ABSTRACT
Akkermansia muciniphila colonizes the mucus layer of the gastrointestinal tract, where the organism can be exposed to the oxygen that diffuses from epithelial cells. To understand how A. muciniphila is able to survive and grow at this oxic-anoxic interface, its oxygen tolerance and response and reduction capacities were studied. A. muciniphila was found to be oxygen tolerant. On top of this, under aerated conditions, A. muciniphila showed significant oxygen reduction capacities and its growth rate and yield were increased compared to those seen under strict anaerobic conditions. Transcriptome analysis revealed an initial oxygen stress response upon exposure to oxygen. Thereafter, genes related to respiration were expressed, including those coding for the cytochrome bd complex, which can function as a terminal oxidase. The functionality of A. muciniphila cytochrome bd genes was proven by successfully complementing cytochrome-deficient Escherichia coli strain ECOM4. We conclude that A. muciniphila can use oxygen when it is present at nanomolar concentrations.
IMPORTANCE This article explains how Akkermansia muciniphila, previously described as a strictly anaerobic bacterium, is able to tolerate and even benefit from low levels of oxygen. Interestingly, we measured growth enhancement of A. muciniphila and changes in metabolism as a result of the oxygen exposure. In this article, we discuss similarities and differences of this oxygen-responsive mechanism with respect to those of other intestinal anaerobic isolates. Taken together, we think that these are valuable data that indicate how anaerobic intestinal colonizing bacteria can exploit low levels of oxygen present in the mucus layer and that our results have direct relevance for applicability, as addition of low oxygen concentrations could benefit the in vitro growth of certain anaerobic organisms.
INTRODUCTION
The gastrointestinal (GI) tract harbors a rich and diverse microbial community, which has proven to play a role in host health and physiology (1). This microbial community is not in direct contact with epithelial cells; a thin layer of host-derived mucus separates them. The outer layer of mucus is colonized with microbes that differ in composition from the luminal microbiota (2, 3). The mucin glycans are used by some bacteria as growth substrates, resulting in the production of short-chain fatty acids (SCFAs) (4). To the host, the SCFAs are important modulators of gut health (4). To the microbial community, SCFAs are a necessary waste product, and the process of SCFA production is required to maintain the redox balance in the cell, as it can restore the NAD+/NADH ratio (5).
One member of the mucosa-associated microbiota is Akkermansia muciniphila, a mucin-degrading specialist that can use mucin as a sole carbon and nitrogen source (6). A. muciniphila is associated with a healthy GI tract, as its abundance is inversely correlated with several GI tract-related disorders (7). Moreover, it has been shown that A. muciniphila has immune-stimulatory capacities, stimulates host mucin production, increases the mucus layer thickness (6, 8–10), and possibly strengthens the intestinal barrier function (8, 11). On top of this, a causal role of A. muciniphila in protection against high-fat-diet-induced obesity in mice was reported previously (9), and its abundance has been identified as potential prognostic marker for predicting the success of dietary interventions for diabetes (12).
A. muciniphila was initially described as a strict anaerobe (6). However, more recently, it was reported that A. muciniphila can tolerate small amounts of oxygen (10). The oxygen that diffuses from the gastrointestinal epithelial cells is thought to be among the factors that keep strictly anaerobic commensal microbiota at a distance (14, 15). However, several mucosa-associated bacteria have developed strategies to cope with low levels of oxygen (16).
Many microorganisms have to build up mechanisms to protect themselves against oxidative stress, with enzymes such as catalase and superoxide dismutase, small proteins such as thioredoxin and glutaredoxin, and molecules such as glutathione (17). Some molecules are constitutively present and help to maintain an intracellular reducing environment or to scavenge chemically reactive oxygen species (ROS). Among these molecules are nonenzymatic antioxidants such as NADPH and NADH. However, enzymes such as superoxide dismutases (SOD), catalases, and hydroperoxidases are under transcriptional regulation and can decrease the steady-state levels of ROS.
Since adaptation of A. muciniphila to the oxygen levels in the mucus layer has not been studied, we used an approach integrating physiological, genetic, and biochemical analyses to characterize the oxygen response of this mucosal symbiont. Here, we show that A. muciniphila is able to survive and grow at nanomolar levels of oxygen, exposes a complex transcriptional response to oxygen, and contains a functional cytochrome bd complex that could be used as a terminal oxidase.
MATERIALS AND METHODS
A. muciniphila growth conditions.
A. muciniphila MucT (CIP 107961T) was grown in a bicarbonate-buffered basal medium (6) with a pH of 6.5 to 7.0, supplemented with 0.5% (wt/vol) hog gastric mucin (type III; Sigma-Aldrich, St. Louis, MO, USA), as described previously (18). On plates, A. muciniphila was grown on the same mucin-based medium supplemented with 0.8% agar (Oxoid, Basingstoke, United Kingdom), referred to here as mucin-based plates (6). The correlation of optical density at 600 nm (OD600) to CFU counts on plates was determined to be 4.0 × 108 CFU per ml for an OD600 of 1.0.
Oxygen survival and tube experiment.
A fully grown culture was exposed to ambient air and incubated at 37°C while shaking. Survival rates were determined in 4-fold over a period of 48 h. Series of 10-fold dilutions were made, and 2 μl of each dilution was spotted on mucin-based plates.
To test for growth at different oxygen concentrations, gas tube experiments were performed, essentially as described previously (15). For this purpose, a 1% inoculum of a mucin-grown culture was mixed with 20 ml of either of the two molten mucin-based media supplemented with 0.8% agar (∼40°C). Medium was poured into glass tubes under strict anaerobic conditions and sealed with a metal cap through which ambient air could diffuse. Thereafter, the tubes were placed in ambient air. The oxygen penetration was measured by the pink-to-colorless turning point of the resazurin color indicator (Sigma-Aldrich), while growth was observed by the turbidity in the tube. The experiments were repeated at least 3 times.
Fermentor growth of A. muciniphila.
Bacterial cultures were grown in two parallel 1.2-liter fermentors (Bio Console ADI1025 and Bio Controller ADI 1010; Applikon Biotechnology B.V., Delft, The Netherlands) using 0.67 liters of medium. The pH was controlled at 7.2, stirring was performed at 50 rpm, the initial N2/CO2 gas flow (80%/20%) was 2 liters/h, and the temperature was set at 37°C. The mucin medium used was slightly altered, replacing the phosphate buffer with a 40 mM MOPS (morpholinepropanesulfonic acid) buffer (Carl Roth GmBH, Karlsruhe, Germany) and supplementing with 0.25% hog gastric mucin (type III; Sigma-Aldrich, St. Louis, MO, USA) as a sole carbon and nitrogen source (19). Thereafter, the medium was sparged with N2/CO2 to remove all oxygen from the medium. A 0.2 liters/h gas flow of ambient air was applied to the medium to measure the oxygen uptake rate of the medium without A. muciniphila, after which the medium was made anaerobic again using N2/CO2 at 2 liters/h. A 1% A. muciniphila inoculum was used to inoculate the fermentors, after which growth was monitored by measuring the OD600 over a period of 27 h. At an optical density of 0.1, one of the fermentors was switched to 0.2 liters/h of ambient airflow while the other remained in 2 liters/h N2/CO2 flow. Samples for transcriptome and metabolic analyses were taken as described below.
To determine heme-dependent oxygen reduction, A. muciniphila was grown in a heme-deprived synthetic medium using five parallel fermentors (Dasgip-Eppendorf). Hemin (Sigma-Aldrich) was added to three fermentors at a concentration of 2 μg/ml. Oxygen was introduced at 0.2 liters/h at an OD600 of 1 to one fermentor with hemin-containing medium and to one fermentor with hemin-deprived medium. The remaining fermentors were used as controls.
