Abstract
Background
Rotaviruses with G6P[14] specificity are mostly isolated in cattle and have been established as a rare cause of gastroenteritis in humans. This study reports the first detection of G6P[14] rotavirus strain in Ghana from the stool of an infant during a hospital-based rotavirus surveillance study in 2010.
Methods
Viral RNA was extracted and rotavirus VP7 and VP4 genes amplified by one step RT-PCR using gene-specific primers. The DNA was purified, sequenced and genotypes determined using BLAST and RotaC v2.0. Phylogenetic tree was constructed using maximum likelihood method in MEGA v6.06 software and statistically supported by bootstrapping with 1000 replicates. Phylogenetic distances were calculated using the Kimura-2 parameter model.
Results
The study strain, GHA-M0084/2010 was characterised as G6P[14]. The VP7 gene of the Ghanaian strain clustered in G6 lineage-III together with artiodactyl and human rotavirus (HRV) strains. It exhibited the highest nucleotide (88.1 %) and amino acid (86.9 %) sequence identity with Belgian HRV strain, B10925. The VP8* fragment of the VP4 gene was closely related to HRV strains detected in France, Italy, Spain and Belgium. It exhibited the strongest nucleotide sequence identity (87.9 %) with HRV strains, PA169 and PR/1300 (Italy) and the strongest amino acid sequence identity (89.3 %) with HRV strain R2775/FRA/07 (France).
Conclusion
The study reports the first detection of G6P[14] HRV strain in an infant in Ghana. The detection of G6P[14], an unusual strain pre-vaccine introduction in Ghana, suggests a potential compromise of vaccine effectiveness and indicates the necessity for continuous surveillance in the post vaccine era.
Electronic supplementary material
The online version of this article (doi:10.1186/s12985-016-0643-y) contains supplementary material, which is available to authorized users.
Keywords: Rotavirus, Gastroenteritis, Genotyping, Ghana
Background
Group A rotaviruses (RVAs) are the leading cause of severe, dehydrating, acute diarrhoea in children <5 years of age worldwide and are estimated to be responsible for 453,000 deaths annually, with a disproportionate number of these deaths occurring in sub-Saharan Africa and South East Asia [1]. The World Health organization (WHO) recommended global use of rotavirus vaccines as one of the effective interventions to reduce RVA-associated diseases [2]. Currently, two RVA vaccines, RotaTeq (Merck & Co., Inc., United States) and Rotarix (GlaxoSmithKline Biologicals, Belgium) have been licensed and introduced into routine childhood immunisation programmes.
Rotavirus, a genus of the Reoviridae family, contains a genome of 11 segments of double-stranded RNA (dsRNA) and are classified in a binary system based upon the main neutralization antigens, namely, the spike protein (VP4) and the major outer capsid glycoprotein (VP7) [3]. VP7 and VP4 nucleotide sequences define G and P genotypes, respectively, and carry independent neutralization-specific epitopes [3]. Recently, the Rotavirus Classification Working Group (RCWG) proposed an extensive RVA classification system that takes into account all 11 rotavirus genome segments [4]. Prior to the present study, at least 27 G genotypes and 37 P genotypes have been identified globally and approximately 73 G/P genotype combinations of rotaviruses have been reported to infect humans [5]. Among these, only six G/P-genotype combinations; G1P[8], G2P[4], G3P[8], G4P[8], G9P[8] and G12P[8] account for most of the HRV strains detected globally [6]. However, several epidemiological studies have reported the sporadic detection of unusual rotavirus G-genotypes (G5-G6, G8, G10, G11 and G20) and P-genotypes (P[1]-P[3], P[5], P[7], P[9]-P[11], P[14], P[19] and P[25]) in humans [3, 6, 7]. Many of these unusual rotavirus genotypes believed to be of animal origin have been introduced into the human population through interspecies transmission and/or reassortment events [3, 8].
Rotaviruses with G6 specificity are recognised as common serotype in cattle and other ruminants and have been found to be an infrequent cause of human disease. Bovine-like human G6 rotavirus strains PA151 and PA169 were first identified in 1987 and 1988 respectively from two Italian children hospitalised with acute gastroenteritis [9]. These two strains were characterised as G6P[9] (strain PA151) and G6P[14] (strain PA169). Subsequently, an increasing number of G6 HRV strains have been detected in combination with other P genotypes across the globe (Hungary, Australia, United States and India) [10–13]. G6 HRV strains are reported to be found mainly in combination with P[14] rotaviruses (commonly detected in antelope, cattle, rabbits, goats, sheep and guanacos) [14]. Previous reports in Africa, documented the isolation of G6 rotavirus strains only in animals [15]. The first detection of G6P[14] HRV strain in Africa was in 2004 from the stool of a child from Egypt [7].
