Abstract
Background
Previously, we demonstrated that bacteria reside in apparently healed alveolar bone, using culture and Sanger sequencing techniques. Bacteria in apparently healed alveolar bone may have a role in peri-implantitis and dental implant failure.
Objective
To compare bacterial communities associated with apical periodontitis, those colonising a failed implant and alveolar bone with reference biofilm samples from healthy teeth.
Methods and results
The study consisted of 196 samples collected from 40 patients undergoing routine dental implant insertion or rehabilitation. The bacterial 16S ribosomal DNA sequences were amplified. Samples yielding sufficient polymerase chain reaction product for further molecular analyses were subjected to terminal restriction fragment length polymorphism (T-RFLP; 31 samples) and next generation DNA sequencing (454 GS FLX Titanium; 8 samples). T-RFLP analysis revealed that the bacterial communities in diseased tissues were more similar to each other (p<0.049) than those from the healthy reference samples. Next generation sequencing detected 13 bacterial phyla and 373 putative bacterial species, revealing an increased abundance of Gram-negative [Prevotella, Fusobacterium (p<0.004), Treponema, Veillonellaceae, TG5 (Synergistetes)] bacteria and a decreased abundance of Gram-positive [(Actinomyces, Corynebacterium (p<0.008)] bacteria in the diseased tissue samples (n=5) relative to reference supragingival healthy samples (n=3).
Conclusion
Increased abundances of Prevotella, Fusobacterium and TG5 (Synergistetes) were associated with apical periodontitis and a failed implant. A larger sample set is needed to confirm these trends and to better define the processes of bacterial pathogenesis in implant failure and apical periodontitis. The application of combined culture-based, microscopic and molecular technique-based approaches is suggested for future studies.
Keywords: oral bacterial communities, apical periodontitis, alveolar bone, dental implant failure, T-RFLP, pyrosequencing
Apical periodontitis is an infection of the tooth root apex resulting from the inflammatory response to bacteria that have migrated through the dental pulp (1, 2). When left untreated, the inflammatory response typically manifests in walling off the bacterial invaders by formation of a periapical lesion (granuloma) (Fig. 1). The treatment of apical periodontitis commonly involves tooth extraction, curettage and irrigation, followed by installation of a dental implant. Peri-implantitis, or the destructive inflammation of soft and hard tissues around dental implants and associated microbial challenge, is a cause of dental implant failure (3, 4). Current 20-year follow-up studies suggest that 99% of implants remain osseointegrated after 1 year, but this drops to 93% after 5 years (5).
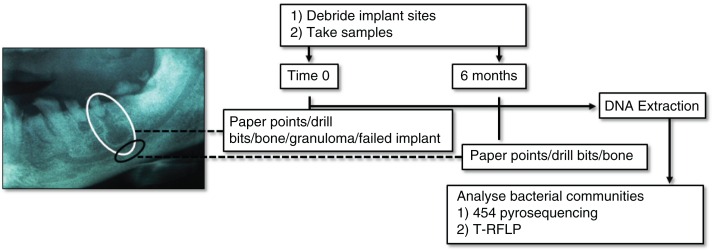
Fig. 1.
Sampling areas and debridement summary at time 0 (white circle) and 6 months (black circle). Of the two periapical lesions (granulomas) pictured in the example radiograph, one features in the white circle. Where possible, sampling was made below the periapical lesion, into trabecular bone (black circle). A reference buccal supragingival biofilm sample from tooth 27 is not shown in the sampling scheme.
Complex bacterial populations exist in the healthy human oral cavity, hosting 500–700 different bacterial species, with 100–200 different species present in any individual, mostly classified within six phyla (6, 7). Such complexity in membership makes it difficult to distinguish the roles of specific bacterial species during health and disease. Nevertheless, certain bacteria are overrepresented at specific sites during various pathologies of the oral cavity (8–10). For example, the bacterial genera Prevotella, Porphyromonas and Fusobacterium tend to be overrepresented in infections of the pulp (7, 10). Slight increases of Prevotella on failed implants with respect to healthy teeth have been detected (9). It has been suggested that bacterial colonisation through the internal cavity of dental implants is a way in which peri-implantitis can begin (11).
Previously, it has been demonstrated that viable bacteria reside in alveolar bone adjacent to the former tooth root apex, including Actinomyes and Prevotella spp (12). The presence of low numbers of bacteria in a normally sterile site (alveolar bone) raised the possibility that these bacteria may later proliferate, contributing to peri-implantitis and early failure of osseointegrated dental implants (12). Given the difficulties in culturing bacteria of the oral cavity, this study aimed to compare the bacteria associated with alveolar bone, apical periodontitis and dental implant failure with reference biofilm samples from healthy teeth via a molecular profiling approach, specifically, comparison of bacterial communities using terminal restriction fragment length polymorphism (T-RFLP) and 454 pyrosequencing of 16S rDNA in diseased samples (root canal, granulomas, alveolar bone and a failed implant).