Metabolic analysis.
To determine the production of metabolites, 1.5 ml of bacterial cultures was centrifuged and the supernatant was stored at −20°C. Levels of short-chain fatty acids (SCFAs) were determined by high-performance liquid chromatography (HPLC) as previously described (20). Total protein concentrations were determined by the use of a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Rockford, IL, USA) and Qubit (Life Technologies, Eugene, OR, USA) according to the manufacturer's protocols. The concentration of proteins was found to be 32 mg/liter at an OD600 of 0.1. Metabolic data were analyzed with the Student t test and the Mann-Whitney test. Two-tailed P values of <0.05 were considered statistically significant.
RNA sequencing and transcriptome analysis.
Cells were collected under N2 flow and were immediately centrifuged (4,800 × g, 5 min, 4°C). Cell pellets were directly suspended into TRIzol reagent (Ambion, Life Technologies, Carlsbad, CA, USA) and stored at −80°C until RNA was purified.
Total RNA was isolated by a method combining TRIzol reagent and an RNeasy minikit (Qiagen GmbH, Hilden, Germany), essentially as described previously (21, 22). Genomic DNA was removed by an on-column DNase digestion step during RNA purification (DNase I recombinant, RNase-free; Roche Diagnostics GmbH, Mannheim, Germany). Yield and RNA quality were assessed using an Experion RNA StdSens analysis kit in combination with an Experion system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Depletion of rRNA was performed using a Ribo-Zero kit for bacteria (Epicentre, Madison, WI, USA) according to the manufacturer's instructions. The success of the rRNA depletion step was checked using an Experion RNA StdSens analysis kit in combination with an Experion system. Library construction for whole-transcriptome sequencing (RNA-Seq) was done by the use of a ScriptSeq v2 RNA-Seq library preparation kit in combination with ScriptSeq Index PCR primers (Epicentre, Madison, WI, USA) according to the manufacturer's instructions.
The barcoded cDNA libraries were sent to BaseClear (Leiden, The Netherlands), where they were pooled, and 50-bp sequencing (single-end reads) was performed on two lanes using an Illumina HiSeq 2500 platform in combination with TruSeq Rapid SBS and TruSeq Rapid SR Cluster kits (Illumina, San Diego, CA, USA).
Reads were mapped to the genome of A. muciniphila with Bowtie2 v2.2.1 (23) using default settings, and BAM files were converted with SAMtools v0.1.19 (24). BEDTools v2.17.0 was used to determine the read count for each protein-coding region (25). Only reads that had a minimum 30% length overlap and were mapped on the correct strand were counted. Differential gene expression was assessed using edgeR (26) with a default trimmed mean of M-value (TMM) settings. Sequences have been deposited in ArrayExpress (see below).
Cloning of A. muciniphila cytochrome bd.
The cytochrome bd genes (Amuc_1694 and Amuc_1695) were amplified from genomic DNA using the following primers: Rev_1694 (AGATCACTCGAGAAAGGATCCTCAGTAACTGTGTTCGTTGAGCTGGA) and FW_1695 (TGAACTAGATCTTTTAAGAAGGAGATATACATATGGACGATCCGGTCTTATTATCCC). The PCR was done with Phusion polymerase (Thermo Scientific, Waltham, MA, USA). The PCR conditions were as follows: denaturation at 96°C for 5 min followed by 35 cycles of a denaturation step at 96°C for 10 s, an annealing step at 59°C for 30 s, and an extension step at 72°C for 1 min, and one final extension step at 72°C for 8 min. The PCR product was cloned into plasmid pBbA5c (Addgene plasmid 35281) containing an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible PlacUV5 promoter, a terminator, a p15a origin of replication, and a chloramphenicol resistance marker (27). For this purpose, the PCR product and the destination vector were digested with BglII and XhoI (FastDigest, Thermo Scientific, Waltham, MA, USA), after which they were ligated with T4 ligase (Thermo Scientific, Waltham, MA, USA) overnight at 15°C. A 2-μl volume of the ligation mixture was used directly to transform Escherichia coli NEB5α (New England BioLabs, Ipswich, MA, USA) chemically competent cells according to the manufacturer's instructions. Transformants were selected on plates with LB agar and 25 μg/ml chloramphenicol (Oxoid, Basingstoke, United Kingdom) and incubated overnight at 37°C. The resulting vector (pBbA5c-AmuCytbd) was confirmed by Sanger sequencing.
Transformation of E. coli ECOM4.
Electrocompetent cells of the cytochrome-deficient, kanamycin-resistant E. coli ECOM4 strain (28), kindly provided by Bernhard Ø. Palsson, were prepared by inoculation with 1% of an overnight E. coli ECOM4 preculture (OD600 of 0.6 to 0.7) in 1 liter of M9 medium (Sigma-Aldrich Chemie, Steinheim, Germany) with 25 μg/liter kanamycin (Carl Roth GmBH, Karlsruhe, Germany) and grown at 37°C shaking at 180 rpm until the exponential phase (OD600 of 0.3 to 0.4). The bacterial culture was cooled on ice for 15 min and centrifuged (5,000 × g, 15 min, 4°C); cells were washed twice with 400 ml of cold demineralized water (demi-water) and once with 10 ml of ice-cold 10% (vol/vol) glycerol and resuspended in 2 ml of 10% glycerol. Finally, 80-μl aliquots were prepared and stored at −80°C for later transformation.
Competent E. coli ECOM4 cells were transformed with pBbA5c-AmuCytbd using electroporation in a 2-mm-gap electroporation cuvette (ECM630 Precision Pulse; Harvard Apparatus, Inc., Holliston, MA, USA) at the following settings: 2,500 V, 200 Ω, and 25 μF. Immediately after, 1 ml of SOC medium (0.5 g/liter NaCl, 20 g/liter tryptone, 5 g/liter yeast extract, 2.5 ml/liter KCl, 2.03 g/liter MgCl2·6H2O, 3.60 g/liter glucose) was added and cells were allowed to recover for 1 h at 37°C. The transformed cells were then plated on LB agar plates with appropriate antibiotics (20 μg/ml kanamycin and 25 μg/ml chloramphenicol) to select for ECOM4-AmuCytbd cells.
Growth of E. coli ECOM4-AmuCytbd and metabolic profiling.
E. coli ECOM4 and ECOM4-AmuCytbd strains were grown on M9 medium (Sigma-Aldrich Chemie, Steinheim, Germany) with 20 μg/ml kanamycin and 25 μg/ml chloramphenicol at 37°C with shaking at 180 rpm. For metabolic analysis, cells were grown in precultures, after which equal cell amounts were inoculated into M9 medium containing 18 mM glucose, either induced with the tested optimal concentration (data not shown) of 25 μM IPTG (Fisher Scientific, Fair Lawn, NJ, USA) or without inducer. Samples were taken over a period of 50 h, growth was determined by measuring optical density (OD600), and HPLC analysis was done as described for the A. muciniphila samples.
Spectra of cytochrome bd.
