Abstract
The present study aimed to evaluate the association between rs11136000 in clusterin (CLU) and late-onset Alzheimer's disease (LOAD) by meta-analysis. Several databases including PubMed, EMbase, CBMdisc and CMCC were searched for relevant case-control studies based on defined selection criteria. Odds ratios (OR) and 95% confidence interval (CI) of the rs11136000 genotype and allele distribution were analyzed with RevMan and Stata software. The control population and heterogeneity between populations were examined in the selected studies using the Hardy-Weinberg equilibrium. Overall OR among the frequencies of the genotype and allele in both patients with AD and controls was estimated using fixed or random effect models. The summary of the OR and 95% CI were then analyzed to obtain the effects across the studies. Publication bias was examined using a funnel plot, Egger's test and Begg's test, and a Fail-safe Number (Nfs). A total of 20 reports were used. The summary OR for studies in the Caucasian population with a frequency of TT+TC/CC genotype and T/C allele at rs11136000 locus in CLU were 0.79 (95% CI, 0.73–0.86; P<0.00001) and 0.87 (95% CI, 0.85–0.90; P<0.00001). The summary OR for the studies conducted in the Asian population were 0.90 (95% CI, 0.81–0.99; P=0.04) and 0.87 (95% CI, 0.81–0.93; P<0.0001). The summary OR in other mixed ethnic groups with regards to the frequency of T/C allele was 0.82 (95% CI, 0.68–0.99; P=0.04). These results demonstrated the presence of a statistically significant difference in LOAD susceptibility between individuals with the T allele CLU rs11136000 polymorphism and those without. The studies conducted in populations of African descent or Hispanics showed no statistically significant difference. Negligible publication bias was present, with Nfs being 750.604. In summary, polymorphism rs11136000 in the CLU gene may contribute to susceptibility to LOAD, and the presence of the T allele may reduce the risk of LOAD in Caucasian and Asian populations. However, no definitive association was found between the presence of the CLU rs11136000 polymorphism and LOAD in populations of African or Hispanic descent.
Keywords: clusterin, rs11136000, Alzheimer's disease, meta-analysis
Introduction
Alzheimer's disease (AD) is a neurodegenerative disorder with progressive cognitive impairment. Molecular genetic studies have demonstrated the presence of complex genetic heterogeneity in AD (1–3). Previous investigations reported that certain gene mutations are associated with familial early-onset AD, such as amyloid-β precursor protein, presenilin I and presenilin II (3–5). In addition, it was demonstrated that there are likely numerous gene polymorphisms that are risk factors for late-onset AD (LOAD), but to date, only the apolipoprotein E (APOE) ε4 allele has been widely confirmed as a susceptibility gene for LOAD (1). However, the APOE gene polymorphism may only contribute to 50% of genetic susceptibility (1). This suggests there may be other genes that could influence susceptibility to the onset of LOAD. The investigation of other susceptibility genes would facilitate the research on LOAD pathogenesis.
In 1996, the clusterin (CLU) gene was suggested to be a potential AD susceptibility gene (2). The CLU gene is located at p21-p12 of chromosome 8 (5). The protein expressed by CLU is known as apolipoprotein J which is secreted by neurons and astrocytes, and being the second-largest glycoprotein in the brain following APOE, it has similar biological functions to APOE (5). CLU is expressed in cerebrospinal fluid and amyloid plaques in brain tissues, is able to combine with β-amyloid peptide to participate in the processes of metabolism, transportation and clearance of amyloid-β (Aβ), and has an important role in the cell cycle regulation and lipid metabolism of AD (3–5). A previous study reported that the CLU gene may be associated with AD (2). Unfortunately, no gene locus identified in the earliest study regarding the correlation between CLU gene variants and AD (2).
With the advance of molecular genetic technology, it became possible to include significantly greater numbers of individuals in the disease-association studies, thereby allowing the identification of more potential disease susceptibility genes. In October 2009, ‘Nature Genetics’ simultaneously published two AD-associated large-scale genome-wide association studies (GWAS) completed by Lambert et al (6) and Harold et al (7), which both confirmed that the CLU gene single nucleotide polymorphism (SNP) rs11136000 was associated with LOAD. Another meta-analysis of genome-wide detection also suggested that the p22-p21.1 of chromosome 8, namely the CLU chromosomal locus, was a LOAD-associated susceptibility region (8). Since then, the association between AD and CLU variants has once again become the focus of numerous studies, in which various experiments were carried out in an attempt to demonstrated the correlation (9–26). However, the results of these studies were not concordant.
The present investigation reviewed and conducted a meta-analysis of previously published case-control studies on the correlation between the CLU gene polymorphism rs11136000 and LOAD both in China and abroad. The increased sample size could improve the significance of the results, and subgroup analyses of various ethnicities could eliminate possible ethnic differences. The aim of the present study was to provide objective evidence-based results for the association between CLU gene polymorphisms and AD susceptibility.
Materials and methods
Subjects
The previously published case-control studies regarding the association between CLU gene variants and LOAD in various ethnic groups were reviewed. The patients with LOAD in these studies were selected as study cases, and the CLU rs11136000 polymorphism as the locus of interest, which in the previous studies was demonstrated to be significant although controversial. The control groups in these studies were healthy participants of the same ethnicity as the cases studied. The genotype frequencies of TT, TC and CC, as well as the allele frequencies of T and C, in the case and control groups of these studies were obtained for meta-analysis. The control groups in this study were healthy participants with the same ethnicity as the cases studied.
Literature search
A systematic search was conducted using PubMed (www.ncbi.nlm.nih.gov/pubmed), EMbase (store.elsevier.com/embase), CBMdisc (www.cnki.com.cn), CMCC, Wanfang (www.wanfangdata.com.cn), and other databases in English or Chinese (the last search was updated on August 31, 2014), with ‘Clusterin AND Alzheimer disease’ as MeSH terms, and ‘Clusterin AND Alzheimer’ and ‘rs11136000’ as free terms. Through literature retrospection, the references of the retrieved studies were further reviewed to ensure that a comprehensive selection of the relevant studies was carried out. According to the abstracts provided by the databases, the original sources of the studies were located in order to obtain full texts and complete data.
Inclusion criteria and quality control
All the participanting authors evaluated the quality of the selected studies and reached a consensus regarding the appropriate inclusion criteria and quality control. Studies meeting the following criteria were included in the meta-analysis: 1) Case-control studies involving the polymorphism rs11136000; 2) LOAD cases diagnosed according to the internationally accepted standards, DSM-IV and NINCDS-ADRDA (27), with the controls being local matched healthy individuals; 3) sample size was explicit, genotype and allele frequency data could be obtained, original data was provided directly or could be used to calculate the odds ratio (OR) and 95% confidence intervals (CI), and information was sufficient to compare the statistical differences between the groups; 4) experimental methods for detecting gene polymorphisms were scientific and rigorous; and 5) allele frequency met the Hardy-Weinberg equilibrium (HWE). The articles published with incomplete data were excluded from the meta-analysis. Among repeatedly reported studies, those with the most complete information were used.
Statistical analysis
RevMan5.2 version 5.2 (The Nordic Cochrane Centre, The Cochrane Collaboration, Oxford, UK) and Stata 11.0 software version 11.0 (StataCorp LP, College Station, TX, USA) were used for statistical analysis.
