SUMMARY
MicroRNAs have been associated with many different biological functions but little is known about their roles in conditioned behavior. We demonstrate that Drosophila miR-980 is a memory suppressor gene functioning in multiple regions of the adult brain. Memory acquisition and stability were both increased by miR-980 inhibition. Whole cell recordings and functional imaging experiments indicated that miR-980 regulates neuronal excitability. We identified the autism susceptibility gene, A2bp1, as an mRNA target for miR-980. A2bp1 levels varied inversely with miR-980 expression; memory performance was directly related to A2bp1 levels. In addition, A2bp1 knockdown reversed the memory gains produced by miR-980 inhibition, consistent with A2bp1 being a downstream target of miR-980 responsible for the memory phenotypes. Our results indicate that miR-980 represses A2bp1 expression to tune the excitable state of neurons, and the overall state of excitability translates to memory impairment or improvement.
INTRODUCTION
MicroRNAs (miRNAs) are small (21–23nt), non-coding RNAs that repress gene expression to regulate cellular development and physiology (Ambros, 2004). A short seed sequence (6–8nt) located at the 5’ end of miRNAs binds to complementary sequences in the 3’-untranslated region of target mRNAs to repress mRNA expression by blocking translation and/or promoting degradation of the mRNA target (Lee et al., 1993; Wightman et al., 1993; Bartel and Chen, 2004; Bartel, 2009, McNeill and Van Vactor, 2012). Thus, miRNAs offer a relatively rapid, analog, and cell-type specific control mechanism for the epigenetic expression of genomic information in both time and space (Kosik, 2006; McNeill and Van Vactor, 2012).
One aspect of miRNA function that remains understudied concerns the roles for these molecules in learning and memory, a primary adaptive function of the CNS. Prior studies revealed that broad insults to the miRNA processing pathway impairs memory formation in both Drosophila and the mouse (Ashraf et al., 2006; Konopka et al., 2010; Schaefer et al., 2010; Bredy et al., 2011). Although eukaryotic genomes encode hundreds of distinct miRNAs and they are generally expressed at high levels in the CNS, only handful of specific miRNAs have been studied and implicated in memory formation through roles in neuronal maturation, connectivity and synaptic plasticity (Bredy, 2011; McNeill and Van Vactor, 2012; Li et al., 2013; Saab and Mansuy, 2014).
To identify the miRNAs that participate in the biology of memory formation, we conducted a large scale, comprehensive screen using a transgenic approach to systematically inhibit 134 different miRNAs (Busto et al., 2015), using a “microRNA sponge” technique (Ebert et al., 2007; Loya et al., 2009). We surveyed the influences of 134 miRNAs for effects on intermediate term (ITM, i.e., at 3 hr after conditioning), olfactory aversive memory. From this screen, we identified several new miRNAs that function to inhibit or promote memory formation at this timepoint (Busto et al., 2015). MiR-980, when inhibited, was shown to enhance memory formation. Thus, MiR-980, a member of the miR-22 family of vertebrate miRNAs (Ruby et al., 2007), was classified as having a memory suppressor function.
Here, we characterize the memory suppressing function of miR-980. Among the mRNA targets for miR-980, we demonstrate that the autism-susceptibility gene, A2bp1 (Ataxin2 binding protein 1, also known as Rbfox-1, Fox-1) is a primary target responsible for miR-980 directed memory suppression. A2bp1 is a known RNA binding protein involved in alternative splicing of a network of critical neuronal genes during development and in adults (Lee et al., 2009; Fogel et al., 2012) and in addition to autism (ASD), is associated with intellectual disability and epilepsy (Bhalla et al., 2004; Martin et al., 2007; Sebat et al., 2007; Mikhail et al., 2011; Davis et al., 2012). Opposite to the role for miR-980, we identify A2bp1 as a memory-promoting gene. Our combined data advance our understanding of the miR-22 family of miRNAs, showing that in Drosophila the magnitude of memory formation is a direct function of miR-980 abundance and of its primary mRNA target for this function, A2bp1.
RESULTS
MiR-980 inhibition enhances olfactory learning and memory stability
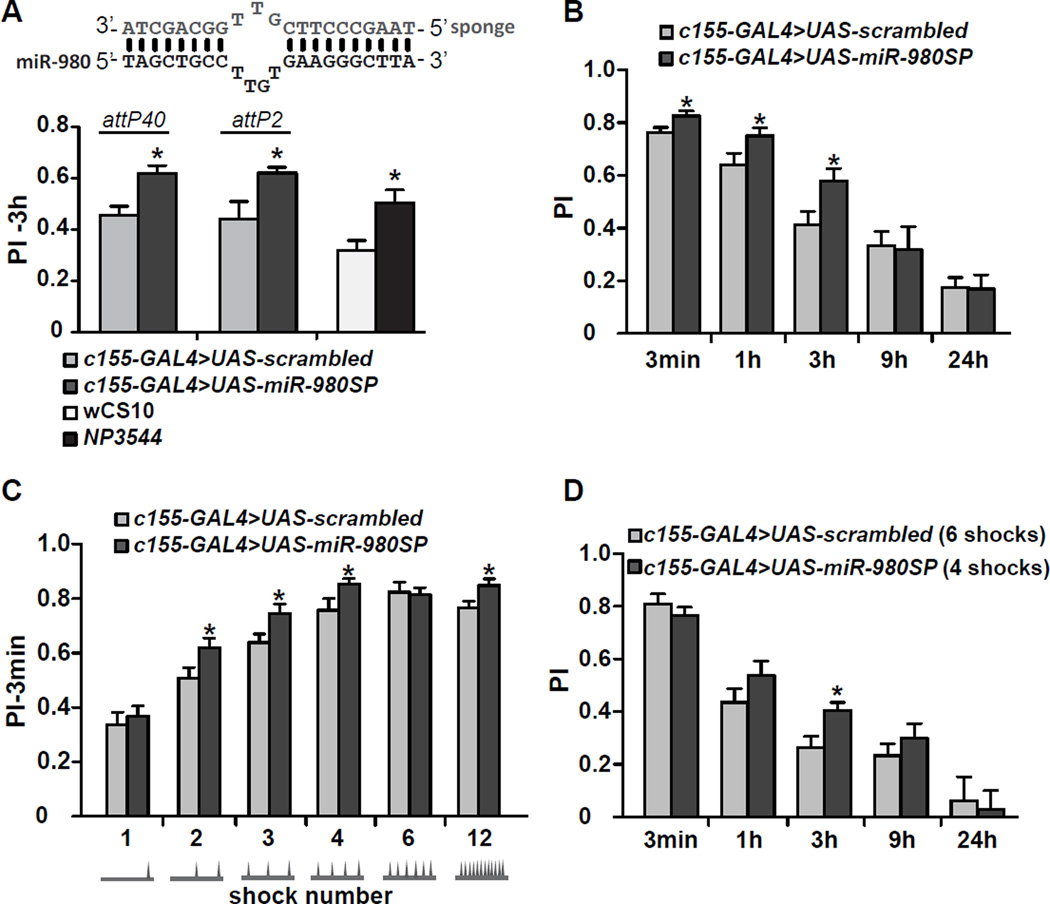
We recently tested 3-hour (3h) olfactory memory of 134 Drosophila miRNA sponge lines (Fulga et al., 2015) using the pan-neuronal driver c155-GAL4 (Busto et al., 2015). Expression of the miR-980 targeting sponge (UAS-miR-980SP) surprisingly enhanced the memory performance index (PI; Figure 1A) by ~40% compared to the UAS-scrambled control flies, without significantly altering odor or shock avoidance (Figure S1A). Two separate miR-980SP transgenes, one inserted at the attP40 locus (2nd chromosome) and the other at the attP2 locus (3rd chromosome), both significantly enhanced 3h PIs compared to their respective UAS-scrambled controls (Figure 1A). We also tested NP3544 flies, which have a P-element in the miR-980 gene reducing its expression by 64% in fly heads as measured by qRT-PCR (Marrone et al., 2012; and data not shown, p<0.01, n=3). The NP3544 hypomorph also enhanced 3h memory compared to the wCS10 control flies without altering odor and shock avoidance (Figure S1B); reinforcing the conclusion that miR-980 normally functions in memory suppression (Figure 1A).
Figure 1. MiR-980 inhibition enhances olfactory memory by potentiating memory acquisition and stability.
(A) MiR-980 inhibition enhances 3h aversive memory. The miR-980 targeting sponge contains sequence mismatches to prevent RNA interference-mediated degradation of the sponge RNA (Ebert et al., 2007). Two independent UAS-miR-980SP insertions, in the attP40 (2nd chromosome) and attP2 (3rd chromosome) sites, and the hypomorphic miR-980 mutant NP3544 improve 3h memory. The UAS-miR-980 sponge expression was driven by the pan-neuronal c155-GAL4 element; transgenic lines containing scrambled sequences (UAS-scrambled) were used as controls. PI: performance index. Statistics: Experimental and control groups were compared by the two-tailed, two-sample Student’s t-test. p<0.01 for attP40 and NP3544 line, p<0.05 for attP2 insert. PIs are the mean ± SEM (standard error of the mean) with n≥6.
