Abstract
Protein ubiquitylation is a widespread post-translational modification, regulating cellular signalling with many outcomes, such as protein degradation, endocytosis, cell cycle progression, DNA repair and transcription. E3 ligases are a critical component of the ubiquitin proteasome system (UPS), determining the substrate specificity of the cascade by the covalent attachment of ubiquitin to substrate proteins. Currently, there are over 600 putative E3 ligases, but many are poorly characterized, particularly with respect to individual protein substrates. Here, we highlight systematic approaches to identify and validate UPS targets and discuss how they are underpinning rapid advances in our understanding of the biochemistry and biology of the UPS. The integration of novel tools, model systems and methods for target identification is driving significant interest in drug development, targeting various aspects of UPS function and advancing the understanding of a diverse range of disease processes.
Keywords: cancer, functional genomics, neurodegeneration, proteomics, proteostasis, ubiquitin
Ubiquitin proteasome system
Conjugation of the small protein modifier ubiquitin (Ub) is one of the most abundant post-translational modifications in cellular signalling. Ubiquitylation regulates a wide range of cellular processes, including protein degradation [1,2], endocytosis [2–4], cell cycle progression [5,6], DNA repair [7,8], transcription [9,10], translation [11] and immune function [12]. The fate of individual substrates following ubiquitylation is determined by ubiquitin chain topology, which acts as a molecular code for substrate fate [13]. Deregulation of the tightly regulated ubiquitin proteasome system (UPS) has been implicated in various human diseases, including cancer, autoimmunity, neurodegeneration and infectious diseases [14–17]. Hence, there is significant interest in defining the biochemistry of this pathway — both to better understand pathophysiology and to underpin the development of novel therapies. While efforts to understand other post-translational modifications — such as phosphorylation and acetylation — and exploit them therapeutically — have progressed rapidly, identifying cellular targets/substrates of the UPS has been hindered by the complex hierarchical biochemistry of the system and significant technical challenges.
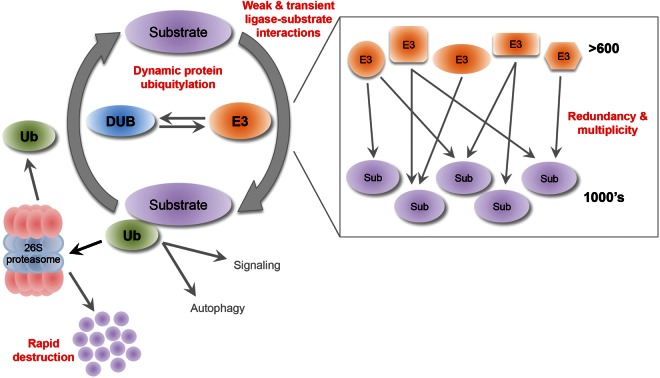
The ubiquitylation cascade (Figure 1) requires the action of three enzymes — Ub-activating enzyme (E1), Ub-conjugating enzyme (E2) and Ub protein ligase (E3 ligase) — in an energy-dependent hierarchical cascade [18]. Initially, the C-terminus of Ub is activated by a two-step ATP-dependent reaction in which a thioester bond is formed with a cysteine residue of E1. Ub is subsequently transferred to a cysteine residue of E2 via a thioester transfer reaction. E3s recruit Ub-loaded E2 enzymes and then either directly catalyze the ubiquitylation of the substrate lysine residue or facilitate the transfer of Ub to the substrate from the E2 via HECT (homology to E6-Ap carboxyl terminus), RING (really interesting new gene) or RBR (RING-in-between-RING) domains [19–21]. Recently, the SidE effector protein family of Legionella pneumophila was shown to directly ubiquitylate substrates via an mART motif. This mechanism operates without the need for E1/E2 enzymes or ATP, and activates ubiquitin using nicotinamide adenine dinucleotide [22]. Interestingly, the mART motif is also present in a family of mammalian proteins, indicating the potential for this all-in-one enzyme activity to exist in mammals [22]. Ubiquitin-specific proteases (USPs) [also known as deubiquitinases (DUBs)] act as reciprocal regulators of ubiquitlyation by catalyzing the removal of Ub from substrate proteins. Substrate ubiquitylation can signal many cellular fates, depending on chain topology, but one of the best-characterized outcomes is degradation by the proteasome.
