously reported (39). This high tolerance to phenolic acids may be somewhat related to the nutritional regimens associated with honeybees and abundance of phenolics in pollen (33).
LAB may metabolize phenolic acids by strain-specific decarboxylase and/or reductase activities. In particular, caffeic and p-coumaric acids may be respectively reduced to dihydrocaffeic and phloretic acids or decarboxylated to the corresponding vinyl derivatives (vinyl catechol and p-vinylphenol, respectively) (38). Gallic acid may be decarboxylated to pyrogallol, whereas no data have been previously reported regarding syringic acid metabolism by LAB (38). First, this study provides a comprehensive overview on the degradation of phenolic acids for FLAB species. Only the presence of the pad gene coding for a phenolic acid decarboxylase in Lactobacillus florum has been previously reported (5). Under the experimental conditions of this study, all FLAB strains consumed p-coumaric acid and caffeic acid. Reductase activities mainly emerged from the analysis of the metabolism of phenolic acids, thus suggesting a specific physiological significance for FLAB species. The only exception was found for L. kunkeei BIII59, which decarboxylated p-coumaric acid to p-vinylphenol. Syringic and gallic acids were also partially metabolized, but related metabolites were not identified, needing more in-depth analysis. These findings are the first data reported in relation to syringic acid degradation by LAB.
Although the degradation of phenolic acids is the main route to detoxify these compounds, the physiological significance was not yet fully elucidated. FLAB require mandatory additional external electron acceptors to regenerate the reduced cofactor NADH. Do FLAB use phenolic acids as external electron acceptors and to enhance glucose dissimilation to counteract the stressful conditions generated by phenolic compounds? As a major component of pollen grains, p-coumaric acid is ubiquitous in the natural diet of honeybees. The metabolism and physiology of FLAB strains, showing phenolic acid reductase activity toward p-coumaric acid, was further investigated in GYP and FYP broth under anaerobic conditions to exclude oxygen as a potential external electron acceptor. Despite the inevitable interference due to the high number of variables involved (e.g., growth conditions and strain-specific variations), the phenome-wide analytical technique and the model systems used allow us to highlight a clear trend. As determined by Omnilog PM technology, the metabolic fingerprint of FLAB strains incubated in GYP (glucose-based medium) supplemented with p-coumaric acid almost matched or approached profiles found in FYP broth, in which fructose acts as an electron acceptors, more than those found in GYP broth. Thirty-nine carbon sources were differentially consumed under the different culture conditions, but the consumption of certain substrates was strain dependent, with a high metabolic variability in each biotype. Among these, several substrates are involved in pathways affecting the NAD+/NADH ratio, such as glycerol and pyruvic acid. Glycerol may have a role in NAD+ regeneration by reduction of 3-hydroxypropionaldehyde to 1,3-propanediol. Glucose has a substantial impact on glycerol metabolism (56). Pyruvate may be used as an external acceptor for reoxidation of NADH from hexose oxidation (57). For instance, glycerol was most widely used by L. kunkeei strain B7 cultured in GYP broth rather than FYP broth and GYP broth supplemented with p-coumaric acid. L. kunkeei BVI52 preferentially used larger amounts of pyruvic acid and methyl pyruvate if cultured in GYP broth not supplemented with p-coumaric acid. The same strain showed also a higher NAD+/NADH ratio in GYP broth supplemented with p-coumaric acid compared to GYP broth. Taken together, the metabolic responses observed in this study suggested that FLAB may use p-coumaric acid as an external electron acceptor, enhancing glucose dissimilation but less efficiently than other acceptors of electrons (e.g., fructose and pyruvic acid) (Fig. 7).
FIG 7.
