ABSTRACT
In Saccharomyces cerevisiae Atg8 coupled to phosphatidylethanolamine is a key component of autophagosome biogenesis. Atg21 binds via 2 sites at the circumference of its β-propeller to PtdIns3P at the phagophore assembly site (PAS). It recruits and arranges both Atg8 and Atg16, which is part of the E3-like ligase complex Atg12–Atg5-Atg16. Binding of Atg8 to Atg21 requires the FK-motif within the N-terminal-helical domain of Atg8 and D146 at the top of the Atg21 β-propeller. Atg16 binds via D101 and E102 within its coiled-coil domain to Atg21.
KEYWORDS: Atg8 lipidation, Atg16, Atg21, PROPPIN, PtdIns3P
In S. cerevisiae the Atg8 lipidation machinery is crucial for autophagosome biogenesis. In brief, after its carboxy terminal processing by the Atg4 proteinase, ubiquitin-like Atg8 is activated by the E1-like enzyme Atg7. Subsequently, Atg8 is transferred to the E2-like Atg3 and finally coupled to the membrane lipid phosphatidylethanolamine (PE). In parallel, ubiquitin-like Atg12 is also activated by Atg7, transferred to the E2-like Atg10 and covalently coupled to Atg5. The Atg12–Atg5 conjugate then interacts with Atg16. Atg16 dimerizes via its coiled-coil domain, and a protein complex containing 2 Atg12–Atg5-Atg16 modules is formed. The Atg12–Atg5-Atg16 complex acts as an E3-like ligase and activates Atg3-mediated Atg8 lipidation. Obviously, efficient Atg8 lipidation requires correct spatial arrangement of the E3 complex and Atg8 at the target membrane.
Remarkably, Atg8 lipidation depends on the presence of PtdIns3P at the phagophore assembly site (PAS), but none of the abovementioned proteins contain a typical PtdIns3P-binding site. In fact, the presence of PtdIns3P is deciphered by a family of β-propellers that bind phosphoinositides (PROPPINs). This highly conserved protein family consists in yeast of the homologous proteins Atg18, Atg21 and Hsv2; the 4 human homologs are termed WIPI1 to WIPI4. In previous studies, we solved the crystal structure of a yeast member of this protein family and demonstrated that these proteins bind PtdIns3P with 2 independent binding sites at the circumference of their propeller and thus are oriented perpendicular to membranes. We and others previously reported that in atg21Δ cells Atg8 lipidation is retarded and that Atg8 is not normally recruited to the PAS in growing atg21Δ cells. We speculated that Atg21 could be the missing component, which senses PtdIns3P at the PAS and recruits in correct orientation Atg8 and the Atg12–Atg5-Atg16 E3 complex.
Atg8 lipidation is thought to take place at the PAS. So far, however, Atg21 was only detected at endosomes. In a first step we could show that Atg21 also colocalizes in a PtdIns3P-dependent manner with the PAS marker precursor Ape1. Next we could demonstrate using the split-ubiquitin system (which is similar to the 2-hybrid system, but better suited for analyses of membrane or membrane-associated proteins), co-immunoprecipitations and affinity isolation experiments a direct interaction between Atg21 and Atg8. Atg8 can interact with other proteins via different sites. Within its ubiquitin-like fold Atg8 exhibits a site that mediates interaction with an Atg8-interacting motif (AIM, or LC3-interacting region/LIR in mammals). These AIM motifs are typically found in cargo receptors or components of the lipidation machinery. In addition to its ubiquitin-fold domain, Atg8 contains an N-terminal helical domain (NHD). In previous work we identified within this NHD a FK-motif also required for Atg8 interactions. Using a set of Atg8 mutants, we found in vivo with fluorescence microscopy, or in vitro with whole cell extracts that both the FK-motif in the NHD-domain and the AIM-binding site is required for interaction with Atg21. In contrast, with recombinant proteins isolated from E. coli the interaction requires only the FK-motif, but not the AIM-binding site. This finding suggests that the direct interaction mainly depends on the FK-motif, but that in vivo further interactions via the AIM-binding sites with other components, most likely the E2-enzyme Atg3, strengthen complex formation.
A routine approach to determine the Atg21 site interacting with Atg8 would be the use of truncated versions. In our case this seemed problematic, since this would most certainly affect the native fold of the propeller. As an alternative, we used a library of synthetic peptides corresponding to the loops of the propeller, and screened for peptides competing with binding of Atg21 and Atg8 in affinity isolation experiments. Because ubiquitin has been reported to bind to the smaller top side of β-propellers, we started with a peptide collection only covering the propeller top. Indeed, a peptide corresponding to loop 2D to 3A significantly competes with the binding. Using mutated variants of the peptide, D146 was identified as a crucial residue. In line with this observation, Atg21D146K shows in fluorescence microscopy a reduced recruitment of Atg8 to the PAS.
Together, these data suggest that Atg8 interacts via its FK motif with the top of the Atg21 propeller, where D146 of Atg21 is required for the interaction. Because in vitro the AIM-binding site of Atg8 is dispensable for Atg21 binding we propose the following model (Fig. 1): Atg8 binds via its NHD-domain to the Atg21 propeller in an orientation such that the AIM-binding site of Atg8 is exposed away from the propeller. This conformation would allow further interactions via the AIM-binding site and is in line with the orientation in which ubiquitin has been reported to bind to β-propellers.
Figure 1.
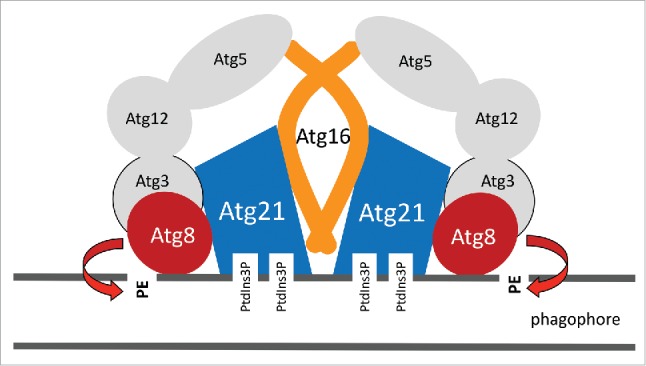
Schematic illustration for how Atg21 acts as a scaffold for Atg8 lipidation. Further details are given in the text.
Having established an interaction between Atg8 and Atg21, we next analyzed interactions between Atg21 and components of the Atg12–Atg5-Atg16 complex. In a series of experiments we detected no interactions between Atg21 and Atg12 or Atg5. However, we could identify a direct interaction between Atg21 and Atg16. In the study of another lab, a set of residues within the coiled-coil domain of Atg16 were uncovered to mediate dimerization. This study further identified 2 residues of this domain to be essential for Atg16 activity, but not for dimerization. We could show with the split-ubiquitin system, co-immunoprecipitations and fluorescence microscopy that indeed these 2 residues, D101 and E102 of Atg16, are required for the interaction with Atg21, without affecting Atg16 dimerization. In parallel with our work, an elegant study showed that mammalian ATG16L1 interacts with the bottom of WIPI2B in a way reminiscent of the yeast situation. Together we propose a model, where Atg8 binds on top and Atg16 of the Atg12–Atg5-Atg16 complex at the bottom of the propeller (Fig. 1). In this way Atg21 arranges Atg8 and its E3-like complex in a functional conformation at membranes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was funded by the “Deutsche Forschungsgemeinschaft” within the CRC860 “Integrative Structural Biology of Dynamic Macromolecular Assemblies.”


