SUMMARY
The design of immunogens that elicit broadly reactive neutralizing antibodies (bnAbs) has been a major obstacle to HIV-1 vaccine development. One approach to assess potential immunogens is to use mice expressing precursors of human bnAbs as vaccination models. The bnAbs of the VRC01-class derive from the IGHV1-2 immunoglobulin heavy chain and neutralize a wide spectrum of HIV-1 strains via targeting the CD4 binding site of the envelope glycoprotein gp120. We now describe a mouse vaccination model that allows a germline human IGHV1-2*02 segment to undergo normal V(D)J recombination and, thereby, leads to the generation of peripheral B cells that express a highly diverse repertoire of VRC01-related receptors. When sequentially immunized with modified gp120 glycoproteins designed to engage VRC01 germline and intermediate antibodies, IGHV1-2*02-rearranging mice, which also express a VRC01-antibody precursor light chain, can support the affinity maturation of VRC01 precursor antibodies into HIV-neutralizing antibody lineages.
Short Paragraph/eTOC
Sequential immunization using modified HIV envelope glycoproteins supports the maturation of HIV-neutralizing antibody lineages in mouse models that have diverse antibody repertoire and therefore are closer to physiological immunization settings than conventional transgenic models.
INTRODUCTION
A major hurdle to the development of an effective HIV-1 vaccine has been the difficulty in designing vaccine immunogens, and immunization strategies, that induce antibodies able to neutralize the majority of diverse HIV-1 strains. Cross-reactive neutralizing antibodies arise over several years in about half of HIV-1 infected subjects, and highly potent broadly cross-reactive antibodies (bnAbs) arise in a subset of this group. Their broad neutralizing activity stems from their ability to recognize relatively conserved epitopes on the HIV-1 envelope (Env) protein and to accommodate natural variations within such epitopes (Burton and Hangartner, 2016; Kwong and Mascola, 2012; Mascola and Haynes, 2013). The VRC01-class of bnAbs targets the CD4-binding site of HIV-1 (Scheid et al., 2011; Wu et al., 2010; Zhou et al., 2010). Because the CD4-binding site is essential for HIV-1 entry into host T cells, this region of HIV-1 envelope protein (Env) is conserved despite the overall diversity of HIV-1 strains. Given its broad and potent neutralization activity, induction of VRC01-class bnAbs is a prime goal for vaccine development.
VRC01-class antibodies share common characteristics (Zhou et al., 2010; Zhou et al., 2015; Zhou et al., 2013). The immunoglobulin (Ig) heavy (IgH) chain variable region is encoded by an exon assembled from different germline VH, D and JH gene segments during B cell development (Alt et al., 2013). Among the 38–46 functional VH segments in the human IgH locus, IGHV1-2 is used exclusively in VRC01-class antibodies, because IGHV1-2 can act as a structural mimic of CD4 (Zhou et al., 2010). VRC01 Ig light (IgL) chains make less important contributions to antigen contact, and different IgL chains are compatible with VRC01 binding activity (Wu et al., 2011). However, all VRC01 light chains have a short complementarity-determining region 3 (CDR3) antigen-binding loop, which is encoded by the junctional region of V and J gene segments. VRC01 IgL chain CDR3s (CDR L3s) are invariably 5-amino acid long, which is far below the average human CDR L3 length. The short VRC01 CDR L3 avoids steric clashes between VRC01 and the Env gp120 V5 loop (Zhou et al., 2010; Zhou et al., 2015; Zhou et al., 2013). A third remarkable feature of VRC01-class antibodies is that up to 32% of their variable region exon sequences arise via somatic hypermutation (SHM) (Scheid et al., 2011; Wu et al., 2010; Wu et al., 2011), a process that takes place in activated B cells within germinal centers in peripheral lymphoid tissues and improves antigen-binding affinity (Victora and Nussenzweig, 2012).
Given the unusual VRC01-class characteristics, inducing such antibodies by vaccination is a daunting task. The immunogen must engage B cells that express a precursor antibody (termed “umutated common ancestor” or “UCA”) composed of an unmutated IGHV1-2 heavy chain and an IgL chain with a 5-amino acid CDR L3; given the low frequency of 5-amino acid CDR L3s such B cells are expected to be very rare (Jardine et al., 2015; Jardine et al., 2016). In contrast to the broad and potent neutralization activity of mature VRC01-class antibodies, the inferred VRC01 UCA does not interact appreciably with gp120 or the native Env trimer (Hoot et al., 2013; McGuire et al., 2013; Zhou et al., 2010). Thus, it has been proposed that induction of such bnAbs would require targeting with an immunogen that binds the UCA with sufficient affinity to prime the corresponding B cells, followed by subsequent sequential immunizations with modified Env-based proteins designed to engage VRC01 intermediates that stimulate SHM and affinity maturation into bnAbs (Haynes et al., 2012; Jardine et al., 2013; Liao et al., 2009; McGuire et al., 2013; Xiao et al., 2009).
Based on the above considerations, HIV gp120 has been engineered into forms that interact with germline VRC01-class antibodies (Jardine et al., 2013; Jardine et al., 2016; McGuire et al., 2016; McGuire et al., 2013). The efficacy of these germline VRC01 binders has been evaluated in immunization experiments in mouse models where the precursor form of VRC01 IgH or IgL genes have been integrated into the corresponding mouse Ig loci (Dosenovic et al., 2015; Jardine et al., 2015; McGuire et al., 2016). In such “knock-in” mouse models, B cells express the rearranged VRC01 IgH or IgL precursor antibody and serve as target cells for test immunogens. It has been shown that a high affinity germline VRC01 binder, eOD-GT8 60mer (Jardine et al., 2015), selectively activates B cells expressing a V(D)J exon consisting of a germline IGHV1-2*02 fused to a CDR H3 sequence from a mature VRC01-class antibody (Dosenovic et al., 2015; Jardine et al., 2015). Moreover, eOD-GT8 60mer immunization expanded B cells that were enriched for antibodies in which the human IgH chain was paired with mouse IgL chains containing 5-amino acid CDR L3s. However, since the human VRC01 IgH chain employed in this knock-in model contained a CDR H3 from a mature VRC01 antibody, the response did not arise from germline variable regions. Moreover, IGHV1-2*02 would be associated with diverse CDR H3s in a human population; thus, an effective vaccine would need to function in such a context. Finally, no prior immunization studies with engineered gp120 immunogens have elicited HIV-1 neutralizing activities (Dosenovic et al., 2015; Jardine et al., 2015; McGuire et al., 2016).
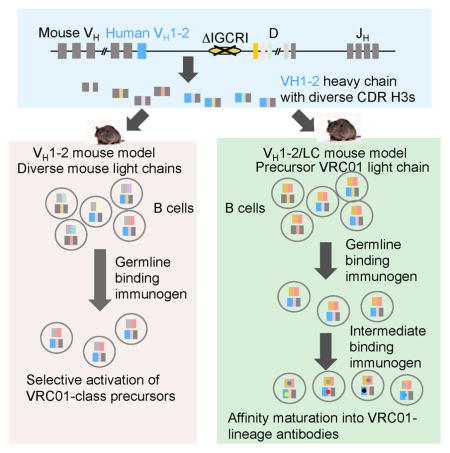
To circumvent some potential shortcomings of standard knock-in models, we developed VRC01 vaccine models based on findings from our prior studies of V(D)J recombination regulation. Although all mouse VHs are represented at varying frequencies in the VH(D)JH repertoire of mature B lymphocytes, the most D-proximal VH (“VH81X”, IGHV5-2 in IMGT nomenclature) is utilized frequently for primary VH(D)JH rearrangements. Nevertheless, VH81X rarely contributes to antibodies expressed by peripheral B cells, due to inability to pair properly with IgL or surrogate IgL chains and a propensity to encode auto-reactive antibodies (Alt et al., 2013). Rearrangement of VH81X is under the control of a major V(D)J recombination regulatory element, termed intergenic control region 1 (IGCR1) (Guo et al., 2011). When IGCR1 is inactivated, VH81X is used in the vast majority of VH to DJH rearrangements, despite integrity of the remaining IgH locus (Guo et al., 2011; Hu et al., 2015). Based on these observations, we hypothesized that, in place of VH81X, the human IGHV1-2*02 would similarly dominate VH usage in the context of IGCRI deletion. Moreover, as IGHV1-2*02 is well represented in the human antibody repertoire (DeKosky et al., 2016; Lin et al., 2016), it may not be subject to negative selection, allowing high representation in the primary VH repertoire to translate into prevalent expression in mature B cells. In such a model, IGHV-1-2*02 would recombine with various mouse D and JH segments, with junctional diversification mechanisms (Alt et al., 2013) creating a diverse range of CDR H3’s with this single human VH. Relative to mouse models where IGHV1-2*02 is linked to a fixed CDR H3, such a model could provide a more physiological setting to test the efficacy of candidate immunogens.
We now report that, when subjected to a sequential immunization strategy, these VH1-2*02-rearranging vaccination mouse models allow maturation of VRC01 precursor antibodies into HIV-1 neutralizing antibody lineages.
RESULTS
A mouse model that expresses IGHV1-2*02 in association with diverse CDR3s
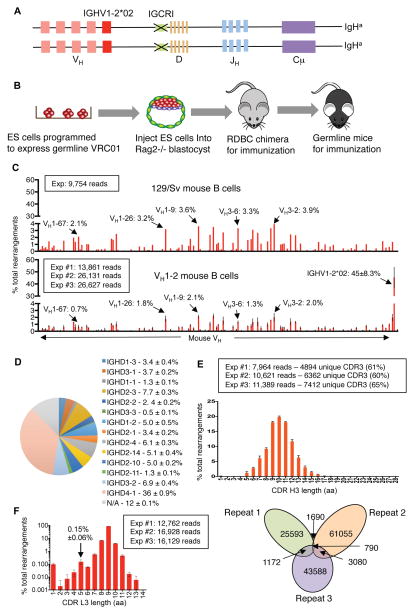
We replaced VH81X with IGHV1-2*02 and deleted IGCRI on one IgH allele in mouse embryonic stem (ES) cells (Figure 1A, Figure S1A and B). To assay these ES cells, we employed Rag2-deficient blastocyst complementation (“RDBC”), which involves injecting test ES cells into Rag2-deficient blastocysts to generate chimeric mice (Chen et al., 1993). Because Rag2 is essential for V(D)J recombination, all B and T cells in the chimeric mice originate from the injected ES cells (Figure 1B). Therefore, such RDBC chimeras can be used directly for immunization experiments. To maximize the frequency of B cells expressing IGHV1-2*02 for some experiments, we also bred RDBC chimeras for germline transmission and generation of mice homozygous for the IGHV1-2*02 replacement and IGCRI deletion mutations (Figure 1A, Figure S1A and B). We refer to these RDBC chimeras and germline mice as the VH1-2 mouse model.
Figure 1. Generation and characterization of VH1-2 mouse model.
