Abstract
A general analytical method for the detection of target ligands has been developed, based on a special class of self-replicating aptazymes. These “autocatalytic aptazymes” are generated by linking an aptamer domain to the catalytic domain of a self-replicating RNA enzyme. Ligand-dependent self-replication of RNA proceeds in a self-sustained manner, undergoing exponential amplification at a constant temperature without the assistance of any proteins or other biological materials. The rate of exponential amplification is dependent on the concentration of the ligand, thus enabling quantitative ligand detection. This system has the potential to detect any ligand that can be recognized by an aptamer, including small molecules and proteins. The instability of RNA in biological samples due to the presence of ribonucleases can be overcome by employing the enantiomeric L-RNA form of the self-replicating enzyme. Methods for real-time fluorescence monitoring over the course of exponential amplification are currently being developed.
Keywords: Aptamer, aptazyme, exponential amplification, ligand detection, L-RNA, self-replicating RNA
Introduction
An analytical method for quantitative ligand detection should be general for various types of ligand molecules, sensitive and specific for the target ligand, robust in the context of biological and environmental samples, inexpensive, and simple to operate. The two most prominent methods in this regard are the enzyme-linked immunosorbent assay (ELISA; Engvall & Perlman, 1971) and the quantitative polymerase chain reaction (qPCR; Wang, Doyle & Mark, 1989). ELISA has broader generality, being applicable to any ligand for which one can generate a corresponding high-affinity antibody. Quantitative PCR is specific for nucleic acid targets, but has greater sensitivity due to its exponential signal amplification compared to the linear amplification of ELISA. Both methods are inexpensive and simple to operate and are carried out routinely in a wide variety of settings.
The discovery of synthetic and naturally-occurring riboswitches (Tang & Breaker, 1997; Winkler, Nahvi, Roth, Collins & Breaker, 2004) has inspired application of this class of molecules to ligand detection, exploiting the ability of RNA to recognize both small molecule and macromolecule targets, and harnessing the ligand-dependent structural rearrangement of RNA to generate a measurable signal. For example, the ligand-recognition (aptamer) domain of a riboswitch can be linked to an RNA enzyme (ribozyme) such that the enzyme is active only when the ligand is bound (Soukup & Breaker, 1999). The resulting “aptazyme” can be used to generate a signal that reflects the abundance of the corresponding ligand.
This chapter describes a special class of aptazymes for which the catalytic component is an RNA enzyme that catalyzes its own replication. As with qPCR, these “autocatalytic aptazymes” undergo exponential amplification with a growth rate that reflects the concentration of the ligand. Unlike qPCR, however, RNA self-replication proceeds at a constant temperature and does not require the assistance of proteins. Ligand-dependent, self-replicating RNA enzymes have not yet been applied outside the research setting, but are now sufficiently robust that they can be designed and utilized by non-specialists for many applications.
The self-replicating RNA enzyme was derived from the R3C ligase, a simple RNA motif that catalyzes the templated ligation of two RNA substrates (Rogers & Joyce, 2001). If those substrates, when joined together, form another copy of the enzyme, then self-replication is achieved (Paul & Joyce, 2002; Lincoln & Joyce, 2009). In the aptazyme format, the enzyme is active only in the presence of the target ligand, this being the case for both the parental enzyme and its copies that are generated through replication (Lam & Joyce, 2009). Thus ligand-dependency is maintained throughout the course of exponential amplification.
The self-replicating RNA enzyme is linked to the aptamer domain via a short stem region, enabling installation of various aptamers that form a closed stem in the presence of their cognate ligand. Closure of the stem results in activation of the conjoined enzyme, which in turn results in replication. Even if the difference in activity is less than absolute for the ligand-bound compared to ligand-free state, its effect is felt as an exponential growth parameter, resulting in signal generation that is strongly ligand dependent.
This chapter discusses the construction and operation of autocatalytic aptazymes, using the well-studied theophylline aptamer as an example (Jenison, Gill, Pardi & Polisky, 1994). Procedures are described for conducting quantitative exponential amplification assays. Also discussed is a nuclease-resistant version of the amplification system, based on L-RNA molecules that can replicate in biological samples (Olea, Horning & Joyce, 2012). Finally, a brief overview is given of efforts to develop a real-time fluorescence assay for ligand detection.
