Abstract
Toll-like receptor 4 (TLR4), a recently identified vertebrate receptor, serves a pivotal role in immune responses. The aim of the present study was to investigate the association between the human TLR4 gene and recurrent spontaneous miscarriage (RSM). A total of 306 RSM patents and 306 age-matched controls were genotyped for four single-nucleotide polymorphisms (SNPs) of the human TLR4 gene (rs1927914, rs1927911, rs4986790 and rs4986791). Data were analyzed for Uygur and Han women separately using a haplotype-based case-control study. There were significant differences between the distributions of rs1927914, rs1927911 and rs4986790 SNPs between RSM patients and the controls (P=0.001, P<0.001 and P=0.015, respectively) were identified in Uygur women, and significant differences between the distributions of the rs1927914 and rs1927911 SNPs between RSM patients and the controls (P<0.001 and P<0.001, respectively) were identified in Han women. Results of the logistic regression analysis indicated that rs1927914, rs1927911 and rs4986790 SNPs were significantly higher in the RSM patients compared with the control individuals (P=0.012, P=0.024 and P=0.035, respectively) in Uygur women. Furthermore, significantly higher frequency was noted for the A-G-G haplotype (SNP1-SNP2-SNP3) (P=0.016) in RSM patients compared with the controls in Uygur women. The results indicate that rs1927914, rs1927911, rs4986790 and the A-G-G haplotype (SNP1-SNP2-SNP3) of the human TLR4 gene may be genetic markers for RSM in Uygur women, while rs1927914 and rs1927911 SNPs of the human TLR4 gene are most likely associated with RSM in Han women in Xinjiang.
Keywords: TLR4, recurrent spontaneous miscarriage, single-nucleotide polymorphism, case-control study
Introduction
Recurrent spontaneous miscarriage (RSM), defined as three or more consecutive losses before 20 weeks of gestation with the same partner (1), comprise a multifactorial condition enhanced by a variety of genetic, endocrine, anatomic, immunological, infectious, environmental, thrombophilic and metabolic factors (2). Regardless of extensive research undertaken in this field, the underlying cause of the majority (≤50%) of RSM cases remains unknown and hence poses a clinical challenge (3).
Toll-like receptors (TLRs), a recently identified group of vertebrate receptors, serve a pivotal role in immune responses. There are 10 TLRs expressed in humans (4), and Toll-like receptor 4 (TLR4) is one of the most actively investigated TLRs. TLR4 serves an essential role in the activation and regulation of the innate and acquired immune responses through the recognition of specific molecular patterns of pathogens and endogenous peptides (5).
Single-nucleotide polymorphisms (SNPs) are present in a number of TLR genes and have been implicated in the loss of self-tolerance, which may lead to autoimmunity and chronic inflammation (6). A number of studies have suggested that TLR4 polymorphisms are associated with the development of chronic inflammatory conditions (7,8), chronic granulomatous conditions (9) and cardivascular diseases (10).
To the best of our knowledge, there have been no previous studies on the association between the human TLR4 gene and RSM in Uygur women in Xinjiang. The aim of the present study was to investigate the association between the human TLR4 gene and RSM using a haplotype-based case-control study that employed SNPs in conjunction with separate analyses that examined the data with regard to Uygur and Han women.
Materials and methods
Subjects
Subjects diagnosed with RSM were recruited at the First Affiliated Hospital of Xinjiang Medical University in Urumqi, China, between 2012 and 2014. A total of 306 RSM patients were enrolled in the present study, with a Uygur/Han ratio of 1.01. RSM was diagnosed based on the following criteria: Patients who had experienced the spontaneous loss of three or more consecutive pregnancies prior to 20 weeks of gestation were included in the RSM group. Patients diagnosed with chromosomal abnormalities, uterine abnormalities, hereditary thrombophilia, endocrinologic disorders, immunologic factors and infections were excluded. A total of 306 age-matched healthy individuals (Uygur/Han ratio, 1.03) were enrolled as controls. There was no history of abortions or fertility treatments and no normal menstrual cycles for the controls. Informed consent was obtained from each of the subjects in accordance with the protocol approved by the Human Studies Committee of Xingjiang Medical University.