(A) Illustration of genetic modifications in the VH1-2 mouse mode (see text, Figure S1A and S1B for details). (B) Illustration of Rag2 deficient blastocyst complementation (See text for details). (C) HTGTS-rep-seq analysis of VH usage in VH1-2 mouse model and in control 129/Sv mouse. The X-axis represents VH locus from the distal to the D-proximal ends; a subset of mouse VHs are labeled for comparison between 129/Sv control and VH1-2 mice. The histogram displays the percent usage of each VH of all productive VH(D)JH rearrangements. Data for the VH1-2 mouse model were average of three experiments with error bars representing standard deviations; data for control 129/Sv mouse is consistent with prior studies (Lin et al. 2016). (D) Pie chart Illustration of D segment usage in productive IGHV1-2*02 rearrangements in VH1-2 mouse model, average D usage frequency with standard deviation for three biological repeats is shown in adjacent panel. (E) Length distribution of IGHV1-2*02 CDR H3 in VH1-2 mouse model derived from data in (C). The number of total reads and unique reads for IGHV1-2*02 associated CDR H3’s are shown and Venn diagram, which reveals tremendous CDR3 complexity since there is little overlap in CDR H3 sequences in three technical repeats of a single B cell sample (see Lin et al. 2016). (F) Length distribution of CDR L3 of mouse IgL chains in VH1-2 mice based on three biological replicates with error bars displayed and frequency of 5-amino acid CDR L3 indicated. Other details are in Methods.
Based on FACS analysis of the expression of several cell surface markers, splenic B and T cell populations of VH1-2 mice appeared comparable to control mice (Figure S1C). To assess frequency of IGHV1-2*02 and D usage, as well as CDR3 length and diversity among peripheral B cells in the VH1-2 model, we employed the recently described HTGTS-rep-seq method (Lin et al., 2016). This assay revealed that 45% of splenic B cells express IGHV1-2*02 heavy chains (Figure 1C). Moreover, IGHV1-2*02-expressing B cells employed all mouse D segments, but with a higher frequency of DQ52 (IGHD4-1 in IMGT nomenclature) and a shorter peak CDRH3 length relative to B cells in normal mice (10 versus 11 amino acids) (Figure 1D and E)(Lin et al., 2016). Due to use of normal junctional diversification mechanisms in their assembly, IGHV1-2*02 heavy chain variable regions in VH1-2 mice collectively contain a tremendously diverse range of CDR H3 sequences (Figure 1D and E, bottom)(Lin et al., 2016).
We did not introduce the VRC01 IgL chain into the VH1-2 mouse model, because the model was designed to test whether a high affinity antigen for the germline VRC01 antibody could engage B cells in which diverse IGHV1-2*02-containing V(D)J IgH chains were paired with different mouse IgL chains containing 5-amino acid CDR L3s. This model could better approximate the challenge of germline-targeting in humans compared to the knock-in mice with a V(D)J IgH chain with a single CDR H3 used in prior tested models. We also used HTGTS-rep-seq to assess the frequency of Igκ chains with 5-amino acid CDR L3s in VH1-2 model B cells. As in normal mice (Lin et al., 2016), the majority VH1-2 model Igκ variable region exons encoded a 9-amino acid long CDR3, with 5-amino acid CDR L3s found in only about 0.15% of expressed Igκ chains (Figure 1F). Thus, the VH1-2 mouse model provided a stringent setting to evaluate ability of candidate immunogens to selectively prime B cells expressing antibodies with VRC01 signatures.
Selective activation of B cells expressing VRC01 type antibodies
A wild-type gp120 or Env trimers would not be effective antigens for immunization experiments with the VH1-2 model since they do not interact appreciably with germline VRC01 antibody (Hoot et al., 2013; McGuire et al., 2013; Zhou et al., 2010). Therefore, we employed a recently described gp120-based immunogen, referred to as engineered outer domain-GT8 (eOD-GT8), which has been engineered to bind with high affinity to germline VRC01-class antibodies (Jardine et al., 2016). This protein was expressed as part of a 60-subunit self-assembling nanoparticle (60mer). Based on binding assays, eOD-GT8 60mer stably binds to predicted germline precursors from different donors with diverse CDR H3s and IgL chains.
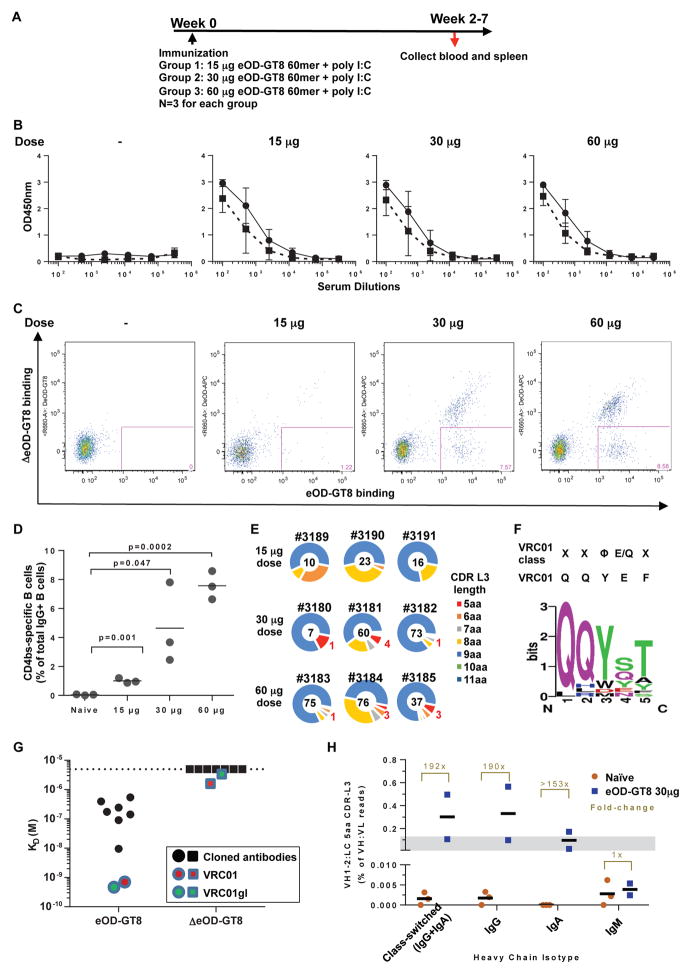
We immunized the VH1-2 mouse model with 15, 30, or 60 μg of eOD-GT8 60mer along with poly I:C adjuvant (Figure 2A). The immunization appeared to elicit antibodies that target the CD4 binding site, as demonstrated by the detection of higher titers of antibody against eOD-GT8 than against ΔeOD-GT8 (the CD4bs-KO mutant) in all mice two weeks after immunization (Figure 2B). Correspondingly, the frequency of IgG+ B cells specific for the gp120 CD4 binding site increased in a dose-dependent manner (Figure 2C and 2D). Similar to immunization studies with germline VRC01 heavy chain knock-in mouse models (Dosenovic et al., 2015; Jardine et al., 2015), a substantial fraction of the immune response was not specific for the CD4 binding site, as evidenced by binding to the ΔeOD-GT8 (Figure 2B), as well as by the population of B cells that bind equally to eOD-GT8 and ΔeOD-GT8 (Figure 2C and Figure S2A). Given that the mice were immunized only once with eOD-GT8, additional boosts with heterologous gp120 antigens may help focus the immune response toward the CD4 binding site. To isolate and characterize CD4 binding site-specific antibodies, we sorted single splenic B cells that bound eOD-GT8, but not ΔeOD-GT8 (Figure S2A) and cloned IgH and IgL chains. Among B cells sorted from mice immunized with 30μg and 60μg eOD-GT8 60mer, some expressed the IGHV1-2*02 V-gene in association with IgL chains containing a 5-amino acid CDR L3, with enrichment for the conserved VRC01-class QQY motif (Figure 2E; Figure 2F). Given the extremely low frequency of IgL chains with a 5-amino acid CDR3s in the pre-immune repertoire (Figure 1F), these results indicated strong selection for B cells encoding antibodies with VRC01 IgL signatures.
Figure 2. Elicitation of VRC01-class germline antibodies in VH1-2 mouse model.
(A) Immunization Schema. (B) Sera binding to eOD-GT8 (solid line) and its CD4bs-knock-out (Δ) mutant (dashed line) assessed by ELISA (mean with standard deviation is shown). (C, D) Sorting and frequency of CD4bs-specific eOD-GT8+/ΔeOD-GT8− splenic IgG+ B-cells. Statistical comparisons were performed using a two-tailed t-test. (E) Distribution of CDR L3 amino acid (aa) length of the cloned VH1-2 antibodies from CD4bs-specific B-cells. Mouse IDs are shown on top of each pie chart and the number of total sequences is shown at the center. VRC01-class germline antibodies were defined as antibodies containing a VH1-2 heavy chain and a light chain with a 5aa CDR L3 (in red). (F) Sequence conservation at each position of the 5aa CDR L3 in the cloned VRC01-class germline antibodies compared to known VRC01-class antibodies and VRC01. (G) Binding affinity of synthesized antibodies compared to VRC01 and its germline-V-gene-revertant (VRC01gl) as assayed with Biolayer Interferometry (BLI) Octet. Dashed line indicates the detection limit at 5μM. (H) Heavy and light chain paired sequencing of the B-cell repertoire of naïve and eOD-GT8 60mer-immunized mice. The bar represents the mean of the samples in C and H.
Among mouse IgL chains, certain Vκ4 family members were over-represented (Table S1), potentially because these V segments encode the QQY motif conserved in the CDR L3 of VRC01-class antibodies. To test binding specificity of the isolated antibodies composed of IGHV1-2*02 IgH chains and mouse IgL chains with 5-amino acid CDR L3s, we produced them as recombinant antibodies. All of these antibodies were highly specific for a functional CD4 binding site, based on binding affinities to eOD-GT8 and ΔeOD-GT8 (Figure 2G and Figure S2B). However, none of these antibodies, which resulted from a single immunization, exhibited HIV neutralizing activity (Duan, H, Cheng, C., Chen, X., and Mascola, J., unpublished data). Of note, among the isolated VRC01 type antibodies, IGHV1-2*02 is associated with different CDR H3’s (Table S1). In contrast, when we immunized VH1-2 mice with immunogens that had lower germline-binding affinity to VRC01-class antibodies than eOD-GT8 60mer, including eOD-GT6 60mer, 426c-Ferritin and C13-Ferritin particles, not a single IgL chain with 5aa CDR L3 was amplified from 473 sorted CD4bs-specific B cells (Table S2). Together, these results indicate that it is feasible to selectively activate B cells expressing diverse precursors of VRC01 type antibodies using a high affinity germline VRC01 binder.
To complement the single-cell based analysis, we used a high throughput paired sequencing method (DeKosky et al., 2015) to assess the effect of immunization on the enrichment of IgL chain transcripts with 5-amino acid CDR L3s at the population level. These analyses demonstrated that immunization with eOD-GT8 60mer substantially elevated the frequencies of 5-amino acid CDR L3 IgL chains paired with IGHV1-2*02 IgH chains. The increase was observed only in B cells that had undergone class switching to IgG or IgA, indicating the enrichment was a consequence of B cell activation (Figure 2H; Figure S2C and Table S3). In conjunction with single-cell analyses described above, these results suggest that eOD-GT8 60mer can serve as an effective priming immunogen to elicit VRC01-type antibodies, even in the context of complex IgH and IgL chain repertoires.