In our previous study, rotavirus was detected in the stool of a four month old child hospitalised with severe, acute dehydrating gastroenteritis at Maamobi Polyclinic in Accra, Ghana in August 2010. This sample exhibited the classical human rotavirus group A migration pattern (4-2-3-2) with a long electrophoretype by Polyacrylamide Gel Electrophoresis (PAGE) (data not shown). However, the amplified VP7 and VP8* fragment of the VP4 gene could not be genotyped using the routine HRV genotype specific primers. In this study, the G and P genotypes of strain GHA-M0084/2010 were identified by sequencing.
Results
The study strain GHA-M0084/2010 was characterised as G6P[14]. Phylogenetic analysis showed the VP7 gene of the Ghanaian G6P[14] strain clustered in lineage III together with randomly selected artiodactyl and human G6 strains (Fig. 1). It clustered in G6 lineage-III together with HRV strains isolated in Italy (RVA/Hu/PA5/89, RVA/Hu-tc/ITA/PA169/1988), Belgium (RVA/Hu/BEL/B10925/1997), and with artiodactyl strain, (RVA/Caprine/CAP455) isolated in South Africa (Fig. 1). The VP7 gene of the study strain shared the strongest nucleotide (88.1 %) and amino acid (86.9 %) sequence identity with the Belgian HRV strain, B10925 detected in 1997. However, it shared very low nucleotide (74.9 %) and amino acid (74.9 %) sequence identity with West African isolates BA356 (Cameroon) and 48-BF (Burkina Faso) (Table 1).
Fig. 1.
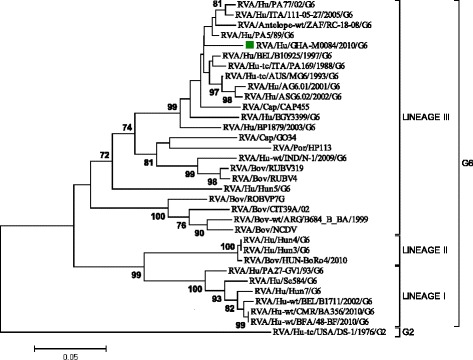
Phylogenetic tree constructed from nucleotide sequence of Ghanaian G6 strain GHA-M0084/2010 and reference G6 strains by the maximum likelihood method using MEGA 6.06 software and rooted with the G2 human rotavirus strain, DS-1. Variation scale (nucleotide substitution per site) is indicated below the phylogenetic tree. Percentage bootstrap support is indicated by values at each node, and values ≥70 % are shown. Study strain GHA-M0084/2010 is indicated by a filled green square. Reference sequences used in the analysis were obtained from the GenBank database. Phylogenetic distance was measured by Kimura two-parameter model. Phylogenetic tree was supported statistically by bootstrapping with 1000 replicates
Table 1.