Genotyping
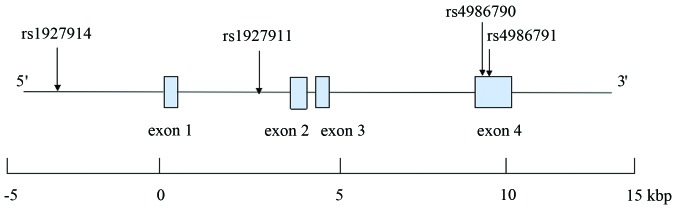
The human TLR4 gene is located on chromosome 9q33.1, spans ~17 kilobase pairs and contains four exons. There are 920 SNPs for the human TLR4 gene listed in the National Center for Biotechnology Information SNP database Build 130 (http://www.ncbi.nlm.nih.gov/SNP). Data for tag SNPs was screened using the International HapMap Project (http://www.hapmap.org/index.html.ja) using a cutoff level of r2 ≥0.5. According to the above criteria, rs1927914 (SNP1), rs1927911 (SNP2), rs4986790 (SNP3) and rs4986791 (SNP4) were selected for the present study. The structure of the human TLR4 gene is presented in Fig. 1.
Figure 1.
Structure of the human TLR4 gene. The gene consists of four exons (boxes) separated by three introns. Lines connecting the boxes indicate introns and intergenic regions. The filled boxes show the coding region. Arrows indicate the locations of single-nucleotide polymorphisms. Kbp, kilobase pairs.
Blood samples were collected from each participant and genomic DNA was extracted from peripheral blood leukocytes by phenol and chloroform extraction. DNA sequencing using the Sanger dideoxy method was performed by Polymorphic DNA Technologies (Alameda, CA, USA). The four SNPs that were used for complete sequencing of the TLR4 region included a portion of the upstream region, introns and exons, and these were analyzed in patients and controls from the cohort. Sequencing reactions were performed by O&E High-Tech Corporation in Beijing, China, and sequencing primers were designed using Primer Premier 5.0 software (Palo Alto, CA, USA).
Genotyping of novel SNPs
The detailed of genotyping of novel SNP have been previously described (11). Briefly, DNA sequencing of the complete TLR4 region was performed using a polymerase chain reaction direct sequencing method, which identified a heterozygous variant in subjects with RSM and controls. The sense primer for SNP1 was 5′TAGCATGAGAAATGAGGAAGTAAGGG3′ and the antisense primer was 5′GAGCTATGATGAGGATTGAAAATGTGG3′. The sense primer for SNP2 was 5′TAGCTGGCTTCTGCAAGGAAT3′ and the antisense primer was 5′GGAGACTGTCTGGCAGTCAAGATGT3′. The sense primer for SNP3 was 5′GTCCCTGAACCCTATGAACTTTATCC3′ and the antisense primer was 5′TCCCACCTTTGTTGGAAGTGAAAG3′. The sense primer for SNP4 was 5′CTGGTGAGTGTGACTATTGAAAGGG3′ and the antisense primer was 5′TTAGTACCTGAAGACTGGAGAGTGAG3′. Genotypes were determined automatically by the signal processing algorithms of Genotyping Console software 4.1 (Cleveland, OH, USA). To aid in the comparison of the results, each scoring method were saved in two separate output files.
Biochemical analysis
Serum concentrations of cholesterol and glucose were measuring using the standard methods employed by the Clinical Laboratory Department of the First Affiliated Hospital of Xinjiang Medical University.
Statistical analysis
All continuous variables were expressed as the mean ± standard deviation. The Mann-Whitney U test was used to compare differences in continuous variables between RSM patients and controls. The Fisher's exact test was used to calculate differences in categorical variables. The Hardy-Weinberg equilibrium was analyzed using χ2 analysis. The Fisher's exact test was used to calculate differences in the distribution of genotypes and alleles between the RSM patients and controls. According to the genotype data resulting from the genetic variations, linkage disequilibrium analysis and haplotype-based case-control analysis was performed using the expectation maximization algorithm and SNPAlyze version 3.2 (Dynacom Co., Ltd., Yokohama, Japan). The four SNPs were used to perform pair-wise linkage disequilibrium analysis. D >0.5 was used to assign SNP locations to one haplotype block. SNPs with r2 <0.5 were selected as tagged. A permutation test was used to calculate the frequency distribution of the haplotypes and P-values in the haplotype-based case-control analysis. The contribution of major risk factors was assessed using logistic regression analysis. P<0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using SPSS software for Windows (version 12; SPSS Inc., Chicago, IL, USA).
Results
Clinical characteristics
Table I presents the clinical characteristics of the study participants. In Uygur women, the age of menarche, systolic blood pressure, diastolic blood pressure and spontaneous miscarriages were significantly higher in RSM patients compared with the controls (P<0.05). In Han women, systolic blood pressure, number of pregnancies, spontaneous miscarriages and the frequency of drinking were significantly higher in RSM patients compared with the controls (P<0.05). There were no significant differences noted between RSM patients and the controls with regards to age, body mass index, pulse rate, serum cholesterol, serum glucose and the frequency of smoking in Uygur and Han women.