Erlenmeyer flasks containing 500 ml M9 medium were inoculated with E. coli ECOM4 and E. coli ECOM4-Amucytbd with or without IPTG. Cells were collected at the end of the exponential phase by centrifugation (4,800 × g, 10 min). A. muciniphila cells were grown in 200-ml cultures in anaerobic bottles. Pellets of E. coli ECOM4 (Amucytbd) and A. muciniphila were dissolved in 1 ml lysis buffer (50 mM Tris-HCl [pH 8], 1 mM EDTA, 150 mM NaCl, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1% dodecyl maltoside) and lysed by 2 passages through a French press (model SPHC; Stansted Fluid Power Ltd., Essex, United Kingdom) at 16,000 lb/in2 or by four 30-s sonications at an output level of 3.5. Cell debris was removed by centrifugation (10,500 × g, 10 min). Spectra were measured by the use of a UV-2501PC instrument (Shimadzu Corporation, Kyoto, Japan) at 380 to 700 nm, and solutions were reduced using 50 mM sodium dithionite.
Accession number(s).
Sequences determined in this work have been deposited in ArrayExpress under accession no. E-MTAB-5111.
RESULTS
Oxygen tolerance and oxygen reduction of A. muciniphila.
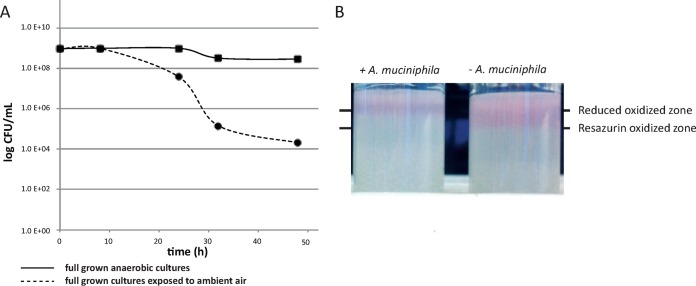
To test oxygen tolerance, survival of A. muciniphila cells was measured after exposure of the cells to ambient air. Based on CFU counts, A. muciniphila cells were able to survive for up to 48 h in ambient air. The viability had dropped to 25% after 24 h and was only 1% after 48 h (Fig. 1A). To test the oxygen reduction capacity of A. muciniphila, the penetration depth of oxygen was measured in a gas tube assay based on the turning point of a redox indicator (resazurin). The oxygenated zone was deeper in noninoculated tubes (10.0 ± 1.0 mm) than in the tubes where A. muciniphila was growing (5.0 ± 1.0 mm) (Fig. 1B).
FIG 1.
(A) Survival of A. muciniphila after exposure to oxygen over time. A survival rate of 1% was measured during 48 h of exposure to ambient air (dashed line), while >90% were still viable in anaerobic cultures (solid line). Points depict averages of results of 4 replicates. (B) Growth of A. muciniphila in the tube assay. Left: an A. muciniphila-inoculated tube. Right: a control tube. The lower line indicates the resazurin-oxidized zone in the control tube; the upper line indicates the reduced oxidized zone in the tube where A. muciniphila was growing. A picture representative of 4 repeats is shown.
A. muciniphila shows enhanced growth under aerated conditions.
The growth of A. muciniphila under aerated conditions was investigated in a fermentor system controlled for temperature and pH. Both dissolved oxygen concentration (expressed in percent dissolved oxygen [dO2]) and redox potential (expressed in millivolts) were measured throughout the experiment. Before A. muciniphila was grown in the fermentor system, a control fermentor (negative control) was run to measure the increase in redox potential and oxygen diffusion by switching the gas flow from an N2 flow of 2.0 liters/h to an ambient airflow of 0.2 liters/h (Fig. 2C). Thereafter, experiments were run in two parallel fermenters. At T1 (the beginning of the exponential phase, at an OD600 of ∼0.1 [gray-shaded area in Fig. 2]), one of the two fermentors was switched to an ambient airflow of 0.2 liters/h (aerated fermentor) and the other fermentor was maintained under strict anaerobic conditions (anaerobic fermentor). After T1, the redox potential showed an increase upon the introduction of ambient air (Fig. 2B); the increased redox potential then declined, however, and the redox potential was lowered again until A. muciniphila reached stationary phase, pointing to the potential use of oxygen as a final electron acceptor. The oxygen concentration remained under our detection level of 0.1% dO2 during the complete period of growth of A. muciniphila and increased only in the stationary phase (Fig. 2B).
FIG 2.
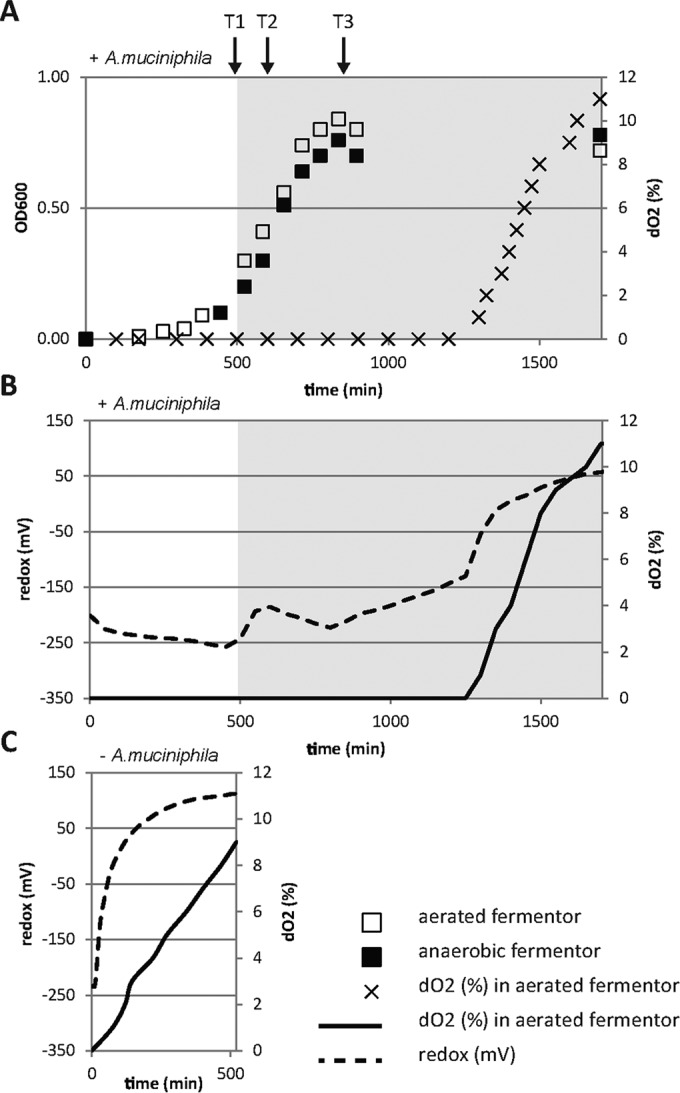
Growth, oxygen concentration, and redox potential of A. muciniphila in aerated fermentor system. (A) Growth of A. muciniphila measured by OD600 in the aerated and anaerobic fermentor and the dissolved oxygen concentration [dO2 (%)] in the aerated fermentor. The gray-shaded area indicates a switch to an ambient airflow of 0.2 liters/h. (B) Redox potential (in millivolts) (dashed line) and the oxygen concentration [dO2 (%)] (solid line) during the growth of A. muciniphila in the aerated fermentor. The gray-shaded area indicates a switch to an ambient airflow of 0.2 liters/h. (C) Redox potential (mV) (dashed line) and the oxygen concentration [dO2 (%)] (solid line) of the negative control (without A. muciniphila). Data are solely from one experiment but are representative of all four experiments performed.