(B) MiR-980 inhibition retards memory decay. MiR-980 inhibition enhances 3min, 1h, and 3h memory. Three min, 1, 3, 9 and 24h memory was tested for UAS-miR-980SP and UAS-scrambled flies containing the c155-GAL4 driver. Statistics: The c155-GAL4>UAS-miR-980SP PI at each time point was compared to the c155-GAL4>UAS-scrambled PI on different days and compared using a two-tailed, two-sample Student’s t-tests for each time point. p<0.05. PIs are the mean ± SEM with n≥6.
(C) MiR-980 inhibition enhances memory acquisition. Three min memory of c155-GAL4>UAS-scrambled and c155-GAL4>UAS-miR-980SP flies was tested after 1, 2, 3, 4, 6 and 12 shock training with a 1min CS+ odor presentation. Shock delivery is schematized below each condition. Statistics: PIs for each shock treatment were tested on different days and were analyzed by two-tailed, two-sample Student’s t-tests for each condition. p<0.05 for 2, 3, 4 and 12 shock treatments. PIs are the mean ± SEM with n=8.
(D) Mir-980 memory retention with a normalized initial PI. Three min PIs of scrambled and miR-980SP flies were normalized with 6 or 4 shocks, respectively during CS+ odor exposure. Memory was tested at 3min, 1, 3, 9 and 24h on different days. Statistics: PIs were compared using two-tailed, two-sample Student’s t-tests for each time point. p<0.05 for 3h memory. PIs are the mean ± SEM with n=8.
Memory time-course experiments showed that expression of miR-980SP enhanced 3-min, 1h and 3h memory but not 9h or 24h memory (Figure 1B). The effect on immediate performance after conditioning suggested that miR-980SP expression might enhance learning. To test acquisition, flies were trained with an increasing number of shock pulses during a 1min CS+ odor presentation followed by a 1min CS- odor (Figure 1C). Memory tested immediately after training with 2, 3, or 4 shocks was enhanced, consistent with the conclusion that inhibition of miR-980 improves the acquisition of the odor:shock contingency (Figure 1C). We also normalized initial PI scores for scrambled and miR-980SP groups by training the former with 6 and latter with 4 shocks (Figure 1D). Although initial performance was similar between the two groups, memory expressed by miR-980SP flies was significantly enhanced at 3h, revealing an additional role in the suppression of memory stability (Figure 1D).
MiR-980 enhances memory when inhibited during adulthood in multiple areas of the CNS
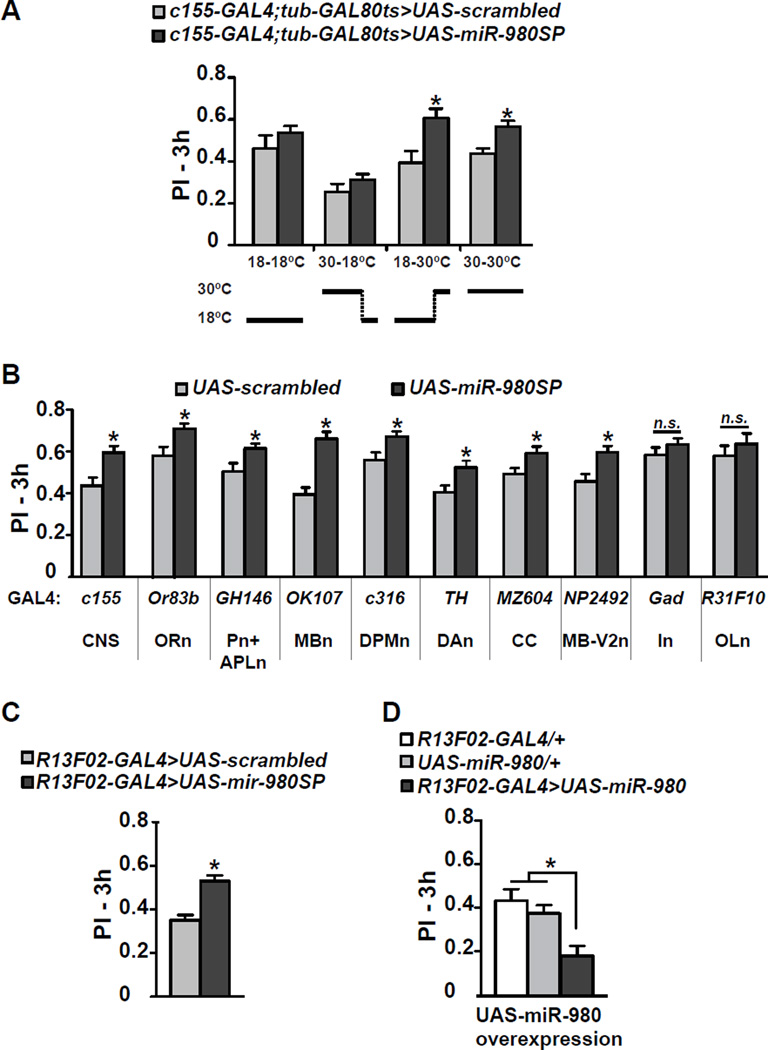
To distinguish whether miR-980 inhibition enhances memory due to developmental changes or roles in adult physiology we restricted the temporal expression of the miR-980SP transgene using the TARGET system (McGuire et al., 2003). With this system, GAL4 function is repressed in the presence of a temperature sensitive GAL80 protein (tub-GAL80ts) at the permissive temperature (18°C) and derepressed at the restrictive temperature (30°C). There was no difference in memory scores between control and experimental flies maintained at 18°C throughout the experiment (Figure 2A). Similarly, miR-980 inhibition only during development produced no difference between control and experimental groups (Figure 2A). In contrast, miR-980 inhibition throughout development and adulthood – or only during adulthood – produced the enhanced memory phenotype (Figure 2A). Therefore, the enhancement of memory occurs from miR-980SP expression during adulthood.
Figure 2. Memory enhancement occurs from inhibiting miR-980 in multiple areas of the CNS during adulthood.
(A) Memory enhancement occurs from miR-980 inhibition during adulthood. The c155-GAL4 expression was modulated during development and adulthood using tub-GAL80ts, with temperature shifts schematized below the bar graph. Flies expressing the scrambled control sequence using c155-GAL4; tub-GAL80ts were used as the control for 3h aversive memory. Statistics: PIs were analyzed by two-tailed, two-sample Student’s t-tests for each condition. p<0.01 for flies kept at 18°C during development and 30°C during adulthood and for flies kept at 30°C during both development and adulthood. PIs are the mean ± SEM with n≥10.
(B) MiR-980SP spatial mapping. The UAS-scrambled and UAS-miR-980SP flies were crossed to a battery of GAL4 lines that drive expression in specific populations of neurons. The c155-GAL4 driver was used as the positive control. The GAL4 drivers used and their abbreviated expression domains are shown below the graph. CNS: central nervous system, ORn: olfactory receptor neurons, Pn: projection neurons, APLn: anterior paired lateral neuron, MBn: mushroom body neurons, DPMn: dorsal paired medial neuron, DAn: dopaminergic neurons, CC: central complex, MB-V2n: mushroom body V2 neuron, In: inhibitory neurons, OL: optic lobe. Statistics: PIs were analyzed by two-tailed, two-sample Student’s t-tests for each driver compared to the scrambled crosses. p<0.05 for miR-980SP crossed to GH146-GAL4, c316-GAL4, TH-GAL4, MZ604-GAL44; p<0.01 for c155-GAL4, Or83b-GAL4, NP2492-GAL4; and p<0.0001 for OK107-GAL4. PIs are the mean ± SEM with n≥16.
(C) Mir-980SP expression in the MB using another GAL4 driver, R13F02-GAL4, improved 3h memory consistent with OK107-GAL4 results. Flies were conditioned with 3 shocks during the 1min CS+ odor representation to avoid ceiling scores. Statistics: PIs were analyzed by two-tailed, two-sample Student’s t-test. p<0.05. PIs are the mean ± SEM with n=8.
(D) Overexpression of miR-980 in MB impairs 3h memory. Statistics: Three hour olfactory memory of R13F02-GAL4>UAS-miR-980 flies was compared to GAL4-only and UAS-only controls using one-way ANOVA followed by Bonferroni’s post-hoc tests. PIs are the mean ± SEM with n=12. p<0.01.
Many types of neurons within the Drosophila olfactory nervous system mediate memory acquisition, consolidation, forgetting and retrieval (Guven-Ozkan and Davis, 2014). Olfactory receptor neurons (ORn) detect the odorants (CS+/CS−) and transmit this olfactory information to the antennal lobe (AL). Projection neurons (Pn) originating within the AL then convey the information to the mushroom body neurons (MBn). Neuromodulatory neurons, like dopamine neurons (DAn) are thought to convey the US (shock) stimulus to MBn. The CS and US stimuli are integrated in the MBn, one “center” for olfactory memory (Davis, 2005, 2011). Other MB extrinsic neurons that modulate memory formation include the anterior paired lateral neurons (APLn), dorsal paired medial neurons (DPMn), neurons in the central complex (CC) and MB-V2n (Guven-Ozkan and Davis, 2014). We employed GAL4 lines that promote expression in these and other neurons to identify the sets of neurons that respond to miR-980 inhibition by enhancing memory. Surprisingly, miR-980 inhibition improved 3h memory using all GAL4 lines tested except for Gad-GAL4, which drives expression in GABAergic inhibitory neurons, and R31F10-GAL4, which drives expression in optic lobe neurons (Figure 2B). To avoid a possible masking effect due to a ceiling on memory performance, MiR-980 inhibition using Gad-GAL4 and R31F10-GAL4 was tested using a milder training protocol, but this failed to improve memory scores (Figure S2A–B). These data indicate that miR-980 functions as a memory suppressor gene in multiple areas of the adult brain and more specifically, in excitatory neurons that generally are part of the olfactory nervous system.