Figure 1. Challenges in the identification of E3 ligase substrates.
1. The dynamic nature of protein ubiquitylation. 2. Weak and/or transient interactions between ligase and substrate. 3. Significant degrees of redundancy and multiplicity. 4. Rapid destruction of many ubiquitylated proteins.
One E1 enzyme (UBA1) is encoded in the human genome, with two known isoforms (nuclear and cytoplasmic localizations) [23,24]. Thirty-eight E2s have been identified (reviewed by ref. [25]), and there are ∼100 known DUBs [26]. However, substrate specificity is largely determined by >600 putative E3s identified to date [19,27]. It should be noted that the catalytic activities of many putative E3s and DUBs are yet to be validated. A key feature of the UPS is a significant degree of redundancy and multiplicity, where individual protein substrates may be targeted by multiple E3s (or DUBs), and a single E3 (or DUB) may have multiple protein substrates [28]. An emerging theme in understanding the regulation of protein ubiquitylation comes from the observation that E3/DUB pairs can act in a highly co-ordinated manner to regulate ubiquitylation (and hence activity) of each other and downstream substrates, although only a few examples have been observed to date [29–31]. The E3 ligase:DUB pair of Rsp5:Ubp2 acts in a co-ordinated fashion, with Ubp2 antagonizing Rsp5 E3 ligase activity by forming a complex together with Rsp5 and Rup1 that deubiquitylates Rsp5 substrates [29]. The DUB USP9X has been shown to stabilize the E3 ligase SMURF1 in MDA-MB-231 cells [31], and another interesting example is the reciprocal regulation of abundance and activity of the E3 ligase:DUB pair of UBR5 and DUBA, which controls IL-17 production in T cells. UBR5 destabilizes DUBA through ubiquitylation, whereas DUBA stabilizes UBR5 in activated T cells by attenuating degradative auto-ubiquitylation, triggering UBR5-mediated ubiquitylation of the transcription factor RORyt in response to TGF-β [30].
Most components of the UPS — from E1 through to the proteasome — are under active investigation as potential therapeutic targets in cancer and other indications [32–35]. The best-known compound targeting the UPS is the dipeptide boronic bortezomib (velcade) [36,37], a 26S proteasome inhibitor approved for clinical use in multiple myeloma [38]. Thalidomide and its analogues (lenalidomide-CC-5013 and pomalidomide-CC-4047) are immunomodulatory drugs (IMiDs) approved for clinical use in the treatment of multiple myeloma (reviewed in ref. [39]). Although the precise molecular mechanism remains unclear, IMiDs are thought to exert their teratogenic and anti-tumour effects by targeting cereblon (CRBN) [40]. The interaction disrupts the function of the E3 ligase complex constituted by CRBN, DDB1 and Cul4, causing down-regulation of fibroblast growth factor genes [40,41]. Apcin (APC inhibitor) is a novel small molecule inhibitor of another E3 ligase complex: anaphase-promoting complex/cyclosome (APC/C) [42]. Apcin binds to and competively inhibits Cdc20, a substrate recognition co-receptor of APC/C [42]. This effect is amplified by the co-addition of another small molecule inhibitor — tosyl-l-argine methyl ester — which also blocks the APC/C–Cdc20 interaction [42].
Another strategy is to target the UPS by inhibiting DUB activity. Currently, there are several small molecule inhibitors in the pipeline, such as HBX 19818 and P050429, which selectively target USP7/HAUSP (reviewed in ref. [34]). More recently, ML323 has been reported as a potent inhibitor of the USP1–UAF1 deubiquitinase complex, revealing USP1–UAF1 to be an important regulator of the DNA damage response and a target for overcoming platinum-based chemotherapy resistance [43].
Several studies have shown that ubiquitin variants (UbVs) can also function as highly specific inhibitors or activators for various enzymes, including E3s and DUBs. For example, UbVs have been developed to either inhibit HECT E3s by binding to the E2-binding site or activate by targeting an ubiquitin-binding exosite [44]. Furthermore, exploitation of the low-affinity interactions between Ub and enzymes of the ubiquitin system by engineered optimization has produced potent and selective modulators of UPS components, including DUBs, E2s and E3s [45]. These UbVs will be very useful tools to elucidate the function of E3s by modulating their activity.