A mouse model with diverse IGHV1-2*02 IgH chains and a VRC01 precursor IgL chain
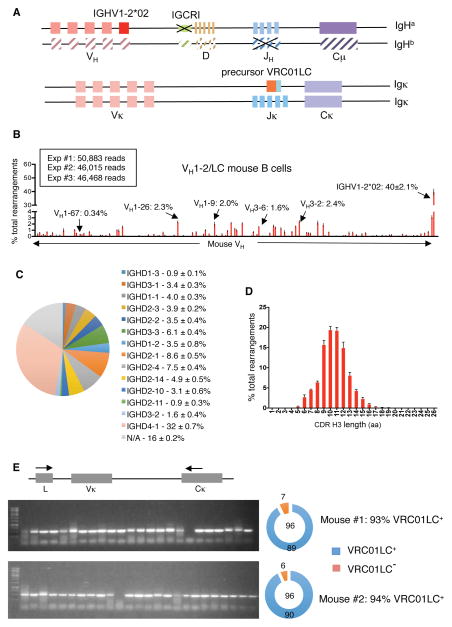
The paucity of IgL chains with 5-amino acid CDR L3s would lead to a similarly low frequency of potential VRC01 precursors in VH1-2 mice, making it difficult to test the efficacy of immunogens to mature precursor antibodies. Mouse Vκ segments also may not provide an optimal context for affinity maturation of IGHV1-2*02. Thus, we generated a related, but distinct, mouse model to facilitate VRC01 affinity maturation studies by integrating a rearranged version of a VRC01 Igκ variable region exon into the mouse Jκ locus of IGHV1-2*02/ΔIGCRI ES cells (Figure 3A, Figure S3A and B). This Igκ variable region exon is composed of germline human IGKV3-20*01, which is used in many VRC01-class antibodies, and includes a 5-amino acid CDR L3 derived from the mature VRC01 antibody (Figure S3B). The use of a CDR L3 from the mature VRC01 antibody was based on two considerations. First, the mature CDR L3 is not only 5-amino acids long, but also contains an E residue that is conserved in VRC01-class antibodies (Zhou et al., 2013; Zhou et al., 2015). Second, the CDR L3 of the bona fide VRC01 UCA is unknown. Thus, the VRC01 IgL chain in this second mouse model is essentially a hybrid of germline IGKV3-20 and the mature CDR L3 containing human Jκ1. We refer to this IgL chain as the “precursor VRC01 IgL”. We also deleted the JH region from the IgH allele that does not contain IGHV1-2*02, thereby limiting IgH chain expression to the IGHV1-2*02/ΔIGCR1 allele (Figure 3A, Figure S3A and B). We refer to these multiply modified ES cells as VH1-2/LC ES cells.
Figure 3. Generation and characterization of VH1-2/LC model.
(A) Illustration of genetic modifications involved in the VH1-2/LC model. See text, Figure S3A and S3B for details. (B) HTGTS-rep-seq analysis of VH usage in splenic B cells from VH1-2/LC mice performed as in Fig. 1C. (C) D segment usage in productive IGHV1-2*02 rearrangements in VH1-2/LC mouse model. (D) Length distribution of IGHV1-2*02-associated CDR H3s in VH1-2/LC mouse model. (E) Single cell analysis of VRC01 IgL chain expression in splenic B cells of VH1-2/LC mouse model. Primers for VRC01LC cDNA in the leader exon (L) of IGKV3-20*01 and the Cκ exon are represented by arrows. Gel images are representative of results from two different VH1-2/LC mice. Numbers of VRC01LC+ and VRC01LC− cells are indicated on the pie chart. Other details are in Experimental Methods.
We employed VH1-2/LC ES cells for RDBC and used the chimeras for the next sets of immunization experiments. In this case, use of RDBC chimeras was advantageous over conventional germline breeding, during which the genetic modifications on three different chromosomes would independently segregate. Similar to VH1-2 mice, splenic lymphocyte populations of VH1-2/LC mice appeared normal based on expression of several cell surface markers (Figure S1C). HTGTS-rep-seq analysis revealed that, in VH1-2/LC mice, approximately 40% of splenic B cells express IGHV1-2*02 IgH chains (Figure 3B). Moreover, due to junctional diversification mechanisms, the IGHV1-2*02 segments in these IgH chains were associated with CDR H3s that employed all D segments, covered a broad length range, and were highly diverse (Figure 3C and D). HTGTS-rep-seq also revealed that that the precursor VRC01 IgL chain dominated the Igκ repertoire of VH1-2/LC B cells (Figure S3C). Finally, single-cell RT-PCR showed that 94% IgM+IgDhi mature splenic B cells expressed immature VRC01 Igκ chains (Figure 3E). Thus, the knock-in precursor VRC01 IgL effectively excluded the rearrangement and expression of endogenous mouse IgL. We conclude that the VH1-2/LC model contains a large and diverse pool of precursor VRC01 IgH- and IgL-expressing B cells that could serve as potential targets for immunization.
Induction of neutralizing antibodies through stepwise immunization
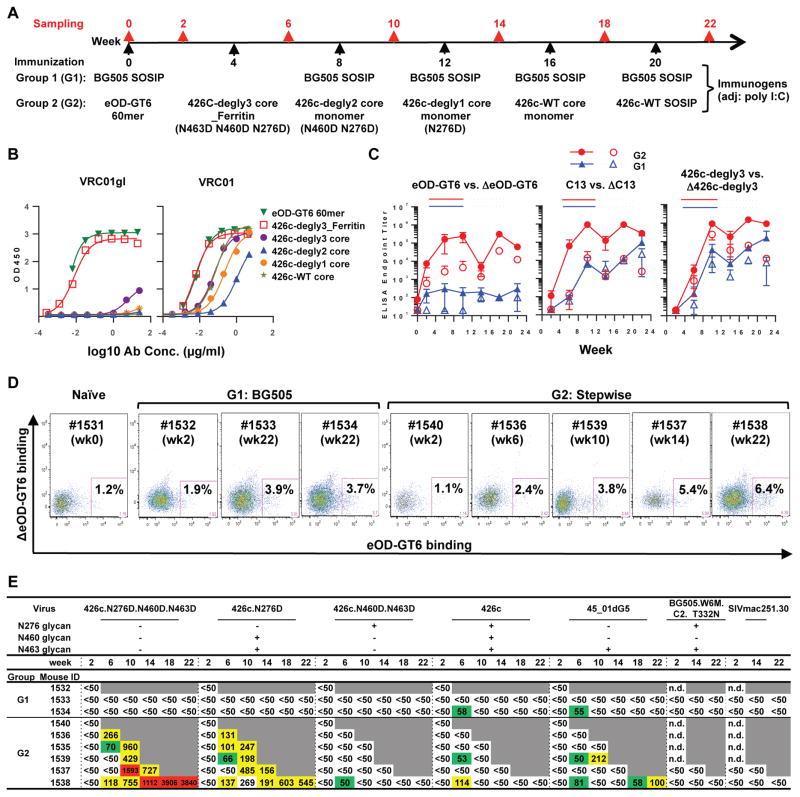
We employed stepwise immunization to help guide affinity maturation of VH1-2/LC B cells towards the end point of neutralizing VRC01-class antibodies (Figure 4A). The overall strategy was to prime the immune response with high affinity germline VRC01 binder and boost with a series of immunogens displaying progressively decreased affinity toward germline relative to mature VRC01 antibodies. Specifically, we primed with eOD-GT6 60mer, which was available and is closely related to eOD-GT8 60mer, but has weaker and less broad reactivity to VRC01-class inferred precursors (Jardine et al., 2016). In subsequent boosts we used a modified gp120 core protein derived from HIV-1 strain 426c (McGuire et al., 2013). For the first boost, we used 426c degly-3_Ferritin; in this 24mer nanoparticle form of gp120 core, three N-linked glycosylation sites near the CD4 binding site, which interfere with the binding of germline VRC01 antibody, were mutated and the variable domains 1,2, and 3 were deleted (McGuire et al., 2016). Although eOD-GT6 60mer and 426c degly-3_Ferrritin are both designed to bind germline VRC01-class antibodies, they are derived from different gp120 backbones, namely the gp120s from clade B and clade C viruses, respectively. In addition, relative to eOD-GT6, a minimal outer domain of gp120, the 426c degly-3 core contains both the outer and inner domains, and is, thus, closer to the native form of gp120. Sequential immunization with these two antigens may help focus the immune response toward the conserved CD4 binding site. The effect of glycosylation at N276, N460 and N463 on germline VRC01 binding is well-characterized for the 426c gp120 (McGuire et al., 2013). In subsequent boost immunizations, we used 426c gp120 core monomers retaining one, two or three of these glycosylation sites. With the gradual restoration of glycosylation sites, these antigens display decreasing affinity for the germline VRC01-class antibodies, but retain similar binding to fully matured VRC01-class antibodies (Figure 4B). Our final boost used a native trimer form of 426c (426c-WT SOSIP). By immunizing with this series of antigens, we aimed to select for antibodies with sufficient affinity maturation to recognize more native forms of gp120. For comparison, we also immunized VH1-2/LC mice with BG505 SOSIP (Sanders et al., 2013), a stabilized native Env trimer that interacts poorly with germline VRC01-class antibodies.
Figure 4. Stepwise immunization of the VH1-2/LC mice elicited VRC01-class CD4bs-specific antibodies with neutralizing activity.
(A) Immunization Schema. (B) Characterization of immunogens (varied symbols) for ELISA binding to VRC01 and its germline reverted variant (VRC01gl). (C) ELISA endpoint titers of post-immune sera to three germline-binding VRC01-class probes and their respective CD4bs-knock out (Δ) mutants. Error bars indicate standard deviations. (D) Splenocytes were sorted with eOD-GT6 and ΔeOD-GT6 probes to isolate CD4bs-specific IgG+ B cells. (E) Neutralizing activity of post-immune sera to various HIV-1 Env-pseudoviruses (ID50 titers). n.d., not determined.
We monitored the immune response by serum ELISA (Figure 4C). In mice treated with stepwise immunization we detected higher titers of VRC01-class antibodies against engineered versions of gp120 outer domain or core with a functional CD4 binding site (eOD-GT6, C13, 426c-dgly3) relative to gp120 with a defective CD4 binding site (ΔeOD-GT6, ΔC13, Δ426-degly3), suggesting that stepwise immunization elicited antibodies specific for the CD4 binding site of gp120. By comparison, immunization with BG505 SOSIP induced lower overall antibody titers for gp120 (Figure 4C). Similarly, flow cytometry revealed that immunization induced B cells that appeared to target the CD4 binding site (Figure 4D). Comparing the two immunization protocols, stepwise immunization yielded more CD4bs-specific B cells with overall higher eOD-GT6 staining signals. In parallel with the induction of CD4bs-specific antibodies, serum from stepwise immunized mice exhibited neutralizing activities against 426c viruses that lack all three (N276D, N460D, N463D) or lacked only the N276 glycosylation (N276D) sites (Figure 4E). Additionally, the immunized serum showed neutralization activity against a wild type HIV-1 strain that was not part of the immunization scheme, 45_01dG5--a virus naturally lacking the N276 glycan (Wu et al., 2012), indicating some heterologous neutralization of elicited antibodies. On the other hand, the immunization did not effectively or consistently elicit antibodies that could neutralize viruses with intact N276 glycosylation (i.e., 426c and 426c.N460D.N463D, BG505)(Figure 4E). Comparing the two immunization methods, the stepwise immunization scheme was superior to immunization with BG505 SOSIP, which failed to elicit detectable serum neutralization activities, as observed previously in related knock-in mouse models (Dosenovic et al., 2015; Jardine et al., 2015).