Exponential Amplification of RNA Enzymes
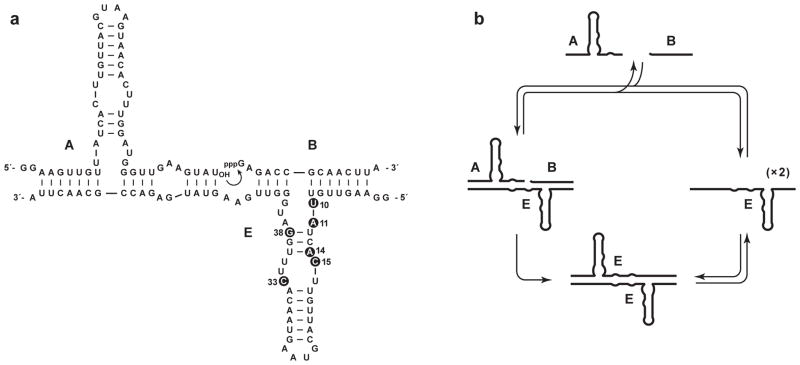
The self-replicating RNA enzyme (E) catalyzes the ligation of two RNA substrates (A and B) to form another copy of itself (Figure 1a). The ligation reaction involves attack of the 3′-hydroxyl of substrate A on the 5′-triphosphate of substrate B, forming a phosphodiester linkage and releasing inorganic pyrophosphate. The resulting E•E complex dissociates in a non-rate-limiting manner at a constant temperature of 40–50 °C, competing the replication cycle (Figure 1b). Replication is self-sustaining, requiring only the starting RNA enzyme, RNA substrates, MgCl2, and a suitable buffer (Lincoln & Joyce, 2009).
Figure 1.
Self-replicating RNA enzyme. (a) Sequence and secondary structure of the enzyme (E), which catalyzes ligation of two substrates (A and B) to form a new copy of E. Curved arrow indicates the site of ligation. Mutations in the improved compared to original form of the enzyme are numbered and highlighted by black circles. These six mutations are: G10→U, C11→A, G14→A, A15→C, G33→C, and A38→G. (b) Self-replication cycle involving binding of A and B to E, ligation of A and B to form of a new copy of E, and dissociation of the E•E complex.
Based on insight from kinetic studies (Ferretti & Joyce, 2013), an improved version of the self-replicating RNA enzyme was recently developed (Robertson & Joyce, 2014). This improved enzyme has an exponential growth rate of 0.14 min−1, corresponding to a doubling time of 5 min. The sequence of the improved enzyme is shown in Figure 1a, with the six mutations relative to the original version noted in the figure legend. Both the original and improved versions of the enzyme also can undergo cross-replication, whereby two different RNA enzymes catalyze each other’s synthesis from a total of four component substrates (Kim & Joyce, 2004). In cross-replication, the templating regions of the paired enzymes are non-identical. This makes it possible for many distinct replicating pairs of enzymes to operate in the same reaction mixture, each drawing upon its corresponding substrates, and thus enabling multiplexed ligand-dependent amplification.
Materials
The RNA enzymes and substrates are prepared in the usual manner by in vitro transcription of corresponding synthetic DNA templates. The substrates also can be prepared by chemical synthesis using D-RNA phosphoramidites (Glen Research, Sterling, VA) or L-RNA phosphoramidites (ChemGenes, Wilmington, MA). Note that preparation of the synthetic B substrate necessitates chemical triphosphorylation of the 5′-hydroxyl, which is a technique that requires expertise in synthetic organic chemistry (Ludwig & Eckstein, 1989). Thus, unless purchased from a commercial vendor (e.g. TriLink, San Diego, CA), the B substrate is normally prepared (in the D-RNA form) by in vitro transcription. Following transcription or synthesis, the enzymes and substrates are purified by denaturing polyacrylamide gel electrophoresis (PAGE) and desalted.
The 3′-terminal homogeneity of the transcribed A or B substrate can be enhanced by processing these materials using either RNase P or a cis-cleaving hammerhead ribozyme, respectively. In the presence of a suitable guide RNA (Forster & Altman, 1990), the RNA subunit of RNase P cleaves at a defined position within an extended-length A substrate to give the desired product, having a reactive 3′-hydroxyl at its terminus. A hammerhead ribozyme located within an extended-length B substrate cleaves at a defined position to remove that extension and yield the desired product, having a 2′,3′-cyclic phosphate at its terminus. The RNA enzyme (E) does not require processing and can be prepared by either run-off transcription or E-catalyzed ligation of A and B.
Procedure for RNA Self-replication
Standard conditions for self-replication employ 0.01–0.25 μM E, 5–10 μM each of A and B, 25 mM MgCl2, and 50 mM EPPS (pH 8.5) at 42 °C. Different forms of the self-replicating enzyme have somewhat different temperature optima, ranging from 40–50 °C (Robertson & Joyce, 2014). The original replicator generally performs best at the lower end of this range, whereas the improved replicator generally performs best at the higher end.
Mix water, EPPS buffer, and RNA in a 97.5-μL volume; heat at 70 °C for 2 min, then hold on ice.