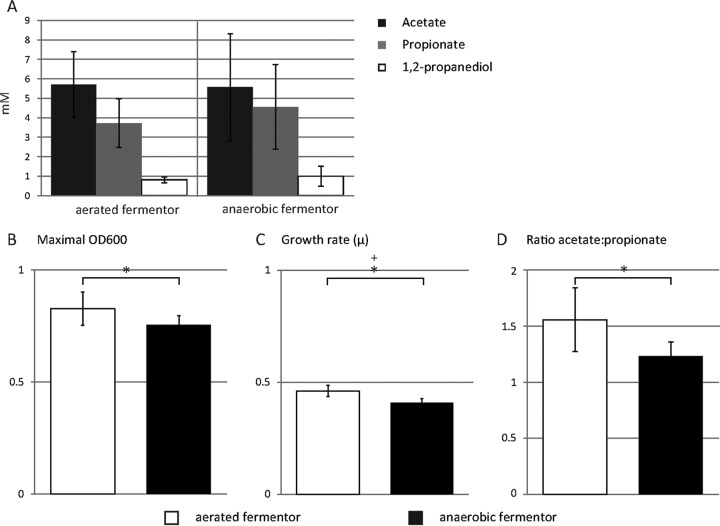
Under aerated conditions, a significantly higher growth rate (P = 0.02) and significantly higher OD600 (P = 0.05) at the end-exponential phase were measured (Fig. 3B and C). The fermentation profile of A. muciniphila consisted of acetate, propionate, and small amounts of 1,2-propanediol, typical for growth in mucin-based medium (Fig. 3A) (29). Under aerated conditions, however, the ratio of acetate to propionate was altered from 1.2 to 1.5 (P = 0.03) (Fig. 3D), suggesting an altered fermentation pathway.
FIG 3.
Growth and physiology of A. muciniphila in the aerated fermentor system. (A) SCFA production profile of A. muciniphila at the stationary phase, including acetate, propionate, and 1,2-propanediol. (B) Maximal OD600 reached during growth in the aerated fermentor and the anaerobic fermentor. (C) Growth rate (μ) based on the OD600 during the exponential-growth phase in the aerated fermentor and the anaerobic fermentor. (D) Ratio of the production of acetate to the production of propionate per fermentor run in the aerated fermentor and the anaerobic fermentor. Bars represent means of n = 4; error bars represent the standard deviations (SD). An asterisk (*) indicates a significant difference determined using the two-tailed Student t test. For panel B, P = 0.05; for panel C, P = 0.02; for panel D, P = 0.03. A plus sign (+) indicates a significant difference determined using the one-tailed Mann-Whitney test (P = 0.03).
The minimal oxygen-reducing capacity of A. muciniphila under the given fermentor conditions was calculated based on the oxygen uptake rate in the negative control and the protein concentration of A. muciniphila cultures. The increase in the oxygen concentration of the negative control was calculated to be 69.4 ± 31.9 mU/liter (mU is defined as nanomoles of substrate per minute). This calculation is based on the difference between the increase in oxygen concentration of the negative control (dO2% in Fig. 2C) and the increase in oxygen concentration of the A. muciniphila-grown fermentor (dO2% in Fig. 2B) for all four experiments. The oxygen reduction capacity of A. muciniphila was determined to be 2.26 ± 0.99 mU/mg total protein (n = 4).
The initial transcriptional response of A. muciniphila to aerated conditions is dominated by the oxygen stress response.
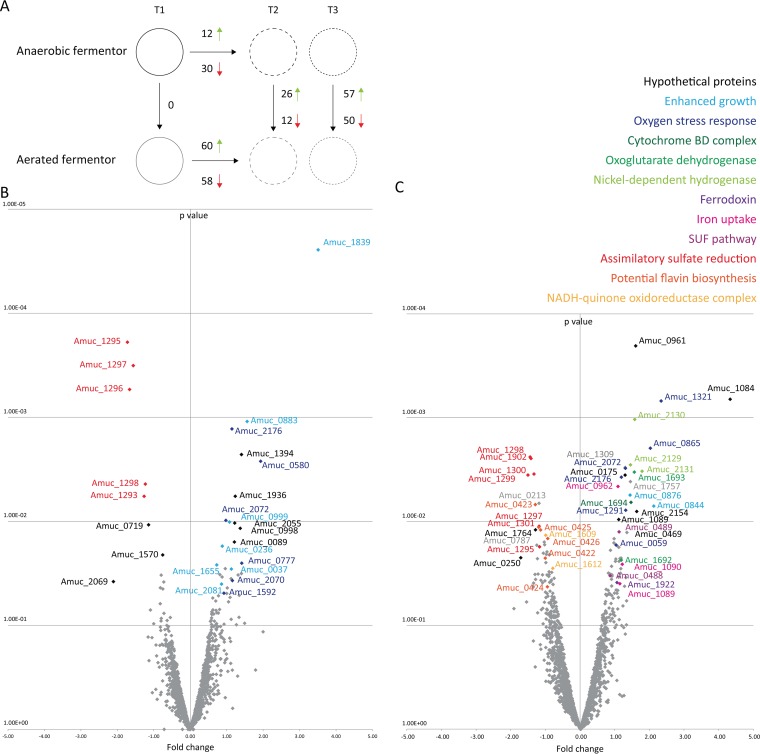
The transcriptome of A. muciniphila was determined by RNA-Seq analysis under both aerated and anaerobic fermentor conditions at the beginning of the exponential phase of growth (before aeration) (T1), during the mid-exponential phase (T2), and at the end-exponential phase (T3) (black arrows in Fig. 2A). All genes that differed significantly between the conditions are listed in Table S1 in the supplemental material. At T1 (before aeration), the two parallel fermentors had identical gene expression profiles (Fig. 4A). Comparing the aerated fermentor to the nonaerated fermentor at T2 (mid-exponential phase) revealed 38 genes that were expressed significantly differently (Fig. 4B). During this initial transcriptional response, a total of 26 genes were significantly upregulated due to the presence of oxygen. Only 18 of the genes are annotated, and 6 of the 18 were annotated as encoding potential oxygen stress-related products: superoxide dismutase; hydroperoxidase; entericidin (EcnAB), homologous to the E. coli EcnAB protein, which is involved in the stress response (30); rubrerythrin, previously implicated in protection from oxidative damage in sulfate-reducing bacteria (31); excinuclease, controlling for potential ROS-mediated DNA damage; and a family 2 glycosyl transferase, part of the capsular polysaccharide biosynthesis pathway upregulated to potentially protect the cells against the presence of oxygen (Table 1). Also, one potential oxidoreductase-encoding gene and six genes that are broadly categorized as potentially growth phase dependent were significantly upregulated (Table 1). Twelve genes were significantly downregulated under aerated conditions. Five of these genes encode the assimilatory sulfate reduction pathway (Fig. 4B and Table 1). Expression of the oxygen stress-related genes, after the initial oxygen response, was assessed by comparing the aerated fermentor results obtained at T2 and T3. Expression of these genes did not further increase significantly, except for the gene encoding hydroperoxidase (see Table S1 in the supplemental material).
FIG 4.
Genes of A. muciniphila differentially expressed under aerated conditions. (A) Schematic representation of the genes significantly differentially expressed under aerated and nonaerated conditions at the different time points. Red arrows indicate downregulated genes; green arrows indicate upregulated genes. (B) Volcano plot of the aerated fermentor versus the nonaerated fermentor at T2. (C) Volcano plot of the aerated fermentor versus the nonaerated fermentor at T3. The P values were determined using edgeR (26) with settings that included the default trimmed mean of M values (TMM) (data corresponding to significant differences in gene expression under all conditions can be found in Table S1 in the supplemental material).
TABLE 1.