Table I.
Characteristics of study participants.
| Uygur | Han | |||||
|---|---|---|---|---|---|---|
| Characteristic | RSM patients | Controls | P-value | RSM patients | Controls | P-value |
| Number of subjects | 154 | 155 | – | 152 | 151 | – |
| Age (years) | 35.2±3.7 | 35.1±4.5 | 0.859 | 35.6±4.1 | 35.7±3.8 | 0.643 |
| Body mass index (kg/m2) | 23.2±2.9 | 22.8±2.7 | 0.261 | 22.5±4.4 | 21.6±3.0 | 0.066 |
| Menarche (years) | 11.8±2.6 | 12.4±3.9 | 0.002a | 12.3±3.9 | 12.5±4.2 | 0.127 |
| Systolic blood pressure (mmHg) | 110.8±15.2 | 119.3±14.3 | <0.001a | 113.1±14.7 | 120.8±13.6 | <0.001a |
| Diastolic blood pressure (mmHg) | 71.8±12.5 | 75.2±13.9 | <0.001a | 75.5±9.7 | 76.6±11.4 | 0.232 |
| Pulse rate (beats/min) | 75.8±8.1 | 75.1±7.2 | 0.356 | 75.1±6.3 | 74.6±5.8 | 0.438 |
| Cholesterol (mmol/l) | 5.3±1.1 | 4.7±0.9 | 0.061 | 5.2±1.0 | 4.8±1.1 | 0.132 |
| Glucose (mmol/l) | 5.2±0.6 | 5.1±0.7 | 0.677 | 5.4±0.3 | 5.3±0.8 | 0.547 |
| Number of pregnancies | 4.5±0.7 | 4.1±0.5 | 0.125 | 4.2±0.8 | 1.8±0.9 | <0.001a |
| Spontaneous miscarriages | 4.1±0.9 | 0±0.0 | <0.001a | 3.8±0.7 | 0±0.0 | <0.001a |
| Smoking (%) | 1.9 | 2.6 | 0.500 | 3.3 | 3.3 | 0.621 |
| Drinking (%) | 12.3 | 13.5 | 0.321 | 21.1 | 14.6 | 0.046a |
P<0.05. Continuous variables are expressed as the mean ± standard deviation. P-values of continuous variables were calculated by Mann-Whitney U test. P-values of categorical variables were calculated by Fisher's exact test. RSM, recurrent spontaneous miscarriage.
Distribution of genotypes
Table II presents the distribution of the genotypes and alleles of the four SNPs. For each SNP, the genotype distributions in the controls were not significantly different from the Hardy-Weinberg equilibrium values (data not shown). In Uygur women, there were significant differences between the distributions of the rs1927914, rs1927911 and rs4986790 SNPs between the RSM patients and the controls (P=0.001, P<0.001 and P=0.015, respectively). In Han women, there were significant differences between the distributions of the rs1927914 and rs1927911 SNPs between the RSM patients and the controls (P<0.001 and P<0.001, respectively).
Table II.
Genotype and allele distributions in RSM patients and controls.