Materials and methods
Patient selection and sample collection
This study was conducted on 40 routine private practice patients presenting for dental implant insertion or rehabilitation. Approval for surgery and sampling was obtained from the University of Sydney Human Research Ethics Committee. Surgical procedures and sampling were performed by a calibrated clinician, following a previously described strictly aseptic two-stage open-flap surgical technique (12, 13) summarised in Fig. 1.
Immediately prior to surgery, a reference sample of normal microflora [supragingival buccal biofilm scraping of tooth 27 (UL7)] was made. Following biofilm sampling, during the first ‘preparatory’ stage (time point 0), microorganisms were removed and soft granulation tissue was curetted using spoon excavators and a Mitchell Osseo Trimmer (Medident, Braeside, Australia). Some bone samples were taken at the height of the former periapex, which was determined by measurements of the extracted calibrated lengths from digital radiographs and radiographic confirmation with the preparation drill in situ. Overlying bone was debrided with a guide drill to the height of the former tooth root apex (also at time point 0). A twist drill was used to penetrate the former tooth root apex into trabecular bone. Debridement of overlying trabecular and cortical bone was performed with a Brånemark System round guide drill equipped with a shaft extender under chilled, sterile saline. Extreme care was taken to prevent contamination of drill bits in sample sites from other areas of the oral cavity. In the second stage (6 months after initial debridement), the previously apparently healed debrided sites were re-debrided in preparation for abutment connection (implant insertion). After measuring the depth of the implant site, implants were mechanically inserted. During curettage and debridement procedures, blood and tissue fluids were absorbed on a size 80 endodontic paper point (Dentsply Maiellefer, Ballaigues, Switzerland), which was left in the sample site for 15 sec. Entire drill bits (with retained bone fragments) used to debride bone or bone filter particles suctioned out of the sample site were retained for analyses, whereas present, intact granulomas, a failed implant or infected tooth were sampled directly. Samples were added to chilled TE buffer (0.5 ml, 10 mM Tris, 1 mM EDTA, pH 7.6) and transported to the molecular laboratory on ice, before storage at −80°C, until DNA extraction. Where patients fell within the study time constraints, at 6 months, apparently healed bone of the implant site was re-debrided and sampled by paper point, bone directly retained on drill bits or bone filter particles suctioned out of the sample site. A total of 196 samples were taken from 104 sites. The failed titanium implant (a 5×7 mm WP Branemark MK 111 TiUnite) presented with bone loss 8 months after prosthetic connection. Other relevant sample details are documented in Supplementary Table 1.
DNA extraction
DNA was extracted by bead-beating samples at 5.5 ms−1 for 30 sec (Fast Prep FP-120, Bio101), followed by purification as described previously (14). Samples were bead-beat to aid lysis of Gram-positive bacteria. One extraction blank, consisting of sterile TE buffer, was included for every 10 samples. DNA quality and quantity was verified by agarose gel electrophoresis.
Terminal Restriction Fragment Length Polymorphism
The extracted DNA was polymerase chain reaction (PCR) amplified using bacterial domain-specific primers, targeting variable regions V1–V5 of the bacterial 16S rRNA gene, using primers 27F (5′-AGAGTTTGATYMTGGCTCAG) and 907R (5′-CCGTCAATTCMTTTRAGTTT), which were modified at the 5′ end with 5-FAM and 6-HEX fluorophores, respectively. Each 25 µl PCR reaction contained 1×Thermopol buffer, 1 µM of forward and reverse primers, 200 µM dNTPs and 1.25 U Taq DNA polymerase. All reagents for PCR amplification and digestion were supplied by New England Biolabs (Ipswich, MA, USA). PCR conditions included an initial denaturation of 94°C for 4 min, then 30 cycles of 94°C for 30 sec, 51°C for 45 sec and 72°C for 60 sec, followed by a final extension at 72°C for 10 min on a Mastercycler S machine (Eppendorf, Hamburg, Germany).
PCR amplicons were separately digested with HaeIII (10U) and MspI (20U), and the resultant terminal restriction fragments (T-RFs) were analysed by capillary electrophoresis using an ABI PRISM 3730 DNA Analyser (AGRF, Westmead, Australia). Genemapper v3.7 (Applied Biosystems, Foster City, CA, USA) was used to determine peak length, height and area using the Local Southern algorithm. T-RFs <50bp or ≤1% of the total fluorescent area were excluded from analysis. For the four combinations of fluorophore and restriction enzyme, remaining T-RFs were rounded to the nearest integer and Primer VI (v. 6.1.6, Primer-E Ltd, Auckland, New Zealand) was used to convert TRFs into a presence and absence matrix. Non-metric multidimensional scaling (NMDS) using the Bray–Curtis coefficient (15), with a restart value of 200 and a one way analysis of similarities (ANOSIM) with a maximum permutation of 9,999, was carried out (16). Samples were separated based on association with health and disease.
