Abstract
The mammalian immune system communicates with skin-resident microbes, and some of these microbes provide benefits to the host. In a recent paper in Nature, Naik et al. (2015) provide evidence that the murine epidermis permits S. epidermidis, a skin-specific bacterium, to shape the immune response.
Mammalian skin is a highly complex semipermeable filtration system that we now know to have many dynamic interactions with the microbes that reside on this surface. Similar to the actions of the microflora within the gut, it was hypothesized that bacteria that reside on the surface of mammalian skin have a mutually beneficial relationship with the host (Cogen et al., 2008). Several studies have emerged to support this hypothesis and have shown multiple ways that skin bacteria can benefit the host. The most direct benefit is seen by observations that some bacteria residing on the skin surface produce selective antimicrobial peptides, lantibiotics, and other molecules that act to resist co-colonization by pathogens (Cogen et al., 2010). A significant fraction of the organisms identified on the skin surface also enter the deeper dermal stroma where they can interact with a variety of host cell types (Nakatsuji et al., 2013). Bacteria present in human skin thus have the opportunity to control cell behaviors below the surface. Examples of beneficial functions induced by specific skin bacteria include the capacity to regulate inflammation after injury (Lai et al., 2009), enhance expression of host innate antimicrobial peptides, and mature T cell responses (Lai et al., 2010; Li et al., 2013; Naik et al., 2012; Wanke et al., 2011). In contrast, Staphylococcus aureus applied to the surface of mouse skin can initiate harmful events that exacerbate allergic responses (Nakamura et al., 2013). Thus, the normal epidermis is not an impermeable barrier to surface microbes but a filter to control entry.
In a recent publication, Naik et al. add to the growing list of experimental evidence in mice that the specific type of bacteria residing on the skin will influence the mammalian immune response (Naik et al., 2015). Their approach followed prior strategies by concentrating on a candidate organism to test the functions of resident human skin bacteria. They chose Staphylococcus epidermidis, one of the most common species of bacteria cultured from normal human skin but also an important opportunistic pathogen in humans. Surprisingly, although S. epidermidis is not commonly detected on mouse skin, the investigators report that after application of a high concentration of live bacteria, they could culture S. epidermidis from mice up to 180 days later. In contrast, parallel measurements of the relative abundance of bacterial phyla by DNA sequencing analysis detected S. epidermidis at 14 days but not at 180 days, a contradictory result that emphasizes the technical challenges inherent in these studies. Importantly however, exposure to S. epidermidis resulted in an increase in CD8 β+ T cells while exposure to other bacteria did not. Other bacterial species tested included Staphylococcus xylosus, Propionibacterium acnes, and Staphylococcus aureus. Each of these organisms induced a different and broader T cell profile that also included cells expressing IL-17A and IFN-γ.
The response to S. epidermidis afforded the investigators the opportunity to study how murine skin detects this specific bacterial species. They show that a subset of dermal dendritic cells, but not epidermal Langerhans cells, is responsible for initiating the unique T cell response. To evaluate the functional significance of these specific CD8 β+ T cells, the study evaluated the capacity of mice to resist infection by Candida albicans. Neutralization of IL-17A or depletion of CD8+ T cells is shown to increase C. albicans abundance. Although the mechanism of resistance to Candida albicans is unclear, mouse skin associated with S. epidermidis was shown to express more of the alarmins S100a8 and S100a9. These proteins have been implicated in a variety of functions including the capacity to induce neutrophil migration.
The authors speculate that many other specific skin commensals may have individual roles in educating the host adaptive immune response. Although by definition this interaction is evidence of mutualism, not commensalism, their finding is consistent with previous demonstrations of specific microbe interactions in the skin. For example, S. epidermidis was uniquely capable of inducing TRAF1 in keratinocytes (Lai et al., 2009) and S. aureus uniquely activated mast cells to exacerbate allergic responses (Nakamura et al., 2013). Furthermore, this study not only demonstrates the potential importance of specific bacteria on skin but also reinforces the idea that bacteria can act across the epidermis and interact with cells in the dermis.
