ABSTRACT
Gut dysbiosis may result in various diseases, such as metabolic and neurobehavioral disorders. Exposure to endocrine disrupting chemicals (EDCs), including bisphenol A (BPA) and ethinyl estradiol (EE), especially during development, may also increase the risk for such disorders. An unexplored possibility is that EDC-exposure might alter the gut microbial composition. Gut flora and their products may thus be mediating factors for the disease-causing effects of these chemicals. To examine the effects of EDCs on the gut microbiome, female and male monogamous and biparental California mice (Peromyscus californicus) were exposed to BPA (50 mg/kg feed weight) or EE (0.1 ppb) or control diet from periconception through weaning. 16s rRNA sequencing was performed on bacterial DNA isolated from fecal samples, and analyses performed for P0 and F1 males and females. Both BPA and EE induced generational and sex-dependent gut microbiome changes. Many of the bacteria, e.g. Bacteroides, Mollicutes, Prevotellaceae, Erysipelotrichaceae, Akkermansia, Methanobrevibacter, Sutterella, whose proportions increase with exposure to BPA or EE in the P0 or F1 generation are associated with different disorders, such as inflammatory bowel disease (IBD), metabolic disorders, and colorectal cancer. However, the proportion of the beneficial bacterium, Bifidobacterium, was also elevated in fecal samples of BPA- and EE-exposed F1 females. Intestinal flora alterations were also linked to changes in various metabolic and other pathways. Thus, BPA and EE exposure may disrupt the normal gut flora, which may in turn result in systemic effects. Probiotic supplementation might be an effective means to mitigate disease-promoting effects of these chemicals.
KEYWORDS: bacteria, california mice, DOHaD, endocrine disrupting chemicals, estrogens, metabolic pathway, Peromyscus
Introduction
Gut dysbiosis is increasingly being recognized to be a key initiator of many disease processes, ranging from metabolic, cardiovascular, reproductive, and neurobehavioral disorders.1-3 Consequently, it has been proposed that various axes linked to the gut and microbiome exist, such as a gut-microbiome-brain axis.1 Dysbiosis linked to changes in the intestinal flora could be due to direct effects of the bacteria, bacterial products, including metabolites and virulence factors (e.g., lipopolysaccharide- LPS), bacterial-induced changes in the host intestines, including increased gut permeability, or through production of hormone-like compounds. It is uncertain whether endocrine disruptors and other environmental chemicals may disrupt normal gut microbial populations.
To date, exposure to arsenic, nanoparticles, and most recently, lead (Pb) are the few environmental chemicals reported to alter the gut microbiome.4-8 Perinatal exposure of mice to Pb alters the gut microbiota, which has been correlated with body weight changes in males but not females.9 Estrogenic compounds can change the composition of the vaginal flora.10,11 Depletion of estrogen due to ovariectomy leads to gut microbial shifts in low and high aerobic capacity rats.12 Women with elevated hydroxylated estrogen metabolites possess a more diverse gut microbiome.13 Thus, EDCs, including BPA and EE might affect the gut and other microbiomes.
BPA is in a wide assortment of household and everyday use items, including plastic storage containers, cardboard products, and credit card receipts. Current BPA production is estimated at ∼15 billion pounds annually and is anticipated to rise dramatically in coming years.14 Its inability to breakdown easily in the environment and prevalence15 ensure long term exposure.16 BPA is measurable in the urine of 93% of the US population,17 and it has also been identified in fetal plasma, placental tissue,18 and breast milk.19 EE is the estrogenic chemical present in birth control pills. The un-metabolized form can be excreted in the urine, and thereby, may also accumulate in various environmental sources.20 BPA and EE exposure, especially during the perinatal period, is linked to a variety of diseases, including cardiovascular, metabolic, reproductive, and neurobehavioral disorders.21-23 To our knowledge, no prior studies have considered the impact of these chemicals on the gut microbiome, even though speculation exists that environmental chemical-induced gut microbiome shifts may serve as the underlying etiology for metabolic and possibly other diseases.24
To address this critical gap in our understanding, we exposed male and female California mice P0 parents (Peromyscus californicus) to BPA or EE through the diet. This species was selected as their genetic outbred state and social organization, monogamous and biparental, might better reflect most human societies. Additionally, we have previously shown that developmental exposure to BPA and EE can lead to various sex-dependent behavioral deficits, including in exploration, territorial marking, parenting ability, and decreased voluntary physical activity.25-27 Further, California mice may serve as a good animal model for human metabolic disorders, in particular T2M.28 When F1 pups were weaned at 30 d of age and prior to all pups being placed on the control diet, fecal samples were collected from P0 parents and F1 sons and daughters to determine whether generational differences in exposure affect the gut microbiota composition. Thus, the fecal samples were collected at the time when the pups were exposed to the varying respective diets (BPA, EE, or control). Figure 1 provides a model of the study design.
Figure 1.
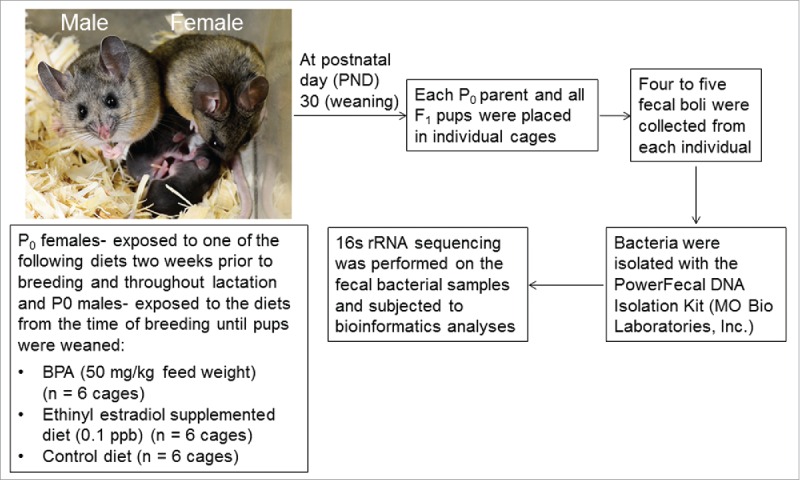
Experimental model design. P0 California mice females were placed on the respective diets 2 weeks prior to breeding to to P0 California mice males, and both parents were maintained on the respective diets throughout the perinatal period. At the time, pups were weaned, each animal was placed in a separate cage and fecal samples were collected from P0 parents and one F1 male and female pup in each litter. Thus, samples were collected prior to all F1 pups being placed post-weaning on the control diet, and fecal samples were only collected at this one time point. Bacteria were then isolated from each sample, 16s rRNA sequencing done, and bioinformatics analyses performed.
Results
Absence of a cage or litter effect on the gut microbiota
To determine whether the P0 and F1 generational microbiota data should consider potential litter effects or whether it was sufficient to consider each individual as the statistical unit, we first determined whether there were any cage or litter effects. Using PERMANOVA of PCoA analyses, we failed to observe a significant effect (p ≤ 0.05) of the cage upon the relationship between samples, either within treatment (Control: p = 0.495, BPA: p = 0.279, EE: p = 0.296) or when all treatments were analyzed together (p = 0.108). Thus, all of the remaining analyses detailed below use the individual animal as the statistical unit.