Just prior to use, incubate this mixture at 42 °C for 3 min.
Initiate replication by adding 2.5 μL of a solution of 1 M MgCl2 that has been pre-warmed at 42 °C; mix quickly and continue incubation at 42 °C.
Samples are removed at various times during the course of replication and quenched by the addition of EDTA in excess of the concentration of Mg2+.
Sampled materials are separated by PAGE, distinguishing unreacted substrates from ligated products.
Typically the A substrate is 5′-labeled using either 32P or a fluorescent dye. The course of amplification is followed by monitoring the increasing concentration of E (and corresponding decreasing concentration of A). The rate of exponential amplification is determined by fitting these data to the logistic growth equation:
where [E]t is the concentration of enzyme at time t, a is the maximum extent of growth, b is the degree of sigmoidicity, and c is the exponential growth rate.
Ligand-dependent Exponential Amplification
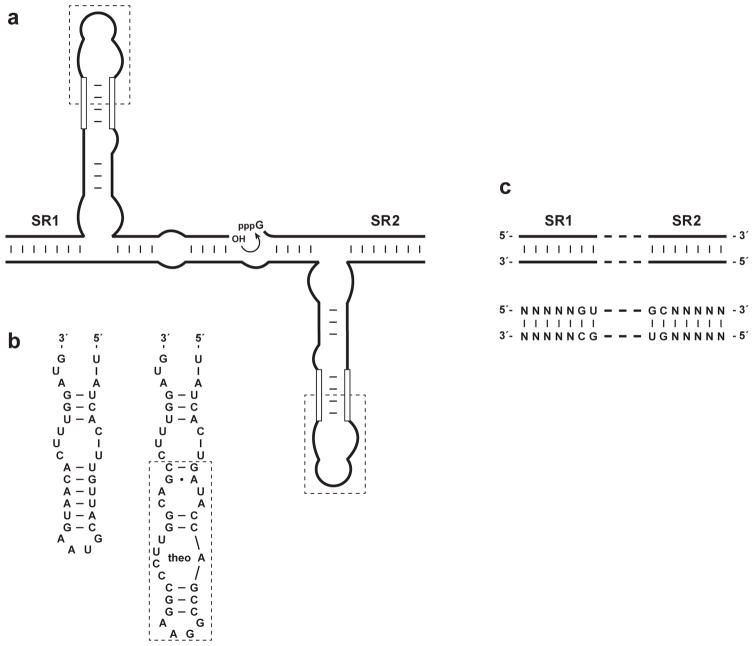
The catalytic domain of the self-replicating RNA enzyme can be linked to an aptamer domain so that amplification occurs in a ligand-dependent manner (Lam & Joyce, 2009). The link is achieved through the central stem-loop of the enzyme, which is made contiguous with a stem that helps define the aptamer motif (Figure 2a). Analogous to naturally-occurring riboswitches, binding of the ligand by the aptamer domain results in a conformational change that stabilizes the joining stem, thereby stabilizing the catalytic center of the enzyme.
Figure 2.
Self-replicating aptazymes. (a) An aptamer domain (dashed box) is linked to the central stem-loop of the self-replicating RNA enzyme via a connecting stem (open lines). (b) Modification of the central stem-loop to include an aptamer that binds theophylline (theo). The connecting stem has been optimized to maximize activity in the presence compared to the absence of theophylline. (c) The two substrate-recognition domains of the enzyme (SR1 and SR2) can be altered in sequence, so long as complementarity is maintained.
Most aptamers contain a stem element that provides a suitable point of attachment to the RNA enzyme. However, some fine-tuning is usually necessary to determine the optimal stability of the connecting stem. As a starting point, the predicted −ΔG° value for the connecting stem should be ~4 kcal/mol. The preferred composition of the stem, both length and sequence, then should be determined empirically. Although the enzyme is a constant, the idiosyncratic nature of various aptamer structures makes it difficult to predict the optimal composition for the connecting stem. In addition, one must choose a balance between minimal activity in the absence of the ligand and maximal activity in the presence of saturating ligand. This can be assessed in a simple ligation assay before testing the preferred composition in a full replication assay.
Ligand-dependent replication proceeds at a constant temperature, with an exponential growth rate that depends on the fractional occupancy of the ligand-binding domain. The maximum extent of amplification is independent of the ligand concentration, being determined by the concentration of the A and B substrates. Thus the concentration of ligand is assessed based on the time required to reach a defined partial extent of amplification. This is analogous to a Ct value for qPCR, which is the number of thermal cycles required to reach a defined threshold of PCR amplification.