HWE test
A χ2 test was used to investigate the HWE of rs11136000 allele distributions among the selected studies, in order to verify the representativeness of the study population. Studies in which the allele frequency of the control group did not satisfy HWE law were removed.
Heterogeneity test
A Q test was performed to examine the heterogeneity among the selected studies. RevMan software provided the P-value of the Q tests, with P<0.10 considered to indicate a statistically significant difference. Different effect models for meta-analysis were selected based on the results of the Q tests.
Selection of statistical models
If there was no statistically significant heterogeneity among the results of the studies, a Peto Mantel-Haenszel fixed-effect model was used to determine the overall OR. If there was statistically significant heterogeneity, a Dersimonian-Laird random-effect model was used. The RevMan software provided the overall OR and its 95% CI.
Test for overall effect
Forest plots were provided by the RevMan software, on which the Z and P-values were shown on the ‘test for overall effect’, with P<0.05 considered to indicate a statistically significant difference.
Bias analysis
Funnel plots were generated by the RevMan software for the preliminary analysis of publication bias. Using the Stata software, Egger's linear regression and Begg's rank correlation tests were performed to objectively evaluate publication bias and to verify the symmetry of the funnel plots, with P<0.10 considered to indicate a statistically significant difference. Lastly, the fail-safe number (Nfs) was calculated to further measure publication bias using the following formula: Nfs0.05 = (ΣZ/1.64)2 - k. The larger the Nfs value, the smaller the bias.
Results
Characteristics of eligible studies
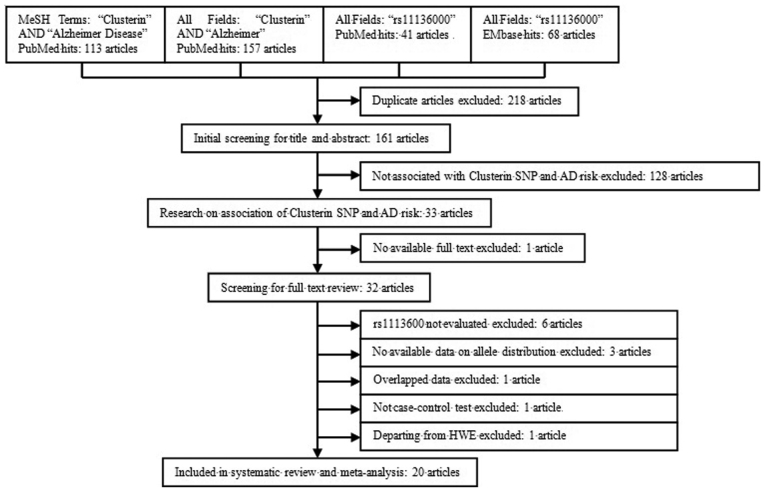
Following a literature review and further screening according to the inclusion criteria, 20 studies were included in the meta-analysis (Fig. 1) (6,7,9–26), all of which were published after 2009 in foreign biomedical journals, but not reported in Chinese journals. The study population of these investigations included Caucasian, Hispanic, and Asian populations, populations of African descent, as well as other ethnicities, of which 12 were from Europe or America, and 8 were from Asia. A total of 32,017 individuals with LOAD were included as cases, whose diagnoses were predominantly based on the clinical criteria, with only a minority diagnosed based on pathological evidence. A total of 43,956 individuals were included as controls, in which the allele distributions were in accordance with the HWE. The characteristics of the eligible studies are shown in Table I. The genotype and allele frequencies of the CLU gene polymorphism rs11136000 are shown in Table II.
Figure 1.
Flowchart for the screening of articles for meta-analysis.
Table I.
Characteristics of the studies included in the meta-analysis.
| LOAD | Control | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Included studies | Population | Results | Cases (% female) | Diagnostic criteria | Average age of onset | Average age | Case (% female) | Average age | Genotyping | References |
| Caucasian | ||||||||||
| Lambert et al, 2009 | Stage 1 (France) | Pos | 2,025 (66%) | C | 68.3±9.0 | 73.7±8.9 | 5,328 (61%) | 73.8±5.4 | Illumina infinium system | (6) |
| Stage 2 (Italy) | Pos | 1,520 (68%) | C | 73.8±8.8 | 76.6±8.7 | 1,291 (55%) | 72.3±8.9 | Illumina infinium system | ||
| Stage 2 (Spain) | Neg | 755 (57%) | C | 72.5±9.4 | 75.3±9.3 | 833 (62%) | 76.9±10.9 | Illumina infinium system | ||
| Stage 2 (Belgium) | Pos | 1,084 (66%) | C | 74.4±8.6 | 78.6±8.1 | 509 (58%) | 67.0±12.9 | Illumina infinium system | ||
| Stage 2 (Finland) | Pos | 619 (67%) | C | 71.4±7.5 | 71.4±7.5 | 664 (60%) | 69.2±6.0 | Illumina infinium system | ||
| Harold et al, 2009 | Stage 1 (USA) | Pos | 1,159 (58%) | M | 73.5 | 80.7 | 1,783 (56%) | 68.1 | Illumina infinium system | (7) |
| Stage 1 (UK, Ireland) | Pos | 2,227 (65%) | M | 72.9 | 79.7 | 5,241 (53%) | 51.2 | Illumina infinium system | ||
| Stage 1 (Germany) | Pos | 555 (64%) | C | 70.5 | 72.9 | 824 (51%) | 56.5 | Illumina infinium system | ||
| Giedraitis et al, 2009 | Sweden (ULSAM) | Neg | 86 (0%) | C | 80.2 | – | 404 (0%) | 81.8 | Illumina GoldenGate assay | (9) |
| Golenkina et al, 2010 | Russian | Neg | 534 (−) | C | – | – | 702 (−) | – | – | (10) |
| Jun et al, 2010 | USA (ADGC-C) | Pos | 5,935 (−) | M | – | – | 7,034 (−) | – | Illumina or affymetrix arrays | (17) |
| Seshadri et al, 2010 | Spain (ACE) | Pos | 1,140 (70%) | C | – | 78.8±7.9 | 1,209 (53%) | 49.9±9.2 | Illumina or affymetrix arrays | (24) |
| Carrasquillo et al, 2010 | USA | Pos | 1,829 (−) | M | – | – | 2,576 (−) | – | TaqMan SNP genotyping assays | (13) |