We tested odor and shock avoidance of miR-980 inhibition using various drivers including GH146-GAL4, OK107-GAL4, 238Y-GAL4 and R13F02-GAL4 (Figure S1C–F and data not shown). Many of the drivers we tested with miR-980SP showed enhanced sensitivity to odors or shock. A MB specific driver, R13F02-GAL4 that also enhances memory (Fig 2C), was the only driver identified other than c155-GAL4 with no significant odor and shock avoidance difference between the control and the sponge-expressing flies (Figure S1F). One explanation for the memory enhancement with broad spatial miR-980 suppression along with increased overall odor and shock sensitivity is that neurons in the experimental flies may be more excitable. This hypothesis was tested with experiments presented below.
MiR-980 overexpression in the mushroom bodies impairs 3h memory
Since reducing miR-980 expression enhanced memory expression, we wondered whether its overexpression might impair expression. MiR-980 overexpression using c155-GAL4 produced pupal lethality. Therefore, we tested the effects of increasing miR-980 expression by driving UAS-miR-980 with the more specific MBn driver, R13F02-GAL4 (Bejarano et al., 2012). Opposite to and consistent with the phenotype observed with miR-980 inhibition, overexpression in the MBn impaired 3h memory relative to the GAL4-only and UAS-only controls (Figure 2D). Memory impairment observed upon overexpression of miR-980 cannot be attributed to impaired odor or shock perception (Figure S1G). Thus, we conclude that MiR-980 expression has a bidirectional influence on memory performance.
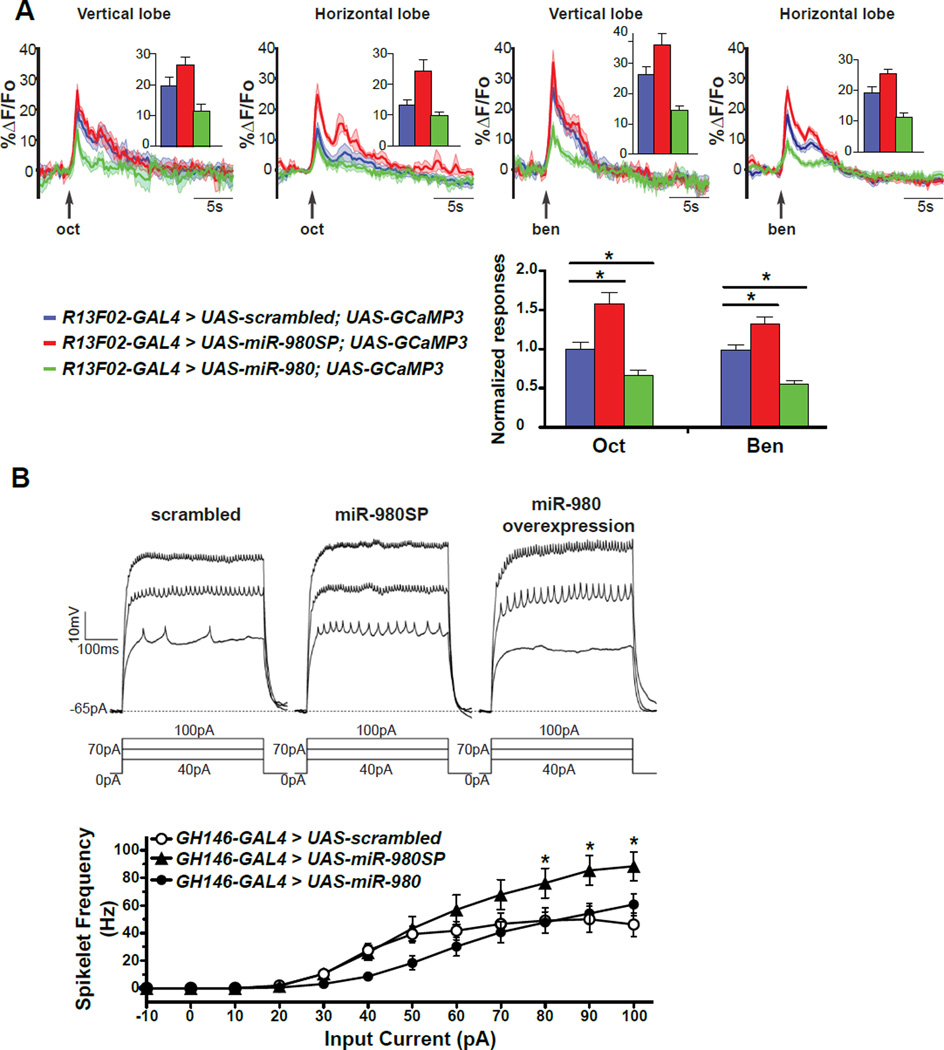
MiR-980 bidirectionally influences naïve odor responses of mushroom body neurons and neuronal excitability
MB responses to odors presented to the fly can be detected by monitoring calcium influx into these neurons (Wang et al., 2001; Yu et al. 2006; Turner et al., 2008). To test whether miR-980 expression might influence the response of MBn when odorants are presented to the fly, we recorded Ca2+ responses using GCaMP3 in naïve flies exposed to octanol or benzaldehyde, the two odorants we used for conditioning. The Ca2+ responses to octanol and benzaldehyde were increased in both the vertical and horizontal lobes of the MBn of the miR-980SP expressing flies, and decreased in the miR-980 overexpressing flies compared to the UAS-scrambled control (Figure 3A). These results reveal that the quantitative representation of odors in the MB is inversely related to miR-980 expression. This relationship is consistent with the hypothesis that miR-980 normally suppresses neuronal excitability.
Figure 3. MiR-980 expression modulates the excitable state of neurons.
(A) Recording of odor evoked Ca2+ influx in the vertical and horizontal MB lobes. GCaMP3 fluorescence was recorded while 3s of octanol- (oct) or benzaldehyde- (ben) laced air was applied to R13F02-GAL4>UAS-scrambled, R13F02-GAL4>UAS-miR-980SP and R13F02-GAL4>UAS-miR-980 overexpressing flies. The graphs show the sliding mean of the ΔF/Fo as a function of time, with the SEM represented as the shaded outline. Group data quantifying the peak response from baseline are shown as insets. Oct and ben responses in the vertical and horizontal lobes were normalized and averaged. The R13F02-GAL4>UAS-miR-980SP flies exhibited elevated odor evoked Ca2+ responses compared to the UAS-scrambled control. The R13F02-GAL4>UAS-miR-980 overexpression flies exhibited impaired odor evoked Ca2+ responses. Statistics: Responses were analyzed using one-way ANOVA followed by Bonferroni’s post-hoc tests. Results are the mean ± SEM with n=12. p<0.0001.
(B) MiR-980 modulates the excitability of projection neurons (Pn) in adult antennal lobes. Representative current clamp recordings of GH146-GAL4>UAS-scrambled control, GH146-GAL4>UAS-miR-980SP and GH146-GAL4>UAS-miR-980 overexpression Pn are shown. Spikelet frequency is plotted as a function of the current step. The firing frequency from miR-980SP Pn was significantly higher than the scrambled control at higher current steps. The mean firing frequency from miR-980 overexpression Pn was not significantly different with the scrambled control PNs, although a strong trend was observed at 40–50 pA. Statistics: Mean firing frequencies were analyzed using two-way ANOVA followed by Bonferroni’s post-hoc tests. Results are the mean ± SEM with n=21 for scrambled, n=19 for miR-980SP and miR-980 overexpression. p<0.001.
To evaluate the effect of miR-980 on neuronal excitability, whole cell recordings were performed using adult brain projection neurons (Pn). Pn of 2-day-old adult female flies expressing scrambled, miR-980SP and miR-980 with the GH146-GAL4 driver were recorded blind with respect to genotype. Depolarizing current injections were used to measure the intrinsic firing properties in the presence of the synaptic blockers curarine and picrotoxin with all cells held at −65 mV. There was no significant difference in holding current, input resistance or cell capacitance in the scrambled control, miR-980SP and miR-980 overexpression flies.
Supra-threshold current injections evoked depolarizations capped by a train of small amplitude spikelets characteristic of sodium-dependent action potentials (Figure 3B). The spikelet frequency in the scrambled control was consistent with that of wild type Pn (Iniguez et al., 2013). The mean firing frequency was significantly different among the scrambled control, miR-980SP and miR-980 overexpression lines. The miR-980SP Pn exhibited a significantly higher firing frequency than the scrambled control at current steps between 80–100 pAs. The mean firing frequency was not significantly different between the miR-980 overexpression and the scrambled control Pn (Figure 3B), although there was a clear difference in the shape of the input-output curve and miR-980 overexpressing Pn exhibited a strong trend for lower mean firing frequency at 40–50 pA, which may indicate a role in inhibiting excitability. Nonetheless, miR-980SP data demonstrate that miR-980 modulates the excitability of Pn with a clearly increased excitability when miR-980 is inhibited, consistent with the GCaMP3 imaging experiments.