Comparison of nucleotide and amino acid sequence identity of Ghanaian G6 and published G6 strains
| Nucleotide identity (%) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RV STRAIN | G-TYPE | PA169 | AG6 | ASG6 | MG6 | PA5 | B10925 | BA356 | 48-BF | PA77 | N-1 | Hun5 | Hun4 | ROBVP7G | NCDV | Hun7 | BoRo4 | PA27 | Hun3 | GHA-M0084 |
| PA169 | G6 | 94.9 | 96.5 | 97.5 | 97.2 | 97.8 | 79.2 | 78.8 | 96.1 | 85.1 | 86.2 | 79.8 | 82.5 | 82 | 79.5 | 80.1 | 79.9 | 79.8 | 87.1 | |
| AG6 | G6 | 96.7 | 97.2 | 97.1 | 94.1 | 94.5 | 76.9 | 76.5 | 93.3 | 83.9 | 85.2 | 78.4 | 81.4 | 81 | 77.2 | 78.8 | 78 | 78.4 | 85 | |
| ASG6 | G6 | 99.1 | 96.7 | 98.7 | 96 | 96.1 | 78.6 | 78.2 | 95.2 | 85.8 | 86.5 | 79.5 | 82.6 | 82.4 | 78.4 | 79.9 | 79.4 | 79.5 | 86.2 | |
| MG6 | G6 | 99.5 | 97.1 | 99.5 | 96.7 | 97.1 | 78.8 | 78.4 | 95.9 | 86.3 | 86.5 | 79.8 | 83.2 | 82.5 | 79.1 | 80.2 | 79.9 | 79.8 | 87.1 | |
| PA5 | G6 | 98.7 | 96.3 | 98.7 | 99.1 | 97.1 | 79.8 | 79.4 | 98 | 85.4 | 86.3 | 79.9 | 83.2 | 82.5 | 79.9 | 80.2 | 80.2 | 79.9 | 87.7 | |
| B10925 | G6 | 98.7 | 96.3 | 98.7 | 99.1 | 99.1 | 78.8 | 78.4 | 96.5 | 86.3 | 86.3 | 79.5 | 83.3 | 82.6 | 79 | 79.7 | 79.5 | 79.5 | 88.1 | |
| BA356 | G6 | 92.2 | 89.3 | 92.6 | 92.2 | 91.4 | 91.4 | 99.5 | 79.7 | 78.8 | 78.6 | 86.2 | 79.8 | 80.2 | 96 | 85.9 | 93.4 | 86.2 | 74.9 | |
| 48-BF | G6 | 92.2 | 89.3 | 92.6 | 92.2 | 91.4 | 91.4 | 100 | 79.2 | 78.4 | 78.7 | 86.3 | 79.7 | 80.1 | 96.1 | 85.8 | 93.5 | 86.3 | 74.9 | |
| PA77 | G6 | 99.1 | 96.7 | 99.1 | 99.5 | 99.5 | 99.5 | 91.8 | 91.8 | 85.4 | 86.5 | 79.9 | 83.1 | 82.8 | 79.8 | 80.1 | 80.1 | 79.9 | 87.6 | |
| N-1 | G6 | 96.7 | 94.2 | 96.7 | 97.1 | 96.7 | 96.7 | 92.6 | 92.6 | 97.1 | 84.4 | 80.1 | 83.6 | 82.6 | 79.9 | 79.8 | 79.9 | 80.1 | 79.2 | |
| Hun5 | G6 | 98.3 | 95.9 | 98.3 | 98.7 | 98.3 | 98.3 | 91.8 | 91.8 | 98.7 | 97.1 | 81.7 | 81.8 | 80.2 | 79.7 | 81.3 | 79.9 | 81.7 | 78.7 | |
| Hun4 | G6 | 92.6 | 89.7 | 92.6 | 92.6 | 91.8 | 92.2 | 93.4 | 93.4 | 92.2 | 92.2 | 92.2 | 81 | 80.5 | 86.1 | 98.6 | 88.2 | 100 | 75.2 | |
| ROBVP7G | G6 | 93 | 90.6 | 92.2 | 92.6 | 92.6 | 92.6 | 89.3 | 89.3 | 93 | 92.2 | 91.8 | 89.7 | 93.7 | 79.9 | 80.6 | 80.3 | 81 | 76.5 | |
| NCDV | G6 | 91.8 | 89.3 | 91 | 91.4 | 91.4 | 91.4 | 88.9 | 88.9 | 91.8 | 91 | 90.6 | 89.3 | 95.9 | 80.3 | 80.2 | 80.7 | 80.5 | 75.8 | |
| Hun7 | G6 | 93 | 90.2 | 92.6 | 93 | 92.2 | 92.2 | 98.7 | 98.7 | 92.6 | 93.4 | 92.6 | 93.8 | 90.2 | 89.7 | 85.4 | 95 | 86.1 | 75 | |
| BoRo4 | G6 | 92.6 | 89.7 | 92.2 | 92.6 | 91.8 | 92.2 | 93 | 93 | 92.2 | 92.2 | 92.2 | 99.1 | 89.7 | 89.3 | 93.8 | 88.2 | 98.6 | 75 | |
| PA27 | G6 | 92.6 | 89.7 | 92.2 | 92.6 | 91.8 | 91.8 | 97.1 | 97.1 | 92.2 | 93.4 | 92.2 | 94.2 | 90.6 | 90.2 | 97.9 | 94.2 | 88.2 | 75.2 | |
| Hun3 | G6 | 92.6 | 89.7 | 92.6 | 92.6 | 91.8 | 92.2 | 93.4 | 93.4 | 92.2 | 92.2 | 92.2 | 100 | 89.7 | 89.3 | 93.8 | 99.1 | 94.2 | 75.2 | |