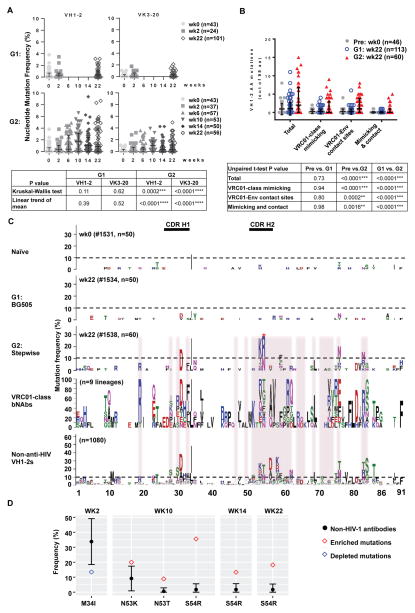
To characterize induced antibodies, we sorted eOD-GT6-specific B cells and cloned IgH and IgL chain pairs from individual B cells (Figure S2A). We focused our analysis on antibodies composed of IGHV1-2*02 heavy chain and IGKV3-20*01 light chains (Table S4), in particular their somatic hypermutation, binding, and neutralization activities. Over the course of 22 weeks of stepwise immunization, the mutation frequency of both the IGHV1-2*02 IgH and VRC01 IgL chains steadily increased, reaching up to 9% and 5%, respectively (Figure 5A). Moreover, the mutation patterns of IGHV1-2*02 after 22 weeks of immunization showed overlap with those of mature VRC01-class antibodies, and there was statistically significant enrichment for mutations at VRC01-Env contact sites (Figure 5B, C, D, Figure S4, S6). As for VRC01 IgL chain, somatic hypermutations appeared primarily focused on CDR L1 of IGKV3-20; a pattern that may be significant in that the mature VRC01 IgL chain contains deletions in CDR L1 (Figure S5). Some of the abundant mutations may reflect intrinsic mutation hotspots of IGVH1-2*02 and substitution bias (for example, G31D and M34I), as they also frequently occur in non-HIV-1 antibodies (Figure 5C). As expected from the poor immune response to BG505, both IGHV1-2*02 IgH chain and IGKV3-20*01 IgL chain accumulated minimal levels of mutation upon BG505 immunization (Figure 5B, C and S4 and 5).
Figure 5. Stepwise immunization of VH1-2/LC mice elicited VRC01-class CD4 binding site-specific antibodies with increased SHM.
(A) Nucleotide mutation frequency in paired VH1-2 and VK3-20 genes, derived from CD4bs-specific IgG+ B cells of the BG505-immunized mice (G1) and stepwise-immunized mice (G2). Each dot represents one VH1-2 or VK3-20 chain in a VH1-2/VK3-20 paired antibody (n, number of VH1-2/VK3-20 pairs in each animal at indicated time points). The median with interquartile range is plotted. (B) Numbers of total, VRC01-class mimicking, Env-contacting, and both mimicking and contacting amino acid mutations in each VH1-2 HC amplified from specified mice plotted with median and interquartile range and statistically assessed using an unpaired t-tests. VRC01-class mimicking mutations are present in at least two of 9 published VRC01-class bnAb lineages. Env-contacting sites are based on VRC01-gp120 or gp160 crystal structures. (C) Amino acid mutations in all VH1-2 HCs from each specified mouse shown in sequence logo profiles. “n” is the number of all VH1-2 chains amplified from each animal. For reference, 9 published VRC01-class HC lineages are represented below panel G2. VRC01-envelope contact sites are highlighted in pink. The mutation profile of IGHV1-2 constructed from 1080 non-HIV-1 neutralizing antibody lineages (from three healthy donors) is also shown. The red asterisk marks significantly enriched S54R mutations in the G2:wk22 sample. (D) Enriched and depleted amino acid substitutions in VH1-2 cloned from the sequential immunization group G2 compared to non-HIV-1 neutralizing antibodies. The calculated mean occurrence and standard deviations of each mutation and 95% CI are depicted in black. The frequency of enriched (red diamond) or depleted (blue diamond) mutations locates outside of the calculated 95% CI.
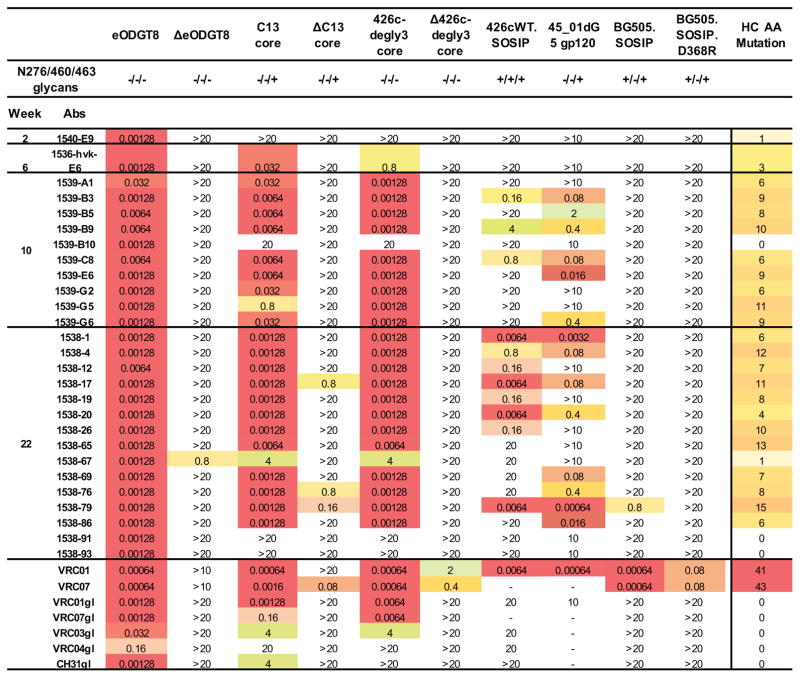
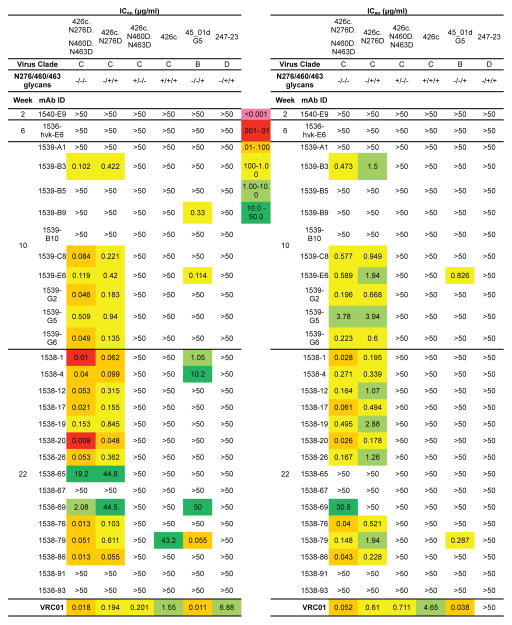
To evaluate binding and neutralization functions of cloned antibodies, we synthesized 27 pairs of IGHV1-2*02 IgH chain and IGKV3-20*01 IgL chain as recombinant monoclonal antibodies (Figure S6). We first tested binding affinities toward various forms of gp120 (Figure 6). Since these antibodies were selected by eOD-GT6, most showed robust binding activity toward three different forms of engineered gp120 outer domain or cores that interact with germline VRC01-class antibodies (eOD-GT8, C13, 426c-degly3), and interaction was strictly dependent on a functional CD4 binding site (Figure 6; compare eOD-GT8 with ΔeOD-GT8, C13 with ΔC13, 426c with Δ426c). Some antibodies were able to bind the 426c-WT SOSIP trimer; one showed detectable binding to BG505 SOSIP. As a more stringent functional test, we performed neutralization assays (Figure 7 and Figure S7). Most of these antibodies potently neutralized a virus that lacked three glycosylation sites (426c.N276D.N460D.N463D) (Figure 7). Some antibodies exhibited substantial neutralization activity against the virus with one glycosylation site mutation (426c.N276D). Consistent with heterologous neutralization activity in sera, a few antibodies neutralized the 45_01dG5 virus, which naturally lacks a glycan at site 276. One antibody, 1538-79, exhibited weak neutralization activity of the wild-type 426c virus with all glycans intact; correspondingly, the same antibody also bound to BG505 SOSIP. Thus the cloned antibody panels largely recapitulated serum binding and neutralization activities (Figure 4E). In summary, these studies demonstrated that our stepwise immunization strategy led to substantial affinity maturation of the germline VRC01 antibody, such that some elicited antibodies recognized more native forms of HIV-1 Env protein.
Figure 6. Selected IGHV1-2*02/IGKV3-20*01 IgGs cloned from the stepwise immunized mice displayed CD4bs-specific binding to multiple Env proteins.
ELISA endpoint binding titers of 27 IGHV1-2*02/IGKV3-20*01 paired IgGs cloned from the stepwise immunized mice 1540 (wk2), 1536 (wk6), 1539 (wk10) and 1538 (wk22) to various Env antigens and their CD4bs-KO mutants (Δ) were compared to the binding titers of VRC01 class antibodies and their germlines (gl). The number of VH1-2 AA mutations in each IgG is listed in the right-most column, and they are reversely correlated with the binding titers to C13 and 426c mutants (p<0.0001).
Figure 7. Selected IGHV1-2*02/IGKV3-20*01 IgGs cloned from the stepwise immunized mice neutralized the same viruses sensitive to the corresponding mouse sera.
Neutralization IC50 and IC80 of the 27 IgGs against Env-pseudoviruses 45-01dG5, 426c, the mutant 426c viruses lacking 1, 2 or all 3 glycans at aa 276, 460 and 463, and 247-23 were listed. The presence (+) or absence (−) of N276/460/463 glycans in each pseudovirus is indicated. The neutralizing titers of VRC01 were shown as positive controls.
DISCUSSION
We have generated two types of mouse models: one designed to test immunization strategies for engaging B cells expressing VRC01-class precursor B cell receptors, and the other for evaluating their affinity maturation. Relative to conventional VRC01 transgenic mouse models, a unique aspect of our models is that IGHV1-2*02 is expressed in association with highly diverse CDR H3s, thus providing a more physiological repertoire on which to test candidate immunogens. In the first mouse model, where only the IGHV1-2*02 IgH chain V exon was introduced, immunization with a high affinity germline VRC01 binder activated B cells that express IGHV1-2*02 in association with a mouse IgL chains containing a 5-amino acid CDR L3, a VRC01-class antibody signature. In the second mouse model where both the IGHV1-2*02 IgH chain and an immature VRC01 IgL chain were expressed, stepwise immunization induced affinity maturation of the VRC01 precursor, and some affinity maturation intermediates exhibited viral neutralization activity. Thus, we have been able to start with mice expressing diverse “germline” VRC01 precursors and move B cell maturation along a path that results in potent neutralization of viruses that lack the 276 glycan, and in one case, of a homologous wild-type virus (426c).
There have been three previous reports of immunization studies aimed at eliciting VRC01 type antibodies in mouse models (Dosenovic et al., 2015; Jardine et al., 2015; McGuire et al., 2016). A major difference between our models and earlier models is that IGHV1-2*02 is expressed in association with diverse CDR H3’s in our models, whereas IGHV1-2*02 was linked to a fixed CDR H3 from mature VRC01-class antibodies in prior models. For mature VRC01-class antibodies, the CDR H3 makes a relatively small contribution to the interface between VRC01-class antibodies and gp120 (Zhou et al., 2010; 2013; 2015); however, in the context of germline IGHV1-2*02, the role of CDR H3 becomes more obvious. In this regard, binding affinities of eOD-GT6 and eOD-GT8 toward different germline reverted VRC01-class antibodies vary over a broad range (Jardine et al., 2013; Jardine et al., 2016); such variation is likely attributable to distinct CDR H3 sequences, as all of these antibodies share the same IGHV1-2*02. In the context of germline IGHV1-2*02, CDR2 and framework 3 (FW3) region, principal contact sites between mature VRC01-class antibodies and gp120, have not been optimized, and contributions from CDR H3 may become relevant.
