Abstract
Environmental stress is among the most important contributors to increased susceptibility to develop psychiatric disorders. While it is well known that acute environmental stress alters gene expression, the molecular mechanisms underlying these changes remain largely unknown. 5-hydroxymethylcytosine (5hmC) is a novel environmentally sensitive epigenetic modification that is highly enriched in neurons and is associated with active neuronal transcription. Recently, we reported a genome-wide disruption of hippocampal 5hmC in male mice following acute stress that was correlated to altered transcript levels of genes in known stress related pathways. Since sex-specific endocrine mechanisms respond to environmental stimulus by altering the neuronal epigenome, we examined the genome-wide profile of hippocampal 5hmC in female mice following exposure to acute stress and identified 363 differentially hydroxymethylated regions (DhMRs) linked to known (e.g., Nr3c1 and Ntrk2) and potentially novel genes associated with stress response and psychiatric disorders. Integration of hippocampal expression data from the same female mice found stress-related hydroxymethylation correlated to altered transcript levels. Finally, characterization of stress-induced sex-specific 5hmC profiles in the hippocampus revealed 778 sex-specific acute stress-induced DhMRs some of which were correlated to altered transcript levels that produce sex-specific isoforms in response to stress. Together, the alterations in 5hmC presented here provide a possible molecular mechanism for the adaptive sex-specific response to stress that may augment the design of novel therapeutic agents that will have optimal effectiveness in each sex.
Keywords: Sex-Specific, Acute Stress, Epigenetics, DNA methylation, 5-hydroxymethylcytosine, Gene Expression
Introduction
Dysregulation of the hypothalamic-pituitary-adrenal (HPA) axis is the most common hallmark across all neuropsychiatric diseases (Bale, 2015; Martin et al., 2010). The HPA axis is a dynamic system that allows an organism to respond to environmental stress in a sex-specific manner by governing the activity of sex-specific endocrine mechanisms including the neuronal epigenome, which can effect gene expression (McCarthy et al., 2009; Meaney, 2001). Genomic analyses of human brain tissue showed that ~2.5% of genes are differentially expressed between males and females (Trabzuni et al., 2013). Thus, fundamental sex differences in the anatomy and the genetic regulatory network of the healthy brain likely underlie pronounced sex differences in susceptibility, progression, symptom severity, and pathology of neurological disorders (Cahill, 2006; Cosgrove et al., 2007; McCarthy et al., 2012). Consistent with this notion, females are more likely than males to develop depression, anxiety, and Alzheimer’s disease, while males are more likely to be diagnosed with attention deficit hyperactivity disorder, autism, and Parkinson’s disease (Balint et al., 2009; Gillberg et al., 2006; Hebert et al., 2013; Nolen-Hoeksema, 1987; Weissman et al., 1996; Wooten et al., 2004). The hippocampus is an important brain structure to study the effects of glucocorticoids and stress on gene expression as studies have shown that known stress-related genes undergo expression changes in the hippocampus following acute and chronic stress exposure (Cirelli et al., 2006; Gray et al., 2014; McGowan et al., 2009; Roth et al., 2009; Rubin et al., 2014). The negative feedback regulation of the HPA axis produced upon binding of glucocorticoids to their receptors in the hippocampus is critical for a healthy stress response. Understanding the biological and molecular underpinnings of sex differences in response to stress is likely to be a useful window into the mechanistic cause of mental illness in both men and women (Altemus et al., 2014; Rutter et al., 2003).
Alterations in environmentally sensitive epigenetic modifications are emerging as important factors in the long-term biological trajectories leading to stress-related psychiatric disorders (Hunter et al., 2009; Maccari et al., 2016). DNA methylation (5-methylcytosine (5mC)) is an epigenetic modification with important roles in chromatin remolding, gene silencing, embryonic development, cellular differentiation, and the maintenance of cellular identity (Chouliaras et al., 2013; Mellen et al., 2012; Tollervey and Lunyak, 2012). Moreover, changes in 5mC have been linked to neurological disorders as well as psychiatric disorders, including depression, anxiety, post-traumatic stress disorders (PTSD) and schizophrenia (Abdolmaleky et al., 2006; Collishaw et al., 2007; Kuratomi et al., 2008; Pidsley et al., 2014; Poulter et al., 2008; Robertson, 2005; Weaver et al., 2004). A recent study found differences in 5mC between males and females across all autosomes, some of which were associated with altered gene expression (Singmann et al., 2015), suggesting the potential for a sex-specific role for this DNA methylation modification. 5mC can be oxidized to 5-hydroxymethylcytosine (5hmC) following exposure to environmental stimuli (e.g., oxidative stress). 5hmC is enriched in neuronal cells and is associated with the regulation of neuronal activity (Szulwach et al., 2011; Yao and Jin, 2014). Related to disease, 5hmC functions independently from 5mC in neurological disorders (e.g., Rett syndrome and Autism) (Mellen et al., 2012; Papale et al., 2015; Zhubi et al., 2014) and neurodegenerative diseases (e.g., Huntington’s and Alzheimer’s) (Chouliaras et al., 2013; Condliffe et al., 2014; Wang et al., 2013). We recently reported a genome-wide disruption of 5hmC in the hippocampus of male mice exposed to acute stress followed by a one-hour recovery (Li et al., 2016). The differential 5hmC pattern significantly resided in known stress-related genes, which serves to validate a role for 5hmC in response to stress and also reveals potentially novel stress-related genes. These findings prompted us to examine the genome-wide profile of hippocampal 5hmC in females following exposure to acute restraint stress to determine the molecular origins of sex-specific vulnerability to stress-related psychiatric disorders. Together, these studies provide new insight into sex-specific functional epigenetic contributions to a stress response and they are intended to serve as a conceptual basis that will facilitate the future study of cellular and brain regional dynamics of 5hmC, especially as it relates to stress and stress recovery. These results demonstrate the power of coupling methylome and transcriptome data to determine the molecular origins of stress-related psychiatric disorders, such as PTSD and anxiety disorders. Here we provide the first genome-wide map of 5hmC in the female mouse hippocampus following an acute stress, which reveals known and potentially novel genes contributing to the stress response in females. These findings establish a sex-specific role for 5hmC in acute stress and provide insights into the immediate genome-wide neuromolecular response to traumatic events.
Methods
Stress paradigm, Tissue acquisition, and DNA/RNA extraction
Mice were purchased from the Jackson laboratories (Bar Harbor, ME) and maintained for several generations on C57BL/6J background prior to experimentation. All mice were housed under uniform conditions in a pathogen-free mouse facility with a 12-hour light/dark cycle with food and water available ad libitum. All experiments were approved by the University of Wisconsin – Madison Institutional Animal Care and Use Committee (M02529). To minimize for the stress of animal handling, all of the following were conducted by a single researcher: animal colony maintenance; administration of the stress paradigm; and behavioral tests.
Notably, all experimental and control (male and female) mice were left undisturbed until weaning day and group housed with same sex littermates. The stress paradigm was administered on both sexes on the same day to ensure that identical procedures were conducted on both sexes. A description of the male mice was described previously (Li et al., 2016). For the female mice, on the day of the experiment seven-week old naïve virgin female C57BL/6 mice were randomly divided into experimental or control (naïve) groups (N = 5 and 3 per group, respectively). Following a published acute stress paradigm that resulted in alterations in epigenetic modifications (including 5hmC in males) and gene expression (Gray et al., 2014; Hunter et al., 2009; Li et al., 2016), the female experimental mice were restrained for thirty minutes (2 hours after lights-on) head first into a 50 ml conical vial that has an 8 millimeter diameter hole at the tip to allow sufficient oxygen flow. Post restraint animals were individually housed in a clean cage for one hour. After this recovery period, animals were briefly anesthetized (isoflurane) to minimize the stress of handling and sacrificed. At the same time that the experimental mice were taken from the cage to begin the stress paradigm, the naïve/control mice also were taken from the cage, briefly anesthetized (isoflurane) prior to sacrifice and tissue dissection. Finally, to minimize the effect of parent-to-offspring interaction per litter, a maximum of 2 pups/litter/sex were randomly selected for molecular analysis.
Whole brains were extracted and immediately flash frozen in 2-methylbutane and dry ice. Whole hippocampal tissue was excised by micropunch (−0.95 to −3.79 mm posterior to bregma) and approximately 30 milligrams of tissue was homogenized with glass beads (Sigma) and DNA and RNA were extracted using AllPrep DNA/RNA mini kit (Qiagen).
5hmC Enrichment of Genomic DNA
Chemical labeling-based 5hmC enrichment was described previously (Li et al., 2016; Song et al., 2011). Briefly, a total of 10ug of hippocampal was sonicated to 300 bp fragments and incubated for 1 hour at 37°C in the following labeling reaction: 1.5 ul of N3-UDPG (2mM); 1.5ulβ of −GT (60uM); and 3ul of 10X β-GT buffer, in a total of 30ul. Biotin was added and the reaction was incubated at 37°C for 2 hours prior to capture on streptavidin-coupled dynabeads (Invitrogen, 65001). Enriched DNA was released from the beads during a 2-hour incubation at room temperature with 100mM DTT (Invitrogen, 15508013), which was removed using a Bio-Rad column (Bio-Rad, 732–6227). Capture efficiency was approximately 5–7% for each sample.
Library Preparation and high-throughput sequencing of genomic DNA
5hmC-enriched libraries were generated for all male and female enriched hippocampal DNA together using the NEBNext ChIP-Seq Library Prep Reagent Set for Illumina sequencing, according to the manufacturer’s protocol. Briefly, 5hmC-enriched DNA fragments were purified after the adapter ligation step using AMPure XP beads (Agencourt A63880). An Agilent 2100 BioAnalyzer was used to quantify the amplified library DNA and 20-pM of diluted libraries (N=6) were used in each lane for sequencing, which yielded approximately 25 to 35 million uniquely mapped 50 bp reads from each library. 50-cycle single-end sequencing was performed by Beckman Coulter Genomics. Image processing and sequence extraction were done using the standard Illumina Pipeline.