Pyrosequencing and sequence analysis
Genomic DNA from eight samples was sent to Research and Testing Laboratory (RTL, Lubbock, TX, USA) for amplification with primers 28F (5′-AGTTTGATCNTGGCTCAG) and 519R (5′-GTNTTACNGCGGCKGCTG) appended with DNA ‘bar codes’ described previously (17). Sequencing of V1–V3 hypervariable regions of the 16S rRNA was performed by the RTL, using a Roche GS-FLX+ Sequencer with Titanium reagents (Roche, Indianapolis, USA) from 28F. Sequences were demultiplexed by RTL, giving 44,211 sequences.
Sequences were processed with mothur following an established 454 pyrosequencing analysis pipleine [(18), described in detail at: www.mothur.org/wiki/454_SOP)]. Sequence barcodes and primers were removed and sequences were truncated if they fell below an average quality threshold of 30 over a sliding window of 50 base pairs. Quality-trimmed sequences were aligned against the mothur-formatted SILVA bacterial reference alignment. Chimeric sequences were removed with uchime and sequences were merged together (‘preclustered’, diffs = 2) if they differed by less than 4 bases (19). Sequences were classified with the mothur-formatted Greengenes 16S rDNA reference taxonomy. Within sequences that passed quality filtering were converted to a percentage of the total sequences within each sample (summarised in Supplementary Table 2). Taxa with less than 0.5% abundance in a given sample are not shown. The average and standard error of the mean were calculated for remaining taxa within each sample type. In preparation for statistical analyses, a similarity matrix was generated using Bray–Curtis coefficient. To avoid bias in analyses, the similarity matrix was subsampled to 1,189 sequences per sample. To analyse variation in community structure, samples were grouped into reference biofilms (n=3) or diseased (n=5) bins and analysis of molecular variance (AMOVA) was used with 1,000 permutations (20). The mothur implementation of metastats (21) was also used to determine if any operational taxonomic units (OTUs) were significantly different in reference biofilm and diseased samples.
Results
PCR amplification of 16S rRNA gene
A total of 196 samples were collected from 40 patients undergoing dental implant preparation and/or insertion. The disease-associated sample set included granulomas, a failed implant, tooth fragments, paper points soaked with extruded fluids and drill bits with attached tissue. Buccal supragingival biofilm scrapings collected from non-diseased teeth were used as references for a normal oral bacterial community.
All dental biofilm samples from healthy teeth yielded strong PCR products, but it proved difficult to amplify 16S rRNA genes from other samples. Attempts to improve the PCR using chemical facilitators (betaine, dimethyl sulfoxide, bovine serum albumin), different polymerases (Kapa ‘Blood Taq’; Kapa Biosystems, Wilmington, USA), different template dilutions, different primer sets or modified thermocycling conditions were not successful (data not shown). Addition of a multiple displacement amplification step (Repli-G mini kit, Qiagen, Hilden, Germany) before the PCR enabled amplification of a few difficult templates, but subsequent T-RFLP and NMDS analyses showed that these samples clustered erroneously, with few terminal fragments; therefore, this approach was not pursued further (data not shown). Further T-RFLP analyses were performed on samples that yielded sufficient PCR products (31 of 196 samples) consisting of intact granulomas (n=2), bone fragments (n=1), an infected tooth (n=1), a failed implant (n=1), paper points (n=5) or healthy tooth biofilms (n=21).
T-RFLP analysis: reference and diseased communities
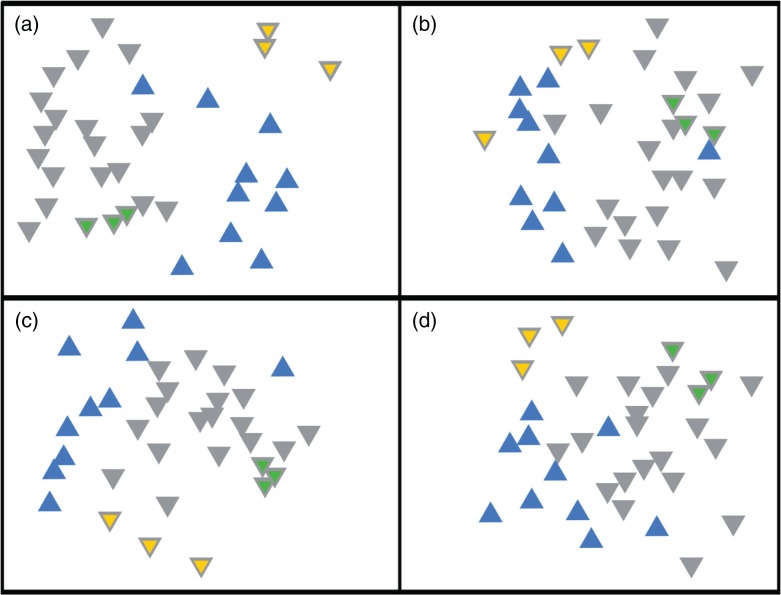
T-RFLP provides a snapshot of community structure based on the size of T-RFs produced from amplified ribosomal RNA genes. For both restriction enzymes (HaeIII, MspI) and fluorophores (5’-FAM, 3’-HEX) used, the same general pattern was observed, that is, the biofilm samples from reference teeth consistently grouped together, separately from diseased tissue samples (Fig. 2), a result confirmed by ANOSIM analysis, in which all four combinations of fluorophore and restriction enzymes were significantly separated (Fig. 2). The number of terminal fragments obtained using Msp1 (17.6±5.7) was similar to that obtained using HaeIII (16.5±5). Taken together, the T-RFLP data indicated that the bacterial communities in the diseased samples were different from that in the reference biofilm samples. Eight of the samples were analysed in greater detail by pyrosequencing of the 16S rDNA gene (regions V1–V3).