| Schjeide et al, 2011 | Germany | Neg | 214 | C | – | – | 211 (−) | – | OpenArray genotyping system | (23) |
| USA | Pos | 2,654 | M | – | – | 1,175 (−) | – | OpenArray genotyping system | ||
| Bettens et al, 2012 | Stage 1 (Belgium) | Pos | 1,057 (66%) | C | 74.9±8.9 | – | 873 (57.4%) | 65.1±14.9 | PCR | (12) |
| Stage 2 (France) | Neg | 1,465 (66%) | C | 69.5±8.2 | – | 717 (62.3%) | 74.0±8.0 | PCR | ||
| Stage 2 (Canada) | Neg | 323 (55%) | C | 75.3±9.7 | – | 250 (60.0%) | 73.0±10.2 | PCR | ||
| Kamboh et al, 2012 | USA | Neg | 1,348 (66%) | M | 72.6±6.4 | – | 1,359 (61%) | 74.7±6.5 | TaqMan SNP genotyping assays | (18) |
| Carrasquillo et al, 2014 | USA | Pos | 54 (76%) | N | – | – | 2,523 (56.7%) | – | TaqMan SNP genotyping assays | (14) |
| Asian | ||||||||||
| Yu et al, 2010 | China | Neg | 324 (56%) | C | – | 76.8±5.5 | 388 (54%) | 75.9±4.6 | MALDI-TOF mass spectrometry | (25) |
| Gu et al, 2011 | Indiana | Neg | 106 (56%) | C | – | 76.7±7.0 | 98 (55.1%) | 76.1±7.1 | PCR | (11) |
| Ohara et al, 2012 | Japan | Pos | 824 (77%) | C | – | 83.2±6.5 | 2,933 (56.0%) | 60.2±11.5 | Invader assay | (22) |
| Lin et al, 2012 | Taiwan | Pos | 268 (−) | C | – | – | 389 (−) | – | – | (19) |
| Chen et al, 2012 | Hong Kong | Neg | 462 (−) | C | – | – | 350 (−) | – | Sequenom platform | (15) |
| Ma et al, 2012 | China | Neg | 127 (58%) | C | – | 73.1±8.6 | 143 (55.2%) | 73.8±6.3 | PCR-RFLP | (20) |
| Miyashita et al, 2013 | Stage 1 (Japan) | Pos | 1,008 (72%) | C | 73.0±4.3 | – | 1,016 (57%) | 77.0±5.9 | Affymetrix GeneChip 6.0 microarrays | (21) |
| Stage 3 (Korean) | Pos | 339 (72%) | C | – | 73.7±9.5 | 1,129 (49%) | 71.0±4.9 | TaqMan assays | ||
| Lu et al, 2014 | Stage 2 (China) | Neg | 499 (55%) | C | – | 70.0±10.0 | 592 (59.3%) | 68.9±9.4 | PCR-RFLP | (26) |
| African descent | ||||||||||
| Jun et al, 2010 | USA (ADGC-AA) | Neg | 462 (−) | M | – | – | 449 (−) | – | Illumina 660Quad | (17) |
| Hispanics | ||||||||||
| Jun et al, 2010 | USA (ADGC-H) | Neg | 549 (−) | M | – | – | 544 (−) | – | Illumina HumanHap 650Y chip | (17) |
| Other/Mixed | ||||||||||
| Jun et al, 2010 | USA (ADGC-Wadi Ara) | Neg | 124 (−) | M | – | – | 142 (−) | – | Illumina 660Quad | (17) |
| Ferrari et al, 2012 | UK | Pos | 342 (59%) | C | 76.8±8.6 | – | 277 (64.6%) | 70.2±8.6 | TaqMan SNP genotyping assays | (16) |
Pos, positive: statistically significant; Neg, negative: non statistically significant; Alzheimer's disease diagnostic criteria: C, clinical criteria; N, neuropathological criteria; M, mixture of clinical and neuropathological criteria; -, no data obtained; PCR, polymerase chain reaction; SNP, single nucleotide polymorphism; RFLP, restriction fragment length polymorphism; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; ADGC-C, the Alzheimer's Disease Genetics Consortium-Caucasian; ADGC-AA, the ADGC-African American; ADGC-H, ADGC-Hispanics; ADGC-Wadi Ara, ADGC-Wadi Arab.
Table II.
Distribution of CLU rs11136000 genotypes and alleles among LOAD cases and controls in the included studies.
| Genotype distribution (%) | Allele distribution (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOAD | Control | LOAD | Control | |||||||||||
| Included studies | Studied population | TT | TC | CC | TT | TC | CC | T | C | T | C | OR | 95% CI | Z-value |
| Lambert et al, 2009 (6) | Stage 1 (France) | 270 | 900 | 869 | 860 | 2,561 | 1,957 | 1,440 | 2,638 | 4,281 | 6,475 | 0.83 | 0.77, 0.89 | −5.0122 |
| (13.2) | (44.1) | (42.6) | (16.0) | (47.6) | (36.4) | (35.3) | (64.7) | (39.8) | (60.2) | |||||
| Stage 2 (Italy) | 197 | 682 | 601 | 211 | 570 | 482 | 1,076 | 1,884 | 992 | 1,534 | 0.88 | 0.79, 0.99 | −2.2241 | |
| (13.3) | (46.1) | (40.6) | (16.7) | (45.1) | (38.2) | (36.4) | (63.6) | (39.3) | (60.7) | |||||
| Stage 2 (Spain) | 99 | 344 | 305 | 112 | 389 | 309 | 542 | 954 | 613 | 1,007 | 0.93 | 0.81, 1.08 | −0.9293 | |
| (13.2) | (46.0) | (40.8) | (13.8) | (48.0) | (38.1) | (36.2) | (63.8) | (37.8) | (62.2) | |||||
| Stage 2 (Belgium) | 155 | 472 | 408 | 79 | 241 | 171 | 782 | 1,288 | 399 | 583 | 0.89 | 0.76, 1.04 | −1.5117 | |
| (15.0) | (45.6) | (39.4) | (16.1) | (49.1) | (34.8) | (37.8) | (62.2) | (40.6) | (59.4) | |||||
| Stage 2 (Finland) | 86 | 286 | 224 | 109 | 323 | 218 | 458 | 734 | 541 | 759 | 0.88 | 0.75, 1.03 | −1.6241 | |
| (14.4) | (48.0) | (37.6) | (16.8) | (49.7) | (33.5) | (38.4) | (61.6) | (41.6) | (58.4) | |||||
| Harold et al, 2009 (7) | Stage 1 (USA) | 163 | 509 | 481 | 328 | 1,085 | 774 | 835 | 1,471 | 1,741 | 2,633 | 0.86 | 0.77, 0.95 | −2.8678 |
| (14.1) | (44.1) | (41.7) | (15.0) | (49.6) | (35.4) | (36.2) | (63.8) | (39.8) | (60.2) | |||||
| Stage 1 | 295 | 1038 | 887 | 787 | 2323 | 1,723 | 1,628 | 2,812 | 3,897 | 5769 | 0.86 | 0.80, 0.92 | −4.1228 | |
| (UK, Ireland) | (13.3) | (46.8) | (40.0) | (16.3) | (48.1) | (35.7) | (36.7) | (63.3) | (40.3) | (59.7) | ||||