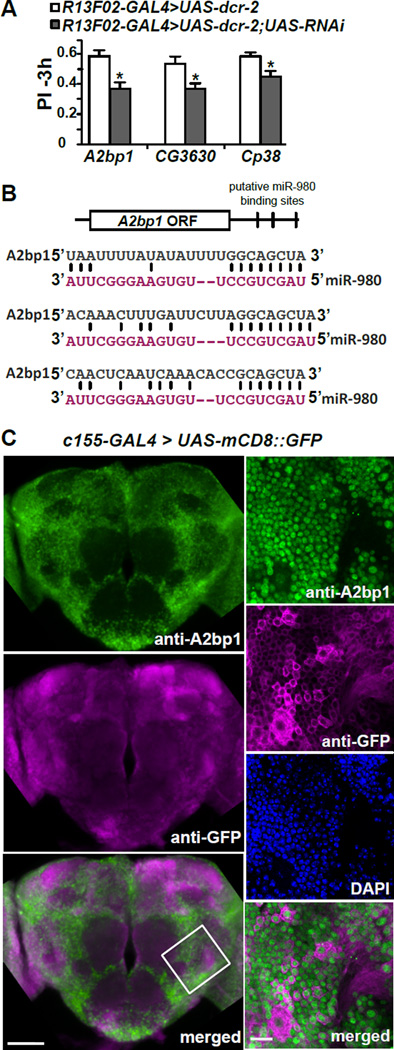
The autism-susceptibility gene, A2bp1, is a target of miR-980
We used bioinformatic prediction software to obtain insights into the potential mRNA targets of miR-980 and then tested the functional relevance of those genes to memory formation, initially using an RNAi knockdown approach. Using Targetscan and microRNA.org bioinformatics tools, we identified 95 mRNA targets with a possible role in the miR-980 phenotype. Fifty-eight of the predicted target genes with an available RNAi line from the VDRC KK library (Dietzl et al., 2007) were previously tested for 3 hr memory using nSyb-GAL4, a pan-neuronal driver (Table S1, Walkinshaw et al., 2015). Given the bidirectional regulation of memory performance with overexpression or inhibition of miR-980, it seemed possible that RNAi knockdown of true targets might expose a memory phenotype. A direct relationship between miRNA expression level, target mRNA expression level, and phenotype predicts that knockdown of a repressive miRNA by a miRNA-SP transgene should produce an elevated level of mRNA target. The phenotype obtained by RNAi knockdown of an authentic mRNA target may thus be opposite to that obtained with a miRNA-SP transgene and in the same direction as miR-980 overexpression.
Eighteen of the lines tested had potential memory functions (Table S2). These lines, along with two additional lines that failed to produce progeny with nSyb-GAL4, were further tested with the MBn driver R13F02-GAL4. This two-step screening strategy, first with a pan-neuronal GAL4 driver followed by a MBn specific GAL4 driver, identified 3 RNAi lines that produced a memory phenotype with MB expression (Figure 4A). The three candidates include A2bp1, a gene encoding a known RNA binding protein involved in alternative splicing (Lee et al., 2009; Fogel et al., 2012); CG3630, a gene encoding a protein containing a Costars domain but with a previously unknown biological function (http://flybase.org); and Cp38, a gene encoding a chorion protein necessary for eggshell formation (Spradling et al., 1980). Among the 3 candidate genes, A2bp1 was the strongest candidate due to its three predicted miR-980 binding sites in its 3’UTR (Figure 4B). Furthermore, logic offered A2bp1 as the most likely choice among the three with a possible authentic role in the biology of memory formation. Memory impairment in A2bp1 RNAi in MBs was not due to defects in odor and shock perception (Figure S3C). We thus focused on A2bp1 as potential target for miR-980 based memory phenotypes.
Figure 4. RNAi screen for potential miR-980 target genes identified the autism-susceptibility gene, A2bp1.
(A) Three of the miR-980 predicted target genes impair 3h memory using an RNAi approach. Predicted miR-980 target genes were screened using RNAi’s expressed in the MB with R13F02-GAL4; UAS-dcr-2. Three hour memory scores for three of the final hits compared to the R13F02-GAL4>UAS-dcr-2 control are shown. Statistics: PIs were analyzed by two-tailed, two-sample Student’s t-tests. p<0.01 for A2bp1 RNAi, p<0.05 for CG3630 and Cp38 RNAi. PIs are the mean ± SEM with n=10.
(B) Schematic diagram of the A2bp1 mRNA showing the location of three predicted miR-980 binding sites in the 3’UTR. Sequences that are complementary between miR-980 and A2bp1 3’UTR are illustrated.
(C) A2bp1 is broadly expressed and primarily localized to nuclei in the Drosophila brain. Representative maximum intensity projection images of anti-A2bp1 (green) and anti-GFP (magenta) immunostaining of the central brain of c155-GAL4>mCD8::GFP flies. The bottom panel shows the merged image for the anti-A2bp1 and anti-GFP images. Scale bar=50µm. The panels to the right show high magnification images of a 3µm single slice of the brain area identified by the white-bordered square in the merged image. Anti-A2bp1 (green), anti-GFP (magenta), DAPI (blue) and the merged image (bottom) indicate that the A2bp1 signal is primarily nuclear. Scale bar=10µm.
A2bp1 is expressed in the nuclei of most brain neurons
Immunostaining of the Drosophila brain with an A2bp1 antisera (Tastan et al., 2010) showed that A2bp1 is expressed broadly in the fly brain (Figure 4C). We marked the cell membranes by expressing mCD8::GFP (Lee and Luo, 1999) and labeled the brains with anti-GFP, anti-A2bp1 and DAPI. High magnification images showed that the A2bp1 and GFP signals were distinct from one another, and that the A2bp1 signal overlapped with DAPI staining. This nuclear localization of Drosophila A2bp1 is consistent with a role for the protein in alternative splicing as demonstrated for other organisms (Jin et al., 2003; Nakahata and Kawamoto, 2005; Underwood et al., 2005; Fogel et al., 2012).
MiR-980 represses A2bp1 protein expression
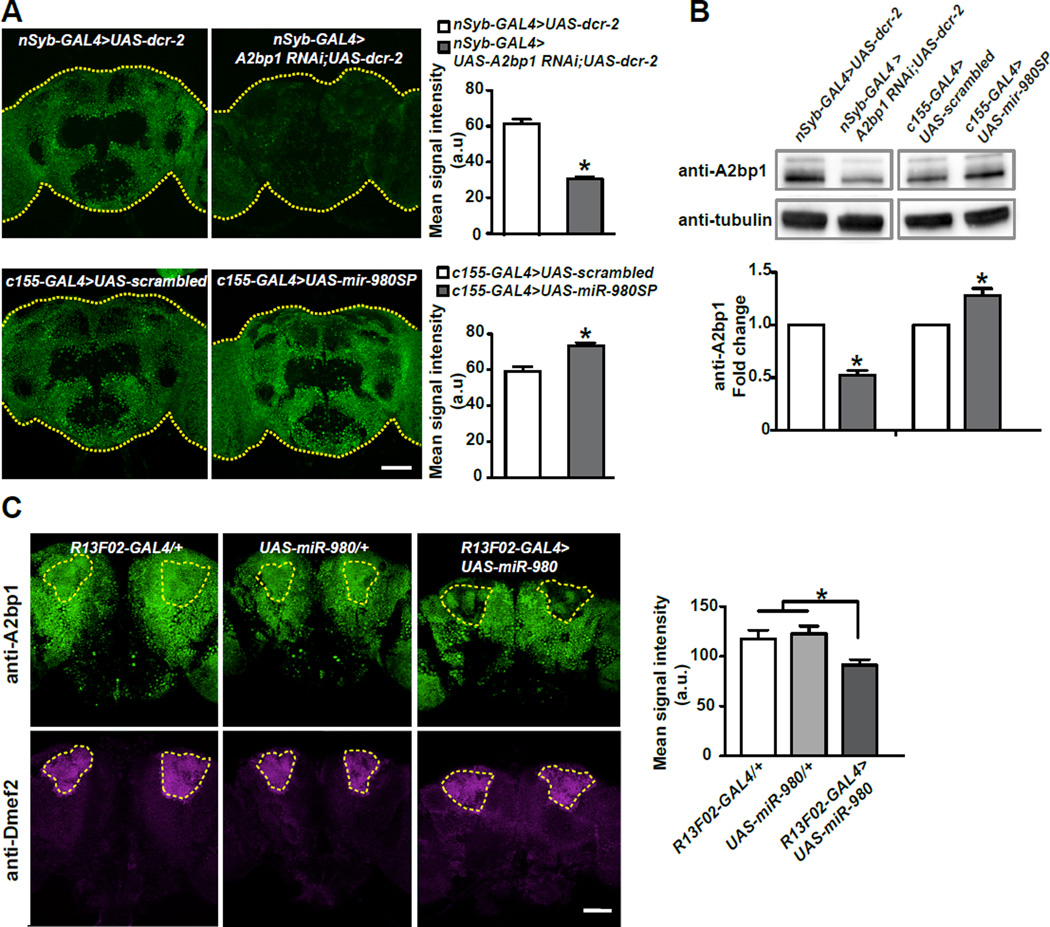
We tested the effect of the A2bp1-RNAi on A2bp1 expression by immunostaining nSyb-GAL4>UAS-A2bp1 RNAi;UAS-dcr-2 brains (Figure 5A) and quantifying the mean signal intensity from the central brain compared to control brains. We measured a ~50% reduction in signal from RNAi knockdown brains compared to the no-RNAi control (Figure 5A). We confirmed this estimate using western blotting of adult heads with the same antibody (Tastan et al., 2010), identifying a protein exhibiting strong immunoreactivity with an apparent mass of ~105kD (Figure S3A). The western blot signal from the ~105kD A2bp1 protein in RNAi knockdown flies was reduced by ~50% compared to control flies (Figure 5B, Figure S4A). The western blots resolved a less abundant isoform of ~125kD that also responded to A2bp1 RNAi expression. The significant decrease in A2bp1 protein upon RNAi knockdown provides molecular support for the effect of the A2bp1 RNAi on behavior (Figure 4A; full size western blot shown in Figure S4). Using the same antibody, we tested whether expression of A2bp1 is altered when miR-980SP is expressed. Inhibiting miR-980 with miR-980SP expression using the pan-neuronal c155-GAL4 driver significantly increased A2bp1 protein by ~25% as detected by both immunostaining (Figure 5A) and western blotting (Figure 5B; Figure S4A).