Oxygen-responsive genes that are significant differentially expressed between aerated and nonaerated fermentors at T2 and T3a
| Gene product or pathway | Gene | Fold change |
Potential mechanism | |
|---|---|---|---|---|
| T2 | T3 | |||
| T2 aerated vs nonaerated fermenter | ||||
| Amino acid permease | Amuc_0037 | 2.19 | 2.13 | Enhanced growth |
| Ion transport 2 domain-containing protein | Amuc_0236 | 1.84 | 0.79 | Enhanced growth |
| Entericidin EcnAB | Amuc_0580 | 3.82 | 0.94 | Response to starvation conditions |
| Short-chain dehydrogenase/reductase (SDR) | Amuc_0777 | 2.66 | 2.30 | Oxidoreductase |
| Cupin barrel domain-containing protein | Amuc_0883 | 1.08 | 1.96 | Enhanced growth |
| Sigma 54 modulation protein/ribosomal protein S30EA | Amuc_0999 | 2.11 | 1.33 | Enhanced growth |
| Superoxide dismutase | Amuc_1592 | 1.90 | 1.22 | 2 O− + 2 H+ → H2O2 |
| Sulfatase | Amuc_1655 | 1.66 | 0.54 | Sulfate cleavage from mucin protein for enhanced growth |
| Heavy-metal-translocating P-type ATPase | Amuc_1839 | 11.39 | 1.68 | Enhanced growth |
| Hydroperoxidase | Amuc_2070 | 2.23 | 1.90 | 2 H2O2 → 2 H2O + O2 |
| Rubrerythrin | Amuc_2072 | 1.97 | 2.48 | Stress response |
| Family 2 glycosyl transferase | Amuc_2081 | 1.81 | 1.18 | Capsular polysaccharide biosynthesis pathway upregulated potentially to protect against oxygen presence |
| Excinuclease | Amuc_2176 | 2.21 | 2.29 | Control for potential ROS-mediated DNA damage |
| Assimilatory sulfate reduction (Amuc_1294–Amuc_1301) | Amuc_1293 | −2.40 | −1.35 | Not functional due to high redox potential |
| Amuc_1295 | −3.30 | −2.25 | ||
| Amuc_1296 | −3.17 | −1.87 | ||
| Amuc_1297 | −2.96 | −2.27 | ||
| Amuc_1298 | −2.35 | −2.67 | ||
| T3 aerated vs nonaerated fermentor | ||||
| Suf pathway (Amuc_0486–Amuc_0489) | Amuc_0488 | 1.20 | 1.82 | Iron uptake [Fe-S] |
| Amuc_0489 | 1.17 | 2.18 | ||
| Oxoglutarate dehydrogenase (Amuc_1692–Amuc_1693) | Amuc_1692 | 1.61 | 2.28 | Potential readthrough of transcript |
| Amuc_1693 | 1.65 | 2.96 | ||
| Cytochrome bd complex (Amuc_1694–Amuc_1695) | Amuc_1694 | 0.91 | 2.77 | Aerobic respiration |
| FeO iron transporter (Amuc_1089–Amuc_1090) | Amuc_1089 | 0.88 | 2.22 | Iron uptake |
| Amuc_1090 | 0.75 | 2.33 | ||
| Nickel-dependent hydrogenase (Amuc_2128–Amuc_2132) | Amuc_2129 | 1.41 | 2.74 | Oxygen-tolerant hydrogenase that might use the H2 present at its ecological niche |
| Amuc_2130 | 1.32 | 2.98 | ||
| Amuc_2131 | 1.78 | 3.46 | ||
| ATPase AAA | Amuc_0059 | 1.26 | 2.05 | UvrB/UvrC protein; ATPase binding domain; part of NER; control for potential ROS-mediated DNA damage |
| Phosphopyruvate hydratase | Amuc_0844 | 2.65 | 4.37 | Involved in glycolysis, enhanced growth |
| Heavy-metal-translocating P-type ATPase | Amuc_0876 | 1.63 | 2.72 | Enhanced growth |
| Alkyl hydroperoxide reductase | Amuc_1321 | 1.25 | 5.06 | ROS scavenger |
| Ferredoxin | Amuc_1922 | 1.01 | 2.09 | Iron uptake |
| Rubrerythrin | Amuc_2072 | 1.97 | 2.48 | Stress response |
| Excinuclease | Amuc_2176 | 2.21 | 2.29 | Control for potential ROS mediated DNA damage |
| Potential flavin biosynthesis (Amuc_0421–Amuc_1426) | Amuc_0422 | −1.15 | −2.00 | Electron sink |
| Amuc_0423 | −1.51 | −2.45 | ||
| Amuc_0424 | −1.26 | −1.92 | ||
| Amuc_0425 | −1.08 | −2.20 | ||
| Amuc_0426 | −1.40 | −1.91 | ||
| Assimilatory sulfate reduction (Amuc_1294–Amuc_1301) | Amuc_1295 | −3.30 | −2.25 | Redox too high to be functional |
| Amuc_1297 | −2.96 | −2.27 | ||
| Amuc_1298 | −2.35 | −2.67 | ||
| Amuc_1299 | −1.74 | −2.83 | ||
| Amuc_1300 | −1.51 | −2.51 | ||
| Amuc_1301 | −1.05 | −2.28 | ||
| NADH-quinone oxidoreductase complex (Amuc_1604–Amuc_1614) | Amuc_1609 | −1.28 | −1.98 | Not essential in the electron transport from NADH to oxygen in a mutant E. coli strain |
| Amuc_1612 | −1.14 | −1.72 | ||
| Menaquinone pathway | Amuc_1017 | −1.56 | −2.43 | Menaquinone pathway |
Only genes that are involved in oxygen metabolism are included in this table; all other differentially expressed genes are listed in Table S1 in the supplemental material.
End-exponential transcriptional response of A. muciniphila to aerated conditions points to respiration using cytochrome bd as a terminal oxidase.
Comparing the aerated fermentor to the nonaerated fermentor at T3 (end-exponential phase) revealed 107 significantly altered genes (Fig. 4C; see also Table S1 in the supplemental material). During this end-exponential transcriptional response, a total of 57 genes were significantly upregulated under aerated conditions. In similarity to the result seen at T2 a series of oxygen stress-related genes was upregulated (Fig. 4C and Table 1). Three major gene clusters were significantly upregulated under aerated conditions: (i) a cluster involved in iron transport, including the FeO iron transporters; (ii) a respiratory gene cluster, including genes encoding subunits of oxoglutarate dehydrogenase and its adjacent gene encoding cytochrome d ubiquinol oxidase subunit II; and (iii) a nickel-dependent hydrogenase cluster, in which 3 of 5 genes were significantly oxygen induced, pointing to the use of cytochrome bd as a terminal oxidase. Cytochrome bd ubiquinol oxidase subunit I, although abundantly present in the transcriptome, showed no transcriptional induction by oxygen. However, comparing the expression levels of the cytochrome bd subunits in the aerated fermentor at T2 and T3, we see that both subunits showed increased expression over time under aerated conditions (see Table S1). At T3, both the sulfur mobilization (SUF) pathway and the ferredoxin gene were significantly upregulated and were potentially involved in electron transfer. In addition, two genes broadly categorized as potentially growth-phase-dependent genes were significantly upregulated under aerated conditions (Fig. 4C and Table 1). A total of 50 genes were significantly downregulated under aerated conditions (see Table S1) and included the following 3 major gene clusters: (i) the assimilatory sulfate reduction genes; (ii) the potential flavin biosynthesis genes; and (iii) the NADH-quinone oxidoreductase complex genes (Fig. 4C and Table 1).
Cytochrome bd genes of A. muciniphila restore respiration in the cytochrome-deficient mutant Escherichia coli ECOM4.