| Stage 1 (Germany) | 66 | 240 | 233 | 144 | 368 | 294 | 372 | 706 | 656 | 956 | 0.76 | 0.65, 0.89 | −3.2328 | |
| (12.2) | (44.5) | (43.2) | (17.5) | (46.8) | (35.7) | (34.5) | (65.5) | (40.7) | (59.3) | |||||
| Giedraitis et al, 2009 (9) | Sweden | 15 | 31 | 33 | 59 | 166 | 140 | 61 | 97 | 284 | 446 | 0.99 | 0.69, 1.41 | −0.0693 |
| (19.0) | (39.2) | (41.8) | (16.2) | (45.5) | (38.4) | (38.6) | (61.4) | (38.1) | (61.1) | |||||
| Golenkina et al, 2010 (10) | Russian | 58 | 262 | 214 | 99 | 341 | 262 | 378 | 690 | 539 | 865 | 0.88 | 0.75, 1.04 | −1.5277 |
| (10.9) | (49.0) | (40.1) | (14.1) | (48.6) | (37.3) | (35.4) | (64.6) | (38.4) | (61.6) | |||||
| Jun et al, 2010 (17) | USA (ADGC-C) | – | – | – | – | – | – | – | – | – | – | 0.91 | 0.85, 0.96 | −3.7532 |
| USA (ADGC-AA) | – | – | – | – | – | – | – | – | – | – | 1.06 | 0.89, 1.28 | 0.6172 | |
| USA (ADGC-H) | – | – | – | – | – | – | – | – | – | – | 1.10 | 0.91, 1.32 | 1.1137 | |
| USA | – | – | – | – | – | – | – | – | – | – | 0.96 | 0.69, 1.32 | −0.2141 | |
| (ADGC-Wadi Ara) | ||||||||||||||
| Seshadri et al, 2010 (24) | Spain (ACE) | 148 | 525 | 467 | 184 | 575 | 450 | 821 | 1459 | 943 | 1475 | 0.88 | 0.78, 0.99 | −2.1150 |
| (13.0) | (46.1) | (41.0) | (15.2) | (47.6) | (37.2) | (36.0) | (64.0) | (39.0) | (61.0) | |||||
| Carrasquillo et al, 2010 (13) | USA | 249 | 848 | 722 | 431 | 1,241 | 893 | 1,346 | 2,292 | 2,103 | 3,027 | 0.85 | 0.77, 0.92 | −3.7725 |
| (13.7) | (46.6) | (39.7) | (16.8) | (48.4) | (34.8) | (37.0) | (63.0) | (41.0) | (59.0) | |||||
| Yu et al, 2010 (25) | China | 2 | 104 | 218 | 12 | 126 | 250 | 108 | 540 | 150 | 626 | 0.83 | 0.64, 1.10 | −1.2983 |
| (0.6) | (32.1) | (67.3) | (3.1) | (32.5) | (64.4) | (16.7) | (83.3) | (19.3) | (80.7) | |||||
| Schjeide et al, 2011 (23) | Germany | – | – | – | – | – | – | – | – | – | – | 0.99 | 0.83, 1.27 | −0.0905 |
| USA | – | – | – | – | – | – | – | – | – | – | 0.84 | 0.73, 0.96 | −3.4529 | |
| Gu et al, 2011 (11) | Indiana | 4 | 72 | 30 | 6 | 67 | 25 | 80 | 132 | 79 | 117 | 0.90 | 0.60, 1.34 | −0.5318 |
| (3.8) | (67.9) | (28.3) | (6.1) | (68.4) | (25.5) | (37.7) | (62.3) | (40.3) | (59.7) | |||||
| Ohara et al, 2012 (22) | Japan | 60 | 295 | 469 | 242 | 1,156 | 1,535 | 415 | 1,233 | 1,640 | 4,226 | 0.87 | 0.77, 0.98 | −2.2324 |
| (7.3) | (35.8) | (56.9) | (8.3) | (39.4) | (52.3) | (25.2) | (74.8) | (28.0) | (72.0) | |||||
| Lin et al, 2012 (19) | Taiwan | 3 | 89 | 176 | 29 | 118 | 242 | 95 | 441 | 176 | 602 | 0.74 | 0.56, 0.97 | −2.1521 |
| (1.1) | (33.2) | (65.7) | (7.5) | (30.3) | (62.2) | (17.7) | (82.3) | (22.6) | (77.4) | |||||
| Chen et al, 2012 (15) | Hong Kong | 15 | 162 | 274 | 24 | 114 | 200 | 192 | 710 | 162 | 514 | 0.86 | 0.68, 1.09 | −1.2616 |
| (3.3) | (35.9) | (60.8) | (7.1) | (33.7) | (59.2) | (21.3) | (78.7) | (24.0) | (76.0) | |||||
| Bettens et al, 2012 (12) | Stage 1 (Belgium) | – | – | – | – | – | – | 676 | 1,232 | 630 | 990 | 0.79 | 0.68, 0.93 | −2.1199 |
| (35.4) | (64.6) | (38.9) | (61.1) | |||||||||||
| Stage 2 (France) | – | – | – | – | – | – | 875 | 1,707 | 452 | 764 | 0.93 | 0.79, 1.10 | −1.9789 | |
| (33.9) | (66.1) | (37.2) | (62.8) | |||||||||||
| Stage 2 (Canada) | – | – | – | – | – | – | 236 | 372 | 179 | 299 | 1.00 | 0.77, 1.31 | 0.4606 | |
| (38.8) | (61.2) | (37.4) | (62.6) | |||||||||||
| Kamboh et al, 2012 (18) | USA | 179 | 623 | 542 | 195 | 636 | 519 | 981 | 1707 | 1,026 | 1,674 | 0.94 | 0.84, 1.05 | −1.1420 |
| (13.3) | (46.4) | (40.3) | (14.4) | (47.1) | (38.4) | (36.5) | (63.5) | (38.0) | (62.0) | |||||
| Ferrari et al, 2012 (16) | UK | – | – | – | – | – | – | 254 | 430 | 242 | 312 | 0.76 | 0.60, 0.96 | −2.3357 |
| (37.1) | (62.9) | (43.7) | (56.3) | |||||||||||
| Ma et al, 2012 (20) | China | 7 | 39 | 81 | 5 | 58 | 80 | 53 | 201 | 68 | 218 | 0.85 | 0.56, 1.27 | −0.8090 |
| (5.5) | (30.7) | (63.8) | (3.5) | (40.6) | (55.9) | (20.9) | (79.1) | (23.8) | (76.2) | |||||
| Miyashita et al, 2013 (21) | Stage 1 (Japan) | – | – | – | – | – | – | – | – | – | – | 0.87 | 0.78, 0.97 | −2.0100 |
| Stage 3 (Korean) | – | – | – | – | – | – | – | – | – | – | 0.79 | 0.63, 0.98 | −2.3006 | |
| Carrasquillo et al, 2014 (14) | USA | 4 | 25 | 25 | 416 | 1,165 | 843 | 33 | 75 | 1,997 | 2,851 | 0.60 | 0.39, 0.92 | −2.2044 |
| (7.4) | (46.3) | (46.3) | (17.2) | (48.0) | (34.8) | (30.6) | (69.4) | (41.2) | (58.8) | |||||
| Lu et al, 2014 (26) | Stage 2 (China) | 18 | 156 | 319 | 23 | 161 | 399 | 192 | 794 | 207 | 959 | 1.12 | 0.90, 1.39 | 1.0224 |
| (3.7) | (31.6) | (64.7) | (4.0) | (27.6) | (68.4) | (19.5) | (80.5) | (17.8) | (82.2) | |||||
LOAD, late-onset Alzheimer's disease; CI, confidence interval; OR, odds ratio; ADGC-C, the Alzheimer's Disease Genetics Consortium-Caucasian; ADGC-AA, ADGC-African American; ADGC-H, ADGC-Hispanics; ADGC-Wadi Ara, ADGC-Wadi Arab.