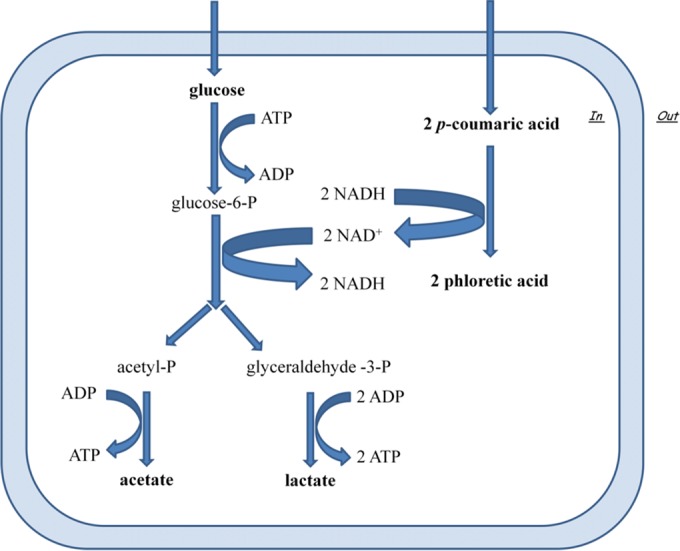
Schematic representation of the presumptive NAD+/NADH recycling mechanism and its links to sugar catabolism by fructophilic lactic acid bacterial strains. The alternative electron acceptor suggested is p-coumaric acid, which can be reduced to phloretic acid.
FLAB are important components in honeybees' GITs, and this study provides more in-depth knowledge on their metabolism. Our findings would be valuable to link unique biochemical features of FLAB and their habitat. These bacteria had to develop specific physiological and biochemical characteristics, such as tolerance to phenolic acids. FLAB may enhance glucose dissimilation through reduction of p-coumaric acid, aiming to counteract the stressful conditions generated by phenolics. The practical relevance of these findings is represented by the potential application in food fermentation. The screening of FLAB strains based on enzyme activities toward phenolic acids may allow the selection of starter cultures with both technological and functional attitudes. Phenolic volatile derivatives resulting from the bioconversion pathways may contribute to the aroma attributes of fermented foods or honeybee products. p-Vinylphenol is one of the permitted food additives and is approved as flavoring agent (38). Phenol derivatives may also exert biological activities. Dihydrocaffeic acid, the reduced derivative of caffeic acid, has well-known higher antioxidant activity than its precursor (58, 59). Thus, the choice of FLAB starter strains with specific enzymatic activities involving phenolic acids may have a promising role in food fermentation.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02194-16.
REFERENCES
- 1.Endo A, Futagawa-Endo Y, Dicks LM. 2009. Isolation and characterization of fructophilic lactic acid bacteria from fructose-rich niches. Syst Appl Microbiol 32:593–600. doi: 10.1016/j.syapm.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Endo A, Futagawa-Endo Y, Sakamoto M, Kitahara M, Dicks LM. 2010. Lactobacillus florum sp. nov., a fructophilic species isolated from flowers. Int J Syst Evol Microbiol 60:2478–2482. doi: 10.1099/ijs.0.019067-0. [DOI] [PubMed] [Google Scholar]
- 3.Endo A, Irisawa T, Futagawa-Endo Y, Sonomoto K, Itoh K, Takano K, Okada S, Dicks LM. 2011. Fructobacillus tropaeoli sp. nov., a fructophilic lactic acid bacterium isolated from a flower. Int J Syst Evol Microbiol 61:898–902. doi: 10.1099/ijs.0.023838-0. [DOI] [PubMed] [Google Scholar]
- 4.Neveling DP, Endo A, Dicks LM. 2012. Fructophilic Lactobacillus kunkeei and Lactobacillus brevis isolated from fresh flowers, bees and bee-hives. Curr Microbiol 65:507–515. doi: 10.1007/s00284-012-0186-4. [DOI] [PubMed] [Google Scholar]
- 5.Mtshali PS, Divol B, Du Toit M. 2012. Identification and characterization of Lactobacillus florum strains isolated from South African grape and wine samples. Int J Food Microbiol 153:106–113. doi: 10.1016/j.ijfoodmicro.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Endo A, Salminen S. 2013. Honeybees and beehives are rich sources for fructophilic lactic acid bacteria. Syst Appl Microbiol 36:444–448. doi: 10.1016/j.syapm.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Endo A, Tanaka N, Oikawa Y, Okada S, Dicks L. 2014. Fructophilic characteristics of Fructobacillus spp. may be due to the absence of an alcohol/acetaldehyde dehydrogenase gene (adhE). Curr Microbiol 68:531–535. doi: 10.1007/s00284-013-0506-3. [DOI] [PubMed] [Google Scholar]
