Figure 4.
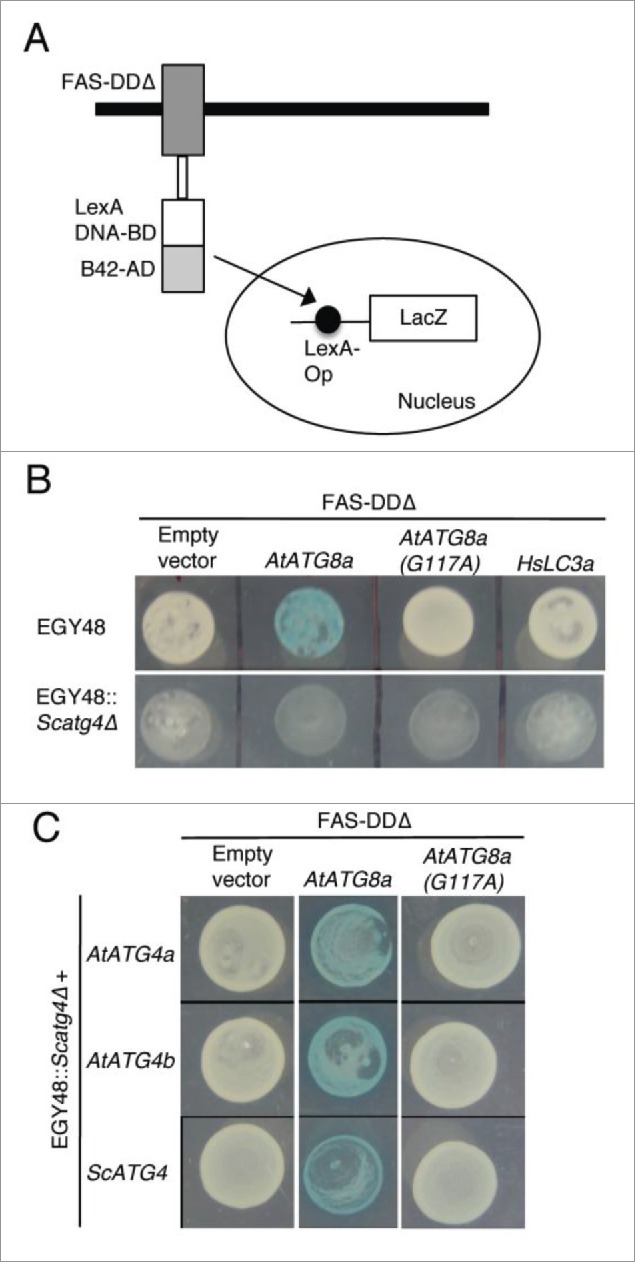
Yeast Atg4 can process plant AtATG8a but not human HsLC3A. (A) Schematics of yeast-based cleavable transcription activator and reporter approach.45 Chimeric transcription activator (TA) comprised of LexA DNA binding domain (LexA DNA-BD) and B42 activation domain (B42-AD) was fused to the C terminus of death domain deleted FAS type I transmembrane receptor (FAS-DDΔ). ATG8 or LC3A is inserted as a translational fusion between FAS-DDΔ and TA. ATG4-mediated processing of ATG8 or LC3A at the Gly residue (not shown) will lead to translocation of TA into the nucleus to induce LacZ expression. (B) FAS-DDΔ-AtATG8a-TA, FAS-DDΔ-AtATG8aG117A-TA, and FAS-DDΔ-HsLC3A-TA plasmids were transformed into EGY48 and Scatg4-deleted EGY48 (EGY48 Scatg4Δ) yeast strains. The selected yeast cells were spotted onto media plates with X-gal substrate. Blue color indicates that endogenous yeast Atg4 can process plant AtATG8a (top panel, column 2). White colony indicates that the endogenous yeast Atg4 cannot process mutant AtATG4aG117A and human HsLC3A (top panel). The deletion of ScATG4 from EGY48 results in lack of processing of AtATG8a (bottom panel, column 2). (C) EGY48 Scatg4Δ yeast cells lacking ScATG4 were transformed with plasmid encoding FAS-DDΔ-AtATG8a-TA and AtATG4a or AtATG4b and FAS-DDΔ-AtATG8a-TA with ScATG4. The transformed cells were grown on media plates with X-gal substrate. Both yeast and plant ATG4s could process plant AtATG8a (middle column). The empty vector and the mutant AtATG8aG117A were used as controls (first and last columns).