Analysis of female 5hmC data: sequence alignment and peak identification
Alignment of sequence data was described previously (Li et al., 2016; Szulwach et al., 2011). Briefly, FastQ files from each sequenced library were separately aligned to mouse NCBI37v1/mm9 references using Bowtie 0.12.9. Each uniquely mapped read (.bed files), with no more than two mismatches in the first 25 bp, was concatenated to separately achieve experimental and control 5hmC peaks of sequence reads. The MACS software was used to identify peaks of 5hmC content using the default parameters, except for the following: effective genome size = 2.7×109; tag size = 85; band width = 300; P-value cutoff = 1e-5.
Identification and annotation of stress-induced female and sex-specific DhMRs
Identification of stress-induced female and sex-specific differentially hydroxymethylated regions (DhMRs) was conducted in R using Bioconductor packages GenomicRanges and DESeq2 (Lawrence et al., 2013; Love et al., 2014). Mapped read data from each female mouse was converted to Granges objects and extended to 275bp, roughly the average fragment length. The peaks of these read data were defined as merged regions that overlap from at least two biological replicates originating from the same condition (e.g., female stress or female control). The peaks from the male read data were previously generated (Li et al., 2016) using the same procedure that was used here to generate the female peak data. Candidate regions for DhMRs were identified by pooling and merging the total peaks from all conditions (i.e., female stress, female control, male stress, and male control). DESeq2 was utilized to perform differential enrichment analysis by implementing a two variable design where sex and treatment (e.g., control or stress) were used as the two factors and sex-specific stress response was used as the interaction term. Each of the following unique contrasting comparisons were performed in DESeq2: female-stress vs female-control, male-stress vs male-control, female-control vs male-control, female-stress vs male-stress, sex-specific stress-response. An alpha level of 0.2 was used when contrasting models in DESeq2. DhMRs were identified by a P-value < 0.05. Annotation of DhMRs to genes (within 10kb) and genomic features was performed using Nebula.curie.fr. A modified function of R package qqman was used to generate a Manhattan plot. DhMRs were termed hyper if stressed animals showed an over-abundance of 5hmC as compared to control animals as determined by the log2 fold change computed by DESeq2. Conversely, DhMRs were termed hypo if stressed animals showed an under-abundance of 5hmC as compared to control animals. When comparing males and females, DhMRs were termed female-specific if female animals showed an over-abundance of 5hmC as compared to male animals. On the other hand, DhMRs were termed male-specific if male animals showed an over-abundance of 5hmC as compared to female animals.
Gene Ontology analysis
Gene symbols of DhMR-associated genes and genes found to be differentially expressed were separately gathered and converted to their Entrez-IDs using R package clusterProfiler (Yu et al., 2012). clusterProfiler was used to perform enrichment analysis of gene ontologies (GO) of biological processes using a p-value cutoff of 0.05. The gene universe for DhMR (GO) analysis consisted of all genes associated with 5hmC peaks from all samples (N= 11,621). The gene universe for differential expression GO analysis consisted of all uniquely tested genes analyzed by the RSEM/Bowtie pipeline (N=21,992).
RNA sequencing
Approximately 100ng of total RNA was used for sequence library construction and all male and female RNA libraries were prepped together following instructions of NuGen mRNA sample prep kit (cat# 0348). In brief, total RNA was copied into first strand cDNA using reverse transcriptase and random primers. This was followed by second strand cDNA synthesis using DNA Polymerase I and RNaseH. These cDNA fragments went through an end repair process, the addition of a single ‘A’ base, and then ligation of the adapters. These products were gel purified and enriched with PCR to create the final cDNA libraries. The library constructs were run on the bioanalyzer to verify the size and concentration before sequencing on the Illumina HiSeq2500 machine where 100-cycle paired-end sequencing was performed by the University of Wisconsin Biotechnology Center. In total, three libraries (N=2 genomes/library, except N=1 for one library; the same mice and combinations that were used to generate the 5hmC data) were sequenced for each experimental condition.
DESeq2 analysis of RNA-seq data
Reads were aligned to NCBI37/mm9 genome. RSEM v1.2.25 which utilized Bowtie v1.1.2 was used to calculate expression of each sample using default parameters with the exception of the addition of the Gencode M1 Release reference genome annotation (http://www.gencodegenes.org/mouse_releases/1.html). Read counts for each sample were concatenated to create a read count matrix using RSEM. As RSEM outputs read counts that may not be whole integers, read counts were rounded to the nearest whole number. In addition, genes originating from sex chromosomes were discarded from further analysis to avoid the calling of DhMRs on chromosome X and Y that are due to karyotype differences rather than stress or sex. Read count matrices were separately generated for the whole-gene and isoform level expression data using RSEM and each matrix was used separately in downstream analyses. Using these generated matrices, R package DESeq2 was utilized to perform differential enrichment analysis (Love et al., 2014) using a two variable design where sex and treatment (i.e., control or stress) were used as the two factors and sex-specific stress response was used as the interaction term. An alpha level of 0.2 was used when contrasting models using DESeq2.
Transcription factor binding site motifs
The DREME suite (Bailey, 2011) was used to identify enrichments of transcription factor sequence motifs in hyper-, hypo-, and potentially functional DhMRs. Motifs that were not found in control-only 5hmC regions, experimental-only 5hmC regions, and all 5hmC regions and that had an E-value <10−3 were considered to be significant. Putative binding factors were predicted using SpaMo directly from the DREME suite software package and are listed in the tables shown with their putative transcription factors.
Statistical analysis
Over- and under-representation of DhMRs (580 peaks): notably, peaks were counted twice if associated with multiple genomic structures. For permutation testing, we randomly selected 580 peaks (from the full set of 61,003peaks) and counted how many peaks were in each genomic structure. We termed this the “permuted number” and generated this number 10,000 times. The permutation P-value is the number of times the set of “permuted numbers” exceeded the “actual number,” divided by 10,000. Over- and under-representation of DhMRs on each chromosome was computed similarly by permutation as described for the genomic structures, except for each chromosome. To correct for multiple hypotheses (i.e., 19 chromosomes), the actual proportion of each chromosome was compared with the permuted proportions of any chromosome for each permutation. Permutations for gene structure: The overlap between the DhMRs and the peaks belonging to a particular gene region (e.g., TSS-1.5K, Exon, etc., called the “actual overlap”) was compared with those obtained by randomly selecting an equal number (i.e., equal in number to the DhMRs) and evaluating the overlap (“permuted overlap”). This was performed for all the DhMRs and the DhMRs that contain both hyper-DhMR and hypo-DhMR peaks.
For the comparison of female DhMR-associated genes and known stress-related genes, we used a chi-square test on genes associated with the following terms: anxiety; stress; depression; psychosis; major depressive disorder; post-traumatic stress disorder; bipolar; fear; schizophrenia; and trauma, which were identified from the GeneCards database; N = 4,348) and the female DhMR-associated genes (N = 363). The gene universe consisted of all the genes associated with peaks from the sequence data of all the mice (N = 13,362). Notably, we also ran an analysis to examine the overlap of stress-induced DhMR-associated genes with known hypoxia-related genes and found a significant overlap in females (N = 53 of 363; Chi-squared P-value < 0.05). However, of the 53 hypoxia-related genes found in this overlap, 42 of them were already accounted for in the initial stress-related gene comparison. Thus, we believe that hypoxia related stress has already been assessed for in our initial comparison.
Results
Acute stress disrupts female hippocampal hydroxymethylation
Finding a disruption in male hippocampal 5hmC following a single acute stress (Li et al., 2016) prompted us to investigate the role of 5hmC in female hippocampal tissue following acute stress. To achieve this goal, we randomly divided seven-week-old C57BL/6 female mice into experimental or control groups, where the experimental mice were restrained for thirty minutes followed by a one-hour recovery period prior to tissue collection (Methods). 5hmC containing DNA sequences were enriched from whole hippocampal genomic DNA, using an established chemical labeling and affinity purification method (Li et al., 2016; Papale et al., 2015; Song et al., 2011; Szulwach et al., 2011), and sequenced using high-throughput sequencing technology. This approach yielded approximately 30 to 55 million uniquely mapped reads from each genome (Methods; Supplementary Fig. 1a). Read density mapping showed no visible differences among the chromosomes, except depletion on the X chromosome, which is consistent with previous observations (Supplementary Fig. 1b, data not shown) (Li et al., 2016; Papale et al., 2015; Szulwach et al., 2011).