Under aerated conditions, A. muciniphila showed both slightly increased growth (Fig. 3) and oxygen reduction capacities of 2.26 ± 0.99 mU/mg total protein, pointing to the potential use of oxygen as a final electron acceptor. Moreover, the A. muciniphila genome contains the genes for cytochrome bd subunits I (Amuc_1695) and II (Amuc_1694) and the gene coding for subunit II was significantly upregulated under aerated conditions (Fig. 4). To test if the A. muciniphila cytochrome bd genes encode a functional cytochrome bd complex that can be used for respiration, we complemented the cytochrome-deficient E. coli ECOM4 strain with pBbA5c-AmuCytbd expressing the genes for cytochrome bd subunits I (Amuc_1695) and II (Amuc_1694) of A. muciniphila, resulting in strain ECOM4-AmuCytbd. As previously shown by Portnoy et al. (28), there was no production of acetate by the cytochrome-deficient E. coli ECOM4 strain under oxic conditions, in contrast to the E. coli MG1655 parental strain, which produces ample amounts of acetate (28). However, in strain ECOM4-amuCytbd, expressing the A. muciniphila cytochrome bd genes, acetate production was partly restored upon induction with IPTG (3.2 ± 0.8 mM) and was slightly lower without induction (2.2 ± 0.4 mM) (Table 2). Moreover, the growth yield increased from an OD600 of 0.18 ± 0.04 to an OD600 of 0.43 ± 0.03 in this complemented strain (Table 2).
TABLE 2.
Acetate production of ECOM4-AmuCytbd and ECOM4a
| Parameter | Values at indicated IPTG concn (μM) |
||
|---|---|---|---|
| Strain ECOM4-AmuCytbd |
Strain ECOM4 |
||
| 25 | 0 | 25 | |
| Acetate (mM) | 3.3 (0.8)A | 2.2 (0.4)A | 0.0 (0.0)B |
| Yield (OD600) | 0.4 (0.0)C | 0.4 (0.0)C | 0.2 (0.0)D |
A negative control of M9 medium without bacteria was included. Values represent means (standard deviations [SD]), n = 3. Data with different superscript letters are significantly different as determined using the 2-tailed Student t test.
Cytochrome bd is present and functional in A. muciniphila.
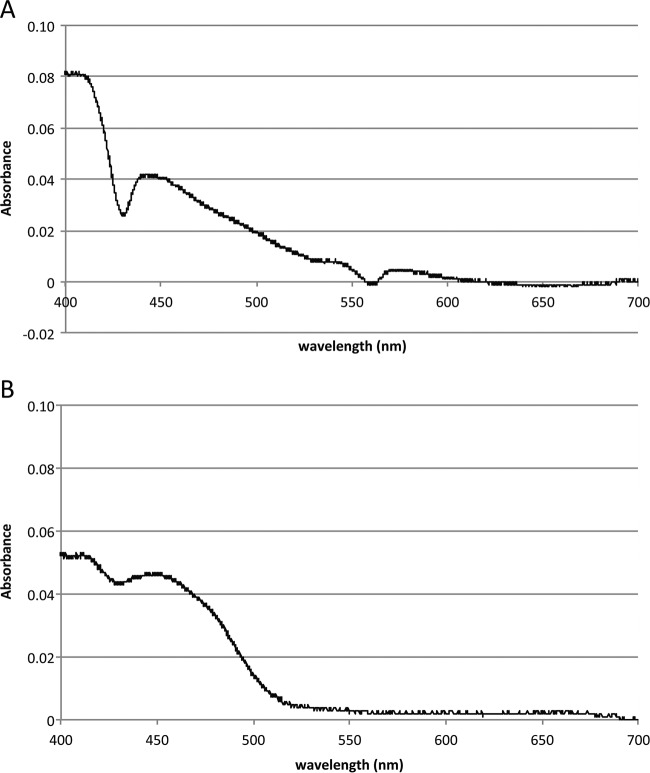
Finally, the presence of the cytochrome bd complex of A. muciniphila was confirmed by spectra showing a small peak at 560 nm, and by a larger peak at 430 nm, indicating the presence of cytochromes (Fig. 5A). The presence of the cytochrome bd complex in the ECOM4-AmuCytbd complemented strain was confirmed using a spectrum measurement that showed a major peak at 430 nm, which is in line with the Soret peak of cytochrome bd (Fig. 5B), as previously described (32, 33). To determine if the oxygen reduction was heme dependent and thus cytochrome bd dependent, three parallel fermentors containing medium with or without hemin were used. Without bacteria, there was no difference observed in the percentage of dO2 between a fermentor with heme and a fermentor without heme (10.5% ± 0.2%). In the presence of A. muciniphila without hemin, the oxygen concentration reached an average saturation of 1.1% ± 0.2% in the first 400 min of aeration. With hemin, the same amount of cells was able to directly and completely reduce the oxygen, preventing accumulation of oxygen in the system. The saturation remained constant at 0.1% ± 0.0% (see Fig. S3 in the supplemental material).
FIG 5.
Cytochrome bd complex of A. muciniphila. (A) Spectrum of A. muciniphila cell extract. The spectrum data show the oxidized spectrum minus the dithionate-reduced spectrum. (B) Spectrum of E. coli ECOM4-AmuCytbd cell extract minus the ECOM4 cell extract spectrum. The baseline is set for absorbance at 700 nm. The two spectra show similar patterns at 430 nm.
DISCUSSION
A. muciniphila is known to colonize the oxic-anoxic interface of the mucus layer. Using an integrated physiological, genetic, and transcriptomic approach, the oxygen response of A. muciniphila was addressed and shown to be highly complex and effective and to involve a functional respiratory complex. Analysis of the global transcriptional response identified 137 genes involved in the reaction to oxygen and other formed ROS (see Table S1 in the supplemental material). This could testify to an adaptation of A. muciniphila to continuous exposure to these products in its natural habitat. Moreover, we also observed the effective use of oxygen in a rudimentary respiratory chain involving a cytochrome bd complex, indicating that A. muciniphila is capable of competing with other strict anaerobes that may colonize the mucosal layer.
The oxygen survival of A. muciniphila is in line with anaerobic gut colonizers such as Bacteroides fragilis and Bifidobacterium adolescentis, which were still viable after 48 h of ambient air exposure (34). As previously described for gut colonizers Faecalibacterium prausnitzii (15) and B. fragilis (35), A. muciniphila had enhanced growth at the oxic-anoxic interphase as shown in the gas tube assay (Fig. 1B). The presence of riboflavin or other vitamins did not affect the observed growth ring (data not shown). Therefore, a riboflavin-dependent extracellular electron shuttle, such as was proposed for the enhanced growth of F. prausnitzii at the oxic-anoxic interface, is probably not present in A. muciniphila. For B. fragilis, cytochrome bd oxidase was shown to be essential for growth at nanomolar oxygen concentrations (35). The genes for cytochrome bd subunits I (Amuc_1695) and II (Amuc_1694) are present in A. muciniphila, and we confirmed their presence by spectral measurements. We also showed the functional relevance of the cytochrome bd complex by revealing the lower oxygen reduction capacity in heme-deprived medium. The bacteria were able to fully reduce the oxygen only when heme was supplemented. Evolutionarily, the A. muciniphila cytochrome bd genes are closest related to those of some aerobic members of the Verrucomicrobia, including Verrucomicrobium spinosum, and Opitutus terrae. The presence of the cytochrome bd genes in A. muciniphila might therefore be conserved during adaptation to the intestinal environment.