Fig. 2.
Non-metric multidimensional scaling of T-RFLP data from reference biofilm versus diseased samples. (a) 5′-FAM label/HaeIII digest (stress = 0.22, p<0.01), (b) 5′-FAM/MspI digest (stress = 0.26, p<0.01), (c) 3′-HEX label/HaeIII digest (stress = 0.20, p<0.01) and (d) 3′-HEX label/MspI digest (stress = 0.23, p<0.02). Reference biofilm and diseased samples are represented by grey and blue triangles, respectively. Technical replicates (amplification and digestion performed in triplicate) of biofilm samples 22A and 23A have yellow or green centres, respectively.
Pyrosequencing analysis: community patterns
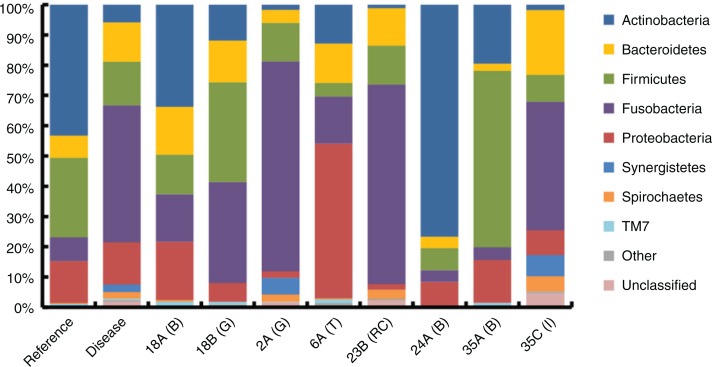
A total of 44,211 sequences were generated from eight samples. Of these sequences, 57% (25,189) passed strict quality control filters (Supplementary Table 2), yielding 11,248 sequences associated with reference biofilm samples (average 3,749±277 per sample) and 11,097 sequences associated with diseased samples (average 2,219±638 per sample). A total of 2,464 unique sequences were clustered at the 97% similarity threshold, giving 373 OTUs across all samples (Supplementary Table 3). The total diversity found in disease-associated samples (278±26 OTUs per sample) was higher than that of healthy tooth biofilms (216±12 OTUs per sample). However, within patients, the abundance of bacteria from reference samples was always higher than corresponding diseased sample (see below and also rarefaction curves in Supplementary Fig. 1). An AMOVA revealed that reference and diseased bacterial populations were significantly different (p<0.049). In the eight patients combined, the most abundant phyla were Actinobacteria (24%), Fusobacteria (23%), Firmicutes (20%), Proteobacteria (18%) and Bacteroidetes (10%) (Fig. 3). Low abundance phyla included Synergistetes (1.7%), Spirochaetes (1.3%), TM7 (0.8%), GN02 (0.1%), SR1 (0.1%), Tenericutes (0.1%), Gemmatimonadetes (0.04%) and AD3 (0.04%). Synergistetes, SR1, Tenericutes, Gemmatimonadetes and AD3 were only detected in disease-associated samples, some of which, such as Synergistetes (TG5), are either not yet culturable or difficult to culture (22).
Fig. 3.
Relative 16S rDNA sequence abundances of bacteria in reference biofilm versus diseased samples. The taxonomic results from 454 pyrosequencing at the phylum-level resolution. Sample types are biofilm (B), granuloma (G), infected tooth (T), root canal (RC) and failed implant (I) from 6 patients (18E/18G, 2A, 6A, 23B, 24A and 35A/35C). The columns labelled ‘Reference’ and ‘Disease’ show the average sequence abundances in these data sets (n=3 and n=5, respectively). ‘Other’ includes GN02, SR1, Tenericutes, Gemmatimonadetes and AD3.
Pyrosequencing analysis: bacterial types associated with disease
The relative abundance of sequences detected in all samples is shown in Table 1 and Fig. 3. Some of the major trends in the diseased samples were decreases in abundance of sequences from Actinomyces (eightfold), Corynebacterium (sixfold) and Streptococcus (fivefold), and increases in abundance of sequences from Fusobacterium and Prevotella (both sevenfold). TG5, a clade within the Synergistetes phylum, was detected only in diseased tissues, comprising an average of 3.4% of sequences in these communities. Of these changes, metastats identified Fusobacterium (OTU 1) and Corynebacterium (OTU 5) as significant (p<0.004 and p<0.008, respectively) between all reference and disease-associated samples.