Current knowledge of the mechanisms by which the microbiome participates in host defense is limited, but improving. The Naik et al. study shows that S. epidermidis participates in several different ways to directly and indirectly add to the innate and adaptive immune barrier to pathogens (Figure 1). Functional specificity seems to exist at both the species and strain level. It remains to be determined how these functions can be reconciled with the great diversity of organisms detected between individuals or how this knowledge can be translated into therapy. Regardless, these studies show again how bacteria participate in control of immunity and can exist in a mutually beneficial relationship with cells of the mammalian immune system.
Figure 1. S. epidermidis Has Several Beneficial Roles in Immune Defense System of the Skin and Interacts with a Complex Innate Cellular Defense System.
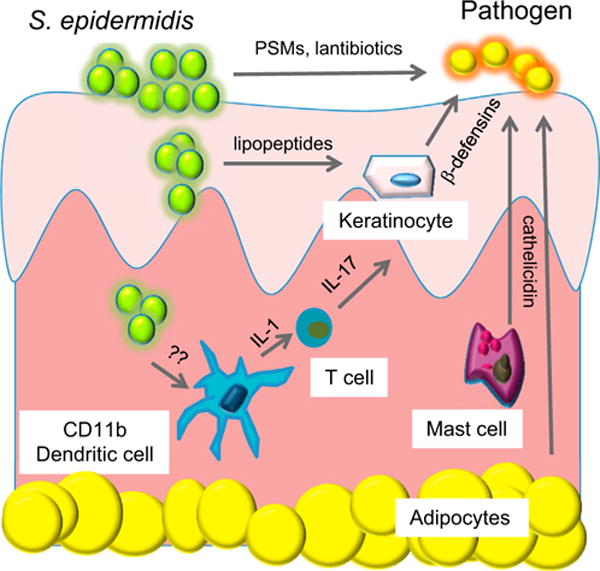
Specific strains of S. epidermidis directly produce antimicrobials including lantibiotics and phenol soluble modulins (PSMs). Lipopeptides produced by S. epidermidis can activate epithelial keratinocytes to increase production of antimicrobial peptides such as β-defensins. S. epidermidis can be detected by dermal dendritic cells to enhance IL-1 production and expansion of T cells producing IL-17. Other resident cells in the skin, including mast cells and adipocytes, produce antimicrobial peptides such as cathelicidin to kill pathogens.
References
- Cogen AL, Nizet V, Gallo RL. Br J Dermatol. 2008;158:442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogen AL, Yamasaki K, Sanchez KM, Dorschner RA, Lai Y, MacLeod DT, Torpey JW, Otto M, Nizet V, Kim JE, Gallo RL. J Invest Dermatol. 2010;130:192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, Ryan AF, Di Nardo A, Gallo RL. J Invest Dermatol. 2010;130:2211–2221. doi: 10.1038/jid.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, Wu ZR, Hooper LV, Schmidt RR, von Aulock S, et al. Nat Med. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Lei H, Li Z, Li H, Wang Y, Lai Y. PLoS ONE. 2013;8:e58288. doi: 10.1371/journal.pone.0058288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, et al. Nature. 2015 doi: 10.1038/nature14052. http://dx.doi.org/10.1038/nature14052. [DOI] [PMC free article] [PubMed]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Muñoz-Planillo R, Hasegawa M, Villaruz AE, Cheung GY, McGavin MJ, Travers JB, et al. Nature. 2013;503:397–401. doi: 10.1038/nature12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. Nat Commun. 2013;4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanke I, Steffen H, Christ C, Krismer B, Götz F, Peschel A, Schaller M, Schittek B. J Invest Dermatol. 2011;131:382–390. doi: 10.1038/jid.2010.328. [DOI] [PubMed] [Google Scholar]


