Abstract
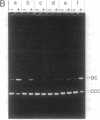
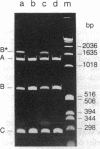
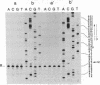
The IncP antibiotic-resistance plasmids transfer to a broad range of bacterial species. The RK2 origin of DNA transfer (oriT) consists of a 250-base-pair segment including the single-stranded cleavage site (nic) needed to generate the DNA strand believed to be transferred. Deletion derivatives and a bank of hydroxylamine-generated oriT mutants were screened for loss of transferability. DNA regions flanking both sides of nic are required for optimal transfer of the oriT clone. Of the chemically induced mutants, critical base-pair changes that dramatically reduced transfer frequency were found in a 10-base-pair region adjacent to nic. Relaxation (nicking) assays performed with these point mutants using protein-DNA complexes reconstituted in vitro revealed a correlation between DNA nicking and transfer frequency. Base-pair changes within the proximal arm of an inverted repeat upstream from the nick site resulted in reduced binding of the essential transfer protein TraJ and correspondingly reduced transfer frequencies. The results support a model of relaxosome formation involving at least two essential proteins: TraI and TraJ. The nick region defined by the point mutants was located in a segment known to be nearly identical in the related plasmid R751. This sequence was also found to be highly conserved in both border junctions of the transfer DNA (T-DNA) of plant tumor-inducing plasmids of Agrobacterium tumefaciens, indicating a relationship between IncP-mediated broad-host-range bacterial conjugation and T-DNA transfer to plants.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albright L. M., Yanofsky M. F., Leroux B., Ma D. Q., Nester E. W. Processing of the T-DNA of Agrobacterium tumefaciens generates border nicks and linear, single-stranded T-DNA. J Bacteriol. 1987 Mar;169(3):1046–1055. doi: 10.1128/jb.169.3.1046-1055.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boe L., Gros M. F., te Riele H., Ehrlich S. D., Gruss A. Replication origins of single-stranded-DNA plasmid pUB110. J Bacteriol. 1989 Jun;171(6):3366–3372. doi: 10.1128/jb.171.6.3366-3372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch M. A., Meyer R. J. A 38 base-pair segment of DNA is required in cis for conjugative mobilization of broad host-range plasmid R1162. J Mol Biol. 1987 Dec 5;198(3):361–369. doi: 10.1016/0022-2836(87)90286-5. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Dürrenberger F., Crameri A., Hohn B., Koukolíková-Nicola Z. Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9154–9158. doi: 10.1073/pnas.86.23.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Ziegelin G., Kröger M., Lanka E. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1771–1775. doi: 10.1073/pnas.86.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Deiss C., Simnad V. Location of the relaxation complex nick site within the minimal origin of transfer region of RK2. Plasmid. 1988 Nov;20(3):259–265. doi: 10.1016/0147-619x(88)90032-7. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Hasegawa P., Davis C. E. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7203–7206. doi: 10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Helinski D. R. The DNA-protein relaxation complex of the plasmid RK2: location of the site-specific nick in the region of the proposed origin of transfer. Mol Gen Genet. 1979 Oct 3;176(2):183–189. doi: 10.1007/BF00273212. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Yakobson E. Location and nucleotide sequence of the transfer origin of the broad host range plasmid RK2. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3595–3598. doi: 10.1073/pnas.80.12.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Heinemann J. A., Sprague G. F., Jr Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989 Jul 20;340(6230):205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- Jen G. C., Chilton M. D. The right border region of pTiT37 T-DNA is intrinsically more active than the left border region in promoting T-DNA transformation. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3895–3899. doi: 10.1073/pnas.83.11.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Balzer D., Kruft V., Lurz R., Lanka E. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6555–6559. doi: 10.1073/pnas.87.17.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Ziegelin G., Lanka E. Covalent association of the traI gene product of plasmid RP4 with the 5'-terminal nucleotide at the relaxation nick site. J Biol Chem. 1990 Jun 25;265(18):10637–10644. [PubMed] [Google Scholar]
- Pansegrau W., Ziegelin G., Lanka E. The origin of conjugative IncP plasmid transfer: interaction with plasmid-encoded products and the nucleotide sequence at the relaxation site. Biochim Biophys Acta. 1988 Dec 20;951(2-3):365–374. doi: 10.1016/0167-4781(88)90108-x. [DOI] [PubMed] [Google Scholar]
- Peralta E. G., Hellmiss R., Ream W. Overdrive, a T-DNA transmission enhancer on the A. tumefaciens tumour-inducing plasmid. EMBO J. 1986 Jun;5(6):1137–1142. doi: 10.1002/j.1460-2075.1986.tb04338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Völker T. A., Showe M. K. Induction of mutations in specific genes of bacteriophage T4 using cloned restriction fragments and marker rescue. Mol Gen Genet. 1980 Feb;177(3):447–452. doi: 10.1007/BF00271483. [DOI] [PubMed] [Google Scholar]
- Wang K., Stachel S. E., Timmerman B., VAN Montagu M., Zambryski P. C. Site-Specific Nick in the T-DNA Border Sequence as a Result of Agrobacterium vir Gene Expression. Science. 1987 Jan 30;235(4788):587–591. doi: 10.1126/science.235.4788.587. [DOI] [PubMed] [Google Scholar]
- Willetts N., Crowther C. Mobilization of the non-conjugative IncQ plasmid RSF1010. Genet Res. 1981 Jun;37(3):311–316. doi: 10.1017/s0016672300020310. [DOI] [PubMed] [Google Scholar]
- Zambryski P. Basic processes underlying Agrobacterium-mediated DNA transfer to plant cells. Annu Rev Genet. 1988;22:1–30. doi: 10.1146/annurev.ge.22.120188.000245. [DOI] [PubMed] [Google Scholar]
- Ziegelin G., Fürste J. P., Lanka E. TraJ protein of plasmid RP4 binds to a 19-base pair invert sequence repetition within the transfer origin. J Biol Chem. 1989 Jul 15;264(20):11989–11994. [PubMed] [Google Scholar]
- van Haaren M. J., Sedee N. J., de Boer H. A., Schilperoort R. A., Hooykaas P. J. Mutational analysis of the conserved domains of a T-region border repeat of Agrobacterium tumefaciens. Plant Mol Biol. 1989 Nov;13(5):523–531. doi: 10.1007/BF00027312. [DOI] [PubMed] [Google Scholar]