Table 1.
Relative sequence abundances (16S rDNA) of bacterial taxa in reference biofilm and diseased dental samplesab
| Abundance (%) | ||
|---|---|---|
|
|
||
| Assigned taxonomy (phylum–family–genus) | Health | Disease |
| Actinobacteria Actinomycetaceae Actinomyces | 26.1±4.3 | 3.3±0.3 |
| Firmicutes Streptococcaceae Streptococcus | 20.2±5.9 | 3.8±0.2 |
| Actinobacteria Corynebacteriaceae Corynebacterium | 13.3±1.3 | 2.2±0.4 |
| Fusobacteria Fusobacteriaceae Fusobacterium | 5.2±1.5 | 38.3±2.6 |
| Bacteroidetes Flavobacteriaceae Capnocytophaga | 4.0±0.6 | 3.8±0.7 |
| Proteobacteria Neisseriaceae Neisseria | 3.5±0.6 | 4.0±0.8 |
| Firmicutes Veillonellaceae Veillonella | 3.0±0.2 | 2.8±0.4 |
| Proteobacteria Burkholderiaceae Lautropia | 2.5±0.4 | 7.0±1.4 |
| Proteobacteria Campylobacteraceae Campylobacter | 2.4±0.5 | 3.5±0.3 |
| Fusobacteria Fusobacteriaceae Leptotrichia | 2.3±0.4 | 0.7±0.1 |
| Firmicutes Carnobacteriaceae Granulicatella | 2.2±0.7 | 0.1±0.0 |
| Proteobacteria Pasteurellaceae Haemophilus | 2.0±0.3 | 1.3±0.3 |
| Firmicutes Veillonellaceae Selenomonas | 1.9±0.5 | 1.3±0.2 |
| Bacteroidetes Porphyromonadaceae Porphyromonas | 1.3±0.4 | 0.1±0.0 |
| Bacteroidetes Prevotellaceae Prevotella | 1.2±0.3 | 8.9±1.1 |
| Proteobacteria Neisseriaceae Kingella | 1.1±0.0 | 0.2±0.0 |
| Proteobacteria Neisseriaceae Eikenella | 1.0±0.3 | 0.5±0.1 |
| TM7 Unclassified | 1.0±0.2 | 0.5±0.1 |
| Actinobacteria Propionibacteriaceae Propionibacterium | 1.0±0.3 | 0.1±0.1 |
| Firmicutes Gemellaceae Gemella | 0.9±0.3 | 0.1±0.0 |
| Actinobacteria Micrococcaceae Rothia | 0.7±0.1 | 0.3±0.1 |
| Proteobacteria Pasteurellaceae Aggregatibacter | 0.5±0.1 | 2.0±0.3 |
| Spirochaetes Spirochaetaceae Treponema | 0.2±0.1 | 2.4±0.3 |
| Proteobacteria Cardiobacteriaceae Cardiobacterium | 0.2±0.0 | 0.6±0.1 |
| Bacteroidetes Porphyromonadaceae Tannerella | 0.2±0.1 | 0.7±0.1 |
| Synergistes Dethiosulfovibrionaceae TG5 | 0.0±0.0 | 3.4±0.5 |
| Actinobacteria Coriobacteriaceae Atopobium | 0.0±0.0 | 0.5±0.1 |
| Firmicutes Peptostreptococcaceae Filifactor | 0.0±0.0 | 0.5±0.2 |
Only OTUs of >0.5% abundance in at least one sample group are shown.
Taxa are arranged from highest to lowest abundance in the reference biofilm data set.
A comparison of one reference sample (biofilm, 18E) and one diseased sample (granuloma, 18G) from the same patient is shown in Table 2 and Supplementary Table 4. The majority of OTUs (75%) from the granuloma corresponded to sequence types in the biofilm, at ≥97% sequence identity, indicating the granuloma bacteria were a subset of this patient's biofilm community. Increases in Prevotella and Fusobacterium and decreases in Actinobacteria (Actinomyces and Corynebacterium) were observed in the granuloma relative to the biofilm control (Sample 18E). Other notable features of the granuloma were increases in Veillonella and Atopobium sequences (6-fold and 29-fold, respectively) and decreases in Capnocytophaga and Neisseria sequences (11-fold and 52-fold, respectively), relative to biofilm.