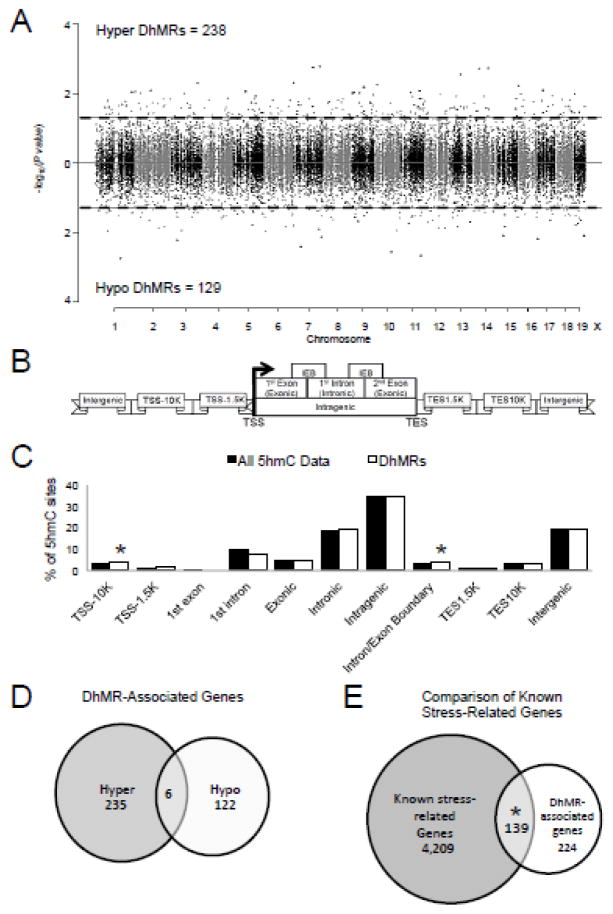
Significant accumulations of uniquely mapped reads represent hydroxymethylated regions throughout the mouse genome. Differentially hydroxymethylated regions (DhMRs) associated with stress were identified in both experimental and control groups (Methods). DhMRs found in experimental mice were classified as hyper-DhMRs and DhMRs absent in experimental mice (present in control mice) were classified as hypo-DhMRs. A total of 238 hyper- and 129 hypo-DhMRs were identified and these loci were distributed across all chromosomes, with a noticeable depletion on chromosome X, which is consistent with previous reports (P-value = 0.05; Fig. 1a; Dataset 1) (Li et al., 2016; Szulwach et al., 2011). Since specific regions of the genome are differentially methylated based on the biological functions of the genes contained within the genomic region, we used permutation testing to determine if the DhMRs are specific to certain regions of the genome by comparing the proportions of DhMRs to the proportions of total detected hydroxymethylated regions in all animals for each chromosome (Methods). This analysis revealed a depletion on chromosome 15 (Permutation P-value = 0.016; Supplementary Fig. 1b; Dataset 1). We next annotated the female DhMRs to the nearest gene structure, meaning the DhMRs were assigned to the following genomic elements: 10 kilobases (kb) upstream of the transcription start site (TSS-10K); 1.5 kb upstream of the TSS (TSS-1.5K); 1st exon; 1st intron; Exonic; Intronic; Intragenic; Intron/Exon boundary (IEB); 1.5 kb downstream of the transcription end site (TES1.5K); 10 kb downstream of the TES (TES-10K); and intergenic regions (Fig. 1b). The distribution of the data indicates that the largest fraction of DhMRs (35%) is in the intragenic regions of the genome (Fig. 1c). To determine the relative abundance of DhMRs in each annotated gene region we compared, via permutation testing, the proportion of DhMRs that reside in or around each gene to the total number of hydroxymethylated regions identified by the enrichment method (Methods). This analysis found a significant abundance of DhMRs in the TSS-10K and at the intron/exon boundaries (P-value < 0.05, respectively; Fig. 1c; Dataset 2). Together, these data indicate that like previously reported in males (Li et al., 2016), female acute stress-related hydroxymethylation primarily resides in genic regions of the genome and suggest that these changes are not randomly distributed throughout the genome.
Figure 1.
Characterization of differential 5hmC across standard genomic structures in acutely stressed females. A. Modified Manhattan plot of female acute stress induced disruptions of 5hmC across the genome. Each point represents a genomic region containing 5hmC (~1kb in size). Points above zero represent increases in 5hmC (hyper) and points below zero represent decreases in 5hmC (hypo). While all regions containing 5hmC alternate between black and gray to indicate each chromosome, significant changes in 5hmC abundance (DhMRs) are found outside the dashed lines (P-value < 0.05). B. Schematic of standard regions of the genome. 5hmC and DhMR data were classified into the following genomic locations: 10 kilobases (kb) upstream of the transcription start site (TSS; TSS-10K); 1.5 kb upstream of the TSS (TSS-1.5K); 1st exon; 1st intron; Exonic; Intronic; Intragenic; Intron/Exon boundary (IEB); 1.5 kb downstream of the transcription end site (TES1.5K); 10 kb downstream of the TES (TES10K); or intergenic regions. Hydroxymethylation data were binned into these groups based on the density of sequence reads. C. Enrichment of 5hmC across genomic structures. The percent distribution (y-axis) of all 5hmC data (black) and DhMRs (white) across each genomic structure is shown with significant enrichments indicated with an asterisk (P-value < 0.05). D. Venn diagram of DhMR-associated genes. Shown is the relative abundance of hyper and hypo DhMR-associated genes and their overlap. E. Venn diagram of DhMR-associated genes compared to stress-related genes. Shown is the significant overlap of DhMR-associated genes and known stress related genes from the literature (GeneCard). An asterisk indicates P-value < 0.05.
Annotation of DhMRs to genes
Annotation of female DhMRs to genes revealed 363 genes with altered hydroxymethylation following acute stress, including 241 and 128 genes that have hyper- and hypo-DhMRs, respectively (Fig. 1d; Dataset 2). Six genes were identified with both hyper- and hypo-DhMRs, meaning these genes contained regions with both increases (hyper) and decreases (hypo) in 5hmC following acute stress (Fig. 1d; Dataset 2). Notably, the average distance between inversely abundant DhMRs was >500 kb, suggesting that when found on the same gene hyper- and hypo-DhMRs are not located near each other and may have unique roles in response to acute stress.
Since we previously found an enrichment of stress- and psychiatric disorder-related genes contained DhMRs following stress in males, we examined whether the female DhMR-associated genes were enriched for stress related genes using the GeneCards database (Methods). Similarly, a significant number of the female DhMR-associated genes are known stress-related genes (N = 139 of 363; P-value < 0.05; Fig. 1e), including genes known to play a role in the adaptation of brain to stress, Nr3c1 and Ntrk2 (Juhasz et al., 2011; Zhang et al., 2013), indicating that stress-related genes harbor DhMRs following exposure to acute stress and suggest that 5hmC has a molecular role in female response to stress. Moreover, these findings also potentially revealed novel genes responding to acute stress (i.e., a subset of the non-stress-related genes, N = 224; Fig. 1e). Notably, unlike in males, the female DhMR-associated genes did not include genes known to function in epigenetic pathways (e.g., Tet2 or Dnmt3a). Together, these data indicate that acute stress disrupts 5hmC on known and potentially novel genes involved in stress response.
To further characterize the genes and pathways altered by acute stress in females, we next examined the gene ontologies of the 241 and 128 female hyper- and hypo-DhMR-associated genes, respectively, and found a significant enrichment of terms with only the hypo-DhMR-associated genes. These terms were significantly enriched for neuronal ontological terms (P-value < 0.01), including regulation of glutamate synthesis, dendrite development, and neurogenesis (FDR P-value < 0.2; Table 1; Dataset 3). Many of the female hypo-DhMR-associated genes that contributed to the overrepresentation of these pathways have been previously implicated in stress-induced psychiatric disorders, including Nr3c1, Ank2, and Nfia (Table 1) (Iossifov et al., 2014; Iossifov et al., 2012; Zhang et al., 2013). Together, these findings suggest that female hippocampal DhMRs are associated with known stress-related neurodevelopmental pathways.
Table 1.