Procedure for Quantitative Ligand Detection
Ligand-dependent exponential amplification is carried out under the same conditions that are employed in RNA self-replication, as described above. The ligand-containing sample is pre-warmed and introduced just prior to the addition of MgCl2 which initiates amplification. A standardized plot is generated by carrying out amplification in the presence of various concentrations of ligand. For each concentration of ligand, the time required to reach a defined threshold of amplification is determined, typically chosen as 25% of the maximum extent. Then a semi-log plot is constructed of time-to-threshold versus ligand concentration. For a sample containing an unknown concentration of ligand, the time-to-threshold is measured and related to the standardized plot to infer the concentration of ligand in the sample.
As an example, the theophylline aptamer (Jenison et al., 1994) was appended to the central stem of the self-replicating RNA enzyme via a short connecting stem that was contiguous with the closing stem of the aptamer (Figure 2b). Shorter connecting stems resulted in lower exponential growth rates even in the presence of saturating theophylline, whereas longer stems resulted in detectable amplification even in the absence of theophylline (Lam & Joyce, 2009).
As an alternative to measuring the time-to-threshold for various ligand concentrations, one can instead follow the entire course of exponential amplification for each ligand concentration, then determine the exponential growth rate by fitting the data to the logistic growth equation, as described above. Then one constructs a saturation plot of exponential growth rate versus ligand concentration. The half-maximal rate corresponds to the apparent Kd of the ligand-binding domain and the maximal rate corresponds to the behavior at saturation. For a sample containing an unknown concentration of ligand, the exponential growth rate is determined and related to the saturation plot to infer the concentration of ligand in the sample. In most cases, however, it is more expedient simply to measure the time required to reach a defined threshold.
Multiplexed Ligand Detection
It is possible to detect multiple ligands simultaneously in a sample, using multiple pairs of cross-replicating RNA enzymes, each containing a different aptamer domain (Lam & Joyce, 2009). Orthogonality is achieved by employing distinct substrate-recognition domains in each pair of cross-replicating aptazymes (Figure 2c). There should be at least two base pairs of discrimination between each combination of paired enzymes. There are five base pairs of variable sequence within each of the two substrate-recognition domains of the enzyme, making it straightforward to design orthogonal combinations. Discrimination is most sensitive for those nucleotides that lie closest to the site of ligation. Highly GC-rich sequences should be avoided because they may impede the rate of dissociation of the E•E complex, which would reduce the maximum rate of exponential amplification.
It also is possible to require the simultaneous detection of two ligands by a cross-replicating pair to achieve exponential amplification (Lam & Joyce, 2009). This requires that each member of the pair contain a different aptamer domain. If only one of the two ligands is present, then only one of the paired enzymes will be active and the system will exhibit linear amplification. If both ligands are present, then the system becomes autocatalytic and exponential amplification is observed.
As an example, the theophylline aptamer was linked to one enzyme and the FMN aptamer (Burgstaller & Famulok, 1994) to the other enzyme of a cross-replicating pair (Lam & Joyce, 2009). The FMN aptamer was linked to the central stem of the RNA enzyme via the same short connecting stem that was used to link the theophylline aptamer (Figure 2b). Exponential growth was observed only in the presence of both ligands. One can construct a plot of either time-to-threshold or exponential growth rate as a function of ligand concentration, doing so for each of the two ligands while maintaining a saturating concentration of the other ligand.
Coupling Ligand Recognition to Ligand-independent Amplification
The potential dynamic range of ligand-dependent exponential amplification, that is, the range of ligand concentrations over which the system exhibits saturation behavior, is determined by the Kd of the ligand-binding domain. Ligand detection is most precise at concentrations that are within 10-fold of the Kd, and becomes imprecise when the ligand concentration is more than 100-fold below the Kd (due to slow amplification) or 100-fold above the Kd (due to saturation). A further constraint is that the Km of the replicating enzyme for the A and B substrates is in the range of 1–10 μM (Ferretti & Joyce, 2013). This means that the substrates must be present at such a concentration to achieve a fast rate of amplification. However, because the aptamer domain lies within the A substrate, the requirement for saturating substrates sets a minimum concentration at which the ligand can be sensed.
Quantitative PCR employs a ligand recognition molecule (PCR primer) that is present at micromolar concentrations, yet can detect subattomolar concentrations of the target nucleic acid. This is because ligand recognition is required to initiate amplification, but subsequent rounds of amplification proceed in a ligand-independent manner. The same principle can be applied to the self-replicating RNA enzyme (Lam & Joyce, 2011). First, a simple aptazyme is used to carry out ligand-dependent joining of two RNA molecules to form seed copies of the self-replicating enzyme. Then these seed copies are used to initiate exponential amplification, which proceeds in a ligand-independent manner. The disadvantage of this approach, like PCR, is that the ligand is not sensed throughout the course of amplification, potentially giving rise to false-positive signals. The advantage is that the dynamic range of the assay is no longer restricted by the Km of the replicating enzyme, instead depending on the number of seed copies of replicating enzyme that are generated in the ligand-dependent ligation reaction.