| GHA-M0084 | G6 | 85.7 | 83.2 | 85.7 | 86.1 | 86.1 | 86.9 | 80.4 | 80.4 | 86.5 | 85.7 | 85.7 | 81.2 | 81.2 | 80 | 81.2 | 81.2 | 80.8 | 81.2 | |
| Amino acid identity (%) | ||||||||||||||||||||
Comparison of the deduced nucleotide (right, upper) and amino acid (left, below) identity of the VP7 gene of the Ghanaian G6 rotavirus strain with G6 reference strains from the GenBank. Study isolate is boldface type
Phylogenetic analysis of the VP8* fragment of the VP4 gene of the Ghanaian strain showed it clustered together with the prototype HRV strain, PA169-88 and published HRV strains from other parts of the world (Fig. 2). It also clustered together with bovine strains, Bov/86 and RUBV81 (India) and lama guanaco strain, Chubut (Argentina) (Fig. 2). The Ghanaian VP4 gene shared the strongest nucleotide sequence identity (87.9 %) with Italian HRV strains PA169 and PR/1300 and the strongest amino acid sequence identity (89.3 %) with R2775/FRA/07 isolated in France (Table 2). However, it shared nucleotide sequence identity of ≥87.0 % ≤89.3 % with randomly selected human and animal rotavirus reference strains detected in Italy (111-05-27), France (R2775/FRA/07), India (Bov/86 & RUBV81) and in Egypt (EGY3399).
Fig. 2.

Phylogenetic tree constructed from nucleotide sequence of Ghanaian P[14] strain GHA-M0084/2010 and reference P[14] strains by the maximum likelihood method using MEGA 6.06 software and rooted with the G4P[6] rotavirus strain RVA/Bov/P14-3/SLOV/G4P6. Variation scale (nucleotide substitution per site) is indicated below the phylogenetic tree. Percentage bootstrap support is indicated by values at each node, and values ≥70 % are shown. Study strain GHA-M0084/2010 is indicated by a filled green square. Reference sequences used in the analysis were obtained from the GenBank database. Phylogenetic distance was measured by Kimura two-parameter model. Phylogenetic tree was supported statistically by bootstrapping with 1000 replicates
Table 2.
Comparison of Amino acid and Nucleotide identity of Ghanaian P[14] and published P[14] rotavirus strains
| Amino acid identity (%) | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RV STRAIN | P-TYPE | MG6 | Hun5 | PA169 | HAL1166 | LRU62150 | B10925 | PR/1300 | GHA-M0084 | 111-05-27 | R2775 | Bov/86 | RUBV81 | EGY3399 | BP1879 | Sp1524 | Sp813 | P14-3 | B137 |
| MG6 | P[14] | 68.9 | 68.1 | 60.9 | 50 | 70.4 | 68.9 | 62.6 | 68.9 | 68.9 | 69.1 | 68.4 | 65.4 | 53.7 | 73.1 | 65.9 | 26.4 | 75.1 | |
| Hun5 | P[14] | 88.6 | 86.3 | 68.4 | 52.9 | 85.6 | 87.8 | 73.1 | 92.4 | 89.3 | 87.9 | 88.7 | 90.2 | 55.9 | 70.1 | 82.5 | 24.2 | 69.9 | |
| PA169 | P[14] | 89.1 | 95.2 | 68.4 | 49.2 | 92.4 | 84 | 73.8 | 83.3 | 93.1 | 83.4 | 84.2 | 82.7 | 55.9 | 73.1 | 84 | 25 | 71.4 | |