Gene ontology pathways of female acute stress hypo DhMR-associated genes
| Gene Ontology Term | **P-Value | *** Gene ontology genes |
|---|---|---|
| * morphogenesis of an epithelium | 0.00018 |
Cxcr4,Enah,Fzd6,Gli2,Nr3c1,Hand1,Foxq1,Sfrp1,Sox2,Tgfb2,Fras1 Arhgap24,Map3k7 |
| tissue morphogenesis | 0.00019 |
Cxcr4,Dscam,Enah,Fzd6,Gli2,Nr3c1,Hand1,Foxq1,Man2a1,Sfrp1,Sox2 Tgfb2,Fras1,Arhgap24,Map3k7 |
| * regulation of glutamate secretion | 0.00047 | Grm7,Nr3c1,Prkg1 |
| * dendrite development | 0.00064 | Dscam,Ezh2,Nr3c1,Sdc2,Prkg1,Abi2,Gsk3b,Tmem106b |
| protein export from nucleus | 0.00070 | Nup214,Ahcyl1,Gsk3b,Park7 |
| * regulation of cellular component movement | 0.00081 |
Ank2,Adra2a,Capn7,Cxcr4,Dscam,Fn1,Itgb3,Prkg1,Sfrp1,Hace1,Tac1 Sema6d,Tgfb2,Gsk3b,Bmper |
| * negative regulation of transport | 0.00081 | Grm7,Adra2a,Fn1,Itgb3,Prkg1,Sfrp1,Tgfb2,Nucb2,Gsk3b,Park7,Ndfip2 |
| * regulation of neurogenesis | 0.00098 |
Cxcr4,Dscam,Ezh2,Fn1,Gli2,Nr3c1,Sdc2,Man2a1,Prkch,Atxn1,Sox2 Ntm,Zfp536,Opcml,Gsk3b |
| * nervous system development | 0.00102 |
Cxcr4,Dscam,Enah,Ezh2,Fn1,Fzd6,Gli2,Nr3c1,Sdc2,Man2a1,Nfia Pikfyve,Prkch,Prkg1,Atxn1,Sfrp1,Sox2,Sema6d,Tgfb2,Ntm,Zfp536 Map3k7,Abi2,Opcml,Gsk3b,Smarca2,Kirrel3,Tmem106b |
| nuclear export | 0.00103 | Atxn1,Nup214,Ahcyl1,Gsk3b,Park7 |
Indicates neuronal related pathways as previously determined by Geigman et al., 2010
All terms shown had an FDR P-Value = 0.138
Bolded genes are known stress related genes from the GeneCards database
Characterization of potentially functional female DhMRs
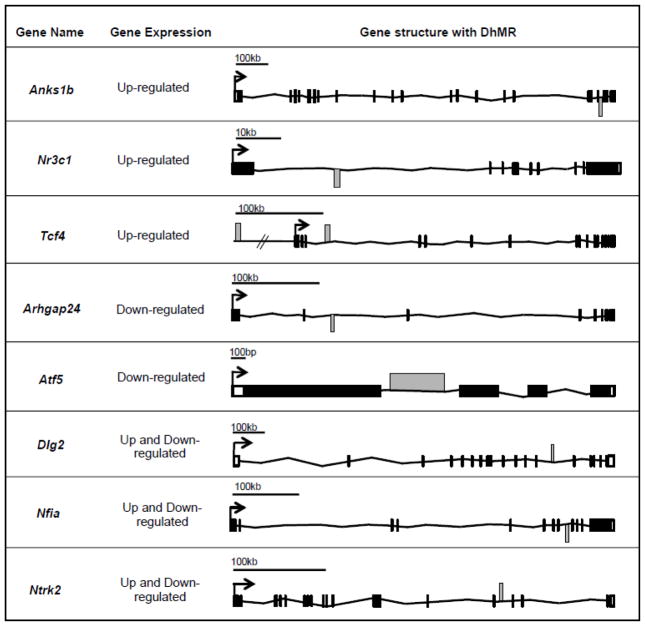
To gain insight into the potential mechanism(s) for female stress-related DhMRs, we used RNA sequencing (RNAseq) to profile gene expression in the hippocampal tissue of the same mice surveyed for 5hmC (see Methods). Comparison of transcript levels in experimental and control mice revealed 2,057 unique genes differentially expressed at the whole-gene and/or isoform level (FDR P-value < 0.05; Dataset 4; Methods). Many of these genes previously were shown to be highly dynamic in response to stress, as we found an up regulation of Nr3c1 and Bdnf, and down regulation of Ntrk2 (Juhasz et al., 2011; Marmigere et al., 2003; Zhang et al., 2013), and other genes that are associated with recent neural activity, including an up regulation of Arc, Irs2, Sgk1, and members of the Fos gene family (Fos, Fosb, and Fosl2) (Dataset 4) (Gray et al., 2014). Examination of the gene ontologies of these 2,057 differentially expressed genes found a significant enrichment of neuronal ontological terms, including corticosteroid signaling pathway, learning and memory and the regulation of dendrite development (FDR P-value < 0.05; Dataset 5; Methods). DhMRs associated with differentially expressed genes represent candidate functional DhMRs that may have a direct role in gene regulation. An overlay of the DhMR data with the differentially expressed (DE) genes revealed 68 potentially functional DhMRs in genes; many with documented roles in stress and/or stress-induced psychiatric related disorders, including Nr3c1, Ntrk2, and Nfia (DE, FDR P-value < 0.05; DhMR, raw P-value < 0.05; Fig. 2; Dataset 4). Notably, these data also revealed potentially functional DhMRs in genes that have an uncharacterized role in stress but have been implicated in mental illness, such as Anks1b, Arhgap24, Ddhd2, Dlg2, and Tcf4 (Fig. 2; Dataset 4) (Gonzalez-Mantilla et al., 2016). Together, these data suggest that 5hmC is correlated to altered gene expression in response to acute stress.
Figure 2.
Potentially functional DhMR-associated genes. Shown are representative DhMR-associated genes that have altered transcript levels following exposure to acute stress. The name of each gene and the direction of expression change is shown to the left of each gene schematic, which depicts the relative location of each transcription start site (TSS; broken arrow), exon (white and black boxes), intron (black horizontal line connecting exons), and hyper- or hypo-DhMR (grey box above or below each gene, respectively).
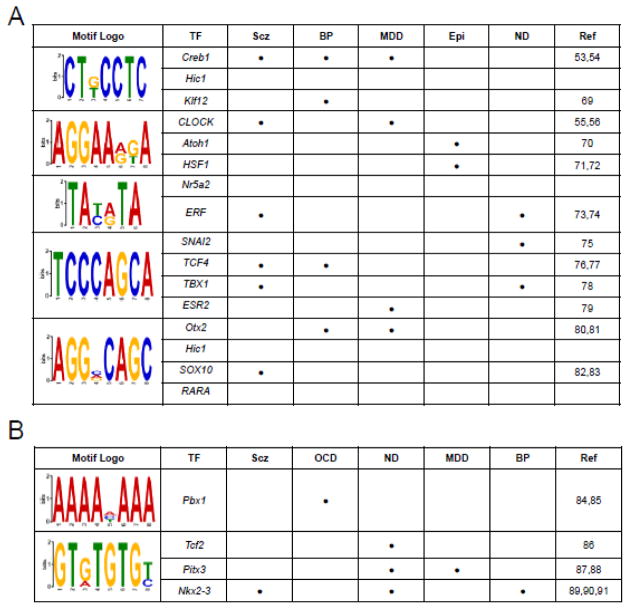
Since it was previously shown that 5hmC in males may regulate the expression of neuronal genes following exposure to acute stress by modulating the binding or function of transcription factors (Li et al., 2016), we investigated the presence of known transcription factor binding sequences within the female DhMRs using the Discriminative Regular Expression Motif Elicitation (DREME) suite software package. Five hyper DhMR-associated motifs and two hypo DhMR-associated motifs were discovered (Evalue < 10e-3; Fig. 3). Many of the transcription factors predicted to bind to the motifs have links to stress-related behaviors and disorders, such as Creb1 and Clock, which are associated with schizophrenia, bipolar disorder, and major depressive disorder (see discussion) (Forero et al., 2016; Johansson et al., 2016; Shi et al., 2016; Wei et al., 2015). Subsequent examination of the sequence data for each DhMR residing in the acute stress-related differentially expressed genes revealed that the majority (>90%) of the potentially functional DhMR-associated genes contained at least one of the enriched transcription factor binding sequence motifs (Dataset 2). Together, these findings are consistent with those found in males, supporting a role for 5hmC in transcription factor binding and functions following exposure to an acute stress.
Figure 3.
Identification of DhMR-associated sequence motifs and their putative transcription factors. The DREME suite was used to predict the sequence motifs in hyper-(A) and hypo-(B) DhMRs. Motifs with E-value <10−3 were considered to be significant. Putative binding factors were predicted using SpaMo directly from the DREME suite software package and are listed in the tables shown with their putative transcription factors. The literature (Ref) links of each TF to psychiatric disorders are shown: Scz (schizophrenia); BP (Bipolar Disorder); MDD (Major Depressive Disorder); Epi (Epilepsy); ND (Neuronal Differentiation); OCD (Obsessive Compulsive Disorder) (Aston et al., 2005; Basmanav et al., 2015; Butts et al., 2014; Castilhos et al., 2014; Chen et al., 2014b; Chiang et al., 2015; Forero et al., 2016; Forrest et al., 2013; Goes et al., 2015; Grados et al., 2014; Gurung and Prata, 2015; Hattori et al., 2014; Johansson et al., 2016; Jukic et al., 2015; Keyes et al., 2015; Kim et al., 2014; Le-Niculescu et al., 2009; Lisowski et al., 2013; Mencarelli et al., 2008; Nestadt et al., 2012; Rivolta et al., 2014; Rotheram-Fuller et al., 2010; Schlaudraff et al., 2014; Shi et al., 2016; Wang et al., 2014; Wei et al., 2015; Wockner et al., 2014).
Naïve male and female 5-hydroxymethylation profiles
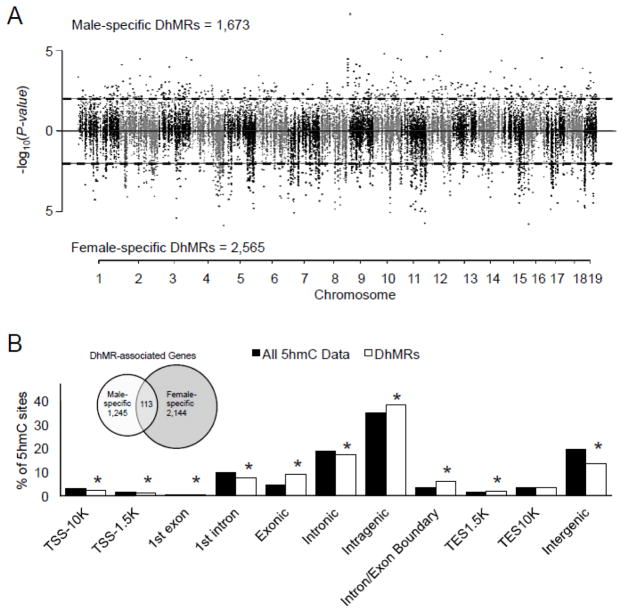
Finding that acute stress disrupts 5hmC in both males and females led us to examine the similarities of these disruptions among the sexes. Prior to this comparison, we first wanted to determine if non-stressed (i.e., naïve) males and females have similar 5hmC distributions in the hippocampus, using the data generated above and previously from seven-week-old C57BL/6 naïve mice (Li et al., 2016). DhMRs enriched or depleted in naïve males compared to naïve females were classified as male-specific or female-specific DhMRs, respectively. A total of 1,673 male-specific DhMRs and 2,565 female-specific DhMRs were found and these loci were distributed across all chromosomes (Fig. 4a; P-value < 0.05; Dataset 6), suggesting that the distribution of 5hmC in naïve animals is sex-specific. Next, we determined whether sex-specific DhMRs were annotated to specific regions of the genome (Methods) and found significant chromosomal enrichments (4, 7, 8, 11, 15, 17, and 19) and depletions (1, 3, 6, 12, 13, 14, and 18; permutation P-value ≤ 0.05; Supplementary Fig. 2), which indicated that specific regions of the genome are differentially hydroxymethylated between the sexes. In addition, examination of DhMRs in relation to gene structures also revealed significant enrichments (exonic, intragenic, intron/exon boundary, and TES1.5K) and depletions (TSS-10K, TSS-1.5K, 1st exon, 1st intron, intronic, and intergenic; all permutation P-value < 0.5; Fig. 4b). Consistent with previous studies, the largest fraction of DhMRs was present in the intragenic regions. Together, these data suggest that 5hmC has sex-specific roles throughout the genome, particularly within gene regions.