Considering ethnic differences, subgroup meta-analyses were conducted according to five ethnicities (Caucasian, Asian, African descent, Hispanic and other/mixed ethnicities)
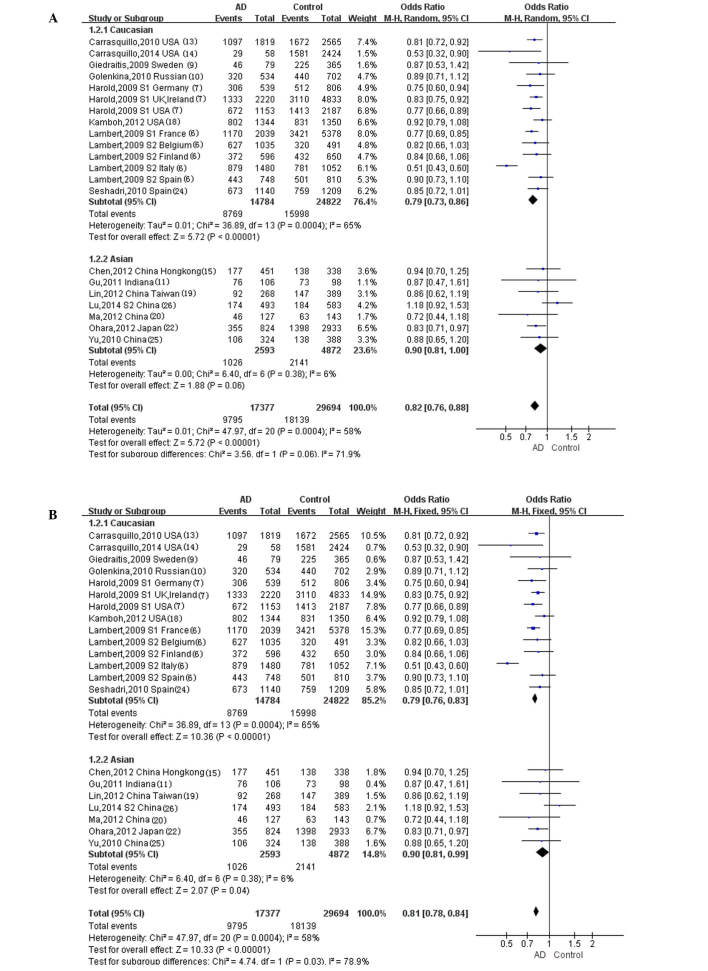
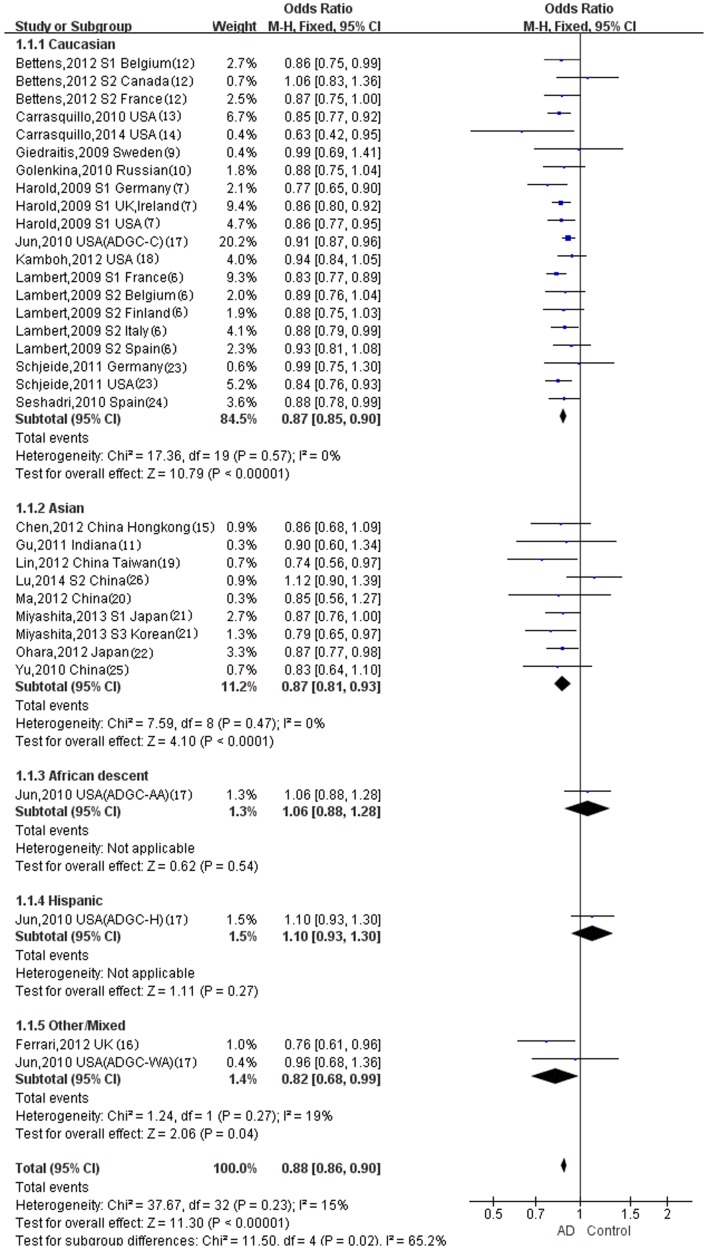
Using the (TT+TC)/CC genotype model (Fig. 2), a significant heterogeneity was observed in the Caucasian population with P=0.0004, although no significant heterogeneity was observed in the Asian population with P=0.38. Using the T/C allele model (Fig. 3), statistically significant heterogeneity was not observed in the Caucasian population with P=0.57, nor in the Asian (P=0.47) or other/mixed ethnicity populations (P=0.27). No heterogeneity tests were required in the ethnic subgroups in which only one study was eligible.
Figure 2.
Forest plot for the meta-analysis of the rs11136000 genotype (TT+TC, vs. CC). (A) Random effect model. (B) Fixed effect model.
Figure 3.
Forest plot for the meta-analysis of the rs11136000 allele (T, vs. C). Fixed effect model. Table I.
Meta-analysis
The random-effect model was used to test for the overall effect of the (TT+TC)/CC genotype model in the Caucasian population (Fig. 2A), in which a statistically significant difference was observed (overall OR, 0.79; 95% CI, 0.73–0.86; P<0.00001). The fixed-effect model was used in the Asian population for the (TT+TC)/CC genotype model (Fig. 2B), and a statistically significant difference was also observed (overall OR, 0.90; 95% CI, 0.81–0.99; P=0.04). The fixed-effect model was used in the Caucasian, Asian and other/mixed ethnicity populations for the T/C allele model (Fig. 3), and statistically significant differences were observed for all ethnicities (Caucasian: Overall OR, 0.87; 95% CI, 0.85–0.90; P<0.00001; Asian: Overall OR, 0.87; 95% CI, 0.81–0.93; P<0.0001; other/mixed ethnicities: Overall OR, 0.82; 95% CI, 0.68–0.99; P=0.04). Statistically significant differences were not however observed in the populations of African descent (overall OR, 1.06; 95% CI, 0.88–1.28; P=0.54) or Hispanics (overall OR, 1.10; 95% CI, 0.93–1.30; P=0.27) for the T/C allele model (Fig. 3).