| HAL_1166 | P[14] | 86 | 88.9 | 89.1 | 48.5 | 69.1 | 67.6 | 64.4 | 64.6 | 69.1 | 69.4 | 70.1 | 68.4 | 72.1 | 65.6 | 66.1 | 23.5 | 63.9 | |
| LRU62150 | P[14] | 80.1 | 80.8 | 79.7 | 79.4 | 50 | 52.2 | 45.9 | 52.9 | 47.7 | 52.5 | 53.2 | 51.4 | 51.4 | 50.3 | 47 | 20.7 | 50.7 | |
| B10925 | P[14] | 89.6 | 94.8 | 97.4 | 89.1 | 79.7 | 84 | 73.8 | 82.5 | 91.6 | 82.7 | 83.4 | 81.9 | 59.7 | 77.6 | 89.3 | 25 | 76.6 | |
| PR/1300 | P[14] | 88.9 | 95.9 | 94.8 | 88.6 | 80.8 | 94.5 | 73.1 | 86.3 | 87.8 | 84.9 | 85.7 | 84.2 | 56.7 | 69.4 | 78 | 24.2 | 68.4 | |
| GHA-M0084 | P[14] | 83.2 | 87.7 | 87.9 | 83.9 | 75.4 | 87.7 | 87.9 | 70.8 | 76.8 | 72.5 | 72.5 | 71.1 | 50.7 | 64.4 | 70.8 | 25.7 | 62.9 | |
| 111-05-27 | P[14] | 88.9 | 97.4 | 94.1 | 87.2 | 80.4 | 93.6 | 95.2 | 87 | 86.3 | 84.9 | 85.7 | 89.4 | 58.2 | 68.6 | 80.3 | 25.7 | 67.6 | |
| R2775 | P[14] | 88.9 | 96.4 | 97.6 | 89.1 | 79.4 | 96.9 | 96.2 | 89.3 | 95.2 | 87.9 | 88.7 | 87.2 | 57.4 | 72.3 | 83.3 | 25 | 71.4 | |
| Bov/86 | P[14] | 88.9 | 95.5 | 94.1 | 89.1 | 80.6 | 93.6 | 94.8 | 87.9 | 94.3 | 95.7 | 99.2 | 85.8 | 59.2 | 71.1 | 75.9 | 24.8 | 70.1 | |
| RUBV81 | P[14] | 88.4 | 95.5 | 94.1 | 89.1 | 81.1 | 93.6 | 94.8 | 87.7 | 94.3 | 95.7 | 99.5 | 86.5 | 60 | 71.8 | 76.6 | 24.1 | 70.8 | |
| EGY3399 | P[14] | 88.2 | 96.2 | 94.3 | 89.3 | 80.8 | 93.8 | 94.5 | 87.2 | 95.5 | 95.5 | 95 | 95 | 59.7 | 67.9 | 79.6 | 25.7 | 66.9 | |
| BP1879 | P[14] | 78.7 | 79.9 | 79.9 | 85.3 | 75.2 | 80.8 | 80.4 | 74.7 | 80.8 | 80.6 | 80.8 | 80.8 | 81.1 | 60.7 | 53.7 | 20.7 | 58.9 | |
| Sp1524 | P[14] | 89.6 | 89.6 | 90.3 | 86.7 | 79 | 91.5 | 89.3 | 84.4 | 88.6 | 90.3 | 89.3 | 89.3 | 88.4 | 80.4 | 73.8 | 21.4 | 91.7 | |
| Sp813 | P[14] | 85.6 | 91.2 | 92.2 | 85.6 | 76.6 | 94.1 | 90 | 85.1 | 90.3 | 91.7 | 88.9 | 88.9 | 90.5 | 76.1 | 88.3 | 25.1 | 72.1 | |
| P14-3 | P[14] | 54.2 | 54.4 | 54.9 | 53.7 | 50.9 | 54.9 | 54 | 53.5 | 55.8 | 54.4 | 54.7 | 54.4 | 54.7 | 50.4 | 52.1 | 52.3 | 22.8 | |
| B137 | P[14] | 90.8 | 90 | 90.5 | 86.7 | 80.4 | 91.9 | 89.8 | 84.1 | 89.1 | 90.8 | 89.8 | 89.8 | 88.9 | 80.6 | 96.2 | 87.7 | 52.8 | |
| Nucleotide identity (%) | |||||||||||||||||||
Discussion
G6 rotaviruses have been found to be an infrequent cause of human disease. Their detection in the human population was suggestive of zoonotic transmission as they have been detected almost exclusively in artiodactyls such as cattle [8, 14]. We report the detection of a G6P[14] rotavirus strain in Ghana from the stool of an infant participating in a hospital-based diarrhea surveillance study conducted in 2010 (pre-vaccine era). This strain was detected in a locality where the human population live in close proximity with their cattle, goats, sheep and pigs. This setting and proximity provide an ideal environment for dual RV infections from human and animal sources which can lead to reassortment of genes and eventual generation of novel strains.