In the VH1-2/LC immunization mouse model, IGHV1-2*02 is associated with diverse CDR H3s, a subset of which may be compatible with high affinity binding to eOD-GT8 60mer and, thus, respond to eOD-GT8 60mer immunization. In this regard, it is significant that all the cloned antibodies with neutralization activities utilized mouse JH1, which is highly homologous to human JH2 and provides the conserved W residue in CDR H3 of VRC01-class antibodies (Table S5). In a recent study, eOD-GT8 was used to isolate B cells from human peripheral blood; strikingly, a substantial fraction of these B cells express antibodies composed of IGHV1-2*02 IgH chain and an IgL chain with 5-amino acid CDR L3’s (Jardine et al., 2016). Together, these studies demonstrate that, even in the context of complex CDR H3, eOD-GT8 60mer is able to effectively engage VRC01 type antibodies, and may, therefore, serve as an effective priming antigen for vaccine development. A potential complicating factor, which remains to be resolved, is that in response to immunization with eOD-GT8 60mer or other germline binders in the context of a complex B cell repertoire, antibodies targeting irrelevant epitopes may eventually dominate the immune response (McGuire et al., 2014). If this were the case, a potential solution would be to use a heterologous Env boost to focus the response to the CD4 binding site. In previous studies, expression of pre-rearranged VRC01 IgH chains largely precluded potential interference from other IgH chains. In our current study, although IGHV1-2*02 IgH chain represents only a fraction of the total repertoire, its frequency is still well above the physiological level, and in the case of VH1-2/LC model, the VRC01 light chain was used in nearly all B cells. Thus, it remains to be tested whether eOD-GT8 or other germline VRC01 binders elicit VRC01 type antibodies when IGHV1-2*02HC and VRC01LC are expressed at physiological frequencies.
With substantial progress at the priming stage of inducing VRC01-class antibodies, the next major challenge was to mature the VRC01-class precursors to attain broad neutralizing activity, and to test such immunogens in the setting of diverse B cell repertoires. None of the isolated antibodies from our current immunizations have attained broad neutralizing status, and additional boosts with appropriate immunogens will be required to further mature the VRC01 intermediates. A major hurdle is N276 glycosylation, which also posed a roadblock to the maturation of VRC01 class antibodies in two HIV-1 infected individuals (Kong et al., 2016; Wu et al., 2012; Wu et al., 2015). Structural studies suggest that overcoming this roadblock will require remodeling of the CDR L1 region. In one case of a VRC01-class antibody (DRVIA7) that failed to attain broad neutralization (Kong et al, 2016), N276 appears to clash with CDR L1; the same problem may have hindered further maturation of VRC01 type of antibodies in our immunization experiments. In mature VRC01-class antibodies, extensive remodeling of CDR L1, including small deletions, established favorable contacts with N276. We found that the CDR L1 of IGKV3-20*01 appeared to be a SHM hotspot region (Figure S5); however, the prevalent mutations may reflect intrinsic hotspot activity rather than antigenic selection, as the mutation patterns are closer to those of non-anti-HIV-1 antibodies than to VRC01-class bnAbs (Figure S5). Thus, further progress in VRC01 maturation would require additional rounds of SHM coupled with effective selection for VRC01 mimetic mutations in CDR L1, including rare deletion events. One potential strategy is to generate mouse models that express affinity maturation intermediates and devise immunization strategies to improve their neutralization breadth and potency. With the use of RDBC, which obviates the need for lengthy and costly mouse breeding, our approach will expedite this reiterative process of model building and immunization.
Some broadly neutralizing antibodies are polyreactive for multiple host molecules or autoreactive with specific antigens, a property that complicates efforts to generate mouse models for broadly neutralizing antibodies (Haynes and Verkoczy, 2014; Liu et al., 2015). In a previously published model of VRC01, which expresses both pre-rearranged IgH and IgL chains of germline 3BNC60, the IgL chain was deleted via receptor editing from the majority of B cells (McGuire et al., 2016). The remaining B cells expressing the germline 3BNC60 HC and LC appeared to take on a marginal zone B cell phenotype. In our VH1-2/LC model, the introduced precursor VRC01LC dominates the peripheral B cell repertoire, with no evidence of receptor editing. Comparing the two mouse models, identities of IgL chains are different: the IgL chain in our current study is composed of germline IGKV3-20 joined to a mature CDR L3 involving Jκ1; the IgL chain in the prior study consists of germline IGKV1-33*01 joined to Jκ3*01. Thus, the different fate of the light chains in the two mouse models might reflect their distinct sequences. Alternatively, the diverse CDR H3’s of IGHV1-2*02 IgH chains in our model may enable the selection for IgH and IgL chain pairs that are compatible with normal B cell maturation. It will be of interest to apply our de novo recombination strategy to the expression of other broadly neutralizing antibodies and test whether our approach could alleviate negative selection against certain polyreactive or autoreactive antibodies.
METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the lead contact.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
The VH1-2 and VH1-2/LC mouse models were generated and housed in the animal facility at Boston Children’s Hospital. The animal work was covered under protocol 14-10-2790R, which was approved by Institutional Animal Care and Use Committee of Boston Children’s Hospital. The VH1-2 mouse model was derived from a F1 hybrid (129/Sv: C57BL/6) ES clone in which human IGHV1-2*02 replaced the mouse VH81X, and IGCRI was deleted on the IgHa allele (129/Sv) (See text and METHODS DETAILS). The ES clone was injected into Rag2−/− blastocysts to generate RDBC chimeras. The chimeric mice were bred with 129/Sv mice for germline transmission of the targeted mutations. Heterozygous VH1-2/IGCRI mice were bred to homozygosity (VH1-2 mouse model). In VH1-2 mouse model, both IgH alleles are of IgHa allotype (129/Sv), but the other chromosomes are of mixed 129/Sv and C57BL/6 origins. The VH1-2/LC mouse model was derived from the F1 ES clone described above that contained the VH1-2 replacement and IGCRI deletion by further gene targeted replacement of the mouse Jκ locus with the precursor VRC01 light chain and targeted deletion of the JHb allele (See text and METHODS DETAILS). The resultant ES clone was injected into Rag2−/− blastocysts to generate chimeric mice (VH1-2/LC mouse model), which are of mixed 129/Sv and C57BL/6 genetic background.
For immunization studies, the VH1-2 and VH1-2/LC mice were housed and cared for in accordance with local, state, federal, and institute policies in an American Association for Accreditation of Laboratory Animal Care-accredited facility at the NIH. All animal experiments were reviewed and approved by the Animal Care and Use Committee of the Vaccine Research Center, NIAID, NIH. The animal work was covered under protocol VRC 14-467 for breeding, and VRC 14-480 for immunization of mice.
METHODS DETAILS
Generation and characterization of VH1-2 and VH1-2/LC mouse models
All the genetic modifications were introduced into ES cells, using standard gene targeting technology (e.g. Chen et al., 1993; Guo et al., 2011). To illustrate the method of genetic modification, the replacement of VH81X with IGHV1-2*02 is described here as an example; all the other genetic modifications followed the same procedure. The targeting construct was based on PGKneolox2DTA.2 (Addgene #13449). In this targeting construct, the IGHV1-2*02 segment, from the start codon in leader exon to the end of VH exon, is flanked with homology arms for the VH81X locus. The 5′ homology arm is 3.2kb long and corresponds to the sequences upstream of the start codon of VH81X coding region; the 3′ homology arm is 5.3kb long and corresponds to the sequences downstream of the VH81X VH exon. A neomycin resistance gene situates between the IGHV1-2*02 segment and the 3′ homology arm to serve as the positive selection marker for transfection; the neomycin selection marker is flanked with two loxP sites to effect the removal of the drug selection marker subsequent to the identification of the correctly targeted clones. The deletion of the drug selection marker is necessary to avoid any potential interference on local transcriptional activity. The vector also contains a DTA gene as a negative selection marker to minimize random integration events. The targeting constructs for the other genetic modifications were configured in a similar manner as described above. For IGCRI and JHb deletion, no extraneous sequences other than those from cloning vectors were introduced in between the homology arms. For expression of the precursor VRC01 light chain, a 1.5kb fragment upstream of mouse IGKV3-1 gene segment was joined directly to the start codon of human IGKV3-20*01 and served as the promoter for transcription of the rearranged Igκ light chain gene. The whole cassette, promoter plus VRC01 light chain, was flanked with 5′ and 3′ homology arms and neomycin selection marker in the targeting construct.
All the genetic modifications were carried out in EF1 ES cell line, which was derived from a F1 hybrid mouse (129/Sv:C57BL/6). The cells were cultured at 37°C with 5% CO2 in DMEM medium supplemented with 15% fetal bovine serum, 20mM HEPES, 1x MEM nonessential amino acids, 2mM Glutamine, 100 units of Penicillin/Streptomycin, 100μM β-mercaptoethanol, 500 units/ml Leukemia Inhibitory Factor (LIF). The ES cells were grown on a monolayer of mouse embryonic fibroblasts (MEF) that have been mitotically inactivated with γ-irradiation. For transfection, 40μg of targeting construct was linearized via a restriction digest and electroporated into 2×107 EF1 cells. Stable transfectants were selected with 0.4mg/ml of G418. 6–7 days after transfection, colonies were picked into 24 well plates that have been coated with MEF. After 3 days, half of the colonies were frozen at −80°C and the other half were expanded for DNA isolation. The genomic DNA was analyzed with Southern blotting to identify clones in which the desired genetic modifications have taken place. After the primary screen, positive clones were further validated via additional Southern blotting analyses, and the modified region was amplified by PCR and sequenced. After a clone has passed all these tests, it was treated with Adeno-cre virus; expression of cre recombinase led to the deletion of the neomycin drug selection marker through flanking loxP sites. After the cre treatment, cells were subcloned and individual clones were screened as above via Southern blotting analyses to identify clones from which the drug selection marker has been eliminated. The correct clones were karyotyped, and clones with a normal karyotype were used for injection into Rag2−/− blastocysts. 3 weeks after the pups were born, blood samples were collected from the mice and stained with anti-B220, anti-IgM, and anti-Thy1.2 antibodies. Mice with good reconstitution of B and T cells in peripheral blood were used directly for immunization experiments (VH1-2/LC mouse model) or for breeding (VH1-2 mouse model). For germline transmission, chimeric VH1-2 mice were bred with 129/Sv mice and the progenies were screened for transmission of IGHV1-2*02 replacement and IGCRI deletion. Heterozygous IGHV1-2*02/ΔIGCRI mice were inter-bred to produce homozygous mice, which were used as the VH1-2 model in the immunization experiments.
To characterize B cells in VH1-2 and the VH1-2/LC mouse models, splenocytes were isolated from 5–8 weeks old mice, and splenocytes from 129/Sv mice of similar age were used as controls. The splenocytes were treated with red blood cell lysis buffer and stained with the following antibodies: PE-Cy5 anti-B220 (eBioscience 15-0452-83); PE anti-Thy1.2 (PharMingen 553006); PE anti-IgM (eBioscience 12-5790-83); PE anti-Igλ (BioLegend 407308); FITC anti-Igκ (SoutnernBiotech 1050-02); FITC anti-IgD (PharMingen 553439).