Figure 4.
Characterization of differential 5hmC across standard genomic structures in naïve animals. A. Modified Manhattan plot of male- and female-specific differences in 5hmC across the genome. Each point represents a genomic region containing 5hmC (~1kb in size). Points above zero represent increases of 5hmC in males (male-specific) and points below zero represent increases of 5hmC in females (female-specific). While all regions containing 5hmC alternate between black and gray to indicate each chromosome, significant changes in 5hmC abundance (DhMRs) are found outside the dashed lines (P-value < 0.05). B. Enrichment of 5hmC across genomic structures. The percent distribution (y-axis) of all 5hmC data (black) and DhMRs (white) across each genomic structure is shown with significant enrichments indicated with an asterisk (P-value < 0.05). Inset: Venn diagram of DhMR-associated genes. Shown is the relative abundance of male-specific and female-specific DhMR-associated genes and their overlap.
Annotation of sex-specific DhMRs to genes found 1,245 male-specific DhMR-associated genes and 2,144 female-specific DhMR-associated genes, and 113 genes that contained both male-specific and female-specific DhMRs associated with the same genes (Fig. 4b, inset; Dataset 7). Again, the large average distance between inversely abundant DhMRs (>500kb) suggested unique roles for DhMRs located proximal to the same gene. To gain further insight into sex-specific genes and pathways, we next examined the gene ontologies of the 1,245 male-specific DhMR-associated genes and 2,144 female-specific DhMR-associated genes and found distinct enrichments for each sex; while male-specific terms were more focused on neuronal pathways (e.g., neuronal generation, differentiation, and development), female-specific terms included broader brain-related terms such as cellular organization and synapse maturation (FDR P-value < 0.05; Dataset 8 and 9). Together, these results suggest that 5hmC may play a role in sex-specific biological functions.
We next used RNAseq to compare transcript levels in the hippocampal tissue of the same mice surveyed for 5hmC and found 512 male-specific and 475 female-specific differentially expressed (DE) at the whole-gene and/or isoform level (FDR P-value < 0.05; Dataset 10; Methods). Since we were most interested in finding potentially functional DhMRs that are sex-specific, we then overlaid the DhMR data with these differentially expressed genes and found 117 male-specific and 140 female-specific potentially functional DhMRs (DE, FDR P-value < 0.05; DhMR, raw P-value < 0.05; Dataset 10; Methods). Together, these data indicate that ~25% of the sex-specific DhMRs are potentially functional, suggesting a role for 5hmC in sex-specific gene expression. These findings led us to further investigate the sex-specific molecular response to acute stress.
Sex-specific disruption of 5hmC and gene expression following acute stress
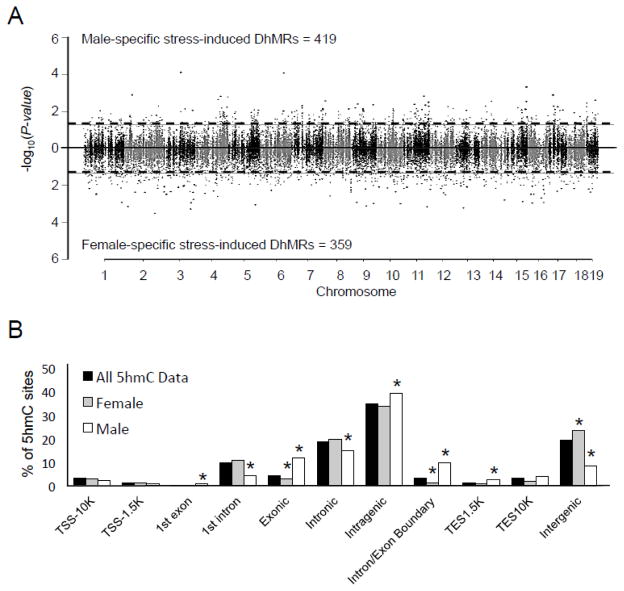
To examine acute stress-related disruptions in 5hmC among males and females we utilized an analysis that allowed for an interaction of both sex and stress (Methods). This approach revealed 778 sex-specific acute stress-induced DhMRs in the mouse hippocampal genome, with 419 and 359 regions having a significantly greater stress induced 5hmC fold change in males and females, respectively (P-value < 0.05; Fig. 5a; Dataset 11). As found above, the sex-specific acute stress-induced DhMRs had significant abundances in specific regions of the genome (Methods), including chromosomal enrichments (7 and 19) and depletions (1 and 18; permutation P-value ≤ 0.05; Supplementary Fig. 3). Examination of sex-specific acute stress-induced DhMRs in relation to gene structures also revealed significant male and female-specific enrichments (1st exon (♂), exonic (♂), intragenic (♂), intron/exon boundary (♂), TES1.5K (♂), and intergenic (♀)) and depletions (1st intron (♂), exonic (♀), intronic (♂), intron/exon boundary (♀), and intergenic (♂); all permutation P-value < 0.5; Fig. 5). Consistent with previous studies, the largest fraction of DhMRs were present in the intragenic regions. Together, these data suggest that 5hmC has sex-specific roles following stress, particularly within genic regions.
Figure 5.
Characterization of differential 5hmC across standard genomic structures related to sex and stress. A. Modified Manhattan plot of sex-specific stress-induced differences in 5hmC across the genome. Each point represents a genomic region containing 5hmC (~1kb in size). Points above zero represent a greater stress induced 5hmC fold change in males (male DhMR) and points below zero represent a greater stress induced 5hmC fold change in females (female DhMR). While all regions containing 5hmC alternate between black and gray to indicate each chromosome, significant changes in 5hmC abundance (DhMRs) are found outside the dashed lines (P-value < 0.05). B. Enrichment of 5hmC across genomic structures. The percent distribution (y-axis) of all 5hmC data (black) and DhMRs (male, white; female, gray) across each genomic structure is shown with significant enrichments indicated with an asterisk (P-value < 0.05).
Annotation of these DhMRs to genes found 424 and 346 DhMR-associated genes with a greater stress induced fold change in males and females, respectively (Dataset 12). Interestingly ten genes contained overlapping inversely abundant sex-specific acute stress-induced DhMRs, suggesting these DhMRs have a sex-specific role in response to stress. To further characterize the genes and pathways altered by acute stress and sex, we examined the gene ontologies of the 424 male and 346 DhMR-associated genes with a greater stress induced fold change in males and females, respectively, and found distinct outputs for each sex; while male-specific terms included axon and neuron projection guidance as well as cholesterol and sterol esterification, a significant enrichment of ontological terms were not found among the female-specific DhMR-associated genes (FDR P-value < 0.2; Dataset 13).
To investigate sex-specific stress induced gene expression changes, we compared RNAseq transcript levels from hippocampal tissue of the same mice surveyed for 5hmC (Methods) and found 1,365 genes and isoforms that had significant sex-specific changes in gene expression, including 807 and 681 genes that have male- and female-specific gene expression, respectively, 123 of which had differential expression of unique isoform transcripts in both sexes (FDR P-value < 0.05; Dataset 14). Again, many of these genes were previously shown to be differentially expressed in response to acute stress, including Bdnf, Ntrk2, and Nr3c2; or have been linked to psychiatric disorders, such as Grip1, Gabrab3, and Kcnq2 (Dataset 14) (Gonzalez-Mantilla et al., 2016). Notably, the sex-specific changes in gene expression also included several genes known to function in epigenetic pathways, including Dnmt3a, Hdac7, and Hdac10. Examination of the gene ontologies of these 807 and 681 differentially expressed genes and isoforms found a significant enrichment of neuronal ontological terms (P-value < 0.01) for both the male- and female-specific differentially expressed genes, including neuronal and axonal differentiation and development (FDR P-value < 0.05; Dataset 15 and 16; Methods). Many of the male- and female-specific genes that contributed to the overrepresentation of these terms have been previously implicated in stress-related disorders, including Ntrk2, Bdnf, Nr3c1, and Nfia. Together, these findings suggest that sex-specific differential gene expression in the hippocampus following acute stress is associated with known stress-related neurodevelopmental pathways.