Influence of exposure to BPA and EE on the gut microbiota of P0 females and males
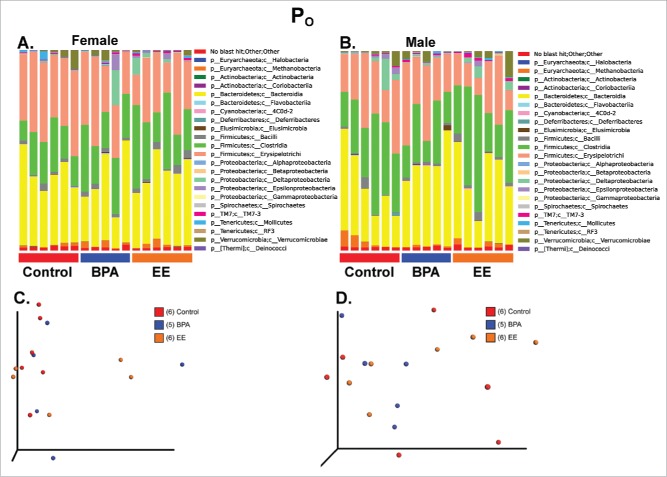
When the 16S rRNA sequencing results were compared using Greengenes Version 13_8 (which is available through QIIME, http://qiime.org/home_static/dataFiles.html ftp://greengenes.microbio.me/greengenes_release/gg_13_5/gg_13_8_otus.tar.gz), no clear distinctions were evident in the various bacterial classes based on treatment in either P0 females or males (Fig. 2A and B). The PCoA analysis revealed no overt differences between the 3 groups in P0 females or males (Fig. 2C and D, PERMANOVA for P0 females = 0.3518 and PERMANOVA for P0 males = 0.5917. Additionally, measures of α-diversity, including Chao1 and Shannon indices as well as rarefaction analysis, were similar among P0 and F1 (Fig. S1).
Figure 2.
Bar plot and PCoA analysis of fecal microbiome data from P0 females and males. A) Bar plot analysis of the most abundant bacterial classes in all 3 treatment groups for P0 females. B) Bar plot analysis of the most abundant bacterial classes in all 3 treatment groups for P0 males. C) PCoA analysis for P0 females. PERMANOVA p value = 0.3518. D) PCoA analysis for P0 males. PERMANOVA p value = 0.5917.
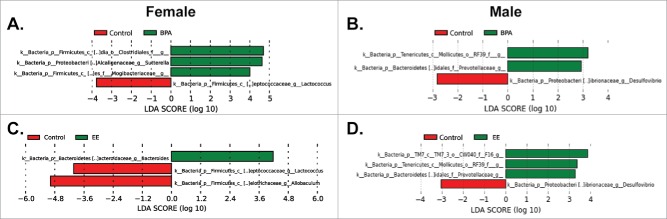
To examine for subtle genera differences, P0 data were analyzed by using linear discriminant analysis effect size (LEfSe) analysis.29 When comparing control females to BPA females, Lactococcus spp. was increased in the control group; whereas, Mogibacteriaceae, Sutterella spp, and Clostridiales were increased in the BPA group (Fig. 3A). Comparison of control males to BPA males, revealed that Mollicutes and Prevotellaceae were characteristic of BPA males; whereas, Desulfovibrio spp was more representative of control males (Fig. 3B). When control females were compared to EE-exposed females, Bacteroides spp was increased in EE females. In contrast, Lactococcus spp and Allobaculum spp were distinctive of control females (Fig. 3C). Mollicutes and Prevotellaceae were representative of EE-exposed males compared to control males, but Desulfovibrio spp was characteristic of control males (Fig. 3D).
Figure 3.
LEfSe analysis of fecal microbiome data from P0 females and males. A) Comparison of BPA females to control females. The linear discriminant analysis (LDA) score of Bacteroides (green bar) was greater in BPA compared to control females. B) Comparison of BPA males and control males. LDA scores that are greater in BPA males are shown in green, whereas the one (Delsufiovibrio) LDA score elevated in control males is depicted in red. C) Comparison of EE females to control females. LDA scores that are greater in EE females are shown in green, whereas the LDA scores elevated in control females are in red. C) Comparison of EE males to control males. LDA scores that are greater in EE males are shown in green, whereas the LDA scores elevated in control males are in red. Only LDA scores ≥ 2 are listed.
Based on the genera that differed between BPA/EE versus control P0 females and males, correlation analyses were performed for various KEGG pathways. Comparison of control females to BPA females revealed that decreased abundance of Sutterella spp. and Clostridiales was inversely associated with histidine metabolism, tryptophan metabolism, lysine degradation, tropane-piperidine- pyridine alkaloid biosynthesis, stilbenoid-diarylheptanoid-gingerol biosynthesis, nitrogen metabolism, NOD-like receptor signaling pathway, antigen processing and presentation, linoleic acid metabolism, arachidonic acid metabolism, riboflavin metabolism, and polycyclic aromatic hydrocarbon degradation; whereas, these bacterial changes in controls were positively associated with glycolysis/gluconeogenesis, starch and sucrose metabolism, butanoate metabolism, pentose-glucoronate interconversions, carbohydrate digestion and absorption, transcription related proteins, and phosphanate and phosphanate metabolism to list a few examples (Fig. S2). While the bacterial differences between control and BPA females was associated with several trends for metabolic and other differences in the BPA group, none of them reached statistical significance (Fig. S2).
Assessments of P0 control vs. BPA males revealed that a decrease in Prevotellaceae abundance in the former group was correlated with several up and down-regulated metabolic pathways; whereas this OTU was only associated with increased glycerophosholipid metabolism in the BPA group (Fig. S3). Alterations in Desulfovibrio and Mollicutes abundance in control males were also correlated with several pathway alterations. In BPA males, increased abundance in Mollicutes positively correlated with changes in secretion.
Comparison of P0 control to EE females revealed that decreased abundance of Bacteroides in the former was positively correlated with changes in amino acid metabolism, citrate cycle (TCA cycle), and lipid biosynthesis proteins; whereas, amino sugar and nucleotide sugar metabolism, peptidases were negatively correlated with reductions in this bacterium (Fig. S4). Allobaculum abundance was increased in control relative to EE females, and this change was negatively associated with flavone and flavonol biosynthesis, and starch and sucrose metabolism. Increased abundance of Bacteroides spp in P0 EE females was positively associated with arginine and proline metabolism, glycine, serine, and threonine metabolism, butirosin and neomycin biosynthesis, citrate cycle (TCA cycle), metabolism of cofactors and vitamins, lipoic acid metabolism, and limonene and pinene degradation to list a few examples (Fig. S4). In contrast, amino sugar and nucleotide sugar metabolism, peptidases, phosphotransferase system (PTS), and nucleotide metabolism were inversely associated with an increase in this bacterium for EE females. Decreased abundance of Allobaculum also led to several pathway associations.
When comparing control to EE males, changes in the abundance of Prevotellaceae, Delsulfovibrio, an uncharacterized bacterium, and Mollicutes in the former were positively and negatively associated with several pathways (Fig. S5). Significant pathway changes, however, were only correlated with an uncharacterized bacterium in EE-exposed males.