| Uygur | Han | |||||
|---|---|---|---|---|---|---|
| RSM patients | Controls | P-value | RSM patients | Controls | P-value | |
| rs1927914 (SNP1) | ||||||
| Genotype | ||||||
| A/A | 88 (57.1%) | 56 (36.1%) | 0.001a | 90 (59.2%) | 54 (35.8%) | <0.001a |
| A/G | 49 (31.8%) | 65 (41.9%) | 48 (31.6%) | 64 (42.4%) | ||
| G/G | 17 (11.1%) | 34 (22.0%) | 14 (9.2%) | 33 (21.8%) | ||
| Allele | ||||||
| A | 225 (73.1%) | 177 (57.1%) | <0.001a | 228 (75.0%) | 172 (57.0%) | <0.001a |
| G | 83 (26.9%) | 133 (42.9%) | 76 (25.0%) | 130 (43.0%) | ||
| rs1927911 (SNP2) | ||||||
| Genotype | ||||||
| G/G | 90 (58.4%) | 55 (35.5%) | <0.001a | 91 (59.9%) | 55 (36.4%) | <0.001a |
| G/A | 47 (30.5%) | 73 (47.1%) | 45 (29.6%) | 70 (46.4%) | ||
| A/A | 17 (11.1%) | 27 (17.4%) | 16 (10.5%) | 26 (17.2%) | ||
| Allele | ||||||
| G | 227 (73.7%) | 183 (59.0%) | <0.001a | 227 (73.7%) | 180 (59.6%) | <0.001a |
| A | 81 (26.3%) | 127 (41.0%) | 81 (26.3%) | 122 (40.4%) | ||
| rs4986790 (SNP3) | ||||||
| Genotype | ||||||
| A/A | 142 (92.3%) | 152 (98.1%) | 0.015a | 148 (97.4%) | 149 (98.7%) | 0.345 |
| A/G | 12 (7.7%) | 3 (1.9%) | 4 (2.6%) | 2 (1.3%) | ||
| G/G | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ||
| Allele | ||||||
| A | 296 (96.1%) | 307 (99.0%) | 0.016a | 296 (98.7%) | 300 (99.3%) | 0.340 |
| G | 12 (3.9%) | 3 (1.0%) | 4 (1.3%) | 2 (0.7%) | ||
| rs4986791 (SNP4) | ||||||
| Genotype | ||||||
| C/C | 154 (100%) | 155 (100%) | – | 152 (100%) | 151 (100%) | – |
| C/T | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
| T/T | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
| Allele | ||||||
| C | 308 (100%) | 310 (100%) | – | 304 (100%) | 302 (100%) | – |
| T | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
P<0.05. P-values for genotype were calculated by Fisher's exact test. RSM, recurrent spontaneous miscarriage; SNP, single nucleotide polymorphism.
Risk factors and SNPs
Parameters in Table I that exhibited significant differences between the RSM patients and the controls were used for the logistic regression analysis. In Uygur women, parameters used included SNPs (rs1927914, rs1927911 and rs4986790), the age of menarche, systolic blood pressure, diastolic blood pressure and spontaneous miscarriages. As shown in Table III, results of the logistic regression analysis indicated that the distributions of rs1927914, rs1927911 and rs4986790 SNPs were significantly different in the RSM patients compared with the control individuals (P=0.012, P=0.024 and P=0.035, respectively). In addition, systolic blood pressure differed significantly between the RSM patients and the controls (P=0.011, P=0.021 and P=0.032, respectively) in Uygur women.
Table III.
Odds ratios and 95% confidence intervals for each risk factor and SNP associated with RSM in Uygur women.
| Uygur | |||
|---|---|---|---|
| Risk factor | Odd ratios | 95% confidence interval | P-value |
| rs1927914 (SNP1) | 6.86 | 1.934–46.950 | 0.012a |
| Menarche (years) | 0.731 | 0.343–1.524 | 0.419 |
| Systolic blood pressure (mmHg) | 1.135 | 1.029–1.249 | 0.011a |
| Diastolic blood pressure (mmHg) | 1.046 | 1.017–1.065 | 0.003a |
| Spontaneous miscarriages | 0.672 | 0.413–1.089 | 0.121 |
| rs1927911 (SNP2) | 4.824 | 1.203–19.672 | 0.024a |
| Menarche (years) | 1.771 | 1.070–2.931 | 0.026a |
| Systolic blood pressure (mmHg) | 2.421 | 1.091–5.297 | 0.021a |
| Diastolic blood pressure (mmHg) | 0.673 | 0.383–1.182 | 0.172 |
| Spontaneous miscarriages | 0.616 | 0.225–1.683 | 0.345 |
| rs4986790 (SNP3) | 8.272 | 1.168–40.358 | 0.035a |
| Menarche (years) | 1.121 | 0.351–3.562 | 0.852 |
| Systolic blood pressure (mmHg) | 2.487 | 1.089–5.745 | 0.032a |
| Diastolic blood pressure (mmHg) | 1.405 | 0.648–3.031 | 0.386 |
| Spontaneous miscarriages | 2.619 | 1.006–6.811 | 0.047a |
P<0.05. RSM, recurrent spontaneous miscarriage; SNP, single nucleotide polymorphism.
In Han women, parameters used included SNPs (rs1927914 and rs1927911), systolic blood pressure, number of pregnancies, spontaneous miscarriages and the frequency of drinking. As shown in Table IV, results of the logistic regression analysis indicated that the distributions of rs1927914 and rs1927911 SNPs were significantly different in the RSM patients compared with the control individuals (P=0.021 and P=0.004, respectively) in Han women.
Table IV.