Table 2.
| Abundance (%) | ||
|---|---|---|
|
|
||
| Assigned taxonomy (phylum–family–genus) | 18E | 18G |
| Fusobacteria Fusobacteriaceae Fusobacterium | 15.7 | 33.4 |
| Actinobacteria Actinomycetaceae Actinomyces | 15.3 | 8.2 |
| Actinobacteria Corynebacteriaceae Corynebacterium | 13.8 | 0.2 |
| Bacteroidetes Flavobacteriaceae Capnocytophaga | 7.8 | 0.7 |
| Firmicutes Veillonellaceae Selenomonas | 5.4 | 8.8 |
| Proteobacteria Campylobacteraceae Campylobacter | 5.4 | 6.0 |
| Proteobacteria Neisseriaceae Neisseria | 5.2 | 0.1 |
| Bacteroidetes Porphyromonadaceae Porphyromonas | 3.8 | 0.0 |
| Actinobacteria Propionibacteriaceae Propionibacterium | 3.2 | 0.0 |
| Firmicutes Streptococcaceae Streptococcus | 3.3 | 3.4 |
| Firmicutes Veillonellaceae Veillonella | 3.2 | 20.7 |
| Proteobacteria Neisseriaceae Eikenella | 3.2 | 0.0 |
| Bacteroidetes Prevotellaceae Prevotella | 3.1 | 12.6 |
| TM7 unclassified | 1.7 | 0.8 |
| Proteobacteria Pasteurellaceae Aggregatibacter | 1.6 | 0.0 |
| Proteobacteria Neisseriaceae Kingella | 1.3 | 0.0 |
| Spirochaetes Spirochaetaceae Treponema | 0.6 | 0.0 |
| Proteobacteria Pasteurellaceae Haemophilus | 0.5 | 0.0 |
| Actinobacteria Coriobacteriaceae Atopobium | 0.1 | 2.9 |
| Tenericutes Erysipelotrichaceae Bulleidia | 0.0 | 0.5 |
Only OTUs of >0.5% abundance in at least one sample group are shown.
Taxa are arranged from highest to lowest abundance in the biofilm data set.
Sequences in a granuloma from a different patient (Sample 2A, Fig. 3 and Supplementary Table 6) confirmed some of the trends observed in granuloma 18G. In Sample 2A, Fusobacterium was the most abundant sequence type (69.4%), Prevotella sequences were more abundant compared with their average abundance in biofilm samples and Actinobacteria decreased in reference to biofilm samples. An unusual feature of the granuloma 2A was that bacteria from phylum TG5 were quite abundant, at 5.5% of all sequences. TG5 sequences were not detected in granuloma 18G, or in any of the biofilm samples, but were detected in the failed implant (Sample 35C, discussed below).
A comparison of the sequences recovered from a failed dental implant (35C) and a biofilm sample (35A) from the same patient (Table 3, Supplementary Table 5 and Fig. 3) revealed similar shifts in bacterial genera as were seen in the granuloma samples, with an increase in Prevotella and Fusobacterium sequences (39-fold and 53-fold, respectively) and a decrease in sequences from Actinobacteria, specifically, complete loss of Corynebacterium, and 10-fold decrease in Actinomyces. Other notable trends in the failed implant community included the appearance of TG5 and Treponema, an increase in Campylobacter sequences (54-fold) and a decrease in Streptococcus (14-fold). A significant portion (4.7%) of the sequences from the failed implant community mapped to the domain Bacteria, but could not confidently be assigned into any known phylum.
Table 3.
| Abundance (%) | ||
|---|---|---|
|
|
||
| Assigned taxonomy (phylum–family–genus) | 35A | 35C |
| Firmicutes Streptococcaceae Streptococcus | 48.4 | 3.4 |
| Actinobacteria Actinomycetaceae Actinomyces | 11.2 | 1.1 |
| Actinobacteria Corynebacteriaceae Corynebacterium | 6.2 | 0.0 |
| Firmicutes Carnobacteriaceae Granulicatella | 5.6 | 0.0 |
| Proteobacteria Burkholderiaceae Lautropia | 4.1 | 0.0 |
| Fusobacteria Fusobacteriaceae Leptotrichia | 3.3 | 0.0 |
| Firmicutes Gemellaceae Gemella | 2.3 | 0.2 |
| Firmicutes Veillonellaceae Veillonella | 1.9 | 0.0 |
| Actinobacteria Micrococcaceae Rothia | 1.3 | 0.0 |
| Bacteroidetes Flavobacteriaceae Capnocytophaga | 1.2 | 0.0 |
| Proteobacteria Neisseriaceae Kingella | 1.0 | 0.0 |
| Fusobacteria Fusobacteriaceae Fusobacterium | 0.8 | 42.4 |
| Bacteroidetes Prevotellaceae Prevotella | 0.5 | 19.3 |
| Proteobacteria Campylobacteraceae Campylobacter | 0.1 | 5.4 |
| Proteobacteria Pasteurellaceae Aggregatibacter | 0.1 | 2.4 |
| Firmicutes Veillonellaceae Dialister | 0.0 | 1.3 |
| Firmicutes Peptococcaceae Peptococcus | 0.0 | 0.9 |
| Actinobacteria Coriobacteriaceae Atopobium | 0.0 | 0.7 |
| Firmicutes Clostridiaceae Mogibacterium | 0.0 | 0.5 |
| Firmicutes Clostridiaceae Eubacterium | 0.0 | 0.5 |
Only OTUs of >0.5% abundance in at least one sample group are shown.