Influence of exposure to BPA and EE on the gut microbiota of F1 females and males
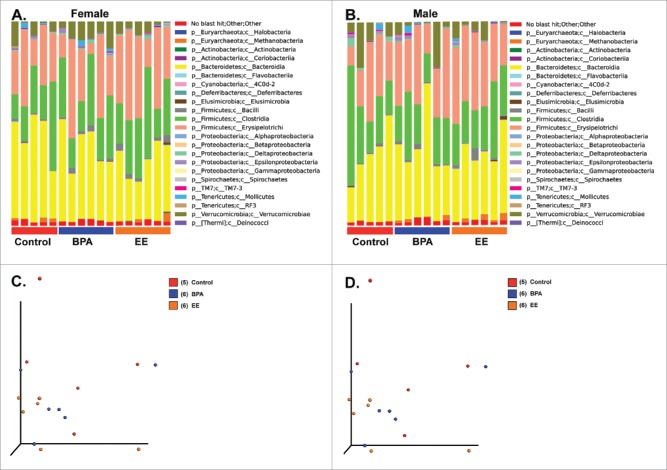
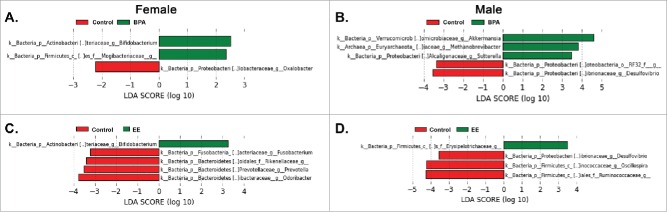
No overt divisions were evident in the various bacterial classes based on treatment in either F1 females or males (Fig. 4A and B). The PCoA analysis revealed no extreme differences between the 3 groups in F1 females or males (Fig. 4C and D, PERMANOVA for F1 females = 0.5492 and PERMANOVA for F1 males = 0.3936). To examine for subtle differences, F1 results were then further analyzed with LEfSe.29 Abundance of Oxalobacter spp was increased in F1 control females relative to F1 BPA females, who showed increased proportion of Bifidobacterium spp and Mogibacteriaceae (Fig. 5A). Comparison of the bacterial genera identified in F1 BPA males and F1 control males revealed several differences. Akkermansia spp, Methanobrevibacter spp, Sutterella spp abundance increased in the former. However, the abundance of a Proteobacteria and Desulfovibrio spp was greater in the latter (Fig. 5B).
Figure 4.
Bar plot and PCoA analysis of fecal microbiome data from F1 females and males. A) Bar plot analysis of the most abundant bacterial classes in all 3 treatment groups for F1 females. B) Bar plot analysis of the most abundant bacterial classes in all 3 treatment groups for F1 males. C) PCoA analysis for F1 females. PERMANOVA p value = 0.5492. D) PCoA analysis for F1 males. PERMANOVA p value = 0.3936.
Figure 5.
LEfSe analysis of fecal microbiome data from F1 females and males. A) Comparison of BPA females to control females. LDA scores that are greater in BPA females are shown in green; whereas, the one (Oxalobacter) elevated LDA score in control females is depicted in red B) Comparison of BPA males and control males. LDA scores that are greater in BPA males are shown in green, whereas LDA scores increased in control males are depicted in red. C) Comparison of EE females to control females. The one LDA score (Bifidobacterium) that is greater in EE females is shown in green, whereas those LDA scores elevated in control females are in red. C) Comparison of EE males to control males. The one LDA score (Erysipelotrichaceae) higher in EE males is shown in green; whereas, those LDA scores elevated in control males are in red. Only LDA scores ≥ 2 are listed.
Bifidobacterium spp abundance was increased in F1 EE females compared to control females, who in contrast had increased abundance of Fusobacterium spp, Rikenellaceae, Prevotella spp, Odoribacter spp (Fig. 5C). Erysiopelotrichaceae abundance was greater in F1 EE males compared to F1 control males, where Delsulfovibrio spp, Oscillospira spp, and Ruminococcaeae levels were elevated (Fig. 5D).
As with the P0 data, correlation analyses were performed for various KEGG pathways based on the microbiota that differed between controls and BPA/EE females and males. Comparison of F1 control to BPA-exposed females revealed that an increased abundance of Oxalobacter in the former was positively linked with galactose metabolism, RIG-I-like receptor signaling pathway, and ether lipid metabolism; whereas, tropane, piperidine, and pyridine alkaloid biosynthesis, other ion-coupled transporters, and nitrogen metabolism were negatively associated with in the abundance of this bacterium (Fig. S6). Increased abundance of Bifidobacterium and Mogibacteriaceae in BPA-exposed females resulted in both positive and negative correlations for various pathways. For instance, fatty acid and arachidonic acid metabolism were positively associated with an elevation in Bifidobacterium abundance in this group.
Comparison of F1 control to BPA-exposed males demonstrated changes in Methanobrevibacter and Desulfiovibrio abundances correlated with several up and downregulated metabolic and other pathways in both groups (Fig. S7). In F1 BPA-exposed males, an increase in Akkermansia abundance associated with several pathways, such as caffeine, insulin signaling pathway, sulfur metabolism, and steroid hormone biosynthesis positively correlated with this bacterium's abundance.
Comparison of F1 EE to control females revealed that an increase in Bifidobacterium abundance in the former was positively associated with renin-angiotensin system but negatively correlated with secretion system (Fig. S8). Increased abundance of Fusobacterium spp, Rikenellaceae, Prevotella spp, Odoribacter spp in control females led to both positive and negative associations. For instance, an increase in Odoribacter spp abundance was negatively correlated with caffeine metabolism, steroid biosynthesis, and fatty acid elongation in mitochondria.
When F1 control male results were compared to EE-exposed males, an increase in Desulfiovibrio abundance in the former was linked to several both positive and negative pathway associations (Fig. S9). In contrast, increased Erysipelotrichaceae abundance in F1 EE-exposed males associated with several negative pathway changes, including penicillin and cephalosporin biosynthesis, caffeine metabolism, ubiquitin system, steroid biosynthesis, and fatty acid elongation in mitochondria.
Comparison of microbiota in BPA and EE exposed groups for P0 and F1 generations
To determine whether there were any microbiota differences between BPA and EE-exposed animals, these 2 groups were directly compared for the P0 and F1 generations. There were no microbiota differences between P0 BPA and P0 EE females. In P0 males, the only difference was that Ruminococcus spp, were greater in the EE compared to BPA group (Fig. S10A). In F1 females, Lachnobacterium spp. and Prevotella spp were increased in the BPA compared to EE group (Fig. S10B). In F1 males, Parabacteroides spp. and Sutterella spp, were more abundant in the BPA compared to EE group (Fig. S10C).
Comparison of generational differences to the different diets
To determine whether the different diets induced generation-dependent differences that varied according to sex, gut microbiota populations were compared between P0 vs. F1 males and females exposed to the same diet. Surprisingly, even the control diet led to generation-dependent genera differences for both females and males. For females in this group, Actinbacteria, Bifidobacterium spp., Streptococcus spp., and Lactococcus spp., were more abundant in the P0 generation; whereas, Fusobacteriaceae, Prevotella spp., and Anaeroplasma spp., were more plentiful in the F1 generation (Fig. S11A). In control males, Coriobacteriaceae and Christensenellaceae were greater in the P0 generation (Fig. S11B). In contrast, Clostridium spp., Prevotellaceae, and Rikenellaceae were more abundant in F1 control males.
When comparing across generations, there were no differences in P0 vs. F1 females in the BPA-supplemented group. However, several genera were different in P0 vs. F1 males in the BPA group (Fig. S11C). For instance, Alphaproteobacteria, Mollicutes, and Cyanobacteria were enriched in P0 BPA males; whereas Fusobacteriales, Sutterella spp., and Akkermansia spp. were greater in F1 BPA males. Comparison of P0 to F1 females exposed to EE revealed that only genera in the P0 group were elevated, and these included Mollicutes and Cyanobacteria (Fig. S11D). In EE-exposed males, Alphaproteobacteria, Oxalobacter spp, and Christensenellaceae were more abundant in the P0 generation; whereas Erysipelotrichaceae was elevated in the F1 generation (Fig. S11E).
Comparison of sex differences in the P0 and F1 generations in response to the different diets
To determine whether there were sex-dependent differences in both generations to the different diets, the gut microbiota of males and females within the same generation and exposed to the same diet were compared with LEfSe analysis.29 In the P0 generation, the control diet resulted in males showing more abundance of Desulfovibrionaceae, Christensenellaceae, and Ruminococcus spp. (Fig. S12A). In contrast, P0 control females possessed greater amounts of Rickenellaceae, Prevotellaceae, and Allobaculum spp. No differences were detected between P0 males and females exposed to BPA. In P0 EE-exposed individuals, Oxalobacter spp. and another Proteobacteria were more abundant in males, but no bacteria were identified as being greater in females compared to males in this group Fig. S12B).