HTGTS-rep-seq analysis of B cell repertoire
2–4μg of DNA from purified splenic B cells was used for generating HTGTS-rep-seq libraries following published procedures (Hu et al., 2016). For analyzing IgH repertoire, two nested primers downstream of JH2 were used: 5′-BiosG/CTCCCAATGACCCTTTCTGAC-3′, 5′-GTCCCTAGTCCTTCATGACCTG-3′. Repertoire analysis with primers downstream of JH1, JH3 and JH4 gave very similar results and conclusions, and were not shown in this paper due to space limitations. The sequence reads from each JH library has been deposited in NCBI data base (see DATA AND SOFTWARE AVAILABILITY) and are available to the reader. Because different JH primers differ in priming efficiencies during library construction, the most accurate view of the overall pattern of VH usage for the four JHs is obtained by analyzing each individually as opposed to pooling primers to generate combined repertoire profiles (e.g. Lin et al., 2016). For analyzing Igκ repertoires, two nested primers downstream of Jκ5 were used: 5′-BiosG/GCCCCTAATCTCACTAGCTTGA-3′, 5′-GTCAACTGATAATGAGCCCTCTCC-3′. Since the Igκ locus in VH1-2 mouse is unaltered, the Igκ repertoire in this mouse model is expected to be essentially identical to that of a normal mouse, which was confirmed via comparison of Igκ repertoire from all four Jκ primers of the VH1-2 mouse with that of normal mice previously described (Lin et al., 2016). All libraries were sequenced via MiSeq and analyzed with the HTGTS-Rep-seq pipeline (Lin et al., 2016).
Single cell RT-PCR
Splenocytes were treated with red blood cell lysis buffer and stained with PE-Cy5 anti-B220 (eBioscience 15-0452-83), PE anti-IgM (eBioscience 12-5790-83), and FITC anti-IgD (PharMingen 553439) antibodies. Single B220+IgM+IgDhi cells were sorted into 96-well plates; each well of the plate contains 5μl of lysis buffer (10mM Tris.Cl pH7.6, 100ng yeast tRNA, 2 units of SUPERasein, 0.5% NP40). After sorting, the plate was heated at 95°C for 3 minutes and chilled to 10°C. To each well was added 5μl of reaction mix containing: 2x First strand buffer, 10mM DTT, 1mM dNTP, 2 units of SUPERasein, 10 units of Superscript III, 5pmol of oligo(dT). The reaction was incubated at 50°C for 1 hour. Following the reverse transcription reaction, 3μl of the cDNA was added to 7μl of PCR reaction mix so that the final reaction contains: 1xCoral Red PCR buffer, 0.5 units of Taq polymerase, 0.2mM dNTP, 10pmol of 5′ IGKV3-20*01 primer (5′-TTC CTC CTG CTA CTC TGG CT-3′) and 3′ Cκ primer (5′-GCC TCA CAG GTA TAG CTG TT-3′). The PCR program was 94°C 30 seconds, 65°C 1 minute, 72°C 1 minute, 35 cycles, 72°C 5 minutes. After the first round of PCR, 1μl of the PCR product was added to 9μl of second PCR reaction mix so that the final reaction contains: 1xCoral Red PCR buffer, 0.5 units of Taq polymerase, 0.2mM dNTP, 10pmol 5′ IGKV3-20*01 primer and nested 3′ Cκ primer (5′-GGA CGC CAT TTT GTC GTT CA-3′). PCR program was the same as the first round of PCR. PCR products were run on agarose gels; PCR products were excised from the gels and sequenced to confirm their identity.
Immunizations
For each immunization, 100 μl of immunogen mix, containing 15–60 μg of specified, filter-sterilized protein immunogen and 60 μg of poly I:C in PBS, was injected to the inner thigh of the two rear legs of each mouse. The VH1-2 mice were immunized only once, and blood was collected 2 weeks after the immunization. Spleens were collected two to seven weeks later. The VH1-2/LC mice were immunized monthly for 6 times. Two weeks after each injection, blood from all animals was collected for serological analyses, and one animal may be sacrificed for spleen collection. Blood and spleen from a naïve mouse were also collected as reference.
Immunogen amino acid sequences
The amino acid sequence of 426c-degly3 gp120 core Ferritin is: MPMGSLQPLATLYLLGMLVASVLAVWKEAKTTLFCASDAKAYEKEVHNVWATHACVPTDPNPQEVVLENVTENFNMWKNDMVDQMQEDVISIWDQSLKPCVKLT(GGS)TLTQACPKVTFDPIPIHYCAPAGYAILKCNNKTFNGKGPCNNVSTVQCTHGIKPVVSTQLLLNGSLAEEEIVIRSK(DLSDNAKI)IIVQLNKSVEIVCT(RPNNGGSGSGGDIRQ)AYCNISGRNWSEAVNQVKKKLKEHFPHKNISFQSSSGGDLEITTHSFNCGGEFFYCNTSGLFNDTISNATIMLPCRIKQIINMWQEVGKAIYAPPIKGNITCKSDITGLLLLRDGG(DTTDNT)EIFRPGGGDMRDNWRSELYKYKVVEIKGGGGSGESQVRQQF[SKDIEKLLNEQVNKEMQSSNLYMSMSSWCYTHSLDGAGLFLFDHAAEEYEHAKKLIIFLNENNVPVQLTSISAPEHKFEGLTQIFQKAYEHEQHISESINNIVDHAIKSKDHATFNFLQWYVAEQHEEEVLFKDILDKIELIGNENHGLYLADQYVKGIAKSRKSGS]. The sequence of 426c-degly3 gp120 core is underlined. The position of deleted V1V2 loop, the D loop, the truncated V3 loop, and the V5 loop are labeled in parentheses, sequentially. The deglycan mutations (N to D) at aa 276, 460 and 463 are in bold. The N-terminal signal peptide is in bold and the ferritin (Helicobacter pylori) portion at the C-terminus is in brackets. The nucleotide sequence of 426c-degly3 gp120 core Ferritin is available from GeneBank: KX462844.
The amino acid sequence of 426c-WT.SOSIP is: MPMGSLQPLATLYLLGMLVASVLAAENLWVTVYYGVPVWKEAKTTLFCASDAKAYEKEVHNVWATHACVPTDPNPQEVVLENVTENFNMWKNDMVDQMQEDVISIWDQSLKPCVKLTPLCVTLNCTNVNVTSNSTNVNSSSTDNTTLGEIKNCSFDITTEIRDKTRKEYALFYRLDIVPLDNSSNPNSSNTYRLINCNTSTCTQACPKVTFDPIPIHYCAPAGYAILKCNNKTFNGKGPCNNVSTVQCTHGIKPVVSTQLLLNGSLAEEEIVIRSKNLSDNAKIIIVQLNKSVEIVCTRPNNNTRRSIRIGPGQTFYATDIIGDIRQAYCNISGRNWSEAVNQVKKKLKEHFPHKNISFQSSSGGDLEITTHSFNCGGEFFYCNTSGLFNDTISNATIMLPCRIKQIINMWQEVGKCIYAPPIKGNITCKSDITGLLLLRDGGNTTNNTEIFRPGGGDMRDNWRSELYKYKVVKIEPLGVAPTRCKRRVVGRRRRRRAVGIGAVFLGFLGAAGSTMGAASMTLTVQARNLLSGIVQQQSNLLRAPEAQQHLLKLTVWGIKQLQARVLAVERYLRDQQLLGIWGCSGKLICCTNVPWNSSWSNRNLSEIWDNMTWLQWDKEISNYTQIIYGLLEESQNQQEKNEQDLLALDGGSGGSGLEVLFQGPGSHHHHHHSAWSHPQFEK. The signal peptide is in bold. The nucleotide sequence of 426c-WT.SOSIP is available from GeneBank: KX462847. The amino acid sequence of the VRC01-class germline-binding probe C13 is: MRVMGIERNYPCWWTWGIMILGMIIICNTAENLWVTVYYGVPVWKDAETTLFCASDAKAYDTEVHNVWATHACVPTDPNPQEIYMENVTEEFNWCRNNMVNQMHTDICSLWDQSLKPCVQLT(PLAGAT)SALTQACPKVTFEPIPIRYCAPAGYAILKCNDKEFAGTGLCKNVSTVQCTHGIRPVVSSQLLLNGSLAEGKVMIRSC(NIKDNAKNI)IVQLNETVTINCT(RPNNGGSGSGGDIRQ)AHCNVSGSQWNRALHQVVGQLREYWNTTIIFKNSSGGDLEITTHWFNCGGEFFYCNTSGLFNSNWTHNDTASMKPNDTITLPCRIKQIINMWCRVGQMIYAPPIQGVIRCESNITGLILTRDGG(DSTNES)QQFRPGGGDMRDNWRSELYKYKVVRIEPLGVAPTKAKRRVVEREKRGSGLNDIFEAQKIEWHELEVLFQGPGHHHHHH. The sequence of C13 chimeric gp120 core is underlined. The truncated V1V2 loop, the D loop, the truncated V3 loop, and the V5 loop are labeled in parentheses, sequentially. D279, which knocks out the CD4bs when mutated to K, is in bold. The N-terminal signal peptide is in bold. The avidin-HRV3c-6xHis tag is at the C-terminus. The nucleotide sequence of C13 is available from GeneBank: KX462845.
Antibody sequences
The sequences of the cloned antibodies from immunized VH1-2 and VH1-2/LC mice have been deposited in GeneBank; their accession numbers are listed in the KEY RESOURCES TABLE.
Protein production
All proteins were produced in transiently transfected Expi293 or 293F cells as previously described (Pancera et al., 2014). The immunogens, eOD-GT6_60mer (lumazine synthase nanoparticle), eOD-GT8_60mer, 426c-degly3 (N276D, N460D, N463D) core-Ferritin, 426c-degly2 (N276D, N460D) core, 426c-degly1 (N276D) core and 426c-WT gp120 core, were purified with Galanthus nivalis (GNA)-lectin gel (EY Laboratories, Inc.) and followed with a gel filtration chromatography. The BG505.SOSIP trimer and the 426c-WT.SOSIP trimer were purified with the 2G12/SEC method as previously described (Pancera et al., 2014). For ELISA and sorting probes, avi-his-tagged eOD-GT6, eOD-GT8, 426-degly3 core, C13 gp120 core, a VRC-developed VRC01-germline-binding UG037.8-based chimeric gp120 core, and their corresponding CD4bs-KO (Δ) mutants, ΔeOD-GT6 (D279K/D368V), ΔeOD-GT8 (D279K/D368R), Δ426-degly3 core (D279K/D368R) and ΔC13 (D279K), were purified with Ni-NTA beads (GE healthcare) and followed by a gel filtration chromatography for monomeric proteins. For antibody production, heavy and light chain plasmids were co-transfected (1:1 ratio) in Expi293 cells using Expi293fectin (Invitrogen) according to the manufacturer’s protocol. The supernatants were harvested six days later, and the IgG was purified using protein G Sepharose (GE Healthcare) and buffer-exchanged into PBS (0.01 M sodium phosphate, pH 7.4, 0.137 M sodium chloride).