Figure 5. MiR-980 inhibits A2bp1 expression.
(A) Maximum projection images of the central brain from nSyb-GAL4>UAS-dcr-2 and nSyb-GAL4>UAS-A2bp1-RNAi;UAS-dcr-2 flies (top row); and c155-GAL4>scrambled and c155-GAL4>miR-980SP flies (bottom row) stained with anti-A2bp1 antisera. Each brain is outlined with a yellow dotted line. The mean signal intensity from the central brain is quantified in the adjacent histogram. Expression of the A2bp1-RNAi reduced the signal by ~50% compared to the no-RNAi control (n≥11, p<0.0001). Expression of miR-980SP increased the signal by ~25% compared to the scrambled control (n≥20, p<0.0001). Statistics: Data were analyzed by two-tailed, two-sample Student’s t-tests. Scale bar=50µm. Results are the mean ± SEM. a.u.=arbitrary unit.
(B) Representative anti-A2bp1 and anti-α-tubulin western blots using: (1) nSyb-GAL4>UAS-dcr-2, (2) nSyb-GAL4>UAS-A2bp1-RNAi; UAS-dcr-2 (3), c155-GAL4>scrambled and (4) c155-GAL4>miR-980SP fly heads. The signal from the A2bp1 band was first normalized to the α-tubulin signal in its own lane, and then the control samples were normalized to 1.0 to calculate the fold change in the experimental groups. A2bp1-RNAi expression reduced protein levels by ~50% compared to the no-RNAi control. Expression of miR-980SP increased A2bp1 by ~25% compared to the scrambled control. For full sized western blots see Figure S4. Statistics: Data were analyzed by one-sample Student t-test. Results are the mean ± SEM with n=8. p<0.0001 for A2bp1 RNAi and p<0.01 for miR-980SP.
(C) Overexpression of miR-980 in MB represses A2bp1 expression. Single section images of the central brain stained with anti-A2bp1 (green) and anti-Dmef2 (magenta) antibodies. R13F02-GAL4/+ and UAS-miR-980/+ brains were used as controls for the R13F02-GAL4>UAS-miR-980 genotype. MBn as defined by Dmef2 immunoreactivity are outlined with yellow dotted lines and mean signal intensity for the region of interest was measured. Overexpressing miR-980 in MBn decreased the anti-A2bp1 ~25%. Statistics: The data were analyzed by one-way ANOVA followed by Bonferroni’s post-hoc tests. Results are the mean ± SEM with n=15–17. p<0.01. Scale bar=50µm.
We also tested the effect of miR-980 overexpression on A2bp1 protein level. MiR-980 overexpression using the pan-neuronal c155-GAL4 driver produced pupal lethality. Therefore, we tested the effects of overexpression using the MBn specific driver R13F02-GAL4 and identified the cell bodies of MBn with Dmef2 co-labeling (Figure 5C). We measured the anti-A2bp1 mean signal intensity in the MBn for R13F02-GAL4>UAS-miR-980, UAS-only and GAL4-only genotypes. Opposite to the results obtained with miR-980 inhibition, elevating miR-980 in MBn significantly decreased A2bp1 expression by ~25% (Figure 5C). These results show that miR-980 represses the expression of the A2bp1 protein.
Overexpression of A2bp1 in the adult mushroom bodies enhances memory
The current annotation of the A2bp1 gene predicts eight different protein isoforms, ranging in size from 547 to 962 amino acids, produced by eight RNAs due to transcription from two transcriptional start sites and alternative splicing (www.flybase.org). It is unknown which of the isoforms dominate expression in the adult head. We designed primers to amplify RNA transcripts from the largest transcriptional unit by PCR using head cDNA and recovered six new splice variants, none of which corresponded to the largest previously annotated isoforms, RH and RL (Figure S3B). We picked one of the two most abundant splice variants and generated UAS-A2bp1 overexpression flies that lack miR-980 binding 3’UTR sites in the transgene. We named the new splice variant RN. It differs from the annotated form, RH, only by the exclusion of exon 6.
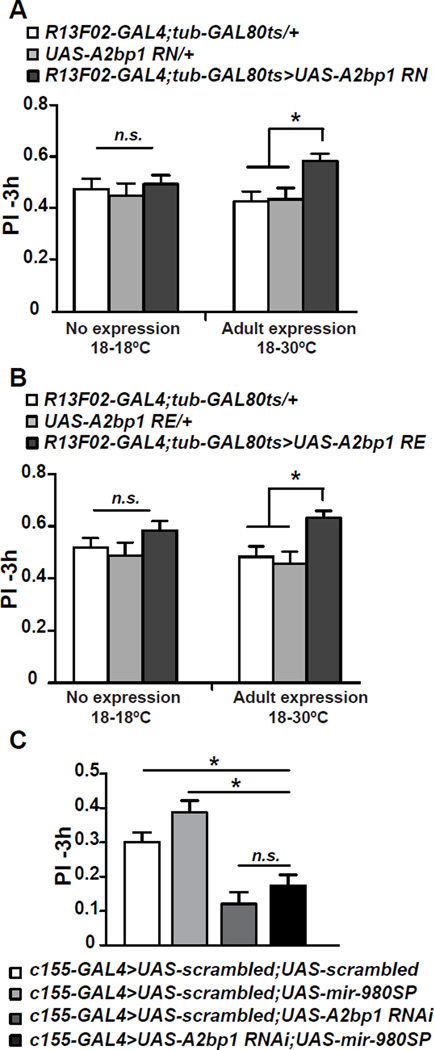
If an increased expression of A2bp1 is the primary reason for the enhanced memory observed in miR-980 inhibited flies due to inadequate repression by miR-980, then overexpressing UAS-A2bp1 should enhance memory. Driving UAS-A2bp1RN with R13F02-GAL4 resulted in embryonic lethality due to unknown developmental defects. We therefore tested whether overexpression, limited to the MB during adulthood, would enhance memory. Restricting A2bp1 expression in adult MB using R13F02-GAL4; tub-GAL80ts resulted in a ~3–4 fold increase in A2bp1 abundance measured by western blotting and immunohistochemistry (Figure S4B–C). Importantly, A2bp1 overexpression in the adult MB without the constraints imposed by miR-980 repression mimicked the effects of miR-980 inhibition, with a significant memory improvement measured at 3 h compared to UAS-only and GAL4-only controls without altering odor and shock perception (Figure 6A, S3D). We also tested a second A2bp1 isoform, RE in adults (Usha and Shashidhara, 2010), and observed the same memory enhancement (Figure 6B, S3D). These results support the model that enhanced memory occurring from miR-980 inhibition results predominantly from the dysregulation in the MBn of the miR-980 target, A2bp1 mRNA. Since overexpression of both isoforms of A2bp1 produced the same memory enhancement, the results also reveal that exons 2, 11, and 13 are unimportant for this function.
Figure 6. Overexpression of A2bp1 potentiates memory; decreasing A2bp1 reverses the memory enhancement due to miR-980SP expression.
(A) Adult-specific overexpression of the A2bp1 RN isoform increases 3h memory. MB specific expression of UAS-A2bp1(RN) was driven by R13F02-GAL4 in the presence of Gal80ts to restrict transgene expression to adulthood. Flies were kept at 18°C throughout development and adulthood (left) or at 18°C during development and switched to 30°C after hatching (right) to induce UAS-A2bp1 overexpression. A2bp1 overexpression in the MB significantly improved memory compared to UAS-only and GAL4-only controls. Statistics: PIs were analyzed using one-way ANOVA followed by Bonferroni’s post-hoc tests. PIs are the mean ± SEM with n=20. p<0.01.