Apart from oxygen tolerance, the oxygen reduction capacity of A. muciniphila was demonstrated in the gas tube assay and was quantified in the fermentor system to be at least 2.26 mU/mg total protein. This lower limit of the oxygen conversion capacity of A. muciniphila is less than that of previously characterized anaerobic bacteria that employ cytochrome bd, including B. fragilis (9 nmol/min/mg total protein) and Moorella thermoacetica (29.8 nmol/min/mg protein) (35, 36). However, A. muciniphila is clearly able to reduce nanomolar concentrations of oxygen. Given the actual concentration of oxygen in the mucus layer, which has been estimated to be 15 mm Hg (∼210 nM) (16), we speculate that A. muciniphila can take advantage of these nanomolar concentrations in its ecological niche to compete with other strict anaerobes. The cytochrome bd complex was described to be essential in intestinal colonization of mice as was reported using respiratory mutants of E. coli (37) and B. fragilis (35). Hence, it is tempting to suggest that A. muciniphila might need cytochrome bd in initial colonization of its host as well.
The mechanism behind the oxygen response was investigated by analyzing the transcriptional response of A. muciniphila under aerated conditions in the fermentor system. Two different responses to oxygen could be distinguished: the detoxification of oxygen and the use of oxygen in aerobic respiration. A variety of mechanisms could explain the upregulation and downregulation of genes after aeration at both T2 (mid-exponential phase) and T3 (end-exponential phase) (Table 1). The detoxification of oxygen is reflected in the initial transcriptional response of A. muciniphila. Both superoxide dismutase (Amuc_1592) and hydroperoxidase (Amuc_2070) are upregulated to scavenge the ROS. Oxygen also induced the transcription of both rubrerythrin genes of A. muciniphila, previously shown to be upregulated as well in the acute murine colitis model where there is an increase of ROS in the GI tract (38). In conclusion, A. muciniphila harbors an arsenal of genes to detoxify oxygen that are significantly upregulated under aerated conditions. However, this does not explain the observed enhanced growth and the shift toward a higher acetate-to-propionate ratio.
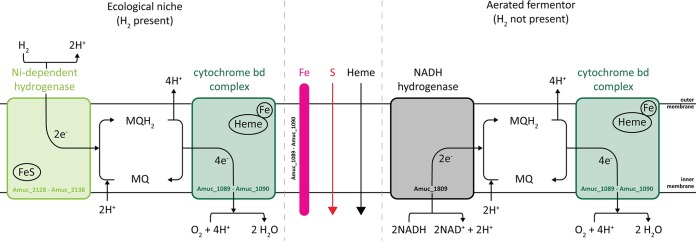
The transcriptome of A. muciniphila at the end-exponential phase points to the use of oxygen in aerobic respiration mediated by cytochrome bd. As the energy yield of aerobic respiration is higher than that of fermentation, this might be the reason for the enhanced growth that we observed under aerated conditions (Fig. 4) (29). Cytochrome d ubiquinol oxidase (subunit II) (Amuc_1694) was significantly upregulated, while subunit I (Amuc_1695), although abundantly present in the transcriptome, showed no transcriptional induction by oxygen (at both T2 and T3). To be functional, the cytochrome bd complex needs to be coupled to an NADH dehydrogenase to recycle its menaquinone and produce NAD+ (Fig. 6). A. muciniphila harbors a NADH hydrogenase complex consisting of the following 14 genes: Amuc_1604 to Amuc_1614, Amuc_1551, Amuc_2157, and Amuc_2158. While the first gene cluster was downregulated under aerated conditions, the other genes were slightly (∼1.5-fold) though not significantly upregulated. Moreover, this NADH hydrogenase complex belongs to NADH dehydrogenase type I (39), which has been shown to be not essential in the electron transport from NADH to oxygen in a mutant E. coli strain (40). Therefore, we screened the genome for another potential NADH dehydrogenase and found that Amuc_1809 might be a candidate. Although abundantly expressed under all tested conditions, Amuc_1809 was upregulated slightly (though not significantly) under aerated conditions at T2 (1.1-fold upregulated) and T3 (1.3-fold upregulated). Hence, this NADH dehydrogenase might operate in conjunction with the cytochrome bd complex to use oxygen as a final electron acceptor.
FIG 6.
Schematic representation of aerobic respiration by A. muciniphila under nanomolar oxygen concentration conditions.
Enhanced growth under aerated conditions was observed together with a marginal but significant shift toward a higher acetate-to-propionate ratio. As cytochrome bd uses oxygen as a final electron acceptor, resulting in the production of H2O and NAD+ (Fig. 6), to maintain the NAD+/NADH ratios, the extra NAD+ production needs to be balanced by additional NADH regeneration. Therefore, the metabolism might shift from propionate, where a net total of 2 NADH are oxidized to 2 NAD+, toward acetate production, where 1 NAD+ is reduced to 1 NADH (a simplified schematic representation is shown in Fig. S2 in the supplemental material). More acetate production leads to more ATP production as well. As acetate is a more highly oxidized fermentation product, more electrons are released from pyruvate and donated to NAD+, leading to the production of more NADH. The NADH can be oxidized and the electrons can be transferred back to oxygen via an NADH dehydrogenase, menaquinones, and cytochrome bd. Due to the change in the acetate-to-propionate ratio, more ATP and NADH can be generated, leading to slightly increased growth rate and yield, as observed in our fermentor experiments. This was confirmed by the increased growth and acetate production of ECOM4-AmuCytbd (Table 2), and its presence in A. muciniphila was confirmed by spectral measurements (Fig. 5).
The cytochrome bd complex is likely to involve iron-containing hemes as cofactors. Both FeO iron transporters (Amuc_1089 and Amuc_1090) predicted to be involved in Fe2+ transport were significantly upregulated under aerated conditions. This might point to increased iron uptake to allow incorporation of the iron-containing heme cofactors in cytochrome bd. All known members of the bd family of oxygen reductases most commonly use ubiquinol or menaquinol as the substrate (41). A. muciniphila is predicted to harbor the complete pathway to produce menaquinones (see Fig. S1 in the supplemental material). However, one gene (Amuc_1017) was significantly downregulated at T3 whereas the other 8 were not significantly altered under aeration conditions.
Taking the data together, we showed that a subunit of cytochrome bd complex is transcriptionally induced under aerated conditions and that its presence in A. muciniphila was confirmed by spectrum measurements. The genes for both FeO iron transporters were significantly upregulated to potentially employ the heme cofactors of cytochrome bd. The oxygen conversion capacity of A. muciniphila is lower than that seen with the previously characterized cytochrome bd of two other bacteria (35, 36), and yet A. muciniphila clearly possesses the ability to reduce oxygen. Finally, the functionality of the cytochrome bd complex of A. muciniphila was confirmed by the increased growth and the acetate production of ECOM4-AmuCytbd. On top of this, A. muciniphila had a lower oxygen reduction capacity in the absence of heme. Therefore, we propose the following mechanism (Fig. 6). A. muciniphila uses the cytochrome bd complex coupled to an unidentified NADH dehydrogenase (possible gene candidate, Amuc_1809) to use oxygen as a final electron acceptor. The use of oxygen as a final electron acceptor by cytochrome bd oxidase shifts the metabolic process toward a higher acetate-to-propionate ratio, resulting in more ATP and NADH and eventually leading to a slightly increased growth rate and yield. In its ecological niche, A. muciniphila might use this additional energy to outcompete strict anaerobes in the mucus layer.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to B. Ø. Palsson for providing the ECOM4 strain and to J. D. Keasling for the pBbA5c plasmid.
The work was supported by ERC Advanced Grant 250172—MicrobesInside from the European Research Council and the Netherlands Organization for Scientific Research (Spinoza Award and SIAM Gravity Grant 024.002.002) to W.M.D.V.