- 8.Endo A, Tanizawa Y, Tanaka N, Maeno S, Kumar H, Shiwa Y, Okada S, Yoshikawa H, Dicks L, Nakagawa J, Arita M. 2015. Comparative genomics of Fructobacillus spp. and Leuconostoc spp. reveals niche-specific evolution of Fructobacillus spp. BMC Genomics 16:1. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porcellato D, Frantzen C, Rangberg A, Umu OC, Gabrielsen C, Nes IF, Amdam GV, Diep DB. 2015. Draft genome sequence of Lactobacillus kunkeei AR114 isolated from honey bee gut. Genome Announc 3:e00144-15. doi: 10.1128/genomeA.00144-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thaochan N, Drew RA, Hughes JM, Vijaysegaran S, Chinajariyawong A. 2010. Alimentary tract bacteria isolated and identified with API-20E and molecular cloning techniques from Australian tropical fruit flies Bactrocera cacuminata and B. tryoni. J Insect Sci 10:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He H, Chen Y, Zhang Y, Wei C. 2011. Bacteria associated with gut lumen of Camponotus japonicus Mayr. Environ Entomol 40:1405–1409. doi: 10.1603/EN11157. [DOI] [PubMed] [Google Scholar]
- 12.Aizen MA, Harder LD. 2009. The global stock of domesticated honey bees is growing slower than agricultural demand for pollination. Curr Biol 19:915–918. doi: 10.1016/j.cub.2009.03.071. [DOI] [PubMed] [Google Scholar]
- 13.Potts SG, Roberts SP, Dean R, Marris G, Brown MA, Jones R, Neumann P, Settele J. 2010. Declines of managed honey bees and beekeepers in Europe. J Apic Res 49:15–22. doi: 10.3896/IBRA.1.49.1.02. [DOI] [Google Scholar]
- 14.Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, Quan PL, Briese T, Hornig M, Geiser DM, Martinson V, van Engelsdorp D, Kalkstein AL, Drysdale A, Hui J, Zhai J, Cui L, Hutchison SK, Simons JF, Egholm M, Pettis JS, Lipkin WI. 2007. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 15.Vanengelsdorp D, Underwood R, Caron D, Hayes JJ. 2007. Estimate of managed colony losses in the winter of 2006–2007: a report commissioned by the Apiary Inspectors of America. Am Bee J 147:599–603. [Google Scholar]
- 16.Kwong WK, Moran NA. 2016. Gut microbial communities of social bees. Nat Rev Microbiol 14:374–384. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell E, Ratnayeke N, Moran NA. 2 August 2016. Strain diversity and host specificity in a specialized gut symbiont of honeybees and bumblebees. Mol Ecol doi: 10.1111/mec.13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz RS, Moran NA, Evans JD. 2016. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc Natl Acad Sci U S A 113:9345–9350. doi: 10.1073/pnas.1606631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vásquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, Szekely L, Olofsson TC. 2012. Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS One 7:e33188. doi: 10.1371/journal.pone.0033188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. 2011. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol 20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 21.Engel P, Martinson VG, Moran NA. 2012. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A 109:11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee FJ, Rusch DB, Stewart FJ, Mattila HR, Newton IL. 2015. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ Microbiol 17:796–815. doi: 10.1111/1462-2920.12526. [DOI] [PubMed] [Google Scholar]
- 23.Moran NA, Hansen AK, Powell JE, Sabree ZL. 2012. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7:36393. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vásquez A, Olofsson TC, Sammataro D. 2009. A scientific note on the lactic acid bacterial flora discovered in the honey stomach of Swedish honey bees—a continuing study on honey bees in the USA. Apidologie 40:26–28. doi: 10.1051/apido:2008063. [DOI] [Google Scholar]
- 25.Vásquez A, Olofsson TC. 2009. The lactic acid bacteria involved in the production of bee pollen and bee bread. J Apic Res 48:189–195. doi: 10.3896/IBRA.1.48.3.07. [DOI] [Google Scholar]
- 26.Forsgren E, Olofsson TC, Vásquez A, Fries I. 2010. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 42:99–108. [Google Scholar]