(B) Adult-specific overexpression of the A2bp1-RE isoform increases 3h memory. MB specific expression of a second A2bp1 isoform, UAS-A2bp1(RE), during adulthood also enhanced 3h memory. Statistics: PIs were analyzed using one-way ANOVA followed by Bonferroni’s post-hoc tests. PIs are the mean ± SEM with n=15. p<0.01.
(C) A2bp1 is genetically downstream of miR-980. The UAS-A2bp1 RNAi and UAS-miR-980SP transgenes were combined with a UAS-scrambled transgene in order to obtain flies with the same number of UAS-elements as in the UAS-A2bp1 RNAi; UAS-miR-980SP experimental group. The UAS-scrambled transgenes inserted at the attP40 and attP2 sites were combined together and used as the control. Expression was driven by c155-GAL4. Three hour PIs of UAS-A2bp1 RNAi;UAS-miR-980 SP expressing flies were significantly lower than the scrambled control and miR-980SP-expressing flies, but not significantly different from A2bp1 RNAi-expressing flies. Statistics: Scores were analyzed using one-way ANOVA followed by Bonferroni’s post-hoc tests. PIs are the mean ± SEM with n=36 for the double scrambled control and n=21 for all other groups. p<0.0001.
MicroRNAs as regulators of mRNA expression are, in essence, upstream of the mRNA targets in the molecular signaling within a cell. To test the predicted genetic interaction between miR-980 and A2bp1, we performed an epistasis experiment combining in the same fly the expression of UAS-miR-980SP that promotes memory, with the expression of the UAS-A2bp1 RNAi transgene that inhibits memory. We compared the memory scores of this experimental group to UAS-A2bp1 RNAi or UAS-miR-980SP controls. The A2bp1 RNAi; miR-980SP double transgenic flies had memory scores significantly lower than both miR-980SP and scrambled controls, indicating that miR-980SP expression loses it normal memory enhancing effect when A2bp1 is reduced (Figure 6C). In addition, the memory scores of the A2bp1 RNAi; miR-980SP double transgenic flies were not different statistically from those of flies expressing A2bp1 RNAi alone. Thus, the observed change in A2bp1 protein abundance by altering miR-980 levels, the improvement of memory with miR-980 inhibition and A2bp1 overexpression, and results from epistasis experiments are consistent with the model that the normal memory suppressing effects of miR-980 occur through its regulation of the memory-promoting gene, A2bp1.
DISCUSSION
MicroRNAs are highly expressed in the vertebrate and invertebrate brain and contribute to fine-tuning of gene expression during development and during physiological events in cells. Nevertheless, their functional roles in the neuronal plasticity underlying learning and memory remains largely unexplored. We previously conducted a behaviorally based ‘miRNA sponge screen’ to systematically identify the miRNAs involved in Drosophila olfactory aversive learning and memory (Busto et al., 2015). The results presented here offer five major advances in our knowledge about the function of this class of regulatory molecules: (1) miR-980 functions to suppress memory formation by acting in multiple types of neurons within the olfactory nervous system. (2) miR-980 works as a suppressor of acquisition and memory stability. (3) miR-980 suppresses the excitability of excitatory neurons, (4) the memory suppressor functions of miRNA-980 are mediated largely by the inhibition of the autism-susceptibility gene, A2bp1. (5) A2bp1, itself, is a memory-promoting gene.
One surprising observation made in our study was that inhibition of miR-980 in multiple neurons within the olfactory nervous system enhances memory performance, as we anticipated finding a single cellular focus for its effects. Initially, it was difficult to understand how a single microRNA could modify behavioral memory when altered in one of many different types of neurons. This was reconciled by showing that excitability of Pns is enhanced with inhibited miR-980 function, offering the explanation that increased signaling, in general, within the olfactory nervous system enhances behavioral memory. This model provides a general explanation for the effects of miR-980 that function in multiple classes of excitable neurons.
We propose that the role of miR-980 in excitability accounts for the increased acquisition when the miRNA is inhibited. An increase in excitable state may simply enhance the signaling through different neuron types within the olfactory nervous system as the organism integrates sensory information into memory. A corollary of this idea is that normal acquisition is a composite effect of multiple neurons within the circuit conveying the sensory information being learned. Although it is possible that increased acquisition also accounts for the increased memory performance observed when immediate performance scores were normalized, an alternative possibility is that miR-980 may have distinct roles in acquisition and memory stability. For instance, although we attribute the increased acquisition to increased neuronal excitability, the increased memory after acquisition may be due to altered regulation of molecules involved in synaptic transmission.
MiR-980 belongs to the miR-22 family of miRNAs found in mammals (Ruby et al., 2007). Within the nervous system, the miR-22 family has been reported to participate in neuroprotection (Yu et al., 2015; Jovicic et al., 2013), neurodegeneration (Lee et al., 2011), neuroinflammation (Parisi et al., 2013; Siegel et al., 2012), neurodevelopment (Volvert et al., 2014; Berenguer et al., 2013), and neuroplasticity (Chen et al., 2013). Thus, although this family appears to have multiple roles in the nervous system and disease, our current studies identify members of this family as specifically involved in the suppression of memory formation. Given the functional association between miR-980 and A2bp1 shown here, it is also tempting to speculate that the miR-980/miR-22 family of miRNAs might be associated with ASD. No evidence for this possibility has yet been reported, but the expression of miR-22 is reduced in ADHD (Kandemir et al., 2014) and is genetically associated with panic disorder and anxiety in humans (Muinos-Gimeno et al., 2011). Thus, there are neuropsychiatric links to miR-22, which could potentially be through a role in excitability. Moreover, miR-22 represses the tumor suppressor gene PTEN in transformed human bronchial epithelial cells (Liu et al, 2010) and PTEN is known to be involved in Cowden syndrome and ASD in humans (Goffin et al., 2001).
Our behavioral, molecular, cellular and genetic data together argue that A2bp1 is a primary target of miR-980 for memory suppression. First, A2bp1 is broadly expressed in the fly brain, consistent with a broad nervous system requirement for miR-980. Second, there are three miR-980 binding sites in A2bp1 3’UTR making it a strong candidate mRNA target for miR-980 regulation. Third, we performed an in vitro mRNA binding experiment using biotinylated mature miR-980 as bait and successfully captured 8× more A2bp1 mRNA using wild-type miR-980 vs a form mutated for the seed region (data not shown). Fourth, A2bp1 shows the precise abundance/behavior relationship predicted as a direct target of miR-980. Overexpression of A2bp1 increases memory; miR-980 suppression increases memory. A2bp1 knockdown impairs memory; miR-980 overexpression impairs memory. Fifth, A2bp1 protein abundance varies as expected by manipulation of miR-980 levels. Overexpression of miR-980 decreases A2bp1 protein abundance and miR-980 suppression increases A2bp1 protein abundance. Finally, reducing A2bp1 levels using RNAi in miR-980 inhibited flies reversed the memory improvement. This finding is consistent with the model that A2bp1 is genetically downstream of miR-980 and a major mediator of the phenotype. However, we cannot exclude the possibilities that there may be additional miR-980 targets that participate in memory suppression and miR-980 regulation of A2bp1 could be indirect. Our simple model for miR-980/A2bp1 interactions and function seem to be at odds with an observation made about A2bp1 using mammalian models. In the mouse (Gehman et al., 2011), neuronal specific knockout of A2bp1 increases excitability in the dentate gyrus, a result opposite of that predicted by our model. This difference might reflect species or cell type differences, the complexity of the gene with its dozens of isoforms, or the multiple layers of regulation on A2bp1 expression. Bioinformatics analyses predict multiple miRNAs as binding to the A2bp1 3’UTR and regulating its expression. Thus, its basal or regulated expression level due to changes in physiological state could be a composite of many different regulatory molecules.
A2bp1 is associated with autism and epilepsy in human patients (Bhalla et al., 2004; Martin et al., 2007; Sebat et al., 2007; Mikhail et al., 2011; Davis et al., 2012) functioning presumably by regulating alternative splicing during both development and in adults (Lee et al., 2009; Fogel et al., 2012). Corominas et al. (2014) proposed that changes in gene-splicing alter the relative abundance of protein isoforms, which remodels protein networks and increases the risk for autism. Consistent with this thought, transcriptome analyses from ASD brains identified A2bp1 as one hub gene that is dysregulated in patients with autism (Voineagu et al., 2011). A2bp1 was originally identified through its interaction with Ataxin-2 (Shibata et al., 2000). Pn specific knockdown of Ataxin-2 impairs long term olfactory habituation associated structural and functional plasticity by regulating the miRNA pathway (McCann et al., 2011). Future studies will shed light on whether memory phenotypes of A2bp1 are dependent on Ataxin-2.
It is intriguing that our studies show that adult stage specific increases in A2bp1 abundance improve aversive olfactory memory, independent of any developmental function for the protein, and that human ASD is a spectrum brain disorder that is associated with poor to extraordinarily robust learning and memory capacities (Grzadzinski et al., 2013). We speculate that the different protein interaction networks that form due to varying levels of A2bp1 function account for the range of intellectual abilities observed in ASD. Drosophila may prove to be a much speedier and simpler system to dissect the specific effect of A2bp1 abundance on the emergence of protein interaction networks and their influence on cognitive abilities.