Taxa are arranged from highest to lowest abundance in the reference biofilm data set.
Pyrosequencing analyses of an infected tooth (Sample 6A, Supplementary Table 7), a root canal paper point sample (23B, Supplementary Table 8) and another reference biofilm sample (24A, Supplementary Table 9) reinforced many of the trends of the failed implant bacterial community, especially the increased abundance of Fusobacterium and the decreased abundance of Actinobacteria in the diseased samples (see also Fig. 3).
Discussion
A molecular profiling approach was used to investigate bacteria associated with a failed dental implant, apical periodontitis and alveolar bone. Several characteristic features of the bacterial communities associated with implant failure and periodontitis were defined, including an increased relative abundance of Prevotella and Fusobacterium and a decreased abundance of Actinomyces and Corynebacterium relative to the biofilm on reference teeth. The reference biofilm and disease-associated bacterial communities were distinguished by community profiling using T-RFLP, and these differences were confirmed by pyrosequencing.
Great difficulty was encountered in amplifying 16S rDNA from the majority of blood and bone-containing samples. Using 454 pyrosequencing, a similar phenomenon in root canal samples has been noted and attributed to competitive inhibition by human DNA (8). In that study, poor PCR amplification was overcome by diluting template DNA, but this approach was not successful for our samples. Two lines of evidence suggest that our PCR amplification difficulties were primarily because of low numbers of bacteria; firstly, biofilm samples and obviously infected samples (such as granulomas) gave strong PCR products, and secondly, human ribosomal sequences (18S rDNA) could be amplified from many samples which did not give positive 16S rDNA PCRs (data not shown). On the other hand, PCRs using a mixture of amplifiable and non-amplifiable templates gave similar results to the non-amplifiable templates alone (data not shown), suggesting the presence of PCR inhibitors was a contributing factor. Calcium ions (23) from bone, or blood factors, such as immunoglobulin G or lactoferrin (24), may have contributed to the problems with PCR amplification and detection of bacteria in many samples.
Previous culture-based work detected an increased abundance of Actinomyces and Prevotella spp. in cases of apical periodontitis and on failed dental implants (12). This trend was not revealed by pyrosequencing analyses performed herein, which showed a decreased relative abundance of Actinobacteria (Actinomyces and Corynebacterium). This apparent discrepancy may represent the ease of culturing facultatives (such as Actinomyces) compared with strict anaerobes (Fusobacterium) (25) which may bias apparent viable counts of these clades. It is notable that the shifts in taxa seen between reference biofilm and diseased samples in this study correlate to a large extent with a shift from facultatives to strict anaerobes. This trend underlines the usefulness of pyrosequencing for studying samples that contain unculturable or fastidious bacteria, where taxa detected in the diseased samples may have been overlooked or underrepresented in a culture-based study.
One of the most striking patterns observed in the pyrosequencing data was the high relative abundance of Fusobacterium sequences in all of the diseased samples. Prevotella populations also increased in diseased samples with an average 20-fold increase in this genus (19.3%) in the failed implant compared to the average detected on reference biofilm. Using pyrosequencing, other investigators confirmed that failed implants are dominated by Gram-negative bacteria (9). However, whilst Prevotella populations associated with failed implants increased with respect to reference biofilm, the same investigators detected the highest populations of Prevotella on healthy implants (9). Other taxa that tended to be more abundant in diseased samples than in reference biofilm samples were Treponema, Veillonellaceae (Selenomonas and Veillonella) and Synergistetes (TG5), all bacteria detected during chronic periodontitis (26, 27).
An increase in sample size is required to draw firm conclusions about bacterial pathogenesis during implant failure and apical periodontitis. Nevertheless, in this pilot molecular study, it is noted that even in the small sample set, clades of bacteria listed above were consistently overrepresented in diseased tissues. Molecular tools were useful here for revealing shifts in bacterial clades in diseased periapical tissues, but it should be noted that DNA-based detection of bacteria in these samples was difficult. Blood- and bone-derived inhibitors and the low numbers of bacterial cells in alveolar samples pose unique restrictions to molecular profiling techniques. Because the numbers of bacteria in alveolar bone were low, application of microscopy-based techniques on intact sections of alveolar bone may overcome discussed restrictions. The combination of molecular, culture and microscopy-based techniques with a larger sample cohort is required to further define the bacterial communities involved in apical periodontitis and determine whether bacteria are able to proliferate in alveolar bone, leading to early dental implant failure.