As E3 ligases primarily determine the substrate specificity of the UPS they may represent more specific targets for inhibition [46,47]. The pursuit of inhibitors that target specific E3 ligase activity or block E3–substrate interactions led to the development of Nutlins (cis-imidazoline compounds), which are currently in clinical trials for various indications. Nutlins prevent binding of the E3 ligase MDM2 to its substrate, the tumour suppressor p53 [19,48–51]. Another compound currently in clinical trials for cancer is the Smac mimetic GDC-0152, which binds to anti-apoptotic proteins (IAPs), inducing IAP self-ubiquitylation and degradation, and consequently apoptosis [52].
Although E3 ligases make promising therapeutic targets, efforts into developing therapeutics targeting E3 ligases have so far been relatively ineffective — with no drugs currently approved for clinical use. This is partly due to limited understanding of ligase–substrate relationships and biological function. A key to the development of effective therapeutics is the identification of E3 ligase substrates and, conversely, the ligases are targeting a specific substrate. These will not only advance understanding of functional roles but also are necessary for the development of functional assays for drug screening and associated discovery and the validation of relevant biomarkers.
E3 ligase substrate identification
Many of the ∼600 E3 ligases identified in the human genome remain relatively uncharacterized [19] and identification of E3 ligase substrates is notoriously difficult. For a few well-characterized E3 ligases, conserved targeting sequences in substrates have been identified, allowing prediction and validation of putative substrates, for example, N-degrons recognized by UBR family ligases, and Nedd4 WW domains, which bind proline-rich domains in target proteins [53–56]. Ligase–substrate recognition can also be modulated by post-translational modifications. For example, the E3 ligase c-Cbl binds to phosphorylated tyrosine motifs in substrates, and ubiquitylation and degradation of phosphoenolpyruvate carboxykinase is mediated via acetylation-dependent recognition by UBR5 under conditions of high glucose [57]. However, in most cases, the identification and validation of substrates of an individual E3 ligase, or the specific ligase(s) targeting a particular substrate, have relied on relatively slow, low-throughput biochemical methods.
Challenges
Many factors present significant technical challenges in identifying E3 ligase substrates or identifying E3 ligases targeting a specific substrate (Figure 1). 1. The dynamic nature of protein ubiquitylation. Ubiquitylation is under tight control of E3 ligases and DUBs acting in a highly co-ordinated manner to edit or remove ubiquitin chains in response to changing cellular environments [58]. 2. Weak and/or transient interactions between ligase and substrate. The weak physical interaction and rapid dissociation rate between some E3–substrate complexes mean that identification by immunoprecipitation, followed by mass spectrometry, is challenging [59]. The low cellular abundance of substrates may also hinder identification [60]. This dynamic equilibrium between E3 ligase and substrate highlights the need for more sensitive techniques and is partly addressed by ‘trapping’ approaches (see below). 3. Significant degrees of redundancy and multiplicity. Any particular substrate may be targeted by multiple E3 ligases at different sites, and a single E3 ligase may target multiple substrates under different conditions or in different cellular compartments. This drives a huge diversity in spatial and temporal control of ubiquitylation (reviewed by ref. [61]). Cellular context is an important consideration, as substrate–ligase pairs identified by biochemical methods may not be expressed or interact in the same sub-cellular compartment. 4. Rapid destruction of many ubiquitylated proteins. The identification of ubiquitylated proteins destined for degradation by the proteasome can be difficult without the use of proteasome inhibitors, and these may produce other confounding biological effects [62]. A complicating factor in E3 ligase substrate identification may be the relative insensitivity of many methods to post-translational modifications of ubiquitin, E3 ligases or substrates — which have the potential to alter activity and substrate binding [63,64].