EXPERIMENTAL PROCEDURES
Fly stocks and behavior
Flies were cultured using standard methods. One- to 4-day-old flies were used for the behavioral experiments. Approximately 30min before training, flies were transferred and maintained in the behavior room (dim red light, 25°C, ~70% humidity). For conditioning, ~50–60 flies were trained using a standard two-odor discriminative aversive conditioning paradigm (Berry et al., 2012) by exposing flies to 1min of CS+ odor paired with 12 electric shock pulses followed by 30s of air and 1min of the CS− odor. Memory was tested using a T-maze, which delivers CS+ from one arm, and CS− from the other. Additional details about the fly stocks utilized in this study and behavioral tests are provided in Supplemental Experimental Procedures.
In vivo Ca2+ imaging
Flies were mounted onto recording chamber as described previously (Berry et al., 2012). Briefly, a single fly was aspirated, without anesthesia, into a custom-designed recording chamber. The head was immobilized by gluing the eyes to the chamber with myristic acid and the proboscis similarly immobilized. A small area of dorsal cuticle was removed to provide optical access to the brain. Fresh saline [103 mM NaCl, 3 mM KCl, 5 mM HEPES, 1.5 mM CaCl2, 4mM MgCl2, 26 mM NaHCO3, 1 mM NaH2PO4, 10 mM trehalose, 7 mM sucrose, and 10 mM glucose (pH 7.2)] was perfused across the brain to prevent desiccation and ensure the health of the fly. We recorded the responses in MB using a 25× water-immersion objective. Odorants were diluted 1:10 in mineral oil and spread on a 1cm2 filter paper in a scintillation vial. Pressurized air was passed through the vial to deliver a three sec pulse of air laced with oct vapor at a rate of 200 ml/min, followed three minutes later by a second pulse of air laced with ben vapor. Images were acquired at 4 frames/s at a resolution of 256×256 pixels from both MB vertical and horizontal lobes. The image data was analyzed as described previously (Yu et al., 2005; Cervantes-Sandoval et al., 2013). For statistical analysis, ΔF/Fo responses from vertical and horizontal lobes were normalized to the scramble control. Significance was determined using one-way ANOVA followed by Bonferroni’s post-hoc tests.
Whole cell recordings from projection neurons in isolated adult brain
Brains were obtained from adult female flies 2 days after eclosion. The entire brain was removed from the head and mounted in the recording chamber with the anterior face of the brain up (Gu and O'Dowd, 2006, 2007). Recordings were made from projection neurons (Pn) in the dorsal neuron cluster using 8–9 MΩ resistance pipettes. All voltages reported refer to pipette potentials at the soma. Depolarization-evoked action potentials were recorded using a pipette solution containing (in mM) 102 potassium gluconate, 0.085 CaCl2, 1.7 MgCl2, 17 NaCl, 0.94 EGTA, 8.5 HEPES, and 4.5 ATP. The pH was adjusted to 7.2 and osmolarity to 234–236 mOsM. The chamber was continuously perfused at 0.5 ml/min with recording solution that contained (in mM) 120 NaCl, 1.8 CaCl2, 0.8 MgCl2, 3 KCl, 5 glucose, 10 HEPES, as well as the synaptic receptor blockers D-turbocurarine (20 µM) and picrotoxin (10 µM). The pH was adjusted to 7.2 and osmolarity to 250–253 mOsM. Data shown were corrected for the 5 mV liquid junction potential generated in these solutions. For examination of the evoked firing properties, the membrane potential was held at −65 mV by injection of hyperpolarizing holding current. Data were acquired with a Patch Clamp L/M-EPC7 amplifier (List Medical), a digidata 1322A D-A converter (Molecular Devices), a Dell computer (Dimension 8200), and pClamp9 software (Molecular Devices).
Bioinformatics and statistical analyses
Putative mRNA targets for miR-980 were predicted using online tools TargetScan and microRNA.org. Targetscan.org predicts 70 mRNA targets for miR-980. We identified 25 additional and non-overlapping candidates from microRNA.org. (Enright et al., 2003; Kheradpour et al., 2007; Ruby et al., 2007). Prism was used for statistical analyses. Two sample, two-tailed Student’s t-tests were used to compare two conditions. To compare one group to a normalized control group, a one sample, two-tailed, Student’s t-test was used. For multiple group comparisons, one-way ANOVA followed by Bonferroni’s post-hoc tests were used.
Immunohistochemistry
Two-to-five day old female fly brains were dissected in 1× PBS and transferred to 1% paraformaldehyde in PBS. We followed the protocol described by Fly Light Project (Jenett et al., 2012). Additional details are found in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We would like to acknowledge David Van Vactor (Harvard) and Tudor Fulga (Oxford) for the miRNA sponge lines. We thank Eric Lai (Sloan Kettering), LS Shashidhara (Indian Institute of Science Education and Research), Stephen Cohen (Institute for Molecular and Cell Biology, Singapore), the Kyoto Drosophila Genetic Resource Center and the Vienna Drosophila RNAi center (VDRC) for RNAi and other fly lines. We are grateful to Michael Buszczak (UT Southwestern) for the anti-A2bp1 antisera, Eric Olson (UT Southwestern) for the anti-Dmef2 antibody, Courtney M. MacMullen and Molee Chakraborty for technical support, and Neelam Shahani for help in obtaining high magnification images. This research was supported by NIH R37 NS19904 and R01 NS052351 to RLD and NIH RO1 NS0830009 to DKO’D.
Footnotes
ACCESSSION NUMBERS
The Genbank accession numbers for the miR-980 and A2bp1 RE and A2bp1 RN sequences reported in this paper are NR_047714NM_001104102 and KU315475 respectively.
AUTHOR CONTRIBUTIONS
TGO, GUB, and RLD designed the behavioral, immunohistochemical, and biochemical experiments. TG performed these experiments. TGO and ICS designed and performed the imaging experiments. SSS and DKO’D designed and performed the whole cell patch clamp recordings. TGO and RLD wrote the initial draft of the manuscript that was then edited by all authors.
REFERENCES
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bejarano F, Bortolamiol-Becet D, Dai Q, Sun K, Saj A, Chou YT, Raleigh DR, Kim K, Ni JQ, Duan H, et al. A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development. 2012;139:2821–2831. doi: 10.1242/dev.079939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenguer J, Herrera A, Vuolo L, Torroba B, Llorens F, Sumoy L, Pons S. MicroRNA 22 regulates cell cycle length in cerebellar granular neuron precursors. Mol Cell Biol. 2013;33:2706–2717. doi: 10.1128/MCB.00338-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JA, Cervantes-Sandoval I, Nicholas EP, Davis RL. Dopamine is required for learning and forgetting in Drosophila. Neuron. 2012;74:530–542. doi: 10.1016/j.neuron.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla K, Phillips HA, Crawford J, McKenzie OL, Mulley JC, Eyre H, Gardner AE, Kremmidiotis G, Callen DF. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Bredy TW, Lin Q, Wei W, Baker-Andresen D, Mattick JS. MicroRNA regulation of neural plasticity and memory. Neurobiol Learn Mem. 2011;96:89–94. doi: 10.1016/j.nlm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Busto GU, Guven-Ozkan T, Fulga TA, Van Vactor D, Davis RL. MicroRNAs that promote or inhibit memory formation in Drosophila melanogaster. Genetics. 2015;200:569–580. doi: 10.1534/genetics.114.169623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Sandoval I, Martin-Peña A, Berry JA, Davis RL. System-like consolidation of olfactory memories in Drosophila. J Neurosci. 2013;33:9846–9854. doi: 10.1523/JNEUROSCI.0451-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Liu H, Guan X. Changes in microRNA expression profile in hippocampus during the acquisition and extinction of cocaine-induced conditioned place preference in rats. J Biomed Sci. 2013;20:96. doi: 10.1186/1423-0127-20-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas R, Yang X, Lin GN, Kang S, Shen Y, Ghamsari L, Broly M, Rodriguez M, Tam S, Trigg SA, et al. Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nature communications. 2014;5:3650. doi: 10.1038/ncomms4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LK, Maltman N, Mosconi MW, Macmillan C, Schmitt L, Moore K, Francis SM, Jacob S, Sweeney JA, Cook EH. Rare inherited A2BP1 deletion in a proband with autism and developmental hemiparesis. Am J Genet A. 2012;158A:1654–1661. doi: 10.1002/ajmg.a.35396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Ann Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- Davis RL. Traces of Drosophila memory. Neuron. 2011;70:8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Belle JS, Heisenberg M. Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science. 1994;263:692–695. doi: 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, Gao F, Parikshak N, Konopka G, Geschwind DH. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet. 2012;21:4171–4186. doi: 10.1093/hmg/dds240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, McNeill EM, Binari R, Yelick J, Blanche A, Booker M, Steinkraus BR, Schnall-Levin M, Zhao Y, DeLuca T, Bejarano F, Han Z, Lai EC, Wall DP, Perrimon N, Van Vactor D. A transgenic resource for conditional competitive inhibition of conserved Drosophila microRNAs. Nature communications. 2015;6:7279. doi: 10.1038/ncomms8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Jr, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nature genetics. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin A, Hoefsloot LH, Bosgoed E, Swillen A, Fryns JP. PTEN mutation in a family with Cowden syndrome and autism. Am J Med Genet. 2001;105:521–524. doi: 10.1002/ajmg.1477. [DOI] [PubMed] [Google Scholar]