Odds ratios and 95% confidence intervals for each risk factor and SNP associated with RSM in Han women.
| Han | |||
|---|---|---|---|
| Risk factor | Odd ratios | 95% confidence interval | P-value |
| rs1927914 (SNP1) | 6.431 | 1.321–31.347 | 0.021a |
| Systolic blood pressure (mmHg) | 1.967 | 1.071–3.589 | 0.037a |
| Number of pregnancies | 1.185 | 0.597–2.358 | 0.634 |
| Spontaneous miscarriages | 0.412 | 0.113–1.514 | 0.186 |
| Drinking (%) | 1.579 | 0.807–3.076 | 0.183 |
| rs1927911 (SNP2) | 2.234 | 1.271–3.917 | 0.004a |
| Systolic blood pressure (mmHg) | 1.127 | 1.029–1.235 | 0.008a |
| Number of pregnancies | 1.156 | 0.933–1.435 | 0.187 |
| Spontaneous miscarriages | 1.046 | 1.022–1.074 | 0.001a |
| Drinking (%) | 1.970 | 0.632–6.145 | 0.249 |
P<0.05. RSM, recurrent spontaneous miscarriage; SNP, single nucleotide polymorphism.
Haplotypes
Since most of the D-values of linkage disequilibrium for the human TLR4 gene in the controls were >0.5 (data not shown), this indicated that the four SNPs were located in one haplotype block. Since all of the r2 values were <0.5 (data not shown), this suggests that every SNP would be useful for the haplotype-based case-control study. Since rs4986791 (SNP4) has no allele distributions in RSM patients and the controls in this study, haplotypes were constructed using SNP1, SNP2 and SNP3. The distributions of the individual haplotypes for these combinations of the three SNPs were analyzed, and the results are presented in Table V. In Uygur women, significantly higher frequencies were noted for the A-G-G haplotype (SNP1-SNP2-SNP3; P=0.016) in RSM patients compared with the controls.
Table V.
Haplotype analysis in RSM patients and controls.
| Haplotype | Frequency in Uygur | Frequency in Han | |||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | SNP1 | SNP2 | SNP3 | RSM patients | Controls | P-value | RSM patients | Controls | P-value |
| H1 | G | A | A | 0.164 | 0.236 | 0.756 | 0.177 | 0.282 | 0.412 |
| H2 | G | G | A | 0.086 | 0.187 | 0.298 | 0.067 | 0.145 | 0.398 |
| H3 | G | G | G | 0.019 | 0.006 | 0.540 | 0.006 | 0.003 | 0.158 |
| H4 | A | A | A | 0.099 | 0.174 | 0.389 | 0.086 | 0.122 | 0.415 |
| H5 | A | G | A | 0.632 | 0.393 | 0.633 | 0.658 | 0.445 | 0.406 |
| H6 | A | G | G | 0.020 | 0.004 | 0.016a | 0.006 | 0.003 | 0.137 |
P<0.05. Haplotypes were estimated using SNPAlyze software. P-values were calculated by the permutation test. RSM, recurrent spontaneous miscarriage; SNP, single nucleotide polymorphism.
Discussion
RSM is a reproductive problem, which affects 1–3% of couples trying to conceive (12). Epidemiological studies have suggested that RSM comprises a multifactorial condition enhanced by a variety of genetic and environmental factors (13). A number of studies have suggested that RSM is associated with genetic disorders that include skewed X chromosome inactivation (14), HLA-G (15), age-related aneuploidy (16), autoimmune disorders (17) and acquired or inherited thrombophilia (18). In addition, several studies have shown that nutritional and lifestyle factors, smoking, and diet (19,20) contribute to RSM pathogenesis. For instance, obesity has a greater incidence in RSM patients compared with controls (21). Furthermore, a woman's medical history (3) is important since the risk of RSM is increased by previous miscarriages (20% after one miscarriage, 28% after two miscarriages) and age (40% risk for 40 year-old women, and 80% for women 45 and older). Considering all these risk factors, the related data (including age, body mass index, number of pregnancies and number of spontaneous miscarriages) has been collected and presented in Table I. Then, the relationship between these risk factors and RSM is analyzed using logistic regression and presented in Tables III and IV.
