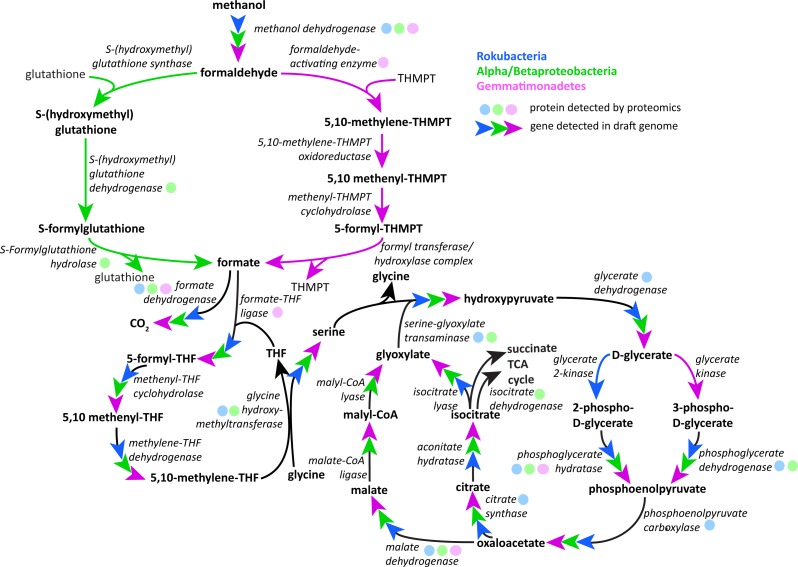
Figure 4. Putative active methylotrophy pathways in Rokubacteria, Proteobacteria, and Gemmatimonadetes.
The colored arrows indicate genetic evidence of proteins that catalyze the reactions that transform methanol into CO2 or cell material and colored dots indicate representation in the proteome. Gemmatimonadetes (purple) and Proteobacteria can oxidize formaldehyde via the tetrahydromethanopterin and glutathione-linked pathways, respectively, while Rokubacteria (blue) cannot. Formate is incorporated by the tetrahydrofolate-linked pathway then converted to serine by glycine hydroxymethyltransferase. Serine and glyoxylate are used by serine-glyoxylate transaminase to produce glycine and hydroxypyruvate, which is then converted to D-glycerate. D-glycerate can be phosphorylated by two different kinases, glycerate 2-kinase in Rokubacteria and glycerate kinase in Gemmatimonadetes. Both phosphoglycerates are converted by different enzymes to phosphoenolpyruvate (phosphoglycerate hydratase or phosphoglycerate dehydrogenase) and then to oxaloacetate by phosphoenolpyruvate carboxylase. Gemmatimonadetes and Proteobacteria (green) contain but Rokubacteria lack the canonical malate-intermediated serine formaldehyde assimilation pathway but do encode citric acid/glyoxylate cycle genes that could assimilate carbon (citrate synthase) and regenerate glyoxylate (isocitrate lyase).