- Grzadzinski R, Huerta M, Lord C. DSM-5 and autism spectrum disorders (ASDs): an opportunity for identifying ASD subtypes. Molecular autism. 2013;4:12. doi: 10.1186/2040-2392-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, O'Dowd DK. Cholinergic synaptic transmission in adult Drosophila Kenyon cells in situ. J Neurosci. 2006;26:265–272. doi: 10.1523/JNEUROSCI.4109-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, O'Dowd DK. Whole cell recordings from brain of adult Drosophila. JoVE. 2007:248. doi: 10.3791/248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T, Davis RL. Functional neuroanatomy of Drosophila olfactory memory formation. Learn Mem. 2014;21:519–526. doi: 10.1101/lm.034363.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez J, Schutte SS, O'Dowd DK. Cav3-type alpha1T calcium channels mediate transient calcium currents that regulate repetitive firing in Drosophila antennal lobe PNs. J Neurophysiol. 2013;110:1490–1496. doi: 10.1152/jn.00368.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Zaldivar Jolissaint JF, Moser R, Silva Santos Mde F, Luthi-Carter R. MicroRNA-22 (miR-22) overexpression is neuroprotective via general anti-apoptotic effects and may also target specific Huntington's disease-related mechanisms. PloS One. 2013;8:e54222. doi: 10.1371/journal.pone.0054222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandemir H, Erdal ME, Selek S, Ay OI, Karababa IF, Kandemir SB, Ay ME, Yilmaz SG, Bayazit H, Tasdelen B. Evaluation of several micro RNA (miRNA) levels in children and adolescents with attention deficit hyperactivity disorder. Neurosci Lett. 2014;580:158–162. doi: 10.1016/j.neulet.2014.07.060. [DOI] [PubMed] [Google Scholar]
- Kheradpour P, Stark A, Roy S, Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M, Kowarsch A, Michaluk P, Dzwonek J, Arnsperger T, et al. MicroRNA loss enhances learning and memory in mice. J Neurosci. 2010;30:14835–14842. doi: 10.1523/JNEUROSCI.3030-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nature Reviews Neuroscience. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Kreher SA, Kwon JY, Carlson JR. The molecular basis of odor coding in the Drosophila larva. Neuron. 2005;46:445–456. doi: 10.1016/j.neuron.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009;23:2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, Park KH, Jung KH, Lee SK, Kim M, et al. Altered microRNA regulation in Huntington's disease models. Experimental neurology. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Li J, Liang S, Yu H, Zhang J, Ma D, Lu X. An inhibitory effect of miR-22 on cell migration and invasion in ovarian cancer. Gynecologic oncology. 2010;119:543–548. doi: 10.1016/j.ygyno.2010.08.034. [DOI] [PubMed] [Google Scholar]
- Li W, Cressy M, Qin H, Fulga T, Van Vactor D, Dubnau J. MicroRNA-276a functions in ellipsoid body and mushroom body neurons for naive and conditioned olfactory avoidance in Drosophila. J Neurosci. 2013;33:5821–5833. doi: 10.1523/JNEUROSCI.4004-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Jiang Y, Zhang H, Greenlee AR, Yu R, Yang Q. miR-22 functions as a micro-oncogene in transformed human bronchial epithelial cells induced by anti-benzo[a]pyrene-7,8-diol-9,10-epoxide. Toxicol In Vitro. 2010;24:1168–1175. doi: 10.1016/j.tiv.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone AK, Edeleva EV, Kucherenko MM, Hsiao NH, Shcherbata HR. Dg-Dys-Syn1 signaling in Drosophila regulates the microRNA profile. BMC Cell Biol. 2012;13:26. doi: 10.1186/1471-2121-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CL, Duvall JA, Ilkin Y, Simon JS, Arreaza MG, Wilkes K, Alvarez-Retuerto A, Whichello A, Powell CM, Rao K, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- McCann C, Holohan EE, Das S, Dervan A, Larkin A, Lee JA, Rodrigues V, Parker R, Ramaswami M. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc the Nat Acad of Sci USA. 2011;108:E655–E662. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- McNeill E, Van Vactor D. MicroRNAs shape the neuronal landscape. Neuron. 2012;75:363–379. doi: 10.1016/j.neuron.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhail FM, Lose EJ, Robin NH, Descartes MD, Rutledge KD, Rutledge SL, Korf BR, Carroll AJ. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Genet A. 2011;155A:2386–2396. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- Muinos-Gimeno M, Espinosa-Parrilla Y, Guidi M, Kagerbauer B, Sipila T, Maron E, Pettai K, Kananen L, Navines R, Martin-Santos R, et al. Human microRNAs miR-22, miR-138-2, miR-148a, and miR-488 are associated with panic disorder and regulate several anxiety candidate genes and related pathways. Biol Psychiatry. 2011;69:526–533. doi: 10.1016/j.biopsych.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Roorda RD, Lima SQ, Zemelman BV, Morcillo P, Miesenböck G. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 2002;36:463–474. doi: 10.1016/s0896-6273(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Parisi C, Arisi I, D'Ambrosi N, Storti AE, Brandi R, D'Onofrio M, Volonté C. Dysregulated microRNAs in amyotrophic lateral sclerosis microglia modulate genes linked to neuroinflammation. Cell Death Dis. 2013;4:e959. doi: 10.1038/cddis.2013.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, Huetteroth W, Burke CJ, Krashes MJ, Lai SL, Lee T, Waddell S. A pair of inhibitory neurons are required to sustain labile memory in the Drosophila mushroom body. Curr Biol. 2011;21:855–861. doi: 10.1016/j.cub.2011.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saab BJ, Mansuy IM. Neuroepigenetics of memory formation and impairment: the role of microRNAs. Neuropharmacology. 2014;80:61–69. doi: 10.1016/j.neuropharm.2014.01.026. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Im HI, Veno MT, Fowler CD, Min A, Intrator A, Kjems J, Kenny PJ, O'Carroll D, Greengard P. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J Exp Med. 2010;207:1843–1851. doi: 10.1084/jem.20100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- Siegel SR, Mackenzie J, Chaplin G, Jablonski NG, Griffiths L. Circulating microRNAs involved in multiple sclerosis. Mol Biol Rep. 2012;39:6219–6225. doi: 10.1007/s11033-011-1441-7. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Digan ME, Mahowald AP, Scott M, Craig EA. Two clusters of genes for major chorion proteins of Drosophila melanogaster. Cell. 1980;19:905–914. doi: 10.1016/0092-8674(80)90082-3. [DOI] [PubMed] [Google Scholar]
- Tastan OY, Maines JZ, Li Y, McKearin DM, Buszczak M. Drosophila ataxin 2-binding protein 1 marks an intermediate step in the molecular differentiation of female germline cysts. Development. 2010;137:3167–3176. doi: 10.1242/dev.050575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GC, Bazhenov M, Laurent G. Olfactory representations by Drosophila mushroom body neurons. J. Neurophysiol. 2008;99:734–746. doi: 10.1152/jn.01283.2007. [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usha N, Shashidhara LS. Interaction between Ataxin-2 Binding Protein 1 and Cubitus-interruptus during wing development in Drosophila. Dev Biol. 2010;341:389–399. doi: 10.1016/j.ydbio.2010.02.039. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volvert ML, Prevot PP, Close P, Laguesse S, Pirotte S, Hemphill J, Rogister F, Kruzy N, Sacheli R, Moonen G, et al. MicroRNA targeting of CoREST controls polarization of migrating cortical neurons. Cell Rep. 2014;7:1168–1183. doi: 10.1016/j.celrep.2014.03.075. [DOI] [PubMed] [Google Scholar]
- Walkinshaw E, Gai Y, Farkas C, Richter D, Nicholas E, Keleman K, Davis RL. Identification of Genes that Promote or Inhibit Olfactory Memory Formation in Drosophila. Genetics. 2015;199:1173–1182. doi: 10.1534/genetics.114.173575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wright NJ, Guo H, Xie Z, Svoboda K, Malinow R, Smith DP, Zhong Y. Genetic manipulation of the odor-evoked distributed neural activity in the Drosophila mushroom body. Neuron. 2001;29:267–276. doi: 10.1016/s0896-6273(01)00196-9. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Yu D, Akalal DB, Davis RL. Drosophila α/β mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron. 2006;52:845–855. doi: 10.1016/j.neuron.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Keene AC, Srivatsan A, Waddell S, Davis RL. Drosophila DPM neurons form a delayed and branch-specific memory trace after olfactory classical conditioning. Cell. 2005;123:945–957. doi: 10.1016/j.cell.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Yu H, Wu M, Zhao P, Huang Y, Wang W, Yin W. Neuroprotective effects of viral overexpression of microRNA-22 in rat and cell models of cerebral ischemia-reperfusion injury. J Cell Biochem. 2015;116:233–241. doi: 10.1002/jcb.24960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.