ELISA
Corning Costar® half-area assay plates were coated with antigens at 100 ng/well at 4°C overnight, blocked with either B3T buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 3.3% fetal bovine serum, 2% bovine albumin, 0.07% Tween 20, 0.02% thimerosal) for mouse sera or 1:10 diluted blocking solution (Immune Technology Corp.) for purified mAbs at room temperature for 1 hr, and incubated with serial diluted sera or antibodies at room temperature for 1 hr. The plates were then washed with PBS with 0.05% Tween (Sigma) 5 times, and 1:5,000 diluted HRP-conjugated secondary antibodies, goat anti-human IgG and/or goat-anti-mouse IgG (Bio-Rad) were added. The plates were washed again and the SureBlue™ TMB Microwell Peroxidase Substrate (Kirkegaard & Perry Laboratories, Inc.) was added to each well. The color reaction was terminated with 1N sulfuric acid and the absorbance at 450 nm (OD450) was recorded. Background absorbance was determined based on wells blotted with a non-related primary antibody and endpoint titer was determined as the largest serum dilution or the lowest primary antibody concentration at which the OD450 is three times over the background level.
TZM-bl Neutralization Assay
Single round replication Env-pseudoviruses were prepared, titered and used for infecting TZM-bl target cells in the presence of different concentrations of mouse sera or purified monoclonal antibodies as described previously (Wu et al., 2010). The neutralizing titer was reported as half-maximum inhibitory concentration (IC50) or 80%-maximum inhibitory concentration (IC80) in μg/ml for mAbs and half-maximum inhibitory dilution (ID50 or ID80) for mouse sera.
Flow cytometry and B-cell sorting
Mouse spleen samples were processed for single B cell sorting based on previously described methods (Tiller et al., 2009). In brief, single cell suspension of splenocytes was stained sequentially with ViVid and a staining mix containing anti-CD3 Cy55PerCP, anti-CD4 Cy55PerCP, anti-CD8 Cy55PerCP, anti-F4/80 Cy55PerCP, anti-B220 TrPE, anti-IgD BV711, anti-IgM Cy7PE, anti-IgG FITC, eOD-GT6(8)-PE and ΔeOD-GT6(8)-APC. Memory B cells were selected for the phenotype B220+, CD3−, CD4−, CD8−, F4/80−, IgM−, IgD−, and IgG+. To make antigen-specific probes, 50 nM of biotinylated Avi-tagged eOD-GT6(8) monomer and its CD4bs-KO variant ΔeOD-GT6(8) (D279K, D368V/R) were coupled to Streptavidin-PE and Streptavidin-APC (Life Technologoies), respectively, in equal molar ratios. Memory B cells with eOD-GT6(8)-PE+ ΔeOD-GT6(8)-APC− were selected and single-cell sorted into 96 well plates containing lysis buffer on a BD FACSAria III sorter and immediately stored at −80°C. Spleen samples without eOD-GT6(8) and ΔeOD-GT6(8) probes and spleen samples from naïve littermates were used as controls and to set up the gate for sorting.
Single B-cell RT-PCR, gene amplification, and cloning
Reverse transcription and subsequent PCR amplification of heavy and light chain variable genes were performed using SuperScript III (Life Technologies) as previously described (Tiller et al., 2009; Wu et al., 2010). All PCR reactions were performed in 25 μl volume with 1–2.5 μl of cDNA transcript using HotStar Taq DNA polymerase master mix (Qiagen). For the VH1-2 single knock-in mice, mixtures of previously described mouse Ig primers (Tiller et al., 2009) were supplemented with human VH1-2 specific primers for 1st and 2nd PCR (Table S6). For the double knock-in mice, specific VH1-2 and VK3-20 5′ primers in combination with designed 3′ primers aligning to the 5′ regions of the mouse gamma (1, 2a, 2b, 2b and 3), mu, or kappa constant segments (Table S6) were used for nested PCR to amplify VH1-2 and VK3-20 heavy and light chains. PCR products were then sequenced using Sanger sequencing and corrected for PCR errors before further analysis and expression. The confirmed heavy or light chain sequences containing the whole V(D)J or VJ segments were synthesized by GenScript and cloned into a mouse IgG1- or mouse Kappa-mammalian expression vector for antibody expression.
Bio-Layer Interferometry Antigenicity Analysis
Kinetics and affinities of antibody-antigen interactions were measured on an Octet.HTX (Forte Bio) using Anti-human IgG Fc and Anti-mouse IgG Fc capture biosensors following the instructions. Briefly, purified VRC01, VRC01gl, and synthesized mouse antibodies were loaded (50 μg/ml) on anti-human or anti-mouse IgG Fc biosensors (AHC, AMC, ForteBio). Following a 1% BSA/PBS buffer wash step, eOD-GT8 or ΔeOD-GT8 monomer at 6 different concentrations was associated (2-fold dilutions starting at 5uM) and then allowed to dissociate into 1%BSA/PBS buffer. A 1:1 ratio of antibody to antigen was assumed for the complex to calculate molar concentration and an irrelevant anti-Influenza antibody was used as a negative control in all experiments. Reference wells containing anti-influenza antibody were subtracted from samples wells. All experiments were performed at 30°C. Octet Analysis Software was used for data analysis, curve fitting and determination of Kd, Kon and Koff. We deemed that no fit was observed if fewer than 3 curves could be fit with an R2 ≥ 0.95.
Mutation profile construction and statistical test for enriched and depleted mutations
The mutation frequency of IGHV1-2 and IGKV3-20 were estimated from 1080 and 4598 antibody clones from three normal donors (Bonsignori et al., 2016). From each clone, one sequence was randomly selected. For enrichment and depletion test, we randomly selected the same number of sequences (equal to the number observed at each time point of vaccination study) from the non-HIV neutralizing IGHV1-2 or IGKV3-20 antibody repertoire for 1000 times. Then the mean occurrence and standard deviations for each mutation was calculated and 95% confidence interval (CI) were decided. Mutations with frequency higher than 95% CI were counted as enriched mutations, and those with frequency lower than 95% CI were counted as depleted mutations. Sequences were aligned using MUSCLE (Edgar, 2004). Consensus sequences for the broadly neutralizing antibody lineages were inferred using SeaView (Gouy et al., 2010).
High-throughput paired heavy-light chain RT-PCR
Splenic mouse B cells were isolated using magnetic bead depletion (EasySep™ Mouse Pan-B Cell Isolation Kit, Stemcell Technologies, Vancouver, CA) and analyzed with a single-cell flow focusing emulsion technology to link paired heavy and light chain antibody genes for high-throughput DNA sequencing (DeKosky et al., 2015). Briefly, paired heavy- and light-chain sequencing was achieved via single-cell isolation in emulsion droplets, cell lysis, and mRNA capture, overlap extension RT-PCR to link heavy and light chains, and Illumina MiSeq sequencing of the VH:VL cDNA product (RT-PCR kit: SuperScript® III One-Step RT-PCR System with Platinum® Taq DNA Polymerase, Invitrogen/Thermo Scientific, Carlsbad, CA, USA). Emulsion linkage RT-PCR primers are provided in Table S7 upper panel, and nested PCR primers are provided in Table S7 lower panel. Paired-end Illumina sequences were quality-filtered to a Q-score of 20 over 50% of the sequence read and annotated using the National Center for Biotechnology Information IgBLAST platform combined with a CDR3 motif identification algorithm (bioinformatic methods/code reported in (DeKosky et al., 2016)). A new IgBLAST V-gene library was used for this analysis containing all IMGT mouse and human VH and Vκ,λ genes; J- and D-gene libraries contained only mouse sequences. Antibody constant region isotype (G/A/M/K/L) was assigned using the nested PCR primer sequences that target the constant region.
QUANTIFICATION AND STATISTICAL ANALYSIS
For statistical comparison of two individual groups, a Student’s t-Test was performed with two tailed distributions and equal variance. For comparison of the mutation frequencies at multiple time points, ANOVA Kruskal-Wallis test was performed using GraphPad Prism 6.05 Software (GraphPad Prism Software, Inc.).
DATA AND SOFTWARE AVAILABILITY
Raw sequence reads for the paired heavy:light DNA sequence data can be downloaded from the NCBI Short Read Archive (SRA) under accession number PRJNA327421 (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA327421). The raw sequence reads for the HTGTS-rep-seq data is available from SPA, accession SRP077660 (http://www.ncbi.nlm.nih.gov/sra/SRP077660).
Supplementary Material
Panels A and B show results of Southern analysis designed to verify genetic modifications in VH1-2 mouse model. The diagrams, not drawn to scale, illustrate the restriction digests and probes that were used to differentiate the region before (germline, GL) and after genetic modifications. (A) Southern analysis of liver DNA from VH1-2 mouse model (lanes 1–2) and wild-type 129/Sv mice (lanes 3–4). The result confirms homozygous replacement of VH81X with IGHV1-2*02 (Vh1-2) in VH1-2 mice. (B) Southern analysis of liver DNA from VH1-2 mouse model (lanes 1–2) and wild-type 129/Sv mouse (lanes 3–4). The result validates IGCRI deletion (ΔIGCRI) on both IgH alleles in VRC01HC mice. (C) FACS analysis of splenic B cells from wild-type 129/Sv, VH1-2 and VH1-2/LC mice. In the left column, splenocytes were stained with anti-B220 and anti-Thy1.2 antibodies. Cells in the lymphocyte gate, as defined by FSC and SSC, were displayed in the FACS plot. The percentage of B cells (B220+) and T cells (Thy1.2+) are indicated on the plot. In the middle column, splenocytes were stained with anti-B220, anti-IgM, and anti-IgD antibodies. B220+ B cells were gated and shown in the plot. The percentage of mature B cells (IgM+IgDhi) is indicated on the plot. In the right column, splenocytes were stained with anti-B220, anti-Igλ, and anti-Igκ antibodies. B220+ B cells were gated and shown in the plot. The percentage of Igκ+ and Igλ+ B cells is indicated on the plot. The data shown in this figure are representative of at least three experiments for each genotype, as summarized in the table below.
(A) Gating strategy for single cell sorting of eOD-GT8-specific memory B cells in naïve and eOD-GT8 60mer-immunized mice. Splenocytes from a naïve mouse were used as a control to set up gating for the analysis of splenocytes from immunized mice. (B) Summary of binding affinity of the cloned antibodies from eOD-GT8 60mer immunized VH1-2 mice compared to VRC01 and its germline revertant. (C) Gene frequency data for naïve and 30 μg eOD-GT8 60mer immunized VH1-2 mice. Data was subdivided by heavy chain isotype prior to analysis.
(A) Southern analysis of wild-type ES cells (lane 1) and VH1-2/LC ES clone (lane 2). The result verifies JH deletion (ΔJHb); additional restriction digest, not shown here, indicated that the deletion occurred on the IgHb allele. (B) Southern analysis of wild-type ES cells (lane 1) and VH1-2/LC ES clone (lane 2). The result verifies the integration of VRC01LC into Jκ locus (VRC01LC KI). The VRC01LC, shown below the diagram, is composed of germline IGKV3-20*01 (sequence in orange) and Jκ1 from mature VRC01 (sequences in blue); two residues in CDR L3, shown in black, cannot be assigned to either IGKV3-20*01 or Jκ1. (C) HTGTS-rep-seq analysis of Vκ usage in 129/Sv control and VH1-2/LC mouse B cells. The analysis was carried out as in Figure 1C and 1F. As mouse Jκ1-5 on one Igκ allele were deleted during the integration of VRC01LC, a primer downstream of Jκ5 was used for the amplification of VRC01LC as well as Vκ-Jκ5 rearrangements on the intact mouse Igκ allele. Given that the human IGKV3-20*01 was inserted at the Jκ region, the slash sign on the X-axis indicates the genetic separation of IGKV3-20*01 from the mouse Vκ segments.
The amino acid mutations in all sequences amplified from each specified mouse are shown in sequence logo graphs. Only residues different from the germline sequences are shown and the size of the letter correlates with its frequency. n: the number of all amplified from each animal and used for the logo graph. As a reference, 36 published VRC01-class HCs were shown below the panel for G2.