Phylogenetic analysis of the VP7 gene of the study strain (GHA-M0084/2010) showed clustering with artiodactyl and human G6 strains in lineage III (Fig. 1). The Ghanaian strain displayed the strongest nucleotide and amino acid sequence identity (74.9–88.1 % and 80.0–86.9 % respectively) to randomly selected non-African reference strains (Table 1). It exhibited the lowest nucleotide identity (74.9 %) to African HRV strains, BA356 (Cameroon) and 48-BF (Burkina Faso) both isolated in 2010 (Table 1). The VP8* fragment of the VP4 gene of the study strain clustered together with HRV strains detected in France (2007), Italy (1988), Spain (2007) and Belgium (1997) (Fig. 2). It displayed the strongest aa (89.3 %) sequence identity to HRV strain R2775 isolated in France and nt (87.9 %) sequence identity to HRV strains PA169 and PR/1300 isolated in Italy (Table 2).
Two paediatric rotavirus vaccines (Rotarix and RotaTeq) are available and have been introduced into national immunization programmes in many countries [16, 17]. The monovalent vaccine, Rotarix, comprises a human G1P[8] rotavirus strain, whereas the pentavalent vaccine, RotaTeq, (a human-bovine reassortant vaccine) comprises human serotypes G1, G2, G3, G4, and P[8] on a bovine strain background. Both vaccines demonstrated broad protection against the most common RVA genotypes [18]. However, their efficacy is low especially on the African continent with diverse pool of strain types [19, 20]. G6P[14] strain detected in this study shares neither G- nor P-genotype with either of the two current vaccines. Though, other rotavirus proteins such as VP6 and NSP4 have been identified to play a role in the protective immunity against rotavirus infection, it is unknown whether Rotarix will provide protection against completely heterotypic G6P[14], an animal-derived human rotavirus strain.
Conclusion
In this study, we determined the VP7 and VP4 genotypes of previously non-typeable rotavirus strain. The study reports for the first time the detection of G6P[14] rotavirus strain in a child in West Africa. Data on the impact of rotavirus vaccination globally has shown that two rotavirus vaccines, RotaTeq™ and Rotarix™, provide good homotypic and heterotypic protection. However, there are concerns about the degree of protection these vaccines provide against rare/unusual rotavirus strains such as G6P[14] that bears G/P genotype specificities not included in the vaccines. The detection of a G6P[14] strain pre-vaccine introduction in Ghana, suggests a potential compromise of vaccine effectiveness and indicates the necessity for continuous surveillance in the post vaccine era.
Methods
Viral RNA extraction and reverse transcription
Viral RNA was extracted from 10 % faecal suspension in phosphate buffered saline using the QIAamp viral RNA Mini kit (Qiagen/Westburg, Leusden, The Netherlands). PCR primer pairs, 9Con1/9Con2 and VP4-F/VP4-R (Additional file 1: Table S1) were used in one step RT-PCR to target the VP7 gene and the VP8* fragment of the VP4 gene to generate 903 and 663 bp respectively [21, 22]. PCR products were purified using the Exo-SAP-IT clean up kit (Affymetrix, Miles Rd Cleveland, OH, USA). Products were directly sequenced for VP7 and VP4 genes using the Bigdye terminator sequencing kit v3.1 (Perkin-Elmer Applied Biosystems, Foster City, CA, USA) on an automated Genetic analyzer ABI PRISM 3130 (Applied Biosystems).
Nucleotide sequence and phylogenetic analysis
Chromatograms were visually inspected and edited. Genotypes were determined using the Basic Local Alignment Search Tool [BLAST] (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and confirmed with RotaC v2.0 genotyping tool [23]. Edited sequences were trimmed and homology table was constructed using Bioedit v.7.0.5. The nucleotide sequences of the study strain were aligned with randomly selected cognate gene sequences available in GenBank using the ClustalW algorithm [24]. Two rotavirus reference strains; HRV serotype G2 (RVA/Human-tc/USA/DS-1/1976/G2) and bovine G4P[6] (RVA/Bov/P14-3/SLOV/G4P[6]) were included in the phylogenetic trees as out-groups for the VP7 and VP4 genes respectively (Figs. 1 and 2). Phylogenetic trees were constructed using the maximum likelihood method in MEGA v6.06 programme [25] and statistically supported by bootstrapping with 1000 replicates. Pyhlogenetic distances were calculated using the Kimura-2 parameter model.
Nucleotide sequence accession number
The nucleotide sequences of the strain reported in this study (GHA-M0084/2010) have been deposited in the GenBank database under accession numbers KJ616414 and KJ616415 respectively.