We declare that we have no competing financial interests.
J.P.O. and K.C.H.V.D.A. designed and performed experiments, analyzed and interpreted results, generated figures and tables, and conceived the manuscript. T.R.F. and N.J.C. designed and performed experiments. M.D. analyzed transcriptomics data. W.M.D.V. and C.B. conceived and supervised the project and contributed to the writing of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01641-16.
REFERENCES
- 1.Flint HJ, Scott KP, Louis P, Duncan SH. 2012. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 2.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 3.Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manach C, Scalbert A, Morand C, Remesy C, Jimenez L. 2004. Polyphenols: food sources and bioavailability. Am J Clin Nutr 79:727–747. [DOI] [PubMed] [Google Scholar]
- 5.van Hoek MJ, Merks RM. 2012. Redox balance is key to explaining full vs. partial switching to low-yield metabolism. BMC Syst Biol 6:22. doi: 10.1186/1752-0509-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 7.Belzer C, de Vos WM. 2012. Microbes inside–from diversity to function: the case of Akkermansia. ISME J 6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derrien M, Van Baarlen P, Hooiveld G, Norin E, Muller M, de Vos WM. 2011. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol 2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, de Vos WM, Satokari R. 2015. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol 81:3655–3662 doi: 10.1128/AEM.04050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, Bae JW. 2014. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63:727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 12.Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, Kayser BD, Levenez F, Chilloux J, Hoyles L, MICRO-Obes Consortium, Dumas ME, Rizkalla SW, Doré J, Cani PD, Clément K. 22 June 2015. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 13.Reference deleted. [Google Scholar]
- 14.Van den Abbeele P, Van de Wiele T, Verstraete W, Possemiers S. 2011. The host selects mucosal and luminal associations of coevolved gut microorganisms: a novel concept. FEMS Microbiol Rev 35:681–704. doi: 10.1111/j.1574-6976.2011.00270.x. [DOI] [PubMed] [Google Scholar]
- 15.Khan MT, Duncan SH, Stams AJ, van Dijl JM, Flint HJ, Harmsen HJ. 2012. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J 6:1578–1585. doi: 10.1038/ismej.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espey MG. 2013. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic Biol Med 55:130–140. doi: 10.1016/j.freeradbiomed.2012.10.554. [DOI] [PubMed] [Google Scholar]
- 17.Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8. [PubMed] [Google Scholar]
- 18.Miller RS, Hoskins LC. 1981. Mucin degradation in human colon ecosystems. Fecal population densities of mucin-degrading bacteria estimated by a “most probable number” method. Gastroenterology 81:759–765. [PubMed] [Google Scholar]
- 19.Plugge CM. 2005. Anoxic media design, preparation, and considerations. Methods Enzymol 397:3–16. doi: 10.1016/S0076-6879(05)97001-8. [DOI] [PubMed] [Google Scholar]
- 20.van Gelder AH, Aydin R, Alves MM, Stams AJ. 2012. 1,3-Propanediol production from glycerol by a newly isolated Trichococcus strain. Microb Biotechnol 5:573–578. doi: 10.1111/j.1751-7915.2011.00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zoetendal EG, Booijink CC, Klaassens ES, Heilig HG, Kleerebezem M, Smidt H, de Vos WM. 2006. Isolation of RNA from bacterial samples of the human gastrointestinal tract. Nat Protoc 1:954–959. doi: 10.1038/nprot.2006.143. [DOI] [PubMed] [Google Scholar]
- 22.Chomczynski P. 1993. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15:532–534, 536–537. [PubMed] [Google Scholar]
- 23.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Handsaker B, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. 2011. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J Biol Eng 5:12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Portnoy VA, Scott DA, Lewis NE, Tarasova Y, Osterman AL, Palsson BO. 2010. Deletion of genes encoding cytochrome oxidases and quinol monooxygenase blocks the aerobic-anaerobic shift in Escherichia coli K-12 MG1655. Appl Environ Microbiol 76:6529–6540. doi: 10.1128/AEM.01178-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lechardeur D, Cesselin B, Fernandez A, Lamberet G, Garrigues C, Pedersen M, Gaudu P, Gruss A. 2011. Using heme as an energy boost for lactic acid bacteria. Curr Opin Biotechnol 22:143–149. doi: 10.1016/j.copbio.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Bishop RE, Leskiw BK, Hodges RS, Kay CM, Weiner JH. 1998. The entericidin locus of Escherichia coli and its implications for programmed bacterial cell death. J Mol Biol 280:583–596. doi: 10.1006/jmbi.1998.1894. [DOI] [PubMed] [Google Scholar]
- 31.Zhou JZ, He Q, Hemme CL, Mukhopadhyay A, Hillesland K, Zhou AF, He ZL, Van Nostrand JD, Hazen TC, Stahl DA, Wall JD, Arkin AP. 2011. How sulphate-reducing microorganisms cope with stress: lessons from systems biology. Nat Rev Microbiol 9:452–466. doi: 10.1038/nrmicro2575. [DOI] [PubMed] [Google Scholar]
- 32.Borisov VB. 2008. Interaction of bd-type quinol oxidase from Escherichia coli and carbon monoxide: heme d binds CO with high affinity. Biochemistry (Mosc) 73:14–22. doi: 10.1134/S0006297908010021. [DOI] [PubMed] [Google Scholar]
- 33.Bloch DA, Borisov VB, Mogi T, Verkhovsky MI. 2009. Heme/heme redox interaction and resolution of individual optical absorption spectra of the hemes in cytochrome bd from Escherichia coli. Biochim Biophys Acta 1787:1246–1253. doi: 10.1016/j.bbabio.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Rolfe RD, Hentges DJ, Campbell BJ, Barrett JT. 1978. Factors related to the oxygen tolerance of anaerobic bacteria. Appl Environ Microbiol 36:306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baughn AD, Malamy MH. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427:441–444. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- 36.Das A, Silaghi-Dumitrescu R, Ljungdahl LG, Kurtz DM Jr. 2005. Cytochrome bd oxidase, oxidative stress, and dioxygen tolerance of the strictly anaerobic bacterium Moorella thermoacetica. J Bacteriol 187:2020–2029. doi: 10.1128/JB.187.6.2020-2029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones SA, Chowdhury FZ, Fabich AJ, Anderson A, Schreiner DM, House AL, Autieri SM, Leatham MP, Lins JJ, Jorgensen M, Cohen PS, Conway T. 2007. Respiration of Escherichia coli in the mouse intestine. Infect Immun 75:4891–4899. doi: 10.1128/IAI.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berry D, Schwab C, Milinovich G, Reichert J, Ben Mahfoudh K, Decker T, Engel M, Hai B, Hainzl E, Heider S, Kenner L, Muller M, Rauch I, Strobl B, Wagner M, Schleper C, Urich T, Loy A. 2012. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J 6:2091–2106. doi: 10.1038/ismej.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baranova EA, Morgan DJ, Sazanov LA. 2007. Single particle analysis confirms distal location of subunits NuoL and NuoM in Escherichia coli complex I. J Struct Biol 159:238–242. doi: 10.1016/j.jsb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 40.Tran QH, Bongaerts J, Vlad D, Unden G. 1997. Requirement for the proton-pumping NADH dehydrogenase I of Escherichia coli in respiration of NADH to fumarate and its bioenergetic implications. Eur J Biochem 244:155–160. doi: 10.1111/j.1432-1033.1997.00155.x. [DOI] [PubMed] [Google Scholar]
- 41.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta 1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.