Supplementary Material
Acknowledgements
Funding for the work and access to samples were provided via the dental surgery business (One Stop Dental Implants, Batemans Bay, NSW, Australia) owned and operated by one of the study authors (SN). We thank Graham Thomas for supervision of SN in related work, and Darryl Nelson for assistance with T-RFLP analyses.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
References
- 1.Kakehashi S, Stanley H, Fitzgerald R. The effects of surgical exposures of dental pulps in germ-free and conventional laboratory rats. Oral Sur Oral Med Oral Pathol. 1965;20:340–9. doi: 10.1016/0030-4220(65)90166-0. [DOI] [PubMed] [Google Scholar]
- 2.Nair PNR. Apical periodontitis: a dynamic encounter between root canal infection and host response. Periodontol. 2000;13:121–48. doi: 10.1111/j.1600-0757.1997.tb00098.x. [DOI] [PubMed] [Google Scholar]
- 3.Lindhe J, Meyle J. Peri-implant diseases: consensus report of the sixth European workshop on periodontology. J Clin Periodontol. 2008;35:282–5. doi: 10.1111/j.1600-051X.2008.01283.x. [DOI] [PubMed] [Google Scholar]
- 4.Mombelli A, Décaillet F. The characteristics of biofilms in peri-implant disease. J Clin Periodontol. 2011;38:203–13. doi: 10.1111/j.1600-051X.2010.01666.x. [DOI] [PubMed] [Google Scholar]
- 5.Ekelund JA, Lindquist LW, Carlsson GE, Jemt T. Implant treatment in the edentulous mandible: a prospective study on Brånemark system implants over more than 20 years. Inter J Prosthodont. 2003;16:602–8. [PubMed] [Google Scholar]
- 6.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–74. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsiao WWL, Li KL, Liu ZQ, Jones C, Fraser-Liggett CM, Fouad AF. Microbial transformation from normal oral microbiota to acute endodontic infections. BMC Gen. 2012;13:345. doi: 10.1186/1471-2164-13-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar PS, Mason MR, Brooker MR, O'Brien K. Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. J Clin Periodontol. 2012;39:425–33. doi: 10.1111/j.1600-051X.2012.01856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zakaria M, Takeshita T, Shibata Y, Maeda H, Wada N, Akamine A, et al. Microbial community in persistent apical periodontitis: a 16S rRNA gene clone library analysis. Inter Endod J. 2015;48:717–28. doi: 10.1111/iej.12361. [DOI] [PubMed] [Google Scholar]
- 11.Jervøe-Storm PM, Jepsen S, Jöhren P, Mericske-Stern R, Enkling N. Internal bacterial colonization of implants: association with peri-implant bone loss. Clin Oral Imp Res. 2015;26:957–63. doi: 10.1111/clr.12421. [DOI] [PubMed] [Google Scholar]
- 12.Nelson S, Thomas G. Bacterial persistence in dentoalveolar bone following extraction: a microbiological study and implications for dental implant treatment. Clin Implant Dent Relat Res. 2010;12:306–14. doi: 10.1111/j.1708-8208.2009.00165.x. [DOI] [PubMed] [Google Scholar]
- 13.Friberg B. Sterile operating conditions for the placement of intraoral implants. J Oral Maxillofac Surg. 1996;54:1334–6. doi: 10.1016/s0278-2391(96)90493-0. [DOI] [PubMed] [Google Scholar]
- 14.Yeates C, Gillings MR. Rapid purification of DNA from soil for molecular biodiversity analysis. Lett Appl Microbiol. 1998;27:49–53. [Google Scholar]
- 15.Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27:325–49. [Google Scholar]
- 16.Rees GN, Baldwin DS, Watson GO, Perryman S, Nielsen DL. Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie van Leeuwenhoek. 2005;86:339–47. doi: 10.1007/s10482-004-0498-x. [DOI] [PubMed] [Google Scholar]
- 17.Dowd S, Callaway T, Wolcott R, Sun Y, McKeehan T, Hagevoort R, et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–98. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson MJ. Permutation tests for univariate or multivariate analysis of variance and regression. Can J Fish Aquat Sci. 2001;58:626–39. [Google Scholar]
- 21.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hugenholtz P, Hooper SD, Kyripides NC. Focus: synergistetes. Environ Microbiol. 2009:1327–9. doi: 10.1111/j.1462-2920.2009.01949.x. [DOI] [PubMed] [Google Scholar]
- 23.Bickley J, Short J, McDowell D, Parkes H. Polymerase chain reaction (PCR) detection of Listeria monocytogenes in diluted milk and reversal of PCR inhibition caused by calcium ions. Lett Appl Microbiol. 1996;22:153–8. doi: 10.1111/j.1472-765x.1996.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 24.Al-Soud WA, Jönsson LJ, Rådström P. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J Clin Microbiol. 2000;38:345–50. doi: 10.1128/jcm.38.1.345-350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wade WG, Spratt DA, Dymock D, Weightman AJ. Molecular detection of novel anaerobic species in dentoalveolar abscesses. Clin Infect Dis. 1997;25(Suppl. 2):S235–6. doi: 10.1086/516215. [DOI] [PubMed] [Google Scholar]
- 26.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–44. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 27.Hutter G, Schlagenhauf U, Valenza G, Horn M, Burgemeister S, Claus H, et al. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiol. 2003;149:67–75. doi: 10.1099/mic.0.25791-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.