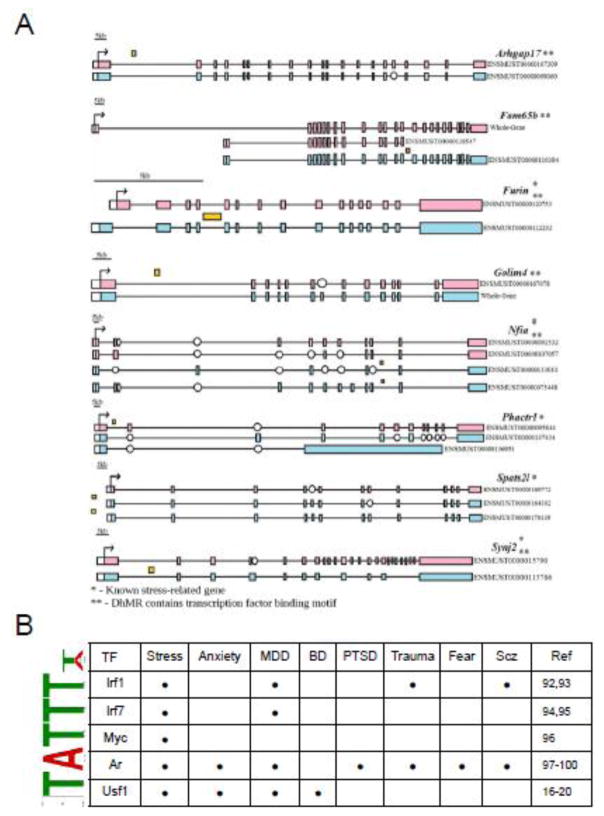
An overlay of the sex-specific DhMR-associated and differentially expressed genes revealed 82 potentially functional sex-specific DhMRs, including 56 and 34 genes with male- and female-specific potentially functional DhMRs, respectively. While it is notable that several of these potentially functional sex-specific DhMRs reside in genes with documented roles in stress and/or stress-induced psychiatric related disorders, including Dst, Grip1, Nfia (Dataset 14) (Gonzalez-Mantilla et al., 2016), it is perhaps most interesting that eight potentially functional DhMRs were associated with genes that produce sex-specific isoforms in response to stress (Figure 6a; Dataset 14). Moreover, the majority of these eight potentially functional DhMRs are located in introns (7/8) and contained a transcription factor binding sequence motif that was found to be significantly enriched in the 82 potentially functional DhMRs (6/8; Figure 6a and 6b). Together, these data demonstrate that sex-specific 5hmC is correlated to altered gene expression in response to acute stress and suggest that differential 5hmC may play a role in stress-induced sex-specific isoform production.
Figure 6.
Sex-specific stress-induced isoform production. A. Shown are the eight potentially functional DhMRs associated with genes that produce sex-specific isoforms in response to stress. The gene schematic contains the gene name on the right along with the relative location of each transcription start site (TSS; broken arrow), exon (white and colored boxes), intron (black horizontal line connecting exons), and hyper- or hypo-DhMR (yellow box above or below each gene, respectively). The sex producing the isoform is depicted as blue (male) or pink (female). Open circle denotes missing exon. Scale bar is present for relative genomic size of the gene. Single asterisk denotes if the gene is known to be associated with stress. Double asterisk denotes if the DhMR contains a transcription factor binding motif. B. Identification of DhMR-associated sequence motifs and their putative transcription factors. The DREME suite was used to predict the sequence motifs in the eight DhMRs shown in A. Motifs with E-value <10−3 were considered to be significant. Putative binding factors were predicted using SpaMo directly from the DREME suite software package and are listed in the tables shown with their putative transcription factors. The literature (Ref) links of each TF to psychiatric disorders are shown: Scz (schizophrenia); BP (Bipolar Disorder); MDD (Major Depressive Disorder); PTSD (Post-Traumatic Stress Disorder) (Chen et al., 2014a; Fenchel et al., 2015; Geng et al., 2007; Hart et al., 2012; Kumar et al., 2005; Liang et al., 2011; Mamdani et al., 2011; Pamment et al., 2002; Shiota et al., 2011).
Discussion
Fundamental differences between sexes in the anatomy and genetic network of the healthy brain are likely to underlie the pronounced sex differences in neurological disorders. Here we present the first genome-wide comparison of 5-hydroxymethylcytosine profiles from the hippocampus of naïve and acute stressed male and female mice and reveal sex-specific molecular responses to environmental stress in known and potentially novel genes in the stress pathway, which suggests that 5hmC is marking biologically relevant plasticity genes in response to acute stress. Perhaps more importantly, 5hmC may contribute to the generation of sex-specific isoforms in response to stress. Together, the DhMRs identified here represent a framework that will facilitate the future study of the complex interactions between the genes involved in stress and stress recovery. A clearer understanding of the molecular mechanisms regulating the expression of these genes may provide potentially modifiable substrates (e.g., 5hmC) that ultimately could be targeted to prevent the onset of some forms of psychiatric-related disorders.
Finding acute stress-related DhMRs in females revealed a role for 5hmC in the female genome at known and potentially novel genes in the stress pathway. For example, female DhMR-associated genes include neurotransmission genes (e.g., glutamate receptors that are a core feature of mental illnesses (Grik1) (Popoli et al., 2012), potassium channels that can induce anxiety-like behaviors in mice (Kcnj15, Kcnj16) (Wallace et al., 2009), and a member of the Kruppel-like factor (KLF) family of transcription factors (e.g., Klf13). These transcription factors are required for late phase neuronal maturation in developing dentate gyrus during adult hippocampal neurogenesis (Scobie et al., 2009). Interestingly, while there are many fewer female DhMRs than male DhMRs, the female DhMR-associated genes generally make up a subset of genes from the same families, suggesting similar neurological pathways are affected in both males an females. Together, these data demonstrate that a remarkable number of known stress-related genes are associated with changes in 5hmC abundance and implicate a role for genes not known to respond to acute stress.
A descriptive analysis of the hydroxymethylation events found in the hippocampus of both male and female (naïve and stressed) mice revealed that the majority of the hydroxymethylation is located in the genic structures (i.e., within the gene body), which is consistent with previous reports of genomic distribution of 5hmC (Li et al., 2016; Szulwach et al., 2011). However, in all genomes and conditions, nearly 20% of the total hydroxymethylation was found in the intergenic region. Despite this somewhat diverse distribution of total 5hmC, the vast majority of DhMRs (sex and/or stress) were found in the genic structures. Notably, while there is more abundance of 5hmC in females, there is more stress-related variation of 5hmC in males, as indicated by the enrichments and depletions of 5hmC in different genomic regions. The female DhMRs only have an overabundance in two genomic regions (Fig. 1), whereas nearly all the male genomic regions investigated (9 of 11) contained significant shifts in their 5hmC distribution following acute stress. These apparently male-specific changes likely drive the significant shifts in the sex specific interaction found in 8 of 11 genomic regions tested (Fig. 5). A common shift in 5hmC among all genomes and conditions was found in the intron/exon boundaries, which is consistent with finding the generation of sex-specific isoforms. Together, these data suggest that acute stress results in a significant shift in 5hmC from the intergenic regions of the genome into the gene body where it may function to regulate gene expression.
Comparison of methylome and transcriptome data found genes with potentially functional DhMRs that are consistent with results from several previous works, further validating this dataset for comparison of genes altered after exposure and recovery to a novel stress. These potentially functional DhMRs also extend previous studies that lacked the identity of specific molecular mechanism(s) that could explain the adaptive response to stress that alters the expression of several genes. Alterations in the expression levels of Nr3c1, an essential transcription factor that mediates stress-induced responses in the hippocampus, are well documented and our data suggests that 5hmC is regulating its expression. We also were encouraged to find that other genes with an uncharacterized role in stress but that have been implicated in mental illness, such as Anks1b, Arhgap24, Ddhd2, Dlg2, Grip1, Nfia, and Tcf4 (Gonzalez-Mantilla et al., 2016) contained potentially functional DhMRs. Together, these findings serve to validate a role for 5hmC in response to stress and also reveal potentially novel stress-related genes. Notably, DhMRs that are not correlated to changes in gene expression may reveal that these methylation levels mediate the action of a noncoding, but functional, RNA found in the genome. Noncoding RNAs are highly expressed in the brain and are involved in neurodevelopment and neuroplasticity-related functions; their dysregulation has been described in numerous neurological diseases(Qureshi and Mehler, 2012). Hence, future studies investigating stress related 5hmC levels need to consider the role of 5hmC in non-coding RNA expression. Since the role of hydroxymethylation in gene regulation is not fully characterized, these data warrant a deeper investigation of the role of 5hmC on individual gene expression changes caused by a novel short stress. It will be important for this future study to also seek to characterize the DNA methylation and expression profiles from distinct hippocampal sub-regions and cell types.
Several of the transcription factors whose binding sites are enriched among the DhMRs have known roles in neurogenesis and neurological activities, suggesting that 5hmC may play a role in transcription factor binding in response to acute stress. For example, PBX/Knotted 1 Homeobox 1 (Pknox1) has been associated with schizophrenia-related intermediate phenotypes and hippocampal volume (Hass et al., 2015). In addition, genetic mutations in the early growth response 2 (EGR2) transcription factor are linked to schizophrenia and bipolar disorder (Hu et al., 2015; Kim et al., 2012). Finally, Sex Determining Region Y-Box 1 (Sox1) is a transcriptional activator that functions as a switch in neuronal development by keeping neural cells undifferentiated until needed. Sox1 is linked to neurogenesis and has a role in epilepsy (Malas et al., 2003). Our data also suggests that 5hmC may have a role in transcript isoform production, as indicated by the potentially functional DhMRs associated with genes exhibiting stress-related sex-specific isoforms. Notably, sex-specific expression following environmental stress has already been documented, including CREB/ATF that are up regulated in males and down regulated in women with PTSD (O’Donovan et al., 2011). Here we found several known stress related genes expressing sex-specific isoforms, such as Furin, Sunj2, and Nfia; the last of which encodes a neuronal transcription factor that also is linked to intellectual disability (ID)/developmental disability (DD), autism spectrum disorders, and bipolar disorder. Together, with the inclusion of non-stress and non-psychiatric related genes producing sex-specific isoforms, these data demonstrate that the production of sex-specific isoforms in response to stress is more common than previously appreciated and may involve the disruption of transcription factor binding by 5hmC. It is of high interest for future studies to determine the unique roles of these sex-specific specific isoforms.