Acknowledgement
The authors greatly appreciate the technical assistance of the staff of the Department of Electron Microscopy and Histopathology, Noguchi Memorial Institute for Medical Research, University of Ghana.
Funding
This work had no funding.
Availability of data and material
The authors have confirmed that the nucleotide sequence data for the VP7 and VP8* portion of the VP4 gene of strain GHA-M0084/2010 for this study have been deposited in the GenBank under the accession numbers KJ616414 and KJ616415.
Authors’ contributions
SD and GEA designed the study. SD conducted the research and co-analysed the data with BL and CA. GEA, TA and MO supervised this project. SD coordinated with BL and FED in drafting the manuscript. All authors read and approved of the final copy of the manuscript.
Authors’ information
SD (PhD) is Chief Research Assistant, BL (PhD candidate) is Principal Research Assistant, CA (PhD candidate) is Principal Research Assistant and FED (PhD) is a Research Fellow in the Department of Electron Microscopy and Histopathology (Regional Rotavirus Reference Laboratory). TA (PhD) is the Head of Medical Virology Laboratory, University of Ghana Medical School. KN (PhD) is a lecturer in the Department of Epidemiology and Disease Control, School of Public Health. MO (PhD) is the Head of Department of Electron Microscopy and Histopathology and GEA (PhD) is the Head of Regional Rotavirus Reference Laboratory.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board, Noguchi Memorial Institute for Medical Research, Legon, Accra, Ghana.
Abbreviations
- aa
Amino acid
- BLAST
Basic local alignment search tool
- bp
Base pair
- EIA
Enzyme immunoassay
- ELISA
Enzyme linked immuno-sorbent assay
- G
Glycoprotein
- HRVA
Human rotavirus group A
- MEGA
Molecular evolutionary genetics analysis
- nt
Nucleotide
- P
Protease-sensitive protein
- PAGE
Polyacrylamide gel electrophoresis
- RNA
Ribonucleic acid
- RRL
Regional reference laboratory
- RT-PCR
Reverse transcription-polymerase chain reaction
- RV
Rotavirus
- RVA
Rotavirus group A
- VP4
Viral protein 4
- VP7
Viral protein 7
- VP8*
Viral protein 8*
Additional file
List of primers used in this study. (DOCX 16 kb)
Contributor Information
Susan Damanka, Email: sdamanka@noguchi.ug.edu.gh.
Belinda Lartey, Email: blartey@noguchi.ug.edu.gh.
Chantal Agbemabiese, Email: cagbemabiese@noguchi.ug.edu.gh.
Francis E. Dennis, Email: fdennis@noguchi.ug.edu.gh
Theophilus Adiku, Email: tekadiku@chs.edu.gh.
Kofi Nyarko, Email: konyarko22@yahoo.com.
Michael Ofori, Email: mofori@noguchi.ug.edu.gh.
George E. Armah, Email: garmah@noguchi.ug.edu.gh
References
- 1.Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD. The WHO-coordinated Global Rotavirus Surveillance Network 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12(2):136–41. doi: 10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- 2.WHO Meeting of the immunization strategic advisory group of experts, April 2009 – conclusions and recommendations. Wkly Epidemiol Rec. 2009;2009(84):220–36. [PubMed] [Google Scholar]
- 3.Estes MK, Kapikian AZ. Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb BA, Roizman B, Straus SE, editors. Fields Virology. 5. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 1917–74. [Google Scholar]
- 4.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Bányai K, Brister JR, et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch Virol. 2011;156:1397–413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trojnar E, Sachsenröder J, Twardziok S, Reetz J, Otto PH, Johne R. Identification of an avian group A rotavirus containing a novel VP4 gene with a close relationship to those of mammalian rotaviruses. J Gen Virol. 2013;94:136–42. doi: 10.1099/vir.0.047381-0. [DOI] [PubMed] [Google Scholar]
- 6.Bànyai K, Làzlò B, Duque J, Steele AD, Nelson EA, Gentsch JR, Parashar UD. Systemic review of Regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30(1):A122–30. doi: 10.1016/j.vaccine.2011.09.111. [DOI] [PubMed] [Google Scholar]