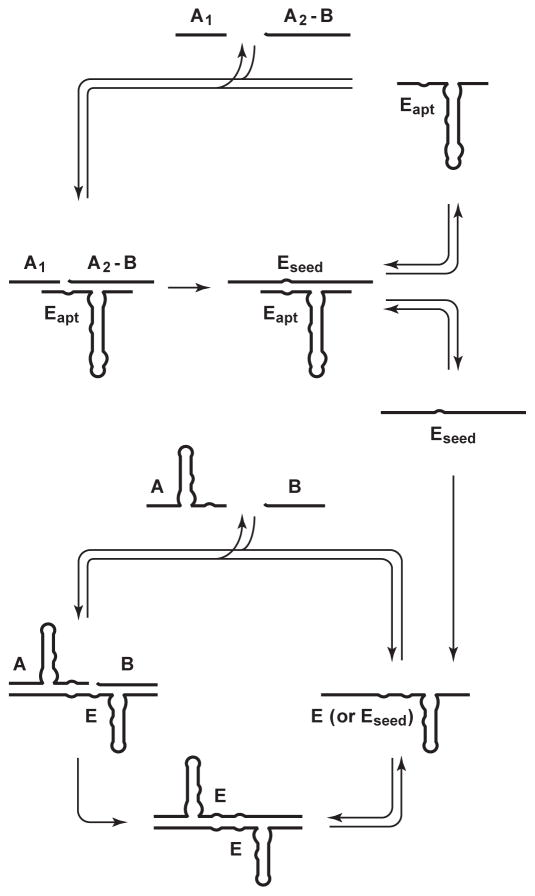
The same R3C ligase motif used in replication also is used to generate seed copies of the replicating enzyme, but is made to perform ligation at a different nucleotide position to avoid interference with the replication cycle (Figure 3). Two substrates are provided: one corresponding to the 5′ half of substrate A (designated A1), and the other corresponding to the 3′-half of substrate A conjoined to substrate B (designated A2-B). The ligated product, A1-A2-B, is equivalent to E, the self-replicating RNA enzyme. The aptamer domain is linked to the central stem of the ligase enzyme in the same manner as for the autocatalytic aptazyme. In principle, ligand-dependent ligation and ligand-independent exponential amplification could be carried out in the same reaction mixture, but it is preferable to carry out these two steps sequentially so that the conditions can be optimized for each reaction. It is not necessary to carry out any purification or processing steps between the two reactions.
Figure 3.
Coupled ligand-dependent ligation and ligand-independent exponential amplification. An aptazyme (Eapt) catalyzes ligand-dependent ligation of two substrates (A1 and A2-B) to form seed copies of the self-replicating RNA enzyme (Eseed), which then initiate exponential amplification utilizing substrates A and B.
Procedure for Coupled Amplification
The reaction mixtures for both ligand-dependent ligation and ligand-independent exponential amplification contain 25 mM MgCl2 and 50 mM EPPS (pH 8.5) at 42 °C. The first reaction employs an excess of the ligase aptazyme and a modest concentration of substrates. The second reaction employs whatever seed enzymes are carried over from the first reaction and a higher concentration of substrates. The RNA substrates for the second reaction can be maintained at dryness until rehydrated by the addition of the completed first reaction mixture.
-
Ligand-dependent generation of seed enzyme:
-
1.1
Mix water, EPPS buffer, 500 pmol ligase aptazyme, 50 pmol A1, and 50 pmol A2-B in a 97.5-μL volume; heat at 70 °C for 2 min, then hold on ice.
-
1.2
Just prior to use, incubate this mixture at 42 °C for 3 min.
-
1.3
Add ligand-containing sample that has been pre-warmed at 42 °C.
-
1.4
Initiate ligation reaction by adding 2.5 μL of a solution of 1 M MgCl2 that has been pre-warmed at 42 °C; mix quickly and continue incubation at 42 °C for 20 min.
-
1.1
-
Ligand-independent exponential amplification:
-
2.1
Prepare a separate aqueous solution containing 200 pmol each of A and B; evaporate to dryness.
-
2.2
Transfer the products of the completed first reaction mixture directly to the second reaction vessel; mix to dissolve and resume incubation at 42 °C.
-
2.3
Samples are removed at various times during the course of replication and quenched by the addition of EDTA in excess of the concentration of Mg2+.
-
2.4
Sampled materials are separated by PAGE, distinguishing unreacted substrates from ligated products.