Evaluation of publication bias
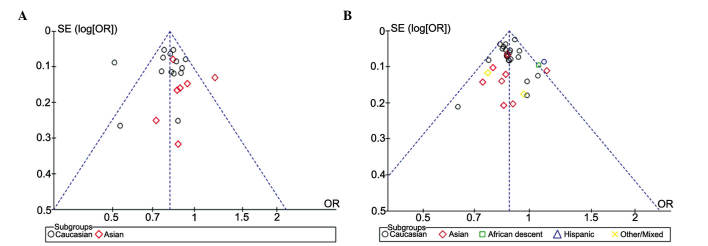
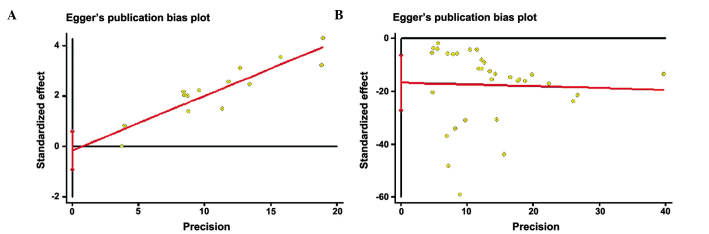
The RevMan funnel plot for the genotype model exhibited relative symmetry, whereas the allele model exhibited incomplete symmetry (Fig. 4). Following Egger's linear regression analysis and Begg's rank correlation test, it was demonstrated that the studies on genotype had no statistically significant publication bias (bias factors, −0.156; P=0.658 of Egger's; P=0.584 of Begg's), whereas those on alleles did have some bias (bias factors, −16.716; P=0.002 of Egger's; P=0.009 of Begg's) (Table III and Fig. 5). Nfs0.05 was equal to 750.604, which was large enough to warrant ignoring the publication bias.
Figure 4.
Funnel plot of the meta-analysis of rs11136000. (A) Genotype (TT+TC, vs. CC); (B) allele (T, vs. C). The funnel plot for the genotype model exhibited relative symmetry, whereas the allele model exhibited incomplete symmetry. OR, odds ratio; SE, standard error.
Table III.
Results of Egger's linear regression analysis and Begg's rank correlation test for publication bias.
| Egger's test | Begg's test | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Std-Eff | Coef.a | SE | t | P>|t| | 95% CI | Adj. Kendall's score (P-Q) | SD of score | Number of studies | Z-value | Pr>|z| | |
| Genotype | Slope | 0.2160835 | 0.0287501 | 7.52 | 0.000 | 0.1534424, 0.2787246 | −11 | 18.27 | 14 | 0.55 | 0.584 |
| bias | −0.1556121 | 0.3430764 | −0.45 | 0.658 | −0.9031114, 0.5918871 | ||||||
| Allele | Slope | −0.0658025 | 0.3326355 | −0.20 | 0.844 | −0.744217, 0.0612612 | −170 | 64.54 | 33 | 2.62 | 0.009 |
| bias | −16.71626 | 5.075586 | −3.29 | 0.002 | −27.06799, −6.364535 | ||||||
Coefficient was the intercept of regression analysis. P>ltl = 0.658, 0.002; Pr>lzl = 0.584, 0.009 (where both stand for no bias). SE, standard error; SD, standard deviation; Std-Eff, standard effect.
Figure 5.
Egger's bias plot for the meta-analysis of rs11136000. (A) Genotype (TT+TC, vs. CC); (B) allele (T, vs. C). The plot for the genotype model exhibited relative symmetry, whereas the allele model exhibited incomplete symmetry.
Discussion
The CLU gene is located on p21-p12 of human chromosome 8, with CLU as its encoded product, which has various physiological functions, including participating in lipid metabolism (28), oxidative stress reaction (29), and cell cycle regulation (30). CLU is highly expressed in cerebrospinal fluid and amyloid plaques in brain tissues, and is involved in the pathogenesis of AD (4,5,31). Yerbury et al (32) demonstrated that the deposition of CLU in senile plaques and neurofibrillary tangles of AD. Howlett et al (33) further reported a correlation between CLU and senile plaque Aβ40 in the brain cortex of patients with AD. Martin-Rehrmann et al (34) demonstrated the presence of dysfunctional neurons with phosphorylated tau protein surrounding the senile plaques in 71% of CLU-positive patients with AD. Furthermore, they also showed that the tau and phosphorylated tau protein were significantly increased in the rat hippocampus, following the injection of a CLU-rich solution (34). It was suggested that CLU may be involved in the formation of Aβ and dysfunctional neurons in the pathological state of AD (5). Wahrle et al (35) reported that CLU regulated the expression and metabolism of APOE in the brain. Indeed, the levels of cholesterol and APOE were decreased in the mouse following CLU gene knocked out. Aβ and clusterin combine together in normal cerebrospinal fluid, which suggested that CLU may be involved in the metabolism, transportation and clearance of Aβ in the brain (30). Bell et al (36) demonstrated that the clearance of Aβ42 through the blood brain barrier was increased by 83% when it was combined with CLU. Thambisetty et al (37) demonstrated the important role of CLU in the pathogenesis of AD. They showed that CLU was associated with atrophy of the entorhinal cortex and rapid clinical progression of AD (37). The higher the plasma concentration levels of CLU, the more significant the entorhinal cortex atrophy, and the more severe the cognitive impairment (37). It was also demonstrated that increased plasma concentration levels of CLU was predictive of greater Aβ burden in the medial temporal lobe (31). In view of the above results, it was hypothesized that gene mutations or specific environmental factors could induce changes in CLU gene expression or function, which have an important role in the pathogenesis of AD.
Since October 2009, association studies between the CLU gene polymorphism and LOAD susceptibility have rapidly progressed worldwide, and have been conducted in numerous populations, including Caucasian, Asian, African descent, and Hispanic populations (6,7,9–26). Notably, SNP rs11136000 in the CLU gene was demonstrated to be significantly associated with LOAD by two GWAS with large sample sizes and two stages, of which one was conducted by Lambert et al (6) (OR, 0.86; 95% CI, 0.81–0.90; P=7.5×10−9) and the other by Harold et al (7) (OR, 0.86; 95% CI, 0.82–0.90; P=8.5×10−10). The meta-analysis carried out by Jones et al (38) also supported the above conclusion (P=1.72 × 10−15). From then on, numerous other studies obtained concordant or non-concordant results (9–26). An issue is that many of the studies differ in their selection of ethnic genetic backgrounds and sample sizes, which leads to inconsistent or contrary conclusions regarding the association between gene polymorphism and diseases. The present study performed a meta-analysis to systematically review the published articles regarding the association between the CLU polymorphism rs11136000 and LOAD susceptibility, with increased sample size to improve and enhance test efficiency and ethnic subgroups to eliminate possible ethnic differences. This investigation aimed to provide objective results of evidence-based medicine for the association between CLU gene polymorphisms and AD susceptibility. The results of the present study demonstrated a positive association between CLU gene polymorphisms and LOAD susceptibility, as expected.