In the F1 generation control group, a Proteobacteria, Ruminococcaeceae, and Oscillospira spp were more abundant in males, but no genera were greater in females relative to males in this group (Fig. S12C). For F1 BPA-exposed individuals, Methanobrevibacter spp. was more abundant in males, whereas, a Cyanobacterium was more plentiful in females (Fig. S12D). No differences were detected between F1 males and females exposed to EE.
Discussion
The main goal of the current study was to examine how generational exposure to BPA and EE affect the gut microbiota at respective doses that have already been shown to lead to later behavioral and metabolic disruptions in F1 offspring.25-27 The 2 attendant goals were to determine 1) whether similar microbiome changes occur in the P0 parents and F1 offspring and 2) whether these chemicals would induce sex-dependent differences in both generations.
In regards to the primary goal, gut microbiota differences were observed in BPA- and EE-exposed P0 and F1 males and females compared to non-exposed control counterparts. To our knowledge, these are the first set of studies to show that parental exposure to environmentally relevant concentrations of BPA and EE causes changes in microbial composition in unexposed offspring. Other environmental chemicals, including lead, arsenic and nanoparticles appear to alter the gut microbiome in rodent models.4-8 Estrogenic compounds and metabolites affect the composition and diversity of the gut and vaginal microbiota.10-13 EDC-induced gut dysbiosis may trigger metabolic and other diseases, as postulated previously.24 By altering the gut microbiome, EDCs may increase the permeability of the intestinal barrier. This pathological change may increase the likelihood that bacterial pathogens, their virulence factors and metabolites will penetrate and enter the systemic circulation, whereupon other target organs, including the brain, may be affected.1,30,31 Perinatal exposure of rats to BPA affects the intestinal barrier function and gut nocioception.32 Gut microbes might also be transmitted to the brain via the enteric nervous system and vagal nerve (Reviewed in33,34). Bacterial-derived metabolites, such as spermidine, urea, short-chained fatty acids (SCFA), and 4-ethylphenylsulfate (4-EPS), might negatively impact various systems, including the central nervous system (Reviewed in1). Other mechanisms by which gut microbes might influence host function are through production of neuroendocrine factors, neurotransmitters, and modulation of the host epigenome (Reviewed in1).
In P0 females, Mogibacteriaceae, Sutterella spp, and Clostridiales were increased in the BPA-exposed individuals relative controls; whereas, Bacteroides levels were elevated in EE-exposed females relative to controls. Abundance of Bacteroides species is negatively correlated with the ratio of hydroxylated estrogen metabolites to parental estrogen compounds (estrone and estradiol) in postmenopausal women.13 In P0 males, both BPA and EE increased the abundance of Mollicutes and Prevotellaceae compared to controls. Mollicutes and Erysipelotrichaceae (abundances increased in EE-exposed F1 males) are more abundant in the gut microbiome of male mice exposed to valproic acid, which is a considered a murine model for autism spectrum disorders.35
F1 females exposed to BPA or EE had increased amounts of Bifidobacterium. This genus is considered a beneficial inhabitant within the gut flora, and based on this premise is in many probiotic formulations, including those administered to pre-term and neonatal infants.36-38 The abundance of Mogibacteriaceae was increased in the gut microbiome of BPA-exposed F1 females. Scant information is available on the potential health implications of the presence of this bacterial family in the gut flora. Mogibacteriaceae tend to cluster with other microorganisms associated with lower body mass index (BMI) in mice and humans39 and non-obese diabetic (NOD) mice fed a cellulose, pectin, and xylan-rich diet.40 For F1 males, BPA- and EE-exposure led to unique changes in the bacterial intestinal flora. Levels of Akkermansia were elevated in the gut microbiome of BPA-exposed males. Akkermansia levels are up-regulated in humans and mouse models stricken with colon cancer,41-43 and this bacterium has been proposed as a target for probiotic treatment.44 Methanobrevibacter levels were also elevated in the gut of F1 BPA-exposed males. These Archaea, such as M. smithii, possess heightened ability to metabolize dietary substrate with resulting increased host energy intake and weight gain.45 Erysipelotrichaceae was the only OTU elevated in the fecal samples of F1 EE-exposed males compared to AIN males. This bacterial family is linked with various diseases and appears to be highly responsive to dietary shifts (Reviewed in46). Comparison of microbiota that were differentially expressed between BPA vs. EE exposed males and females in both generations revealed no differences in P0 females and only isolated bacterial differences in P0 males and F1 females and males. Notwithstanding, the findings may suggest that BPA and EE can induce differential effects on the gut microbiota, even though BPA is considered a weak estrogen.47
In P0 and F1 males and females, various genera are associated with metabolic and other pathway changes that were group-dependent (AIN, BPA, or EE). The bioinformatic analyses only predict which pathways might be affected, but such predictions can be tested by assaying for specific metabolite changes. Alterations in bacterial metabolite and other pathway changes may be another mechanism by which these EDCs can lead to various diseases.
In relation to attendant goal 1, bacterial gut colonization occurs at the time of birth. When neonates pass through the birth canal, they are innoculated with a complex mixture of maternal vaginal microorganisms.48-50 In human infants, the intestinal microbiome resembles that of their mother up until about one year of age, whereupon a distinct microbiome profile develops.51,52 Thus, our prediction was that the gut microbiome changes in BPA or EE-exposed F1 offspring would mirror that of their P0 mothers. However, bacterial changes in the F1 EDC-exposed generation varied from that of their P0 mothers and fathers. The findings support the notion that generational exposure to BPA or EE differentially impacts the gut microbiome relative to adults. The etiology of these differences is unclear. It is interesting to note that even within the control diet, there were genera differences in males and females in the P0 relative to the F1 generation. Divergences in the F1 progeny from their P0 parents and across treatments could be attributed to the in utero environment or occur postnatally due to altered milk composition. In human infants, breastfeeding increases fecal Bifidobacterium compared to supplementation with standard formulas.53 As detailed above, Bifidobacterium was more abundant in BPA- and EE-exposed F1 females. Other studies provide additional evidence that the gut microbiome is vulnerable to infant dietary changes in humans and animal models.54,55
For attendant goal 2, clear sex differences were evident in both generations after exposure to BPA or EE. Even for the control diet, different genera varied between males and females in the 2 generations, suggesting that even a so-called control diet can lead to sex-dependent drift across generations in gut microbial populations. Neonatal NOD mice also show sex differences in the gut microbiome, which governs sex hormone levels and autoimmunity progression.56 In males, resident microbiota increase serum testosterone concentrations and confer protection against type I diabetes. These phenotypes might be transmitted to juvenile females receiving fecal transplantation from adult males.
In summary, the current findings indicate EDC exposure results in gut microbiome changes and potentially accompanying changes in metabolic and other pathways. The genera altered by these treatments are dependent upon generation and sex. While some of the intestinal flora changes were similar in BPA- and EE-exposed females and males in both generations, others differed. The results suggest that even though BPA is a weak estrogen, it can induce effects outside of binding and engaging estrogen receptors. Overall, many of the genera whose abundances were elevated by EDC-exposure are associated with various diseases, including IBD, metabolic disorders, and colorectal cancer. The notable exception was the surge of Bifidobacterium in the fecal samples of BPA- and EE-exposed F1 females. The escalation of this bacterium in the gut flora of these groups might be due to direct effects of generational exposure to one of the EDCs or mediated via changes in the in utero environment/milk composition in the P0 exposed mothers. The F1 males and females used in this study were also generated to examine for gene expression differences at adolescence, which is currently ongoing. Thus, one limitation of the current study is that we cannot link the gut microbiome changes in the P0 and F1 generations to phenotypic or molecular alterations. However, future studies will aim to do such correlation analyses and also test several doses of BPA and EE. Even so, the current study demonstrates that BPA and EE at doses that have previously been shown to result in behavioral and metabolic disturbances in F1 California mice offspring25-27 also leads to generational and sex-dependent changes in the gut microbiome that could initiate subsequent adverse effects to the host.