Mutation profiles of IGKV3-20*01 LCs amplified from each mouse were compared with that of three mature VRC01-class bnAb (VRC01, VRC04 and VRC18) IGKV3-20*01 LCs, as well as that of 1080 non-anti-HIV-1 antibody IGKV3-20*01 LCs from three healthy donors.
(A) Alignment of VH1-2*02 germline sequence with 25 published VRC01-class bNAbs HCs from 9 lineages/donors (Zhou et al. 2015). (B) Based on A, the consensus or representative sequence of each of the 9 lineages were aligned. At each VH1-2 residue position, somatic mutations that appear in at least 2 out of the 9 VRC01-class lineages were defined as VRC01-class mimicking mutations and listed in the yellow-highlighted region corresponding to each VH1-2 position. (C) Amino acid alignment of the heavy chains of the 27 synthesized double knock-in mouse IgGs with human VH1-2*02 genes. (D) Amino acid alignment of the light chains of the 27 synthesized double knock-in mouse IgGs with human VK3-20*01 genes.
The presence (+) or absence (−) of N276, N460 and N463 glycans in each pseudovirus is indicated. The neutralizing titers of VRC01 are shown as positive controls.
Highlights.
Mice that generate diverse primary B cell receptors from a single human VH segment
Efficient generation of mouse models for assessing immunogens for human vaccination
Step-wise immunization for affinity maturation of HIV-neutralizing antibodies in mice
Acknowledgments
We thank Norman Letvin and Connie Gee for stimulating this collaborative study. We also thank Sam Darko for assistance with data analysis. This work was supported by NIH grants R01AI077595 and AI020047 (to FWA), P01 AI094419 U19 AI109632 (to LS) and NIAID, Division of AIDS, Center for HIV/AIDS Vaccine Immunology- Immunogen Discovery (CHAVI-ID) AI100645 (BFH and FWA) and CHAVI-ID 5UM1AI100645 (to FWA); by the International AIDS Vaccine Initiative Neutralizing Antibody Consortium and Center (W.R.S.); CAVD funding for the IAVI NAC Center (W.R.S.); NIH Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) 1UM1 AI100663 (W.R.S.); and the Ragon Institute of MGH, MIT and Harvard (W.R.S.). This work was also supported by the intramural research program of the Vaccine Research Center, NIAID, NIH. M.D. was supported by an HHMI Medical Student Fellowship. FWA is an investigator and ZD a postdoctoral fellow of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
MT, CC, XC, HD, HLC, PK, BH, JM, and FA designed the study and analyzed and interpreted data. MT, CC, XC, HD, HLC, MD, ZS, MK, LW, SL, SS, MGJ, YC, BD, YC, EN, EC, RC, NADR, YZ, WS, WPK, MC, AH, FL, IG, PYH, and SJ performed experiments. AM, WS, DD and LS provided reagents. ZD, BD, and LS performed computational analyses. MT, JM, and FA drafted the manuscript and BH, LS, WS, AM, CC, XC, HD helped revise and polish the manuscript. MT, CC, XC, HD, MD, MK, SL, ZD, BD, SJ generated figures and tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, et al. Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody. Cell. 2016;165:449–463. doi: 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annual review of immunology. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lansford R, Stewart V, Young F, Alt FW. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nature medicine. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- DeKosky BJ, Lungu OI, Park D, Johnson EL, Charab W, Chrysostomou C, Kuroda D, Ellington AD, Ippolito GC, Gray JJ, et al. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E2636–2645. doi: 10.1073/pnas.1525510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell. 2015;161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular biology and evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nature biotechnology. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Verkoczy L. AIDS/HIV. Host controls of HIV neutralizing antibodies. Science (New York, NY) 2014;344:588–589. doi: 10.1126/science.1254990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoot S, McGuire AT, Cohen KW, Strong RK, Hangartner L, Klein F, Diskin R, Scheid JF, Sather DN, Burton DR, et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS pathogens. 2013;9:e1003106. doi: 10.1371/journal.ppat.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, Frock RL. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nature protocols. 2016;11:853–871. doi: 10.1038/nprot.2016.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zhang Y, Zhao L, Frock RL, Du Z, Meyers RM, Meng FL, Schatz DG, Alt FW. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell. 2015;163:947–959. doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science (New York, NY) 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ereno-Orbea J, Kalyuzhniy O, Deresa I, et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science (New York, NY) 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science (New York, NY) 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Ju B, Chen Y, He L, Ren L, Liu J, Hong K, Su B, Wang Z, Ozorowski G, et al. Key gp120 Glycans Pose Roadblocks to the Rapid Development of VRC01-Class Antibodies in an HIV-1-Infected Chinese Donor. Immunity. 2016;44:939–950. doi: 10.1016/j.immuni.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang KK, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. Journal of virological methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SG, Ba Z, Du Z, Zhang Y, Hu J, Alt FW. Highly sensitive and unbiased approach for elucidating antibody repertoires. Proc Natl Acad Sci USA. Jun 27; doi: 10.1073/pnas.1608649113. pii: 201608649. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- Liu M, Yang G, Wiehe K, Nicely NI, Vandergrift NA, Rountree W, Bonsignori M, Alam SM, Gao J, Haynes BF, et al. Polyreactivity and autoreactivity among HIV-1 antibodies. Journal of virology. 2015;89:784–798. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunological reviews. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Dreyer AM, Carbonetti S, Lippy A, Glenn J, Scheid JF, Mouquet H, Stamatatos L. HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science (New York, NY) 2014;346:1380–1383. doi: 10.1126/science.1259206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Gray MD, Dosenovic P, Gitlin AD, Freund NT, Petersen J, Correnti C, Johnsen W, Kegel R, Stuart AB, et al. Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nature communications. 2016;7:10618. doi: 10.1038/ncomms10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. The Journal of experimental medicine. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS pathogens. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science (New York, NY) 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Busse CE, Wardemann H. Cloning and expression of murine Ig genes from single B cells. Journal of immunological methods. 2009;350:183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annual review of immunology. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Wu X, Wang C, O’Dell S, Li Y, Keele BF, Yang Z, Imamichi H, Doria-Rose N, Hoxie JA, Connors M, et al. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. Journal of virology. 2012;86:5844–5856. doi: 10.1128/JVI.07139-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science (New York, NY) 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhang Z, Schramm CA, Joyce MG, Kwon YD, Zhou T, Sheng Z, Zhang B, O’Dell S, McKee K, et al. Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell. 2015;161:470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science (New York, NY) 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, Zhang MY, Longo NS, Dimitrov DS. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochemical and biophysical research communications. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science (New York, NY) 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panels A and B show results of Southern analysis designed to verify genetic modifications in VH1-2 mouse model. The diagrams, not drawn to scale, illustrate the restriction digests and probes that were used to differentiate the region before (germline, GL) and after genetic modifications. (A) Southern analysis of liver DNA from VH1-2 mouse model (lanes 1–2) and wild-type 129/Sv mice (lanes 3–4). The result confirms homozygous replacement of VH81X with IGHV1-2*02 (Vh1-2) in VH1-2 mice. (B) Southern analysis of liver DNA from VH1-2 mouse model (lanes 1–2) and wild-type 129/Sv mouse (lanes 3–4). The result validates IGCRI deletion (ΔIGCRI) on both IgH alleles in VRC01HC mice. (C) FACS analysis of splenic B cells from wild-type 129/Sv, VH1-2 and VH1-2/LC mice. In the left column, splenocytes were stained with anti-B220 and anti-Thy1.2 antibodies. Cells in the lymphocyte gate, as defined by FSC and SSC, were displayed in the FACS plot. The percentage of B cells (B220+) and T cells (Thy1.2+) are indicated on the plot. In the middle column, splenocytes were stained with anti-B220, anti-IgM, and anti-IgD antibodies. B220+ B cells were gated and shown in the plot. The percentage of mature B cells (IgM+IgDhi) is indicated on the plot. In the right column, splenocytes were stained with anti-B220, anti-Igλ, and anti-Igκ antibodies. B220+ B cells were gated and shown in the plot. The percentage of Igκ+ and Igλ+ B cells is indicated on the plot. The data shown in this figure are representative of at least three experiments for each genotype, as summarized in the table below.
(A) Gating strategy for single cell sorting of eOD-GT8-specific memory B cells in naïve and eOD-GT8 60mer-immunized mice. Splenocytes from a naïve mouse were used as a control to set up gating for the analysis of splenocytes from immunized mice. (B) Summary of binding affinity of the cloned antibodies from eOD-GT8 60mer immunized VH1-2 mice compared to VRC01 and its germline revertant. (C) Gene frequency data for naïve and 30 μg eOD-GT8 60mer immunized VH1-2 mice. Data was subdivided by heavy chain isotype prior to analysis.
(A) Southern analysis of wild-type ES cells (lane 1) and VH1-2/LC ES clone (lane 2). The result verifies JH deletion (ΔJHb); additional restriction digest, not shown here, indicated that the deletion occurred on the IgHb allele. (B) Southern analysis of wild-type ES cells (lane 1) and VH1-2/LC ES clone (lane 2). The result verifies the integration of VRC01LC into Jκ locus (VRC01LC KI). The VRC01LC, shown below the diagram, is composed of germline IGKV3-20*01 (sequence in orange) and Jκ1 from mature VRC01 (sequences in blue); two residues in CDR L3, shown in black, cannot be assigned to either IGKV3-20*01 or Jκ1. (C) HTGTS-rep-seq analysis of Vκ usage in 129/Sv control and VH1-2/LC mouse B cells. The analysis was carried out as in Figure 1C and 1F. As mouse Jκ1-5 on one Igκ allele were deleted during the integration of VRC01LC, a primer downstream of Jκ5 was used for the amplification of VRC01LC as well as Vκ-Jκ5 rearrangements on the intact mouse Igκ allele. Given that the human IGKV3-20*01 was inserted at the Jκ region, the slash sign on the X-axis indicates the genetic separation of IGKV3-20*01 from the mouse Vκ segments.
The amino acid mutations in all sequences amplified from each specified mouse are shown in sequence logo graphs. Only residues different from the germline sequences are shown and the size of the letter correlates with its frequency. n: the number of all amplified from each animal and used for the logo graph. As a reference, 36 published VRC01-class HCs were shown below the panel for G2.
Mutation profiles of IGKV3-20*01 LCs amplified from each mouse were compared with that of three mature VRC01-class bnAb (VRC01, VRC04 and VRC18) IGKV3-20*01 LCs, as well as that of 1080 non-anti-HIV-1 antibody IGKV3-20*01 LCs from three healthy donors.
(A) Alignment of VH1-2*02 germline sequence with 25 published VRC01-class bNAbs HCs from 9 lineages/donors (Zhou et al. 2015). (B) Based on A, the consensus or representative sequence of each of the 9 lineages were aligned. At each VH1-2 residue position, somatic mutations that appear in at least 2 out of the 9 VRC01-class lineages were defined as VRC01-class mimicking mutations and listed in the yellow-highlighted region corresponding to each VH1-2 position. (C) Amino acid alignment of the heavy chains of the 27 synthesized double knock-in mouse IgGs with human VH1-2*02 genes. (D) Amino acid alignment of the light chains of the 27 synthesized double knock-in mouse IgGs with human VK3-20*01 genes.
The presence (+) or absence (−) of N276, N460 and N463 glycans in each pseudovirus is indicated. The neutralizing titers of VRC01 are shown as positive controls.