Abstract
Macroautophagy (hereafter referred to as autophagy) is an essential self-digestion process to maintain homeostasis and promote survival in response to starvation. Although the components of autophagy in the cytoplasm have been well studied, little has been known about the fine-tuning mechanism of autophagy through epigenetic regulations. Recently, we identified the histone arginine methyltransferase CARM1 as a new component and followed histone H3R17 dimethylation as a critical epigenetic mark in starvation-induced autophagy. Upon nutrient starvation, CARM1 is stabilized in the nucleus, but not in the cytoplasm, whereas it is constantly degraded under nutrient-rich conditions by the SKP2-containing SCF (SKP1-CUL1-F-box protein) E3 ubiquitin ligase. We further showed that nutrient starvation induces the protein levels and activity of AMPK in the nucleus. Activated AMPK then phosphorylates FOXO3, leading to SKP2 downregulation and increased CARM1 protein levels in the nucleus. Stabilized CARM1 in turn functions as an essential co-activator of TFEB and regulates the expression of autophagy and lysosomal genes. Our findings provide a conceptual advance that activation of specific epigenetic programs is indispensable for a sustained autophagic response, and shed light on a potential therapeutic targeting of the newly identified AMPK-SKP2-CARM1 signaling axis in autophagy-related diseases.
KEYWORDS: AMPK, arginine methylation, CARM1, ellagic acid, epigenetic regulation, FOXO, SKP2, starvation, TFEB, transcription
Epigenetic regulations including histone modifications and DNA methylation affect the overall chromatin structure and function, leading to the various outcomes of cellular processes including autophagy. We asked an intriguing question as to whether epigenetic and transcriptional regulations are actively involved in autophagy to consistently replenish the materials of the autophagy machinery and sustain the process. Further, we wished to identify the role of epigenetic mechanisms in autophagy and understand how their perturbation could lead to autophagy-related diseases.
Among various histone modifications, we found arginine methylation (H3R17me2) as a critical epigenetic mark in autophagic induction. We observed a dramatic increase in H3R17me2 marks following glucose and amino acid starvation, and searched for its function in the process of autophagy. This histone modification is a transcriptional activation mark that is solely mediated by the CARM1 arginine methyltransferase. Interestingly, concomitant with an increase in H3R17me2, an increase in CARM1 protein levels is observed in various cell lines, and the ablation of CARM1 or inhibition of histone arginine methylation significantly impairs starvation-induced autophagy.
Although present in both the cytoplasm and the nucleus, CARM1 induction following glucose starvation occurs primarily in the nucleus due to the exclusive nuclear localization of SKP2, a component of the SCF E3 ubiquitin ligase complex, which is responsible for CARM1 degradation under nutrient-rich conditions. Interestingly, SKP2 expression is downregulated in response to glucose starvation and this regulation turns out to be mediated by AMPK. We found that in response to glucose starvation FOXO3 is phosphorylated by AMPK and functions as a repressor for Skp2 transcription. Reduction of SKP2 expression results in CARM1 stabilization and the stabilized CARM1 then functions as a co-activator of TFEB and transcriptionally activates various TFEB target genes with functions at all stages of the autophagy process. Intriguingly, amino acid starvation or rapamycin treatment also increase CARM1 expression and subsequent H3R17me2 in various cell lines tested. We therefore speculate that an alternative AMPK-independent mechanism might exist to tightly regulate the transcriptional activation through CARM1 following amino acid starvation or rapamycin treatment. Taken together, we reported a previously unidentified AMPK-SKP2-CARM1 signaling axis, in which nutrient starvation-induced AMPK increases CARM1 expression in the nucleus via reduction of SKP2 expression, and provided evidence that CARM1-dependent histone arginine methylation functions as a critical nuclear event of autophagy for epigenetic and transcriptional regulation (Fig. 1).
Figure 1.