Materials and methods
Animal husbandry
Founder captive adult (60-90 d of age) California mouse females and males, free of common rodent pathogens, were purchased from the Peromyscus Genetic Stock Center (PGSC) at the University of South Carolina (Columbia, SC). When they were shipped to the University of Missouri, they were placed in quarantine, along with sentinel mice, at the MU Lab Animal Center (LAC) for a minimum of 8 weeks to ensure that they did not carry any transmittable and zoonotic diseases. No diseases have been identified in any sentinel animals or colony animals. Once the animals were deemed pathogen-free, they were transported from the LAC to the Animal Sciences Research Center (ASRC). At this facility, we have established our own breeding colony established. Additional animals are purchased, though as needed, from the PGSC to maintain the outbred status of the line and similar procedures as previously mentioned are followed. All experiments were approved by University of Missouri Animal Care and Use Committee (Protocol #7753) and performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. To reduce any background BPA exposure, animals were housed in polypropylene cages (Allentown, NJ), provided glass water bottles, and BPA-free water. Two weeks prior to breeding, virgin females, 8 to 12 wks of age were randomly assigned to receive one of 3 diets: 1) a low phytoestrogen AIN 93G diet supplemented with 7% by weight corn oil to minimize potential phytoestrogenic contamination (control), 2) the same diet supplemented with 50 mg BPA/kg feed weight, which we have reported results in internal serum concentrations approximating those measured in pregnant women unknowingly exposed to this chemical,57,58 and 3) AIN93G diet supplemented with 0.1 parts per billion of EE, as the US Food and Drug Administration (FDA) required estrogen positive control for BPA studies.59 The FDA has requested EE be included in BPA studies that may guide policy decisions based on the notion that BPA acts primarily as a weak estrogen.47 Each generation (P0 and F1) included control females and males that were used as the as the base comparison within generation to their counterpart BPA-exposed and EE-exposed females and males, respectively. Treatments were initiated 2 weeks prior to breeding to span the peri-conceptional period. P0 females were maintained on these diets throughout gestation and lactation, as described previously.25-27,57,60 Exposure to these doses has previously been shown to induce behavioral and metabolic alterations in F1 male and female California mice offspring.25-27 Further, the BPA dose falls below the diet-administered maximum nontoxic dose for rodents (200mg/kg of body weight per day), which is within the presumptive NOAEL61-65 and yields serum concentrations comparable to those identified in human populations.58,66-70 P0 males were exposed to the diets at the time from breeding up until weaning the F1 pups. The F1 generation sons and daughters were weaned at 30 d of age and fecal samples collected at this time. They were then placed on the control diet. The study design model is detailed in Figure 1.
Collection of fecal samples and isolation of fecal microbial DNA
Prior to fecal collection at weaning, each animal was placed one per cage without any bedding. Four to 5 fecal boli were collected from each animal and placed in a 7ml polypropylene vial (Fisher Scientific, St. Louis, MO). The fecal microbial DNA was isolated using the PowerFecal DNA Isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA) and in accordance with the manufacture's protocol. The quantity of DNA isolated was measured using Qubit 3.0 Fluorometer (Life Technologies, Grand Island, NY). Fecal samples were only collected once for the P0 and F1 generations. The number of replicates included in the final analysis for the P0 and F1 generations is listed in Supplemental Table 1. The number of replicates tested is comparable to those used in other maternal diet and offspring gut microbiome studies that showed such sample sizes can result in statistical differences between offspring groups.71,72 While we tested both males and females in the P0 pairings, one control male and one BPA female sample did not properly sequence, and thus, these were not included in the final analyses.
16s rRNA sequencing
The University of Missouri DNA Core Facility prepared bacterial 16S rDNA amplicons from extracted fecal DNA by amplification of the V4 hypervariable region of the 16s rDNA with universal primers (U515F/806R) flanked by Illumina standard adapter sequences.73,74 Universal primer sequences are available at proBase (http://www.microbial-ecology.net/probebase/).75 A forward primer and reverse primer with a unique 12-base index were used in each PCR reaction. PCR reactions (50μl) contained 100ng of genomic DNA, forward and reverse primers (0.2μM each), dNTPs (200μM each), and Phusion High-Fidelity DNA Polymerase (1U). PCR amplification was performed as follows: 98°C(3:00) + [98°C(0:15) + 50°C(0:30) + 72°C(0:30)] × 25 cycles + 72°C(7:00). Maximum sample volume was added to each PCR reaction for seminal fluid samples which contained <100 ng of input DNA. Amplified product (5μl) from each PCR reaction was combined and thoroughly mixed to prepare a single pool. Pooled amplicons were then purified by addition of Axygen AxyPrep MagPCR Clean-up beads (50μl) to an equal volume of 50μl of the amplicon library pool and incubated at room temperature for 15 minutes. Products were placed on a magnetic stand for 5 minutes and supernatant (95 μl) was removed and discarded. Each well was washed by addition of 200μl of freshly prepared 80% EtOH, incubation at room temperature for 30 seconds, and removal of supernatant. Wash steps were repeated once and plate was allowed to dry on magnetic stand for 15 minutes. The dried pellet was resuspended in Qiagen EB Buffer (32.5 μl), incubated at room temperature for 2 minutes, and then placed on the magnetic stand for 5 minutes. Supernatant (30 μl) was transferred to low binding microcentrifuge tube for storage. The final amplicon pool was evaluated using the Advanced Analytical Fragment Analyzer automated electrophoresis system, quantified with the Qubit flourometer using the quant-iT HS dsDNA reagent kit (Invitrogen), and diluted according to Illumina's standard protocol for sequencing on the MiSeq.
Bioinformatics and amplicon analyses
Paired-end Illumina MiSeq DNA reads were joined using FLASH.76 Usearch777 was used to clean contigs and remove those with E > 0.5, as explained here: http://drive5.com/usearch/manual/exp_errs.html. Contigs were clustered to 97% identity against DNA sequences in the Greengenes database,78 version 13_5, using the QIIME,79 version 1.8, script pick_closed_reference_otus.py, which obviates chimera and PCR error detection. For α-diversity in P0 and F1 fecal samples, Chao1 (species richness) and Shannon (species diversity) values were calculated and plotted using the phlyoSeq R package.80 Rarefaction metrics were calculated using the α_rarefaction.py script in the Qiime package79 and plotted using Microsoft Excel (Fig. S1).
For the follow-up analyses, we first sought to determine whether the P0 and F1 generational microbiota data should consider potential litter effects or if it was instead appropriate to consider each individual as the statistical unit. The PERMNAVOVA method was employed by using the QIIME script compare_categories.py to measure the significance of differences among cages (litters) used across all treatments or within individual treatments. Based on these analyses, it was determined that no litter effects were evident, and thus, the individual animal was considered the statistical unit for the remaining analyses.