TLR4 is present in numerous cell types and signals the innate immune response. TLR4 expression has been investigated in tumor cells or cell lines, including extranodal marginal zone B-cell lymphomas, gastric carcinoma, hepatocellular carcinoma cells, pituitary epithelial tumor cell lines, colon cancer cells and human prostate epithelial PC3 cells (22). The innate immune system is the first line of defense against microbial infection and TLR4 serves as the first point of defense in the innate immune system. TLR4 is one of the most extensively investigated TLRs and recognizes endotoxic lipopoly saccharide (LPS) in the outer membrane of Gram-negative bacteria with its co-receptor MD-2 (23). TLR4 is critical in the recognition of viruses and bacteria, serving as a key immune system effector. Excessive signaling in response to LPS frequently results in uncontrolled amplification of inflammatory responses, leading to fatal septic shock (24,25).
Dysregulation of TLR4 signalling owing to SNPs may alter the ligand binding and balance between pro- and anti-inflammatory cytokines, thereby modulating the risk of chronic inflammatory and immune disorders. Consequently, several studies have shown that common SNPs of a series of low penetrance alleles of TLR4, modified during the above mentioned processes, are likely to serve an important role in cancer susceptibility (26,27). Two SNPs of the human TLR4, A299G (ref SNP ID: rs4986790, SNP3) and T399I (ref SNP ID: rs4986791, SNP4), have been identified and suggested to be associated with LPS hyporesponsiveness (28,29). Furthermore, two non-synonymous polymorphisms of TLR4 (Asp299Gly and Thr399Ile) have been suggested to alter the function of the receptor (30,31) and have been related to the development of chronic inflammatory conditions (32). In addition, Hirschfeld et al (33) reported that there was no association between fetal TLR4 polymorphisms, Asp299Gly and Thr399Ile, known to blunt LPS responsiveness, and the risk of idiopathic, chromosomally normal miscarriage. However, how the SNPs affect the function of TLR4 remains largely unknown.
The aim of the present study was to assess the association between the human TLR4 gene and RSM in Uygur and Han women in Xinjiang by using a haplotype-based case-control study. The results of the genotype distributions and logistic regression analysis indicated that rs1927914, rs1927911 and rs4986790 are most likely associated with RSM in Uygur women, while rs1927914 and rs1927911 are most likely associated with RSM in Han women. Additionally, logistic regression analysis indicated that systolic blood pressure is one of the risk factors for RSM patients, which is consistent with results of a previous study (34).
It has been demonstrated that haplotypes can provide valuable information that can be used for predicting the severity and prognosis of certain genetic disorders (35). With the dramatic increase in the number of available SNP markers, haplotyping based on genotype data has become an important component of genetic studies in general and has shown that these types of studies have a number of advantages over analyses that are just based on individual SNPs (36). In the present study, haplotypes for the human TLR4 gene were successfully established that were created from the combinations of the three SNPs. In the present haplotype analysis, there was a significantly higher frequency of the A-G-G haplotype (SNP1-SNP2-SNP3) in RSM patients compared with the controls in Uygur women. Therefore, it can be suggested that the result indicates that the A-G-G haplotype (SNP1-SNP2-SNP3) is the susceptibility haplotype for RSM in Uygur women, and it can be proposed that the A-G-G haplotype (SNP1-SNP2-SNP3) should be regarded as the genetic marker for RSM in Uygur women in Xinjiang. In addition, for SNP3, the G allele is the risk allele while the A allele is the safe allele for RSM in Uygur women.
To date, to the best of our knowledge, there have been no previous studies that have reported data on the association between TLR4 and RSM in Uygur women. Therefore, the current results provide novel data on the possibility that the TLR4 gene is most likely associated with RSM, and rs4986790 may be the nationality-specific genetic marker for RSM for Uygur women.
In conclusion, the present study examined the association between the human TLR4 gene and RSM. The present results indicate that rs1927914, rs1927911, rs4986790 and the A-G-G haplotype of the human TLR4 gene may be genetic markers for RSM in Uygur women in Xinjiang. In addition, the results suggest that rs1927914 and rs1927911 of the human TLR4 gene are most likely associated with RSM in Han women. Further studies are required to elucidate the functional association of TLR4 with RSM progression.