In the present study, 20 studies of interest were included in a meta-analysis, covering 33 independent study populations from various regions, ethnicities and stages, all meeting the predetermined inclusion criteria. The results demonstrated that there were some differences in the association between rs11136000 and LOAD among the various ethnicities. Meta-analysis supported the following conclusions: 1) TT+TC genotypes and the T allele of rs11136000 in Caucasian populations are associated with LOAD, as for the overall-effect test P<0.01; 2) TT+TC genotypes and the T allele of rs11136000 in Asian populations is associated with LOAD, as for the overall-effect test both P<0.05; 3) the T allele of rs11136000 in the population of African descent and the Hispanic population was not associated with LOAD, as for the single tests P>0.05; and 4) the T allele of rs11136000 in the other/mixed ethnicity group was associated with LOAD, as for the overall-effect test P<0.05. To summarize, the meta-analysis demonstrated that TT+TC genotypes and (or) the T allele of rs11136000 in Caucasian, Asian, and other/mixed ethnicity populations were associated with LOAD (P<0.05), and the population carrying the T allele would likely suffer lower LOAD risk (OR<1). The results therefore demonstrated that the T allele of rs11136000 had a protective effect on LOAD. The results also demonstrated the impact of racial differences on association studies of gene polymorphisms and disease.
In addition to ethnic differences, sample size also had an important effect on the study results. In the present meta-analysis, there was only one study population of African descent or Hispanic population included (17), with a relatively small sample size, which may not have provided accurate results. For instance, in the studies conducted in the Chinese Han population, Yu et al (25), Chen et al (15) and Ma et al (20) did not report any statistical significance in the T allele distribution of rs11136000 (P=0.21, 0.22, 0.42, respectively). However, when sample size was enlarged through meta-analysis and the three studies were included, a weak association was revealed (P=0.048 in the overall effect test). The majority of the studies and those with the largest sample size were conducted in the Caucasian population in the current meta-analysis, and therefore the results obtained for the Caucasian population are the most reliable. Further studies are required in other ethnic groups with larger sample sizes in order to improve the reliability of the study findings.
It is not only rs11136000 but also other polymorphisms of the CLU gene may contribute to LOAD susceptibility. For instance, the study carried out by Yu et al (25) in the Chinese Han population did not find an association between rs11136000 and LOAD, although the results indicated instead an association between rs9331888 and LOAD, with OR=1.39 (95% CI, 1.13–1.72) and P=0.002. Other studies also demonstrated the association between polymorphisms of rs9331888 (6,11,25), rs2279590 (6,15,21,23), rs7982 (7,17), rs7012010 (7,17), rs9331908 (12), rs1532278 (12,39), and rs9331949 (40) and LOAD, although these studies were few and inconsistent in their results. The rs11136000 polymorphism systematically reviewed in the present meta-analysis is likely one of the factors that influences LOAD susceptibility. The precise forms of the genetic variants require further investigation. The meta-analysis only involved rs11136000, the most well-established CLU gene polymorphism, but was not representative of the entire CLU gene. The results of the meta-analysis supported that the CLU gene polymorphism was associated with LOAD susceptibility in Caucasian and Asian populations, but there was no such definitive association in populations of African descent or Hispanic populations.
In conclusion, although the studies included in the meta-analysis of the present investigation were less than those conducted on the APOE gene, all were published in authorized journals after 2009, which indicated that the research on the CLU gene polymorphism was novel and promising. It is likely that the CLU polymorphism will be the subject of numerous studies on the genetic susceptibility to AD in the future. Further investigations are required in various ethnic population. The present meta-analysis will be regularly updated in order to draw more scientific and reliable conclusions regarding the association between the CLU gene polymorphism and susceptibility to LOAD.
There are clinical significances and wider implications of the findings in the present study. For example, there may be a use in monitoring disease progression in the elderly at risk. Furthermore, understanding the influence of the LOAD risk variants on cognitive decline may provide additional information with regard to their plausible mechanism of action.
References
- 1.Ossenkoppele R, van der Flier WM, Zwan MD, Adriaanse SF, Boellaard R, Windhorst AD, Barkhof F, Lammertsma AA, Scheltens P, van Berckel BN. Differential effect of APOE genotype on amyloid load and glucose metabolism in AD dementia. Neurology. 2013;80:359–365. doi: 10.1212/WNL.0b013e31827f0889. [DOI] [PubMed] [Google Scholar]
- 2.Tycko B, Feng L, Nguyen L, Francis A, Hays A, Chung WY, Tang MX, Stern Y, Sahota A, Hendrie H, Mayeux R. Polymorphisms in the human apolipoprotein-J/clusterin gene: Ethnic variation and distribution in Alzheimer's disease. Hum Genet. 1996;98:430–436. doi: 10.1007/s004390050234. [DOI] [PubMed] [Google Scholar]
- 3.Bertram L, Tanzi RE. Alzheimer's disease: New light on an old CLU. Nat Rev Neurol. 2010;6:11–13. doi: 10.1038/nrneurol.2009.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malkki H. Alzheimer disease: Chaperone protein clusterin is involved in amyloid-β-associated entorhinal atrophy in early AD. Nat Rev Neurol. 2014;10:60. doi: 10.1038/nrneurol.2014.1. [DOI] [PubMed] [Google Scholar]
- 5.Nuutinen T, Suuronen T, Kauppinen A, Salminen A. Clusterin: A forgotten player in Alzheimer's disease. Brain Res Rev. 2009;61:89–104. doi: 10.1016/j.brainresrev.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 7.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler AW, Ng MY, Hamshere ML, Forabosco P, Wroe R, Al-Chalabi A, Lewis CM, Powell JF. Meta-analysis of linkage studies for Alzheimer's disease-a web resource. Neurobiol Aging. 2009;30:1037–1047. doi: 10.1016/j.neurobiolaging.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Giedraitis V, Kilander L, Degerman-Gunnarsson M, Sundelöf J, Axelsson T, Syvänen AC, Lannfelt L, Glaser A. Genetic analysis of Alzheimer's disease in the Uppsala Longitudinal Study of Adult Men. Dement Geriatr Cogn Disord. 2009;27:59–68. doi: 10.1159/000191203. [DOI] [PubMed] [Google Scholar]
- 10.Golenkina SA, Gol'tsov AIu, Kuznetsova IL, Grigorenko AP, Andreeva TV, Reshetov DA, Kunizheva SS, Shagam LI, Morozova IIu, Goldenkova-Pavlova IV, et al. Analysis of clustering gene (CLU/APOJ) polymorphism in Alzheimer's disease patients and in normal cohorts from Russian populations. Mol Biol (Mosk) 2010;44:620–626. doi: 10.1134/S0026893310040072. (In Russian) [DOI] [PubMed] [Google Scholar]
- 11.Gu H, Wei X, Chen S, Kurz A, Müller U, Gasser T, Dodel RC, Farlow MR, Du Y. Association of clusterin gene polymorphisms with late-onset Alzheimer's disease. Dement Geriatr Cogn Disord. 2011;32:198–201. doi: 10.1159/000331276. [DOI] [PubMed] [Google Scholar]