Measurements of β-diversity were facilitated by the QIIME script jackknifed_β_diversity, as implemented in QIIME.79 LEfSe29 was used to identify genera most characteristic of different sample types. LEfSe results were visualized using taxonomy bar-chart and cladogram plots, as implemented on the LEfSe website, http://huttenhower.sph.harvard.edu/galaxy/. Bacterial metabolic characterization of sample types was facilitated with PICRUSt,81 version 1.0.0. To correlate the genera changes with metabolic characteristics of sample types, we used a custom R script provided as a gift from Dr. Jun Ma and Kjersti Aagaard-Tillery, Baylor College of Medicine, Houston, TX. In these figures, the correlation of the abundance of genera (from the OTU table) with their predicted metabolic function (from KEGG pathways as determined by PICRUSt), was calculated with the R stats function cor.test (https://cran.r-project.org/), using the Kendall method, a rank-based measure of association. The cor.test function outputs a matrix of correlation coefficients and a matrix of results of tests of their significance. The matrix of correlation values was visualized using an adaptation of the R package corrplot made by Ma et al.71 The area and intensity change together so that larger, darker, circles represent correlation coefficients that are larger in magnitude. The scale to the right of each figure relates those shades of color to the value of the correlation coefficient. Those values whose correlation coefficients were found to be significant at the 0.05 level have a red square around them.
Supplementary Material
Abbreviations
- BMI
Body mass index
- BPA
Bisphenol A
- EDCs
Endocrine disrupting chemicals
- EE
Ethinyl estradiol
- FDA
Food and drug administration
- GF
Germ-free
- IBD
Inflammatory bowel disease
- LAC
Lab Animal Center
- LPS
Lipopolysaccharide
- NOD
Non-obese diabetic
- OTU
Operational taxonomic unit
- PGSC
Peromyscus genetic stock center
- TCA
Citric acid cycle
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are grateful to Mr. Donald L. Connor for his assistance in preparing the figures.
Funding
The studies were supported by NIH Grant 5R21ES023150 to Cheryl S. Rosenfeld.
References
- [1].Rosenfeld CS. Microbiome disturbances and autism spectrum disorders. Drug Metab Dispos 2015; 43:1557-71; PMID:25852213; http://dx.doi.org/ 10.1124/dmd.115.063826 [DOI] [PubMed] [Google Scholar]
- [2].Hansen TH, Gobel RJ, Hansen T, Pedersen O. The gut microbiome in cardio-metabolic health. Genome Med 2015; 7:33; PMID:25825594; http://dx.doi.org/ 10.1186/s13073-015-0157-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Al-Asmakh M, Stukenborg JB, Reda A, Anuar F, Strand ML, Hedin L, Pettersson S, Söder O. The gut microbiota and developmental programming of the testis in mice. PLoS One 2014; 9:e103809; PMID:25118984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Williams K, Milner J, Boudreau MD, Gokulan K, Cerniglia CE, Khare S. Effects of subchronic exposure of silver nanoparticles on intestinal microbiota and gut-associated immune responses in the ileum of Sprague-Dawley rats. Nanotoxicology 2015; 9:279-89; PMID:24877679; http://dx.doi.org/ 10.3109/17435390.2014.921346 [DOI] [PubMed] [Google Scholar]
- [5].Dheer R, Patterson J, Dudash M, Stachler EN, Bibby KJ, Stolz DB, Shiva S, Wang Z, Hazen SL, Barchowsky A, et al.. Arsenic induces structural and compositional colonic microbiome change and promotes host nitrogen and amino acid metabolism. Toxicol Appl Pharmacol 2015; 289:397-408; PMID:26529668; http://dx.doi.org/ 10.1016/j.taap.2015.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Guo X, Liu S, Wang Z, Zhang XX, Li M, Wu B. Metagenomic profiles and antibiotic resistance genes in gut microbiota of mice exposed to arsenic and iron. Chemosphere 2014; 112:1-8; PMID:25048881; http://dx.doi.org/ 10.1016/j.chemosphere.2014.03.068 [DOI] [PubMed] [Google Scholar]
- [7].Lu K, Abo RP, Schlieper KA, Graffam ME, Levine S, Wishnok JS, Swenberg JA, Tannenbaum SR, Fox JG. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ Health Perspect 2014; 122:284-91; PMID:24413286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lu K, Cable PH, Abo RP, Ru H, Graffam ME, Schlieper KA, Parry NM, Levine S, Bodnar WM, Wishnok JS, et al.. Gut microbiome perturbations induced by bacterial infection affect arsenic biotransformation. Chem Res Toxicol 2013; 26:1893-903; PMID:24134150; http://dx.doi.org/ 10.1021/tx4002868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu J, Wen XW, Faulk C, Boehnke K, Zhang H, Dolinoy DC, Xi C. Perinatal lead (Pb) exposure alters gut microbiota composition and results in sex-specific bodyweight increases in adult mice. Toxicol Sci 2016; 151:324-33; PMID:26962054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Silva C, Rey R, Elena Nader-Macias M. Effects of estrogen administration on the colonization capability of lactobacilli and Escherichia coli in the urinary tracts of mice. Methods Mol Biol 2004; 268:387-99; PMID:15156049 [DOI] [PubMed] [Google Scholar]
- [11].Heinemann C, Reid G. Vaginal microbial diversity among postmenopausal women with and without hormone replacement therapy. Can J Microbiol 2005; 51:777-81; PMID:16391657; http://dx.doi.org/ 10.1139/w05-070 [DOI] [PubMed] [Google Scholar]
- [12].Cox-York KA, Sheflin AM, Foster MT, Gentile CL, Kahl A, Koch LG, Britton SL, Weir TL. Ovariectomy results in differential shifts in gut microbiota in low vs. high aerobic capacity rats. Physiological reports 2015; 3:e12488; PMID:26265751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fuhrman BJ, Feigelson HS, Flores R, Gail MH, Xu X, Ravel J, Goedert JJ. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J Clin Endocrinol Metab 2014; 99:4632-40; PMID:25211668; http://dx.doi.org/ 10.1210/jc.2014-2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].GrandViewResearch Global bisphenol A (BPA) market by appliation (appliances, automotive, consumer, construction, electrical & electronics) expected to reach USD 20.03 billion by 2020. http://www.digitaljournal.com/pr/2009287. Published 2014; Last accessed September 25, 2016. [Google Scholar]
- [15].Environment Canada Screening Assessment for the Challenge Phenol, 4,4′ -(1-methylethylidene)bis-(Bisphenol A) Chemical Abstracts Service Registry Number 80-05-7. In: Ministers of the Environment and of Health, ed., 2008:1-107 [Google Scholar]
- [16].Vandenberg LN, Maffini MV, Sonnenschein C, Rubin BS, Soto AM. Bisphenol-A and the great divide: a review of controversies in the field of endocrine disruption. Endocr Rev 2009; 30:75-95; PMID:19074586; http://dx.doi.org/ 10.1210/er.2008-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. Exposure of the US population to bisphenol A and 4-tertiary-octylphenol: 2003-2004. Environ Health Perspect 2008; 116:39-44; PMID:18197297; http://dx.doi.org/ 10.1289/ehp.10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].vom Saal FS, Akingbemi BT, Belcher SM, Birnbaum LS, Crain DA, Eriksen M, Farabollini F, Guillette LJ, Jr, Hauser R, Heindel JJ, et al.. Chapel Hill bisphenol A expert panel consensus statement: integration of mechanisms, effects in animals and potential to impact human health at current levels of exposure. Reprod Toxicol 2007; 24:131-8; PMID:17768031; http://dx.doi.org/ 10.1016/j.reprotox.2007.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vandenberg LN, Hauser R, Marcus M, Olea N, Welshons WV. Human exposure to bisphenol A (BPA). Reprod Toxicol 2007; 24:139-77; PMID:17825522; http://dx.doi.org/ 10.1016/j.reprotox.2007.07.010 [DOI] [PubMed] [Google Scholar]