The differences in 5hmC profiles in male and female mice shed light on an underlying mechanism that may contribute to variations in the development of altered behaviors. The DhMR-associated genes found here represent targets within the HPA axis that are responding to environmental stress in a sex-specific manner. These results can serve as a benchmark for future comparison of sex-specific responses to different trauma types and durations and it will be important to relate these findings to chronic stress paradigms and to behavioral phenotypes. These future studies of behavior-related 5hmC will improve our understanding of the role of 5hmC in sex-specific susceptibility, progression, symptom severity, and pathology of neurological disorders.
While previous studies have identified differential distributions of 5hmC over a lifespan (Al-Mahdawi et al., 2014; Bradley-Whitman and Lovell, 2013; Chouliaras et al., 2013; Mellen et al., 2012; Villar-Menendez et al., 2013; Wang et al., 2013; Zhubi et al., 2014), here we also show dynamic genome-wide changes in 5hmC after an extremely short amount of time (i.e., 90 minutes). Moreover, these stress-induced rapid changes in 5hmC correlated with altered transcript levels, supporting an environmentally-sensitive functional role for 5hmC. These findings further support the emergence of 5hmC in environmentally-induced psychopathy. The identification of potentially functional sex-specific DhMRs that result in sex-specific isoforms encoded by genes with demonstrated roles in psychiatric disorders provide strong evidence that 5hmC plays a role in the regulation of gene expression in a sex specific manner in response to stress. Indeed, a link between 5hmC and gene regulation was recently postulated in human prefrontal cortex (Gross et al., 2015). Thus, the DhMRs presented here provide a modifiable functional molecular substrate (e.g., 5hmC) that may be harnessed to design novel therapeutic agents that will have optimal effectiveness in each sex.
Supplementary Material
Highlights.
Adult acute stress results in disruptions of female hippocampal 5hmC
Characterization of sex-specific 5hmC profiles in the mouse genome
Correlated sex-specific 5hmC and transcript levels reveal potentially functional5hmC
Stress-induced sex-specific 5hmC profiles are associated with neuronal ontologies
5hmC may mediate transcription factor binding and alternative splicing
Acknowledgments
The authors would like to thank Kim Sorens, the WISPIC animal facility, and the UW biotechnology center. This work was supported in part by the University of Wisconsin-Madison department of Psychiatry, University of Wisconsin Vilas Cycle Professorships #133AAA2989 and University of Wisconsin Graduate School #MSN184352 (all to RSA), NARSAD Young Investigator Grant from the Brain & Behavioral Research Foundation #22669 (LP), National Science Foundation under Grant No. 1400815 (AM), University of Wisconsin Neuroscience training grant T32-GM007507 (SL) and NIH grants HG003747 and U54AI117924 (SK), HG007019 (SK and QZ). The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdolmaleky HM, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15:3132–45. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mahdawi S, et al. The emerging role of 5-hydroxymethylcytosine in neurodegenerative diseases. Front Neurosci. 2014;8:397. doi: 10.3389/fnins.2014.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altemus M, et al. Sex differences in anxiety and depression clinical perspectives. Front Neuroendocrinol. 2014;35:320–30. doi: 10.1016/j.yfrne.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston C, et al. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10:309–22. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–9. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale TL. Epigenetic and transgenerational reprogramming of brain development. Nat Rev Neurosci. 2015;16:332–44. doi: 10.1038/nrn3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint S, et al. Attention deficit hyperactivity disorder (ADHD): gender- and age-related differences in neurocognition. Psychol Med. 2009;39:1337–45. doi: 10.1017/S0033291708004236. [DOI] [PubMed] [Google Scholar]
- Basmanav FB, et al. Investigation of the role of TCF4 rare sequence variants in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2015;168B:354–62. doi: 10.1002/ajmg.b.32318. [DOI] [PubMed] [Google Scholar]
- Bradley-Whitman MA, Lovell MA. Epigenetic changes in the progression of Alzheimer’s disease. Mech Ageing Dev. 2013;134:486–95. doi: 10.1016/j.mad.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butts T, et al. Transit amplification in the amniote cerebellum evolved via a heterochronic shift in NeuroD1 expression. Development. 2014;141:2791–5. doi: 10.1242/dev.101758. [DOI] [PubMed] [Google Scholar]
- Cahill L. Why sex matters for neuroscience. Nat Rev Neurosci. 2006;7:477–84. doi: 10.1038/nrn1909. [DOI] [PubMed] [Google Scholar]
- Castilhos RM, et al. Huntington disease and Huntington disease-like in a case series from Brazil. Clin Genet. 2014;86:373–7. doi: 10.1111/cge.12283. [DOI] [PubMed] [Google Scholar]
- Chen CV, et al. New knockout model confirms a role for androgen receptors in regulating anxiety-like behaviors and HPA response in mice. Horm Behav. 2014a;65:211–8. doi: 10.1016/j.yhbeh.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, et al. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl Psychiatry. 2014b;4:e375. doi: 10.1038/tp.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, et al. Rosiglitazone activation of PPARgamma-dependent signaling is neuroprotective in mutant huntingtin expressing cells. Exp Cell Res. 2015;338:183–93. doi: 10.1016/j.yexcr.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, et al. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiol Aging. 2013;34:2091–9. doi: 10.1016/j.neurobiolaging.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, et al. Changes in brain gene expression after long-term sleep deprivation. J Neurochem. 2006;98:1632–45. doi: 10.1111/j.1471-4159.2006.04058.x. [DOI] [PubMed] [Google Scholar]
- Collishaw S, et al. Resilience to adult psychopathology following childhood maltreatment: evidence from a community sample. Child Abuse Negl. 2007;31:211–29. doi: 10.1016/j.chiabu.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Condliffe D, et al. Cross-region reduction in 5-hydroxymethylcytosine in Alzheimer’s disease brain. Neurobiol Aging. 2014;35:1850–4. doi: 10.1016/j.neurobiolaging.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove KP, et al. Evolving knowledge of sex differences in brain structure, function, and chemistry. Biol Psychiatry. 2007;62:847–55. doi: 10.1016/j.biopsych.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel D, et al. Beyond the HPA-axis: The role of the gonadal steroid hormone receptors in modulating stress-related responses in an animal model of PTSD. Eur Neuropsychopharmacol. 2015;25:944–57. doi: 10.1016/j.euroneuro.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Forero DA, et al. A network of synaptic genes associated with schizophrenia and bipolar disorder. Schizophr Res. 2016;172:68–74. doi: 10.1016/j.schres.2016.02.012. [DOI] [PubMed] [Google Scholar]
- Forrest MP, et al. Knockdown of human TCF4 affects multiple signaling pathways involved in cell survival, epithelial to mesenchymal transition and neuronal differentiation. PLoS One. 2013;8:e73169. doi: 10.1371/journal.pone.0073169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng YG, et al. Comparison of the polymorphisms of androgen receptor gene and estrogen alpha and beta gene between adolescent females with first-onset major depressive disorder and controls. Int J Neurosci. 2007;117:539–47. doi: 10.1080/00207450600773640. [DOI] [PubMed] [Google Scholar]
- Gillberg C, et al. Brief report: “the autism epidemic”. The registered prevalence of autism in a Swedish urban area. J Autism Dev Disord. 2006;36:429–35. doi: 10.1007/s10803-006-0081-6. [DOI] [PubMed] [Google Scholar]
- Goes FS, et al. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am J Med Genet B Neuropsychiatr Genet. 2015;168:649–59. doi: 10.1002/ajmg.b.32349. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mantilla AJ, et al. A Cross-Disorder Method to Identify Novel Candidate Genes for Developmental Brain Disorders. JAMA Psychiatry. 2016;73:275–83. doi: 10.1001/jamapsychiatry.2015.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grados M, et al. Genetic findings in obsessive-compulsive disorder connect to brain-derived neutrophic factor and mammalian target of rapamycin pathways: implications for drug development. Drug Dev Res. 2014;75:372–83. doi: 10.1002/ddr.21223. [DOI] [PubMed] [Google Scholar]
- Gray JD, et al. Hippocampal gene expression changes underlying stress sensitization and recovery. Mol Psychiatry. 2014;19:1171–8. doi: 10.1038/mp.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JA, et al. Characterizing 5-hydroxymethylcytosine in human prefrontal cortex at single base resolution. BMC Genomics. 2015;16:672. doi: 10.1186/s12864-015-1875-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung R, Prata DP. What is the impact of genome-wide supported risk variants for schizophrenia and bipolar disorder on brain structure and function? A systematic review. Psychol Med. 2015;45:2461–80. doi: 10.1017/S0033291715000537. [DOI] [PubMed] [Google Scholar]
- Hart LS, et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest. 2012;122:4621–34. doi: 10.1172/JCI62973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass J, et al. Associations between DNA methylation and schizophrenia-related intermediate phenotypes - a gene set enrichment analysis. Prog Neuropsychopharmacol Biol Psychiatry. 2015;59:31–9. doi: 10.1016/j.pnpbp.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, et al. DISC1 (disrupted-in-schizophrenia-1) regulates differentiation of oligodendrocytes. PLoS One. 2014;9:e88506. doi: 10.1371/journal.pone.0088506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert LE, et al. Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology. 2013;80:1778–83. doi: 10.1212/WNL.0b013e31828726f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TM, et al. Resequencing of early growth response 2 (EGR2) gene revealed a recurrent patient-specific mutation in schizophrenia. Psychiatry Res. 2015;228:958–60. doi: 10.1016/j.psychres.2015.05.035. [DOI] [PubMed] [Google Scholar]
- Hunter RG, et al. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci U S A. 2009;106:20912–7. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson AS, et al. Altered circadian clock gene expression in patients with schizophrenia. Schizophr Res. 2016;174:17–23. doi: 10.1016/j.schres.2016.04.029. [DOI] [PubMed] [Google Scholar]