References
- 1.Rai R, Regan L. Recurrent miscarriage. Lancet. 2006;368:601–611. doi: 10.1016/S0140-6736(06)69204-0. [DOI] [PubMed] [Google Scholar]
- 2.Cramer DW, Wise LA. The epidemiology of recurrent pregnancy loss. Semin Reprod Med. 2000;18:331–339. doi: 10.1055/s-2000-13722. [DOI] [PubMed] [Google Scholar]
- 3.Ford HB, Schust DJ. Recurrent pregnancy loss: Etiology, diagnosis, and therapy. Rev Obstet Gynecol. 2009;2:76–83. [PMC free article] [PubMed] [Google Scholar]
- 4.Akira S, Takeda K. Functions of toll-like receptors: Lessons from KO mice. C R Biol. 2004;327:581–589. doi: 10.1016/j.crvi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 6.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroder NW, Schumann RR. Single nucleotide polymorphisms of toll-like receptors and susceptibility to infectious disease. Lancet Infect Dis. 2005;5:156–164. doi: 10.1016/S1473-3099(05)01308-3. [DOI] [PubMed] [Google Scholar]
- 8.Horie Y, Meguro A, Ota M, Kitaichi N, Katsuyama Y, Takemoto Y, Namba K, Yoshida K, Song YW, Park KS, et al. Association of TLR4 polymorphisms with Behcet's disease in a Korean population. Rheumatology (Oxford) 2009;48:638–642. doi: 10.1093/rheumatology/kep077. [DOI] [PubMed] [Google Scholar]
- 9.Pabst S, Baumgarten G, Stremmel A, Lennarz M, Knüfermann P, Gillissen A, Vetter H, Grohé C. Toll-like receptor (TLR) 4 polymorphisms are associated with a chronic course of sarcoidosis. Clin Exp Immunol. 2006;143:420–426. doi: 10.1111/j.1365-2249.2006.03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frantz S, Ertl G, Bauersachs J. Mechanisms of disease: Toll-like receptors in cardiovascular disease. Nat Clin Pract Cardiovasc Med. 2007;4:444–454. doi: 10.1038/ncpcardio0938. [DOI] [PubMed] [Google Scholar]
- 11.Varshney RK, Kudapa H, Roorkiwal M, Thudi M, Pandey MK, Saxena RK, Chamarthi SK, Mohan SM, Mallikarjuna N, Upadhyaya H, et al. Advances in genetics and molecular breeding of three legume crops of semi-arid tropics using next-generation sequencing and high-throughput genotyping technologies. J Biosci. 2012;37:811–820. doi: 10.1007/s12038-012-9228-0. [DOI] [PubMed] [Google Scholar]
- 12.Sierra S, Stephenson M. Genetics of recurrent pregnancy loss. Semin Reprod Med. 2006;24:17–24. doi: 10.1055/s-2006-931797. [DOI] [PubMed] [Google Scholar]
- 13.Cramer DW, Wise LA. The epidemiology of recurrent pregnancy loss. Semin Reprod Med. 2000;18:331–339. doi: 10.1055/s-2000-13722. [DOI] [PubMed] [Google Scholar]
- 14.Kuo PL, Huang SC, Chang LW, Lin CH, Tsai WH, Teng YN. Association of extremely skewed X-chromosome inactivation with Taiwanese women presenting with recurrent pregnancy loss. J Formos Med Assoc. 2008;107:340–343. doi: 10.1016/S0929-6646(08)60096-0. [DOI] [PubMed] [Google Scholar]
- 15.Pfeiffer KA, Fimmers R, Engels G, van der Ven H, van der Ven K. The HLA-G genotype is potentially associated with idiopathic recurrent spontaneous abortion. Mol Hum Reprod. 2001;7:373–378. doi: 10.1093/molehr/7.4.373. [DOI] [PubMed] [Google Scholar]
- 16.Hodes-Wertz B, Grifo J, Ghadir S, Kaplan B, Laskin CA, Glassner M, Munné S. Idiopathic recurrent miscarriage is caused mostly by aneuploid embryos. Fertil Steril. 2012;98:675–680. doi: 10.1016/j.fertnstert.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 17.Finan RR, Al-Irhayim Z, Mustafa FE, Al-Zaman I, Mohammed FA, Al-Khateeb GM, Madan S, Issa AA, Almawi WY. Tumor necrosis factor-alpha polymorphisms in women with idiopathic recurrent miscarriage. J Reprod Immunol. 2010;84:186–192. doi: 10.1016/j.jri.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Paidas MJ, Ku DH, Langhoff-Roos J, Arkel YS. Inherited thrombophilias and adverse pregnancy outcome: Screening and management. Semin Perinatol. 2005;29:150–163. doi: 10.1053/j.semperi.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Metwally M, Saravelos SH, Ledger WL, Li TC. Body mass index and risk of miscarriage in women with recurrent miscarriage. Fertil Steril. 2010;94:290–295. doi: 10.1016/j.fertnstert.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 20.Dasarathy J, Gruca LL, Bennett C, Parimi PS, Duenas C, Marczewski S, Fierro JL, Kalhan SC. Methionine metabolism in human pregnancy. Am J Clin Nutr. 2010;91:357–365. doi: 10.3945/ajcn.2009.28457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Dwyer V, Monaghan B, Fattah C, Farah N, Kennelly MM, Turner MJ. Miscarriage after sonographic confirmation of an ongoing pregnancy in women with moderate and severe obesity. Obes Facts. 2012;5:393–398. doi: 10.1159/000336253. [DOI] [PubMed] [Google Scholar]