- [20].Laurenson JP, Bloom RA, Page S, Sadrieh N. Ethinyl estradiol and other human pharmaceutical estrogens in the aquatic environment: a review of recent risk assessment data. The AAPS journal 2014; 16:299-310; PMID:24470211; http://dx.doi.org/ 10.1208/s12248-014-9561-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rosenfeld CS. Bisphenol A and phthalate endocrine disruption of parental and social behaviors. Frontiers in neuroscience 2015; 9:1-15; PMID:25653585; http://dx.doi.org/ 10.3389/fnins.2015.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Heindel JJ, Schug TT. The obesogen hypothesis: Current Status and implications for human health. Curr Enviro Health Rpt 2014; 1:33; http://dx.doi.org/ 10.1007/s40572-014-0026-8 [DOI] [Google Scholar]
- [23].Rochester JR. Bisphenol A and human health: a review of the literature. Reprod Toxicol 2013; 42:132-55; PMID:23994667; http://dx.doi.org/ 10.1016/j.reprotox.2013.08.008 [DOI] [PubMed] [Google Scholar]
- [24].Snedeker SM, Hay AG. Do interactions between gut ecology and environmental chemicals contribute to obesity and diabetes? Environ Health Perspect 2012; 120:332-9; PMID:22042266; http://dx.doi.org/ 10.1289/ehp.1104204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Johnson SA, Javurek AB, Painter MS, Peritore MP, Ellersieck MR, Roberts RM, Rosenfeld CS. Disruption of parenting behaviors in California Mice, a monogamous rodent species, by endocrine disrupting chemicals. PLoS One 2015; 10:e0126284; PMID:26039462; http://dx.doi.org/ 10.1371/journal.pone.0126284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Johnson SA, Painter MS, Javurek AB, Ellersieck MR, Wiedmeyer CE, Thyfault JP, Rosenfeld CS. Sex-dependent effects of developmental exposure to bisphenol A and ethinyl estradiol on metabolic parameters and voluntary physical activity. J Dev Orig Health Dis 2015:;6:1-14; PMID:26378919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Williams SA, Jasarevic E, Vandas GM, Warzak DA, Geary DC, Ellersieck MR, Roberts RM, Rosenfeld CS. Effects of developmental bisphenol A exposure on reproductive-related behaviors in California mice (Peromyscus californicus): A monogamous animal model. PLoS ONE 2013; 8:e55698; PMID:23405200; http://dx.doi.org/ 10.1371/journal.pone.0055698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Krugner-Higby L, Shadoan M, Carlson C, Gendron A, Cofta P, Marler C, Wagner J. Type 2 diabetes mellitus, hyperlipidemia, and extremity lesions in California mice (Peromyscus californicus) fed commercial mouse diets. Comp Med 2000; 50:412-8; PMID:11020161 [PubMed] [Google Scholar]
- [29].Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol 2011; 12:R60; PMID:21702898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Alonso C, Vicario M, Pigrau M, Lobo B, Santos J. Intestinal barrier function and the brain-gut axis. Adv Exp Med Biol 2014; 817:73-113; PMID:24997030; http://dx.doi.org/ 10.1007/978-1-4939-0897-4_4 [DOI] [PubMed] [Google Scholar]
- [31].Ait-Belgnaoui A, Durand H, Cartier C, Chaumaz G, Eutamene H, Ferrier L, Houdeau E, Fioramonti J, Bueno L, Theodorou V. Prevention of gut leakiness by a probiotic treatment leads to attenuated HPA response to an acute psychological stress in rats. Psychoneuroendocrinology 2012; 37:1885-95; PMID:22541937; http://dx.doi.org/ 10.1016/j.psyneuen.2012.03.024 [DOI] [PubMed] [Google Scholar]
- [32].Braniste V, Jouault A, Gaultier E, Polizzi A, Buisson-Brenac C, Leveque M, Martin PG, Theodorou V, Fioramonti J, Houdeau E. Impact of oral bisphenol A at reference doses on intestinal barrier function and sex differences after perinatal exposure in rats. Proc Natl Acad Sci U S A 2010; 107:448-53; PMID:20018722; http://dx.doi.org/ 10.1073/pnas.0907697107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stilling RM, Dinan TG, Cryan JF. Microbial genes, brain & behaviour - epigenetic regulation of the gut-brain axis. Genes Brain Behav 2014; 13:69-86; PMID:24286462; http://dx.doi.org/ 10.1111/gbb.12109 [DOI] [PubMed] [Google Scholar]
- [34].Douglas-Escobar M, Elliott E, Neu J. Effect of intestinal microbial ecology on the developing brain. JAMA pediatrics 2013; 167:374-9; PMID:23400224; http://dx.doi.org/ 10.1001/jamapediatrics.2013.497 [DOI] [PubMed] [Google Scholar]
- [35].de Theije CG, Wopereis H, Ramadan M, van Eijndthoven T, Lambert J, Knol J, et al.. Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav Immun 2014; 37:197-206; PMID:24333160; http://dx.doi.org/ 10.1016/j.bbi.2013.12.005 [DOI] [PubMed] [Google Scholar]
- [36].Tojo R, Suarez A, Clemente MG, de los Reyes-Gavilan CG, Margolles A, Gueimonde M, Garssen J, Kraneveld AD, Oozeer R. Intestinal microbiota in health and disease: role of bifidobacteria in gut homeostasis. World J Gastroenterol 2014; 20:15163-76; PMID:25386066; http://dx.doi.org/ 10.3748/wjg.v20.i41.15163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gritz EC, Bhandari V. The human neonatal gut microbiome: a brief review. Frontiers in pediatrics 2015; 3:17; PMID:25798435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Moles L, Escribano E. Administration of Bifidobacterium breve PS12929 and Lactobacillus salivarius PS12934, two strains isolated from human milk, to very low and extremely low birth weight preterm infants: a pilot study. 2015; 2015:538171; PMID:25759843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Goodrich Julia K, Waters Jillian L, Poole Angela C, Sutter Jessica L, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al.. Human Genetics Shape the Gut Microbiome. Cell 2014; 159:789-99; PMID:25417156; http://dx.doi.org/ 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Toivonen R, Emani R, Munukka E, Rintala A, Laiho A, Pietilä S, Pursiheimo JP, Soidinsalo P, Linhala M, Eerola E, et al.. Fermentable fibres condition colon microbiota and promote diabetogenesis in NOD mice. Diabetologia 2014; 57:2183-92; PMID:25031069; http://dx.doi.org/ 10.1007/s00125-014-3325-6 [DOI] [PubMed] [Google Scholar]
- [41].Baxter NT, Zackular JP, Chen GY, Schloss PD. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome 2014; 2:20; PMID:24967088; http://dx.doi.org/ 10.1186/2049-2618-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD. The gut microbiome modulates colon tumorigenesis. mBio 2013; 4:e00692-13; PMID:24194538; http://dx.doi.org/ 10.1128/mBio.00692-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 2013; 8:e70803; PMID:23940645; http://dx.doi.org/ 10.1371/journal.pone.0070803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Thomas LV, Ockhuizen T, Suzuki K. Exploring the influence of the gut microbiota and probiotics on health: a symposium report. Br J Nutr 2014; 112 Suppl 1:S1-18; PMID:24953670; http://dx.doi.org/ 10.1017/S0007114514001275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sweeney TE, Morton JM. The human gut microbiome: a review of the effect of obesity and surgically induced weight loss. JAMA surgery 2013; 148:563-9; PMID:23571517; http://dx.doi.org/ 10.1001/jamasurg.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kaakoush NO. Insights into the Role of Erysipelotrichaceae in the Human Host. Front Cell Infect Microbiol 2015; 5:84; PMID:26636046; http://dx.doi.org/ 10.3389/fcimb.2015.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vandenberg LN, Maffini MV, Sonnenschein C, Rubin BS, Soto AM. Bisphenol-A and the great divide: a review of controversies in the field of endocrine disruption. Endocr Rev 2009; 30:75-95; PMID:19074586; http://dx.doi.org/ 10.1210/er.2008-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends Mole Med 2014; 21:109-17; PMID:25578246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Murgas Torrazza R, Neu J. The developing intestinal microbiome and its relationship to health and disease in the neonate. J Perinatol 2011; 31 Suppl 1:S29-34; PMID:21448201; http://dx.doi.org/ 10.1038/jp.2010.172 [DOI] [PubMed] [Google Scholar]