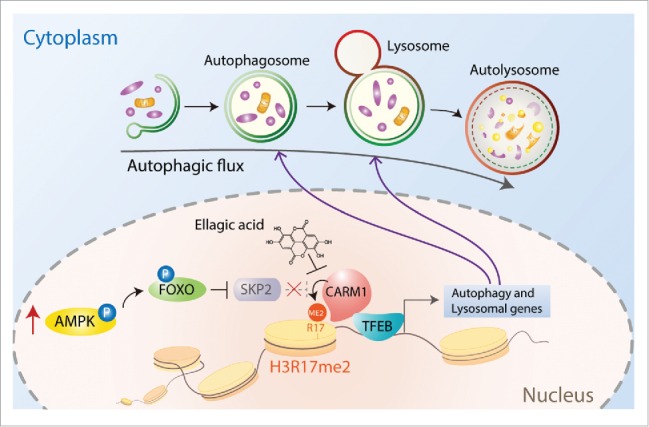
AMPK-SKP2-CARM1 signaling cascade in epigenetic and transcriptional regulation of autophagy. In response to prolonged nutrient starvation, the protein levels and activity of AMPK increase in the nucleus. The FOXO3 transcription factor is then phosphorylated by AMPK and transcriptionally represses SKP2, the component of the SCF E3 ubiquitin ligase complex responsible for CARM1 degradation. Stabilized CARM1 following starvation functions as a co-activator of TFEB and increases H3R17me2 to transcriptionally activate autophagy and lysosomal genes. Ellagic acid treatment impairs CARM1-induced autophagy through selective inhibition of histone H3 arginine 17 dimethylation (H3R17me2). Illustration by Kyung-Ah Lee.
Surprisingly, we found an unexpected nuclear role of AMPK in epigenetic and transcriptional regulation of autophagy. In response to prolonged starvation, AMPK protein levels and activity increase in the nucleus. In our experimental setting, the transcriptional activation of autophagy and lysosomal genes occurs starting at 12 h after glucose starvation. It is noteworthy that the time frame of nuclear AMPK accumulation is concomitant with the induction of CARM1 and subsequent transcriptional activation of autophagy genes. While an acute and rapid response of autophagy occurs primarily in the cytoplasm, prolonged starvation results in the activation of transcriptional programs and changes in the epigenetic network in the nucleus. AMPK has been known to activate autophagy through inactivation of TORC1 and phosphorylation of ULK1, all of which occur in the cytoplasm at early times of starvation. However, our results indicate that when transcription of various autophagy and lysosomal genes are upregulated to sustain autophagy, AMPK accumulates at a later time of starvation in the nucleus and distinctly functions in coordinating the transcription of target genes involved in the outcome of autophagy.
The importance of CARM1 and histone arginine methylation in autophagy was also assessed in vivo. In 8- to 10-wk-old mice, fasting-induced autophagy in the liver is dramatically attenuated and induction of various autophagy and lysosomal genes regulated by CARM1 is impaired in fasted mice pre-injected with ellagic acid. Ellagic acid is a naturally occurring polyphenol, abundant in fruits and vegetables, which was identified through general screening as a specific inhibitor of H3R17me2 modifications. Ellagic acid was also reported to have a beneficial effect against cancer and cardiovascular diseases, which has prompted active research into its potential health benefits. Moreover, ellagic acid shows anti-malarial properties and when combined with chloroquine, a commonly used drug against malaria but also an autophagy inhibitor, improves disease treatment. It would be therefore interesting to study the effect of ellagic acid on autophagy-related diseases and investigate its therapeutic potential. The effects of ellagic acid in the impairment of CARM1-induced autophagy strongly support our finding that H3R17me2 is a major epigenetic mark of autophagy. It is tempting to speculate that there exist readers of methyl-arginine marks able to recognize H3R17me2, and then connect this epigenetic mark to the process of autophagy. It will be interesting to screen for potential readers of methyl-arginine marks and further explore the direct functional link of autophagic induction, histone modifications, and epigenetic regulation.
In summary, our findings demonstrate nutrient signaling through CARM1 and histone arginine methylation as a central nuclear event of autophagy and provide a new signaling axis of AMPK-SKP2-CARM1 that links nutrient sensing to epigenetic regulation in autophagy induction by nutrient starvation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
FUNDING
This work was supported by Creative Research Initiatives Program (Research Center for Chromatin Dynamics, 2009-0081563) to S.H.B.; the Basic Science Research Program (NRF-2014R1A6A3A04057910) to H.K. from the National Research Foundation (NRF) grant funded by the Korea government (MSIP).
Supplementary Material
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