- 27.Kim EB, Tyler CA, Kopit LM, Marco ML. 2013. Draft genome sequence of fructophilic Lactobacillus florum. Genome Announc 1:e00025-12. doi: 10.1128/genomeA.00025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, Leblond-Bourget N, Decaris B, Bolotin A, Delorme C, Dusko Ehrlich S, Guédon E, Monnet V, Renault P, Kleerebezem M. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol Rev 29:435–463. [DOI] [PubMed] [Google Scholar]
- 29.Vogel RF, Pavlovic M, Ehrmann MA, Wiezer A, Liesegang H, Offschanka S, Voget S, Angelov A, Böcker G, Liebl W. 2011. Genomic analysis reveals Lactobacillus sanfranciscensis as stable element in traditional sourdoughs. Microb Cell Fact 10(Suppl 1):S6. doi: 10.1186/1475-2859-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makarova KS, Koonin EV. 2007. Evolutionary genomic of lactic acid bacteria. J Bacteriol 189:1199–1208. doi: 10.1128/JB.01351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adler LS. 2000. The ecological significance of toxic nectar. Oikos 91:409–420. doi: 10.1034/j.1600-0706.2000.910301.x. [DOI] [Google Scholar]
- 32.Irwin RE, Adler LS. 2008. Nectar secondary compounds affect self-pollen transfer: implications for female and male reproduction. Ecology 89:2207–2217. doi: 10.1890/07-1359.1. [DOI] [PubMed] [Google Scholar]
- 33.Ulusoy E, Kolayli S. 2014. Phenolic composition and antioxidant properties of Anzer bee pollen. J Food Biochem 38:73–82. doi: 10.1111/jfbc.12027. [DOI] [Google Scholar]
- 34.Campos M, Markham KR, Mitchell KA, da Cunha AP. 1997. An approach to the characterization of bee pollens via their flavonoid/phenolic profiles. Phytochem Anal 8:181–185. [Google Scholar]
- 35.Mao W, Schuler MA, Berenbaum MR. 2013. Honey constituents up-regulate detoxification and immunity genes in the western honey bee Apis mellifera. Proc Natl Acad Sci U S A 110:8842–8846. doi: 10.1073/pnas.1303884110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manson JS, Otterstatter MC, Thomson JD. 2010. Consumption of a nectar alkaloid reduces pathogen load in bumble bees. Oecologia 162:81–89. doi: 10.1007/s00442-009-1431-9. [DOI] [PubMed] [Google Scholar]
- 37.Martinez JL, Sánchez MB, Martínez-Solano L, Hernandez A, Garmendia L, Fajardo A, Alvarez-Ortega C. 2009. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol Rev 33:430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- 38.Rodrìguez H, Curiel JA, Landete JM, de Las Rivas B, de Felipe FL, Gòmez-Cordovés C. 2009. Food phenolics and lactic acid bacteria. Int J Food Microbiol 132:79–90. doi: 10.1016/j.ijfoodmicro.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 39.Filannino P, Gobbetti M, De Angelis M, Di Cagno R. 2014. Hydroxycinnamic acids used as external acceptors of electrons: an energetic advantage for strictly heterofermentative lactic acid bacteria. Appl Environ Microbiol 80:7574–7582. doi: 10.1128/AEM.02413-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson KE, Sheehan TH, Mott BM, Maes P, Snyder L, Schwan MR, Walton A, Jones BM, Corby-Harris V. 2013. Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS One 8:e83125. doi: 10.1371/journal.pone.0083125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Degrandi-Hoffman G, Eckholm BJ, Huang MH. 2013. A comparison of bee bread made by Africanized and European honey bees (Apis mellifera) and its effects on hemolymph protein titers. Apidologie 44:52–63. doi: 10.1007/s13592-012-0154-9. [DOI] [Google Scholar]