- 12.Bettens K, Brouwers N, Engelborghs S, Lambert JC, Rogaeva E, Vandenberghe R, Le Bastard N, Pasquier F, Vermeulen S, Van Dongen J, et al. Both common variations and rare non-synonymous substitutions and small insertion/deletions in CLU are associated with increased AD risk. Mol Neurodegener. 2012;7:3. doi: 10.1186/1750-1326-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Dickson DW, Graff-Radford NR, et al. Replication of CLU, CR1 and PICALM associations with alzheimer's disease. Arch Neurol. 2010;67:961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrasquillo MM, Khan Qu, Murray ME, Krishnan S, Aakre J, Pankratz VS, Nguyen T, Ma L, Bisceglio G, Petersen RC, et al. Late-onset Alzheimer disease genetic variants in posterior cortical atrophy and posterior AD. Neurology. 2014;82:1455–1462. doi: 10.1212/WNL.0000000000000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen LH, Kao PY, Fan YH, Ho DT, Chan CS, Yik PY, Ha JC, Chu LW, Song YQ. Polymorphisms of CR1, CLU and PICALM confer susceptibility of Alzheimer's disease in a southern Chinese population. Neurobiol Aging. 2012;33:210.e1–210.e7. doi: 10.1016/j.neurobiolaging.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 16.Ferrari R, Moreno JH, Minhajuddin AT, O'Bryant SE, Reisch JS, Barber RC, Momeni P. Implication of common and disease specific variants in CLU, CR1 and PICALM. Neurobiol Aging. 2012;33:1846.e7–1846.e18. doi: 10.1016/j.neurobiolaging.2012.01.110. [DOI] [PubMed] [Google Scholar]
- 17.Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, Buxbaum JD, Ertekin-Taner N, Fallin MD, Friedland R, et al. Meta-analysis confirms CR1, CLU and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67:1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamboh MI, Minster RL, Demirci FY, Ganguli M, Dekosky ST, Lopez OL, Barmada MM. Association of CLU and PICALM variants with Alzheimer's disease. Neurobiol Aging. 2012;33:518–521. doi: 10.1016/j.neurobiolaging.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin YL, Chen SY, Lai LC, Chen JH, Yang SY, Huang YL, Chen TF, Sun Y, Wen LL, Yip PK, et al. Genetic polymorphisms of clusterin gene are associated with a decreased risk of Alzheimer's disease. Eur J Epidemiol. 2012;27:73–75. doi: 10.1007/s10654-012-9650-5. [DOI] [PubMed] [Google Scholar]
- 20.Ma JF, Liu LH, Zhang Y, Wang Y, Deng YL, Huang Y, Wang G, Xu W, Cui PJ, Fei QZ, et al. Association study of clusterin polymorphism rs11136000 with late onset Alzheimer's disease in Chinese Han population. Am J Alzheimers Dis Other Demen. 2012;26:627–630. doi: 10.1177/1533317511432735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, Kawarabayashi T, Shoji M, Tomita N, Arai H, et al. SORL1 is genetically associated with late-onset Alzheimer's disease in Japanese, Koreans and Caucasians. PLoS One. 2013;8:e58618. doi: 10.1371/journal.pone.0058618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohara T, Ninomiya T, Hirakawa Y, Ashikawa K, Monji A, Kiyohara Y, Kanba S, Kubo M. Association study of susceptibility genes for late-onset Alzheimer's disease in the Japanese population. Psychiatr Genet. 2012;22:290–293. doi: 10.1097/YPG.0b013e3283586215. [DOI] [PubMed] [Google Scholar]
- 23.Schjeide BM, Schnack C, Lambert JC, Lill CM, Kirchheiner J, Tumani H, Otto M, Tanzi RE, Lehrach H, Amouyel P, et al. The role of clusterin, complement receptor 1 and phosphatidylinositol binding clathrin assembly protein in Alzheimer disease risk and cerebrospinal fluid biomarker levels. Arch Gen Psychiatry. 2011;68:207–213. doi: 10.1001/archgenpsychiatry.2010.196. [DOI] [PubMed] [Google Scholar]
- 24.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu JT, Li L, Zhu QX, Zhang Q, Zhang W, Wu ZC, Guan J, Tan L. Implication of CLU gene polymorphisms in Chinese patients with Alzheimer's disease. Clin Chim Acta. 2010;411:1516–1519. doi: 10.1016/j.cca.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Lu SJ, Li HL, Sun YM, Liu ZJ, Yang P, Wu ZY. Clusterin variants are not associated with southern Chinese patients with Alzheimer's disease. Neurobiol Aging. 2014;35:2656.e9–2656.e11. doi: 10.1016/j.neurobiolaging.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 27.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/WNL.34.7.939. [DOI] [PubMed] [Google Scholar]
- 28.Leeb C, Eresheim C, Nimpf J. Clusterin is a ligand for apolipoprotein E receptor 2 (ApoER2) and very low density lipoprotein receptor (VLDLR) and signals via the Reelin-signaling pathway. J Biol Chem. 2014;289:4161–4172. doi: 10.1074/jbc.M113.529271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trougakos IP. The molecular chaperone apolipoprotein J/clusterin as a sensor of oxidative stress: Implications in therapeutic approaches-a mini-review. Gerontology. 2013;59:514–523. doi: 10.1159/000351207. [DOI] [PubMed] [Google Scholar]
- 30.Song HB, Jun HO, Kim JH, Yu YS, Kim KW, Min BH, Kim JH. Anti-apoptotic effect of clusterin on cisplatin-induced cell death of retinoblastoma cells. Oncol Rep. 2013;30:2713–2718. doi: 10.3892/or.2013.2764. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Ma Y, Wei X, Li Y, Wu H, Zhuang J, Zhao Z. Clusterin in Alzheimer's disease: A player in the biological behavior of amyloid-beta. Neurosci Bull. 2014;30:162–168. doi: 10.1007/s12264-013-1391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yerbury JJ, Poon S, Meehan S, Thompson B, Kumita JR, Dobson CM, Wilson MR. The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillar structures. FASEB J. 2007;21:2312–2322. doi: 10.1096/fj.06-7986com. [DOI] [PubMed] [Google Scholar]
- 33.Howlett DR, Hortobágyi T, Francis PT. Clusterin associates specifically with Aβ40 in Alzheimer's disease brain tissue. Brain Pathol. 2013;23:623–632. doi: 10.1111/bpa.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin-Rehrmann MD, Hoe HS, Capuani EM, Rebeck GW. Association of apolipoprotein J-positive beta-amyloid plaques with dystrophic neurites in Alzheimer's disease brain. Neurotox Res. 2005;7:231–242. doi: 10.1007/BF03036452. [DOI] [PubMed] [Google Scholar]
- 35.Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, Holtzman DM. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted ApoE. J Biol Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- 36.Bell RD, Sagare AP, Friedman AE, Bedi GS, Holtzman DM, Deane R, Zlokovic BV. Transport pathways for clearance of human Alzheimer's amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J Cereb Blood Flow Metab. 2007;27:909–918. doi: 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thambisetty M, Simmons A, Velayudhan L, Hye A, Campbell J, Zhang Y, Wahlund LO, Westman E, Kinsey A, Güntert A, et al. Association of plasma clusterin concentration with severity, pathology and progression in Alzheimer disease. Arch Gen Psychiatry. 2010;67:739–748. doi: 10.1001/archgenpsychiatry.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones L, Harold D, Williams J. Genetic evidence for the involvement of lipid metabolism in Alzheimer's disease. Biochim Biophys Acta. 2010;1801:754–761. doi: 10.1016/j.bbalip.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 39.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu JT, Ma XY, Wang YL, Sun L, Tan L, Hu N, Tan L. Genetic variation in Clusterin gene and Alzheimer's disease risk in Han Chinese. Neurobiol Aging. 2013;34:1921.e17–1921.e23. doi: 10.1016/j.neurobiolaging.2013.01.010. [DOI] [PubMed] [Google Scholar]