- 7.El Sherif M, Esona MD, Wang Y, Gentsch JR, Jiang B, Glass RI, et al. Detection of the first G6P[14] human rotavirus strain from a child with diarrhea in Egypt. Infect Genet Evol. 2011;11:1436–42. doi: 10.1016/j.meegid.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Martella V, Bányai K, Matthijnssens J, Buonavoglia C, Ciarlet M. Zoonotic aspects of rotaviruses. Vet Microbiol. 2010;140:246–55. doi: 10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 9.Gerna G, Sarasini A, Parea M, Arista S, Miranda P, Brussow H, et al. Isolation and characterization of two distinct human rotavirus strains with G6 specificity. J Clin Microbiol. 1992;30(1):9–16. doi: 10.1128/jcm.30.1.9-16.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banyai K, Gentsch JR, Glass RI, Szucs G. Detection of human rotavirus serotype G6 in Hungary. Epidemiol Infect. 2003;130:107–12. doi: 10.1017/S0950268802007975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palombo EA, Bishop RF. Genetic and antigenic characterization of a serotype G6 human rotavirus isolated in Melbourne, Australia. J Med Virol. 1995;47:348–54. doi: 10.1002/jmv.1890470410. [DOI] [PubMed] [Google Scholar]
- 12.Griffin DD, Nakagomi T, Hoshino Y, Nakagomi O, Kirkwood CD, Parashar UD, et al. Characterization of nontypeable rotavirus strains from the United States: identification of a new rotavirus reassortant (P2A[6], G12) and rare P3[9], G6 strains related to bovine rotaviruses. Virology. 2002;294:256–69. doi: 10.1006/viro.2001.1333. [DOI] [PubMed] [Google Scholar]
- 13.Kelkar SD, Ayachit VL. Circulation of group A rotavirus subgroups and serotypes in Pune, India, 1990–1997. J Health Popul Nutr. 2000;18:163–70. [PubMed] [Google Scholar]
- 14.Matthijnssens J, Potgieter CA, Ciarlet M, Parreño V, Martella V, Bányai K, et al. Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates belonging to the mammalian order of Artiodactyla? J Virol. 2009;83:2917–29. doi: 10.1128/JVI.02246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fodha I, Boumaiza A, Chouikha A, Dewar J, Armah G, Geyer A, et al. Detection of group a rotavirus strains circulating in calves in Tunisia. J Vet Med. 2005;52(1):49–50. doi: 10.1111/j.1439-0450.2004.00810.x. [DOI] [PubMed] [Google Scholar]
- 16.Armah GE, Sow SO, Breiman RF, Dallas MJ, Tapia MD, Feikin DR, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomized, double-blind, placebo-controlled trial. Lancet. 2010;376:606–14. doi: 10.1016/S0140-6736(10)60889-6. [DOI] [PubMed] [Google Scholar]
- 17.Madhi SA, Kirsten M, Louw C, Boss P, Aspinall S, Bouckenoogbe A, et al. Efficacy and immunogenicity of two or three dose rotavirus-vaccine regimen in South African children over two consecutive rotavirus-seasons: a randomized, double-blind placebo-controlled trial. Vaccine. 2012;1:A44–51. doi: 10.1016/j.vaccine.2011.08.080. [DOI] [PubMed] [Google Scholar]
- 18.Patton JT. Rotavirus diversity and evolution in the post-vaccine world. Discov Med. 2012;13:85–97. [PMC free article] [PubMed] [Google Scholar]
- 19.Agbemabiese CA, Nakagomi T, Suzuki Y, Armah G, Nakagomi O. Evolution of a G6P[6] rotavirus strain isolated from a child with acute gastroenteritis in Ghana 2012. J Gen Virol. 2015;96:2219–31. doi: 10.1099/vir.0.000174. [DOI] [PubMed] [Google Scholar]
- 20.Nyaga MM, Stucker KM, Esona MD, Jere KC, Mwinyi B, Shonhai A, et al. Whole-genome analyses of DS-1-like human G2P[4] and G8P[4] rotavirus strains from Eastern, Western and Southern Africa. Virus Genes. 2014;49:196–207. doi: 10.1007/s11262-014-1091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das BK, Gentsch JR, Cicirello HG, Woods PA, Gupta A, Ramachandran M, et al. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994;32:1820–2. doi: 10.1128/jcm.32.7.1820-1822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simmonds MK, Armah G, Asmah R, Banerjee I, Damanka S, Esona M, et al. New oligonucleotide primers for P-typing of rotavirus strains: Strategies for typing previously untypeable strains. J Clin Virol. 2008;42:368–73. doi: 10.1016/j.jcv.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Maes P, Matthijnssens J, Rahman M, VanRanst M. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009;9:238. doi: 10.1186/1471-2180-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]