- 42.De Angelis M, Siragusa S, Berloco M, Caputo L, Ragni A, Burzigotti R, Gobbetti M. 2006. Selection of potential probiotic lactobacilli from pig feces to be used as additives in pelleted feeding. Res Microbiol 157:792–801. doi: 10.1016/j.resmic.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 43.Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Madden TL, Shaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goebel BM, Stackebrandt E. 1994. Cultural and phylogenetic analysis of mixed microbial populations found in natural and commercial bioleaching environments. Appl Environ Microbiol 60:1614–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Cagno R, Minervini G, Rizzello CG, Lovino R, Servili M, Taticchi A, Urbani S, Gobbetti M. 2011. Exploitation of sweet cherry (Prunus avium L.) puree added of stem infusion through fermentation by selected autochthonous lactic acid bacteria. Food Microbiol 28:900–909. doi: 10.1016/j.fm.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Zwietering MH, Jongeberger I, Roumbouts FM, Van't Riet K. 1990. Modelling of bacterial growth curve. Appl Environ Microbiol 56:1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeppa G, Conterno L, Gerbi V. 2001. Determination of organic acids, sugars, diacetyl, and acetoin in cheese by high-performance liquid chromatography. J Agric Food Chem 49:2722–2726. doi: 10.1021/jf0009403. [DOI] [PubMed] [Google Scholar]
- 49.Sanchez-Maldonado AF, Schieber A, Gänzle MG. 2011. Structure-function relationships of the antibacterial activity of phenolic acids and their metabolism by lactic acid bacteria. J Appl Microbiol 111:1176–1184. doi: 10.1111/j.1365-2672.2011.05141.x. [DOI] [PubMed] [Google Scholar]
- 50.Rodrìguez H, Landete JM, Rivas BDL, Muñoz R. 2008. Metabolism of food phenolic acids by Lactobacillus plantarum CECT 748T. Food Chem 107:1393–1398. doi: 10.1016/j.foodchem.2007.09.067. [DOI] [Google Scholar]
- 51.Barthelmebs L, Divies C, Cavin JF. 2000. Knockout of the p-coumarate decarboxylase gene from Lactobacillus plantarum reveals the existence of two other inducible enzymatic activities involved in phenolic acid metabolism. Appl Environ Microbiol 66:3368–3375. doi: 10.1128/AEM.66.8.3368-3375.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aizen MA, Garibaldi LA, Cunningham SA, Klein AM. 2008. Long-term global trends in crop yield and production reveal no current pollination shortage but increasing pollinator dependency. Curr Biol 18:1572–1575. doi: 10.1016/j.cub.2008.08.066. [DOI] [PubMed] [Google Scholar]
- 53.Rangberg A, Diep DB, Rudi K, Amdam GV. 2012. Paratransgenesis: an approach to improve colony health and molecular insight in honey bees (Apis mellifera)? Integr Comp Biol 52:89–99. doi: 10.1093/icb/ics089. [DOI] [PubMed] [Google Scholar]
- 54.Asenjo F, Olmos A, Henríquez-Piskulich P, Polanco V, Aldea P, Ugalde JA, Trombert AN. 2016. Genome sequencing and analysis of the first complete genome of Lactobacillus kunkeei strain MP2, an Apis mellifera gut isolate. PeerJ 4:e1950. doi: 10.7717/peerj.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Linjordet MS. 2016. A comparative analysis of lactic acid bacteria isolated from honeybee gut and flowers, with focus on phylogeny and plasmid profiling. M.S. thesis Norwegian University of Life Sciences, Oslo, Norway. [Google Scholar]
- 56.Lüthi-Peng Q, Dileme F, Puhan Z. 2002. Effect of glucose on glycerol bioconversion by Lactobacillus reuteri. Appl Microbiol Biotechnol 59:289–296. [DOI] [PubMed] [Google Scholar]
- 57.Zaunmüller T, Eichert M, Richter H, Unden G. 2006. Variations in the energy metabolism of biotechnologically relevant heterofermentative lactic acid bacteria during growth on sugars and organic acids. Appl Microbiol Biotechnol 72:421–429. [DOI] [PubMed] [Google Scholar]
- 58.Huang J, de Paulis T, May JM. 2004. Antioxidant effects of dihydrocaffeic acid in human EA.hy926 endothelial cells. J Nutr Biochem 15:722–729. doi: 10.1016/j.jnutbio.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Cavin JF, Barthelmebs L, Diviès C. 1997. Molecular characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum: gene cloning, transcriptional analysis, overexpression in Escherichia coli, purification, and characterization. Appl Environ Microbiol 63:1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.