- [50].Walker WA. Initial intestinal colonization in the human infant and immune homeostasis. Ann Nutr Metab 2013; 63 Suppl 2:8-15; PMID:24217032; http://dx.doi.org/ 10.1159/000354907 [DOI] [PubMed] [Google Scholar]
- [51].Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr 1999; 69:1035s-45s; PMID:10232646 [DOI] [PubMed] [Google Scholar]
- [52].Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol 2007; 5:e177; PMID:17594176; http://dx.doi.org/ 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Liu Z, Roy NC, Guo Y, Jia H, Ryan L, Samuelsson L, Thomas A, Plowman J, Clerens S, Day L, et al.. Human Breast Milk and Infant Formulas Differentially Modify the Intestinal Microbiota in Human Infants and Host Physiology in Rats. J Nutr 2015; 146(2):191-9 [DOI] [PubMed] [Google Scholar]
- [54].Narayan NR, Mendez-Lagares G, Ardeshir A, Lu D, Van Rompay KK, Hartigan-O'Connor DJ. Persistent effects of early infant diet and associated microbiota on the juvenile immune system. Gut Microbes 2015; 6:284-9; PMID:26177107; http://dx.doi.org/ 10.1080/19490976.2015.1067743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Praveen P, Jordan F, Priami C, Morine MJ. The role of breast-feeding in infant immune system: a systems perspective on the intestinal microbiome. Microbiome 2015; 3:41; PMID:26399409; http://dx.doi.org/ 10.1186/s40168-015-0104-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013; 339:1084-8; PMID:23328391; http://dx.doi.org/ 10.1126/science.1233521 [DOI] [PubMed] [Google Scholar]
- [57].Jasarevic E, Sieli PT, Twellman EE, Welsh TH Jr., Schachtman TR, Roberts RM, Geary DC, Rosenfeld CS. Disruption of adult expression of sexually selected traits by developmental exposure to bisphenol A. Proc Natl Acad Sci U S A 2011; 108:11715-20; PMID:21709224; http://dx.doi.org/ 10.1073/pnas.1107958108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sieli PT, Jasarevic E, Warzak DA, Mao J, Ellersieck MR, Liao C, Kannan K, Collet SH, Toutain PL, Vom Saal FS, et al.. Comparison of serum bisphenol A concentrations in mice exposed to bisphenol A through the diet versus oral bolus exposure. Environ Health Perspect 2011; 119:1260-5; PMID:21642047; http://dx.doi.org/ 10.1289/ehp.1003385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].vom Saal FS, Richter CA, Ruhlen RR, Nagel SC, Timms BG, Welshons WV. The importance of appropriate controls, animal feed, and animal models in interpreting results from low-dose studies of bisphenol A. Birth Defects Res A Clin Mol Teratol 2005; 73:140-5; PMID:15751043; http://dx.doi.org/ 10.1002/bdra.20120 [DOI] [PubMed] [Google Scholar]
- [60].Jasarevic E, Williams SA, Vandas GM, Ellersieck MR, Liao C, Kannan K, Roberts RM, Geary DC, Rosenfeld CS. Sex and dose-dependent effects of developmental exposure to bisphenol A on anxiety and spatial learning in deer mice (Peromyscus maniculatus bairdii) offspring. Horm Behav 2013; 63:180-9; PMID:23051835; http://dx.doi.org/ 10.1016/j.yhbeh.2012.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Anderson OS, Nahar MS, Faulk C, Jones TR, Liao C, Kannan K, Weinhouse C, Rozek LS, Dolinoy DC. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A. Environ Mol Mutagen 2012; 53:334-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Pro Natl Acad Sci USA 2007; 104:13056-61; PMID:17670942; http://dx.doi.org/ 10.1073/pnas.0703739104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect 2006; 114:567-72; PMID:16581547; http://dx.doi.org/ 10.1289/ehp.8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cox KH, Gatewood JD, Howeth C, Rissman EF. Gestational exposure to bisphenol A and cross-fostering affect behaviors in juvenile mice. Horm Behav 2010; 58:754-61; PMID:20691692; http://dx.doi.org/ 10.1016/j.yhbeh.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Vandenberg LN, Chahoud I, Padmanabhan V, Paumgartten FJ, Schoenfelder G. Biomonitoring studies should be used by regulatory agencies to assess human exposure levels and safety of bisphenol a. Environ Health Perspect 2010; 118:1051-4; PMID:20444668; http://dx.doi.org/ 10.1289/ehp.0901717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Jasarevic E, Williams SA, Vandas GM, Ellersieck MR, Liao C, Kannan K, Roberts RM, Geary DC, Rosenfeld CS. Sex and dose-dependent effects of developmental exposure to bisphenol A on anxiety and spatial learning in deer mice (Peromyscus maniculatus bairdii) offspring. Horm Behav 2013; 63:180-9; PMID:23051835; http://dx.doi.org/ 10.1016/j.yhbeh.2012.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Padmanabhan V, Siefert K, Ransom S, Johnson T, Pinkerton J, Anderson L, Tao L, Kannan K. Maternal bisphenol-A levels at delivery: a looming problem? J Perinatol 2008; 28:258-63; PMID:18273031; http://dx.doi.org/ 10.1038/sj.jp.7211913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Teeguarden JG, Calafat AM, Ye X, Doerge DR, Churchwell MI, Gunawan R, Graham MK. Twenty-four hour human urine and serum profiles of bisphenol a during high-dietary exposure. Toxicol Sci 2011; 123:48-57; PMID:21705716; http://dx.doi.org/ 10.1093/toxsci/kfr160 [DOI] [PubMed] [Google Scholar]
- [69].Vandenberg LN, Chahoud I, Heindel JJ, Padmanabhan V, Paumgartten FJ, Schoenfelder G. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ Health Perspect 2010; 118:1055-70; PMID:20338858; http://dx.doi.org/ 10.1289/ehp.0901716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Vandenberg LN, Hauser R, Marcus M, Olea N, Welshons WV. Human exposure to bisphenol A (BPA). Reprod Toxicol 2007; 24:139-77; PMID:17825522; http://dx.doi.org/ 10.1016/j.reprotox.2007.07.010 [DOI] [PubMed] [Google Scholar]
- [71].Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Blundell P, Alan Harris R, Frias AE, Grove KL, et al.. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun 2014; 5:3889; PMID:24846660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Thorburn AN, McKenzie CI, Shen S. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. 2015; 6:7320; PMID:26102221 [DOI] [PubMed] [Google Scholar]
- [73].Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 2011; 108 Suppl 1:4516-22; PMID:20534432; http://dx.doi.org/ 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, Knight R. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics 2011; 27:1159-61; PMID:21349862; http://dx.doi.org/ 10.1093/bioinformatics/btr087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Loy A, Maixner F, Wagner M, Horn M. probeBase–an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res 2007; 35:D800-4; PMID:17099228; http://dx.doi.org/ 10.1093/nar/gkl856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011; 27:2957-63; PMID:21903629; http://dx.doi.org/ 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460-1; PMID:20709691; http://dx.doi.org/ 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- [78].DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-72; PMID:16820507; http://dx.doi.org/ 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al.. QIIME allows analysis of high-throughput community sequencing data. Nature methods 2010; 7:335-6; PMID:20383131; http://dx.doi.org/ 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:e61217; PMID:23630581; http://dx.doi.org/ 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al.. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31:814-21; PMID:23975157; http://dx.doi.org/ 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.