-
2.1
The extent of the first reaction should not be allowed to exceed more than 20% relative to the starting amount of substrate. Otherwise the production of seed enzymes would begin to reflect the maximum extent of reaction rather than the concentration of ligand. A saturation plot can be constructed based on the yield of seed enzymes as a function of ligand concentration. The yield is determined either at a fixed time for all samples or at various times (depending on reaction rate) and normalized to a fixed time. These data are fit to the equation:
where [E] is the yield of seed enzyme for a given concentration of ligand, ε is the yield in the absence of ligand, and [E]max is the yield at saturating ligand concentration.
Even if no seed enzyme is produced in the first reaction mixture, exponential amplification can still occur in the second reaction mixture due to the spontaneous initiation of amplification. The A and B substrates can form an A•B•A•B tetramolecular complex that undergoes ligation at a reduced rate to produce E molecules (Ferretti & Joyce, 2013). This is analogous to the spontaneous initiation of PCR amplification that can occur in the absence of a target nucleic acid, largely as a consequence of primer-dimer formation (Rychlik, 1995). Spontaneous initiation of RNA amplification can be reduced by decreasing the stability of the tetramolecular complex relative to the E•A•B complex. Alternatively, it may be preferable to carry out RNA amplification in the cross-replication format to enable the use of more discriminating substrates that are less prone to spontaneous initiation. As with qPCR, the spontaneous initiation of amplification cannot be eliminated entirely, and is instead managed as a control reaction that helps to define the sensitivity of ligand detection.
The ligand-dependent generation of seed enzyme is carried out in the presence of various concentrations of ligand (including no ligand), and each of these reaction mixtures is then used to initiate ligand-independent exponential amplification. For each concentration of ligand, the time required to reach a defined threshold is determined in the subsequent exponential amplification reaction, typically chosen as 25% of the maximum extent. Then a semi-log plot is constructed of time-to-threshold versus ligand concentration. To be more precise, one can plot the time-to-threshold as a function of the yield of seed enzyme, normalized to the yield obtained at saturating ligand concentration (Lam & Joyce, 2011). For a sample containing an unknown concentration of ligand, the time-to-threshold is measured and related to the standardized plot to infer the concentration of ligand in the sample.
Nuclease-resistant Autocatalytic Aptazymes
RNA is a fragile molecule in the presence of biological or environmental samples due to its susceptibility to degradation by ribonucleases. For the detection of small-molecule ligands, such as drugs or metabolites, it is possible to deproteinize the samples by phenol extraction prior to analysis. Ligand-dependent exponential amplification of RNA proceeds readily in human serum that has been pre-treated in this manner (Lam & Joyce, 2009). For the detection of protein ligands, however, a different approach is required. The tactic that is commonly applied to RNA aptamers is to replace the nucleotides that are most susceptible to ribonuclease degradation by nuclease-resistant nucleotide analogs. Unpaired pyrimidine residues are especially vulnerable in this regard, although almost any RNA sequence is susceptible to cleavage by ribonucleases. The challenge when introducing nuclease-resistant nucleotide analogs is to avoid impairing RNA function, which is especially difficult for a highly optimized molecule such as the self-replicating RNA enzyme discussed in this chapter.
An alternative approach that can be applied to the autocatalytic aptazymes is to construct the enzyme and its substrates entirely from non-natural L-ribonucleotides (Olea et al., 2012). The mirror-image enzyme behaves identically as its D-RNA counterpart, but is completely resistant to ribonucleases. The L-RNA molecules must be prepared by chemical synthesis, which poses three special challenges. First, the A substrate cannot be [5′-32P]-labeled using T4 polynucleotide kinase and instead is labeled with a fluorophore following oligonucleotide synthesis. Second, the B substrate must be chemically 5′-triphosphorylated, which requires expertise in synthetic organic chemistry (see Materials). Third, the size of the enzyme plus appended aptamer domain approaches the limit of what can be prepared by chemical synthesis. Thus it may be more expedient to prepare starting copies of the enzyme using the autocatalytic reaction itself, employing the two L-RNA substrates.
The protocols for self-replication, quantitative ligand detection, and coupled amplification are identical for the D- and L-RNA self-replication systems. The aptamer domain that is linked to the enzyme also must be constructed of L-nucleotides. For achiral ligands, such as theophylline, the same sequence can be used for the D- and L-aptamers. For chiral ligands, including proteins, it is necessary to obtain a corresponding L-RNA aptamer, usually referred to as a “Spiegelmer” (Klussmann, Nolte, Bald, Erdmann & Furste, 1996). These compounds are obtained by first selecting D-aptamers against the enantiomer of the target ligand, then preparing a L-RNA of the same sequence that binds the desired target. Protein-binding L-aptamers usually are obtained by selecting D-RNAs that bind a D-peptide corresponding to the enantiomer of a structural epitope within the target protein (Leva, Lichte, Burmeister, Muhn, Jahnke, Fesser et al., 2002).