- Juhasz G, et al. The CREB1-BDNF-NTRK2 pathway in depression: multiple gene-cognition-environment interactions. Biol Psychiatry. 2011;69:762–71. doi: 10.1016/j.biopsych.2010.11.019. [DOI] [PubMed] [Google Scholar]
- Jukic MM, et al. Abnormal development of monoaminergic neurons is implicated in mood fluctuations and bipolar disorder. Neuropsychopharmacology. 2015;40:839–48. doi: 10.1038/npp.2014.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes K, et al. The role of allelic variation in estrogen receptor genes and major depression in the Nurses Health Study. Soc Psychiatry Psychiatr Epidemiol. 2015;50:1893–904. doi: 10.1007/s00127-015-1087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, et al. Pitx3 deficient mice as a genetic animal model of co-morbid depressive disorder and parkinsonism. Brain Res. 2014;1552:72–81. doi: 10.1016/j.brainres.2014.01.023. [DOI] [PubMed] [Google Scholar]
- Kim SH, et al. Genetic association of the EGR2 gene with bipolar disorder in Korea. Exp Mol Med. 2012;44:121–9. doi: 10.3858/emm.2012.44.2.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, et al. Human serum from patients with septic shock activates transcription factors STAT1, IRF1, and NF-kappaB and induces apoptosis in human cardiac myocytes. J Biol Chem. 2005;280:42619–26. doi: 10.1074/jbc.M508416200. [DOI] [PubMed] [Google Scholar]
- Kuratomi G, et al. Aberrant DNA methylation associated with bipolar disorder identified from discordant monozygotic twins. Mol Psychiatry. 2008;13:429–41. doi: 10.1038/sj.mp.4002001. [DOI] [PubMed] [Google Scholar]
- Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:155–81. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- Li S, et al. Genome-wide alterations in hippocampal 5-hydroxymethylcytosine links plasticity genes to acute stress. Neurobiol Dis. 2016;86:99–108. doi: 10.1016/j.nbd.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, et al. Negative regulation of IRF7 activation by activating transcription factor 4 suggests a cross-regulation between the IFN responses and the cellular integrated stress responses. J Immunol. 2011;186:1001–10. doi: 10.4049/jimmunol.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowski P, et al. Stress susceptibility-specific phenotype associated with different hippocampal transcriptomic responses to chronic tricyclic antidepressant treatment in mice. BMC Neurosci. 2013;14:144. doi: 10.1186/1471-2202-14-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccari S, et al. Early-life experiences and the development of adult diseases with a focus on mental illness: The Human Birth Theory. Neuroscience. 2016 doi: 10.1016/j.neuroscience.2016.05.042. [DOI] [PubMed] [Google Scholar]
- Malas S, et al. Sox1-deficient mice suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexcitability. Neuroscience. 2003;119:421–32. doi: 10.1016/s0306-4522(03)00158-1. [DOI] [PubMed] [Google Scholar]
- Mamdani F, et al. Gene expression biomarkers of response to citalopram treatment in major depressive disorder. Transl Psychiatry. 2011;1:e13. doi: 10.1038/tp.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmigere F, et al. Rapid induction of BDNF expression in the hippocampus during immobilization stress challenge in adult rats. Hippocampus. 2003;13:646–55. doi: 10.1002/hipo.10109. [DOI] [PubMed] [Google Scholar]
- Martin EI, et al. The neurobiology of anxiety disorders: brain imaging, genetics, and psychoneuroendocrinology. Clin Lab Med. 2010;30:865–91. doi: 10.1016/j.cll.2010.07.006. [DOI] [PubMed] [Google Scholar]
- McCarthy MM, et al. Sex differences in the brain: the not so inconvenient truth. J Neurosci. 2012;32:2241–7. doi: 10.1523/JNEUROSCI.5372-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, et al. The epigenetics of sex differences in the brain. J Neurosci. 2009;29:12815–23. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–8. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ. Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu Rev Neurosci. 2001;24:1161–92. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- Mellen M, et al. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–30. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencarelli MA, et al. Private inherited microdeletion/microduplications: implications in clinical practice. Eur J Med Genet. 2008;51:409–16. doi: 10.1016/j.ejmg.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Nestadt G, et al. Homeobox genes in obsessive-compulsive disorder. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:53–60. doi: 10.1002/ajmg.b.32001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen-Hoeksema S. Sex differences in unipolar depression: evidence and theory. Psychol Bull. 1987;101:259–82. [PubMed] [Google Scholar]
- O’Donovan A, et al. Transcriptional control of monocyte gene expression in post-traumatic stress disorder. Dis Markers. 2011;30:123–32. doi: 10.3233/DMA-2011-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamment J, et al. Regulation of the IRF-1 tumour modifier during the response to genotoxic stress involves an ATM-dependent signalling pathway. Oncogene. 2002;21:7776–85. doi: 10.1038/sj.onc.1205981. [DOI] [PubMed] [Google Scholar]
- Papale LA, et al. Genome-wide disruption of 5-hydroxymethylcytosine in a mouse model of autism. Hum Mol Genet. 2015;24:7121–31. doi: 10.1093/hmg/ddv411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 2014;15:483. doi: 10.1186/s13059-014-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoli M, et al. The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nat Rev Neurosci. 2012;13:22–37. doi: 10.1038/nrn3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter MO, et al. GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biol Psychiatry. 2008;64:645–52. doi: 10.1016/j.biopsych.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–41. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta D, et al. Source-reconstruction of event-related fields reveals hyperfunction and hypofunction of cortical circuits in antipsychotic-naive, first-episode schizophrenia patients during Mooney face processing. J Neurosci. 2014;34:5909–17. doi: 10.1523/JNEUROSCI.3752-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Roth TL, et al. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–9. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotheram-Fuller E, et al. Social involvement of children with autism spectrum disorders in elementary school classrooms. J Child Psychol Psychiatry. 2010;51:1227–34. doi: 10.1111/j.1469-7610.2010.02289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin TG, et al. Experience and the ever-changing brain: what the transcriptome can reveal. Bioessays. 2014;36:1072–81. doi: 10.1002/bies.201400095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, et al. Using sex differences in psychopathology to study causal mechanisms: unifying issues and research strategies. J Child Psychol Psychiatry. 2003;44:1092–115. doi: 10.1111/1469-7610.00194. [DOI] [PubMed] [Google Scholar]
- Schlaudraff F, et al. Orchestrated increase of dopamine and PARK mRNAs but not miR-133b in dopamine neurons in Parkinson’s disease. Neurobiol Aging. 2014;35:2302–15. doi: 10.1016/j.neurobiolaging.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobie KN, et al. Kruppel-like factor 9 is necessary for late-phase neuronal maturation in the developing dentate gyrus and during adult hippocampal neurogenesis. J Neurosci. 2009;29:9875–87. doi: 10.1523/JNEUROSCI.2260-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SQ, et al. Molecular analyses of circadian gene variants reveal sex-dependent links between depression and clocks. Transl Psychiatry. 2016;6:e748. doi: 10.1038/tp.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota M, et al. Oxidative stress and androgen receptor signaling in the development and progression of castration-resistant prostate cancer. Free Radic Biol Med. 2011;51:1320–8. doi: 10.1016/j.freeradbiomed.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Singmann P, et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenetics Chromatin. 2015;8:43. doi: 10.1186/s13072-015-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–16. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Lunyak VV. Epigenetics: judge, jury and executioner of stem cell fate. Epigenetics. 2012;7:823–40. doi: 10.4161/epi.21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D, et al. Widespread sex differences in gene expression and splicing in the adult human brain. Nat Commun. 2013;4:2771. doi: 10.1038/ncomms3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Menendez I, et al. Increased 5-methylcytosine and decreased 5-hydroxymethylcytosine levels are associated with reduced striatal A2AR levels in Huntington’s disease. Neuromolecular Med. 2013;15:295–309. doi: 10.1007/s12017-013-8219-0. [DOI] [PubMed] [Google Scholar]
- Wallace DL, et al. CREB regulation of nucleus accumbens excitability mediates social isolation-induced behavioral deficits. Nat Neurosci. 2009;12:200–9. doi: 10.1038/nn.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, et al. Genome-wide loss of 5-hmC is a novel epigenetic feature of Huntington’s disease. Hum Mol Genet. 2013;22:3641–53. doi: 10.1093/hmg/ddt214. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. The systemic amyloid precursor transthyretin (TTR) behaves as a neuronal stress protein regulated by HSF1 in SH-SY5Y human neuroblastoma cells and APP23 Alzheimer’s disease model mice. J Neurosci. 2014;34:7253–65. doi: 10.1523/JNEUROSCI.4936-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Wei Y, et al. Association study of three single-nucleotide polymorphisms in the cyclic adenosine monophosphate response element binding 1 gene and major depressive disorder. Exp Ther Med. 2015;9:2235–2240. doi: 10.3892/etm.2015.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman MM, et al. Cross-national epidemiology of major depression and bipolar disorder. JAMA. 1996;276:293–9. [PubMed] [Google Scholar]
- Wockner LF, et al. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl Psychiatry. 2014;4:e339. doi: 10.1038/tp.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten GF, et al. Are men at greater risk for Parkinson’s disease than women? J Neurol Neurosurg Psychiatry. 2004;75:637–9. doi: 10.1136/jnnp.2003.020982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Jin P. Cytosine modifications in neurodevelopment and diseases. Cell Mol Life Sci. 2014;71:405–18. doi: 10.1007/s00018-013-1433-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TY, et al. Epigenetic mechanisms for the early environmental regulation of hippocampal glucocorticoid receptor gene expression in rodents and humans. Neuropsychopharmacology. 2013;38:111–23. doi: 10.1038/npp.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A, et al. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl Psychiatry. 2014;4:e349. doi: 10.1038/tp.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.