- 22.Bulut Y, Faure E, Thomas L, Karahashi H, Michelsen KS, Equils O, Morrison SG, Morrison RP, Arditi M. Chlamydial heat shock protein 60 activates macrophages and endothelial cells through toll-like receptor 4 and MD2 in a MyD88-dependent pathway. J Immunol. 2002;168:1435–1440. doi: 10.4049/jimmunol.168.3.1435. [DOI] [PubMed] [Google Scholar]
- 23.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Defective LPS signalingin C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 24.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: Analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Rangel-Frausto MS. Sepsis: Still going strong. Arch Med Res. 2005;36:672–681. doi: 10.1016/j.arcmed.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Zidi S, Verdi H, Yilmaz-Yalcin Y, Yazici AC, Gazouani E, Mezlini A, Atac FB, Yacoubi-Loueslati B. Involvement of Toll-like receptors in cervical cancer susceptibility among Tunisian women. Bull Cancer. 2014;101:E31–E35. doi: 10.1684/bdc.2014.2037. [DOI] [PubMed] [Google Scholar]
- 27.He C, Tu H, Sun L, Xu Q, Gong Y, Jing J, Dong N, Yuan Y. SNP interactions of Helicobacter pylori-related host genes PGC, PTPN11, IL1B, and TLR4 in susceptibility to gastric carcinogenesis. Oncotarget. 2015;6:19017–19026. doi: 10.18632/oncotarget.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rallabhandi P, Bell J, Boukhvalova MS, Medvedev A, Lorenz E, Arditi M, Hemming VG, Blanco JC, Segal DM, Vogel SN. Analysis of TLR4 polymorphic variants: New insights into TLR4/ MD-2/CD14 stoichiometry, structure, and signaling. J Immunol. 2006;177:322–332. doi: 10.4049/jimmunol.177.1.322. [DOI] [PubMed] [Google Scholar]
- 29.Figueroa L, Xiong Y, Song C, Piao W, Vogel SN, Medvedev AE. The Asp299Gly polymorphism alters TLR4 signaling by interfering with recruitment of MyD88 and TRIF. J Immunol. 2012;188:4506–4515. doi: 10.4049/jimmunol.1200202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, Frees K, Watt JL, Schwartz DA. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 31.Ferwerda B, McCall MB, Verheijen K, Kullberg BJ, van der Ven AJ, Van der Meer JW, Netea MG. Functional consequences of Toll-like receptor 4 polymorphisms. Mol Med. 2008;14:346–352. doi: 10.2119/2007-00135.Ferwerda. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radstake TR, Franke B, Hanssen S, Netea MG, Welsing P, Barrera P, Joosten LA, van Riel PL, van den Berg WB. The toll-like receptor 4 Asp299Gly functional variant is associated with decreased rheumatoid arthritis disease susceptibility but does not influence disease severity and/or outcome. Arthritis Rheum. 2004;50:999–1001. doi: 10.1002/art.20114. [DOI] [PubMed] [Google Scholar]
- 33.Hirschfeld AF, Jiang R, Robinson WP, McFadden DE, Turvey SE. Toll-like receptor 4 polymorphisms and idiopathic chromosomally normal miscarriage. Hum Reprod. 2007;22:440–443. doi: 10.1093/humrep/del377. [DOI] [PubMed] [Google Scholar]
- 34.Almawi WY, Saldanha FL, Mahmood NA, Al-Zaman I, Sater MS, Mustafa FE. Relationship between VEGFA polymorphisms and serum VEGF protein levels and recurrent spontaneous miscarriage. Hum Reprod. 2013;28:2628–2635. doi: 10.1093/humrep/det308. [DOI] [PubMed] [Google Scholar]
- 35.Chen CK, Yu HT, Soong YK, Lee CL. New perspectives on preimplantation genetic diagnosis and preimplantation genetic screening. Taiwan J Obstet Gynecol. 2014;53:146–150. doi: 10.1016/j.tjog.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Stein CM, Elston RC. Finding genes underlying human disease. Clin Genet. 2009;75:101–106. doi: 10.1111/j.1399-0004.2008.01083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]