Real-time Florescence Assays
It is time consuming to use PAGE analysis to follow the course of exponential amplification. As with modern qPCR, it would be preferable to monitor amplification in real-time based on the increasing production of a fluorescent signal. However, unlike PCR which generates new copies of the DNA amplicon from dNTPs, the self-replicating RNA enzyme generates new copies of itself from oligonucleotide building blocks that are already present in the reaction mixture. Thus the detection method must distinguish the ligated products from these unligated substrates. Several approaches are being investigated that would provide such a detection method, but none is yet ready for general application. Three approaches are discussed briefly here.
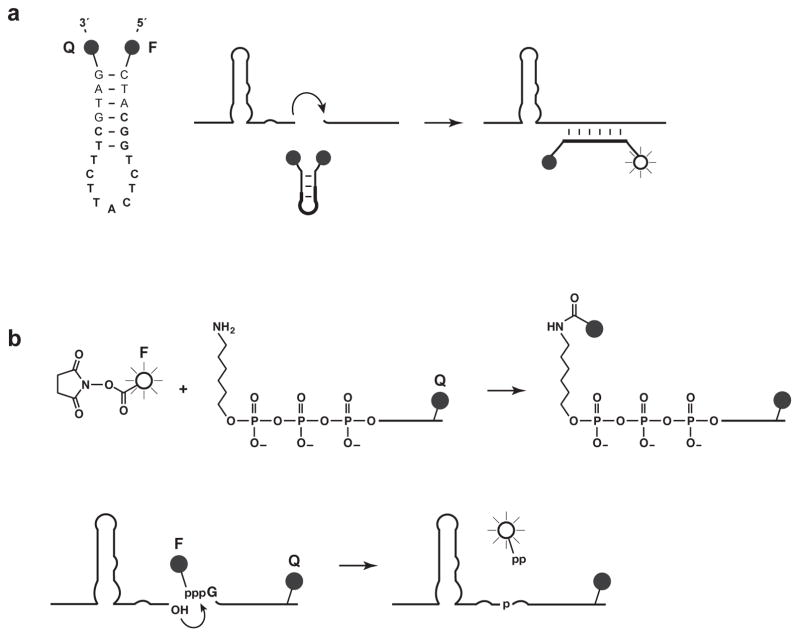
The first approach utilizes a molecular beacon that binds across the ligation junction, hybridizing to E, but not appreciably to A or B (Figure 4a). A molecular beacon is an oligodeoxynucleotide that contains both a fluorescent label and a fluorescence quencher, and adopts either a hairpin conformation in the absence of the target or an extended conformation when hybridized to the target (Tyagi & Kramer, 1996). In the hairpin conformation the label and quencher are in close proximity and fluorescence is quenched, whereas in the extended conformation the label and quencher are well separated and a fluorescent signal occurs. The preferred molecular beacon for the self-replicating RNA binds to seven nucleotides on either side of the ligation junction, thereby separating a 5′-fluorescein label from a 3′-dabcyl quencher. Amplification can be monitored using a standard qPCR instrument, but operating at constant temperature. There are two disadvantages to employing molecular beacons: first, they somewhat inhibit the rate of exponential amplification; and second, they cannot distinguish among multiplexed pairs of cross-replicating enzymes because these molecules all have the same sequence surrounding the ligation junction.
Figure 4.
Two approaches for real-time florescence monitoring of exponential amplification. (a) A molecular beacon contains a fluorescent label (F) and fluorescence quencher (Q) and forms either a non-fluorescent hairpin structure in the presence of unligated substrates or a fluorescent extended structure when hybridized to the ligated product. The sequence of the beacon is shown at the left, with the region of hybridization in bold. (b) A fluorescent label is linked to the γphosphate of substrate B, which also contains a fluorescence quencher. Ligation of the two substrates results in the release of labeled pyrophosphate, now separated from the quencher.
A second approach exploits the fact that with each RNA-catalyzed ligation event a molecule of inorganic pyrophosphate is released. The released pyrophosphate can be used to generate a luminescent signal based on an ATP-regenerative luciferase assay (Ronaghi, Karamohamed, Pettersson, Uhlén & Nyrén, 1996). This approach has been used to monitor the course of ligand-dependent exponential amplification (Lam & Joyce, 2009). However, it was necessary to withdraw samples from the amplification mixture due to differences in the preferred conditions for exponential amplification and luciferase-mediated signaling. Furthermore, this method does not enable multiplexed detection of different ligands.
A third approach also takes advantage of the release of inorganic pyrophosphate, but links a fluorescent dye to the γ-phosphate of the B substrate (Kumar, Sood, Wegener, Finn, Nampalli, Nelson et al., 2005) and incorporates a fluorescence quencher within that substrate. Fluorescence is quenched for the unreacted substrate, but not for the dye-labeled pyrophosphate that is released (Figure 4b). The labeled B substrate is prepared by in vitro transcription in the presence of 5 mM γ-aminohexyl-GTP (Jena Bioscience, Jena, Germany) and 2 mM GTP, followed by reaction with the N-hydroxysuccinimide ester of various fluorophores (e.g. TAMRA, Cy5, Alexa Fluor 488, and Alexa Fluor 610). In principle, different labels could be employed to allow multiplex analysis. At present, however, the method requires further optimization to obtain a rapid rate of amplification with the various dye-labeled substrates.
Conclusions
The autocatalytic aptazymes described in this chapter offer a new tool for the general-purpose, quantitative detection of target ligands. The exponential growth rate is dependent on the concentration of ligand, enabling quantitative detection. Moreover, the system can operate in biological samples by employing the L-RNA form of the replicating enzyme and its substrates. Additional effort is needed to develop a real-time fluorescence assay and to apply the system to a broad range of complex targets.
Acknowledgments
This work was supported by grant no. GM065130 from the National Institutes of Health. C.O. was supported by Ruth L. Kirschstein National Research Service Award no. F32CA165430 from the National Institutes of Health.
References
- Burgstaller P, Famulok M. Isolation of RNA aptamers for biological cofactors by in vitro selection. Angew Chemie. 1994;33:1084–1087. [Google Scholar]
- Engvall E, Perlman P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochem. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- Ferretti AC, Joyce GF. Kinetic properties of an RNA enzyme that undergoes self-sustained exponential amplification. Biochemistry. 2013;52:1227–1235. doi: 10.1021/bi301646n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster AC, Altman S. External guide sequence for an RNA enzyme. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- Kim DE, Joyce GF. Cross-catalytic replication of an RNA ligase ribozyme. Chem Biol. 2004;11:1505–1512. doi: 10.1016/j.chembiol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Klussmann S, Nolte A, Bald R, Erdmann VA, Furste JP. Mirror-image RNA that binds D-adenosine. Nat Biotechnol. 1996;14:1112–1115. doi: 10.1038/nbt0996-1112. [DOI] [PubMed] [Google Scholar]
- Kumar S, Sood A, Wegener J, Finn PJ, Nampalli S, Nelson JR, Sekher A, Mitsis P, Macklin J, Fuller CW. Terminal phosphate labeled nucleotides: synthesis, applications, and linker effect on incorporation by DNA polymerases. Nucleosides Nucleotides Nucleic Acids. 2005;24:401–408. doi: 10.1081/ncn-200059823. [DOI] [PubMed] [Google Scholar]
- Lam BJ, Joyce GF. Autocatalytic aptazymes enable ligand-dependent exponential amplification of RNA. Nat Biotechnol. 2009;27:288–292. doi: 10.1038/nbt.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam BJ, Joyce GF. An isothermal system that couples ligand-dependent catalysis to ligand-independent exponential amplification. J Am Chem Soc. 2011;133:3191–3197. doi: 10.1021/ja111136d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leva S, Lichte A, Burmeister J, Muhn P, Jahnke B, Fesser D, Erfurth J, Burgstaller P, Klussmann S. Chem Biol. 2002;9:351–359. doi: 10.1016/s1074-5521(02)00111-4. [DOI] [PubMed] [Google Scholar]
- Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig J, Eckstein F. Rapid and efficient synthesis of nucleoside 5′-O-(1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J Org Chem. 1989;54:631–635. [Google Scholar]
- Olea C, Jr, Horning DP, Joyce GF. Ligand-dependent exponential amplification of a self-replicating L-RNA enzyme. J Am Chem Soc. 2012;134:8050–8053. doi: 10.1021/ja302197x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul N, Joyce GF. A self-replicating ligase ribozyme. Proc Natl Acad Sci USA. 2002;99:12733–12740. doi: 10.1073/pnas.202471099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson MP, Joyce GF. Highly efficient self-replicating RNA enzymes. Chem Biol. 2014;21:238–245. doi: 10.1016/j.chembiol.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Joyce GF. The effect of cytidine on the structure and function of an RNA ligase ribozyme. RNA. 2001;7:395–404. doi: 10.1017/s135583820100228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyrén P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- Rychlik W. Selection of primers for polymerase chain-reaction. Molec Biotechnol. 1995;3:129–134. doi: 10.1007/BF02789108. [DOI] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc Natl Acad Sci USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Wang AM, Doyle MV, Mark DF. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]