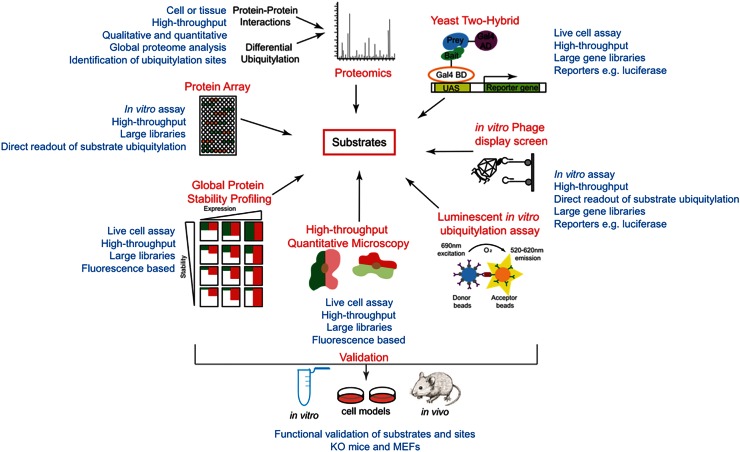
The relatively recent development of systematic approaches to identify E3 ligase substrates (shown in Figure 2) is providing an unprecedented mechanistic insight into ubiquitin signalling. In a very short timeframe, the defined cellular ubiquitome has expanded by orders of magnitude to now include a significant proportion of the total proteome — with >23 000 individual modification sites mapping to >4600 proteins [65,66]. In particular, integrated approaches combining genetic models (i.e. modulating E3 ligase expression or activity) with powerful functional genomics or proteomics have rapidly expanded our understanding of breadth, depth and diversity of the ubiquitin-modified proteome (ubiquitome) in various contexts [67–69]. The various platforms and techniques present their own advantages and disadvantages, depending on the study context and available model systems (Table 1). Differences in the catalytic mechanisms of HECT, RING and RBR classes of E3 ligases also present slightly different challenges to the various methods for the identification of substrates. These include different mechanisms of Ub binding and transfer, variations in regulatory mechanisms and the need for accessory/adaptor proteins. Unfortunately, there is little empirical evidence comparing the relative performance of each method against different E3 classes, making it difficult to draw definitive conclusions.
Figure 2. Diversity of systematic approaches available to identify E3 ligase substrates.
Table 1. Comparison of systematic approaches to identify E3 ligase substrates.
| Method | Advantages | Disadvantages |
|---|---|---|
| Yeast two-hybrid (Y2H) |
|
|
| In vitro ubiquitylation: phage display and luminescent |
|
|
| Protein microarrays |
|
|
| Global protein stability (GPS) profiling |
|
|
| High-throughput quantitative microscopy (HCA) |
|
|
| Differential expression (shotgun) proteomics |
|
|
| Affinity purification proteomics: Ub, TUBEs, PAC-Compass |
|
|
| Ubiquitin ligase trapping and proximity labelling |
|
|
| DiGly remnant affinity proteomics |
|
|
A comprehensive summary of the application of these various strategies to identifying E3 ligase substrates is provided in Table 2. While interacting proteins of E3 ligases represent potential substrates, further validation by orthogonal methods is necessary to attribute status as bona fide substrates.
Table 2. Examples of E3 ligase substrate identification with various approaches.
| Method | E3 ligase | Number of substrates identified | Reference |
|---|---|---|---|
| Yeast two-hybrid (Y2H) | LNX | 64 Interactors and 8 validated substrates | [70] |
| VHL | 100 Interactors, 8 putative substrates and 2 known substrates | [71] | |
| Nedd4 | 4 Interactors. 2 substrates | [72] | |
| SCF (Cdc4) | 4 Putative substrates and 1 known substrate | [73] | |
| NKLAM (RNF19b) | 1 Substrate | [74] | |
| TRIM32 | 1 Substrate | [75] | |
| In vitro ubiquitylation — phage display | MDM2 | 4 Known substrates and 11 validated substrates | [76] |
| In vitro ubiquitylation — luminescent | Rsp5 | 9 Validated substrates | [77] |
| Protein microarray | Rsp5 | 45 Putative substrates, 11 known substrates and 6 validated | [78] |
| Rsp5 | 86 Candidates identified and 28 validated substrates | [79] | |
| Nedd4 | ∼15 Validated substrates | [80] | |
| SMURF1 | 7 Validated substrates | [81] | |
| Praja1 | 14 Putative substrates | [82] | |
| Global protein stability (GPS) profiling | SCF | 359 Putative substrates and 31 validated substrates | [83] |
| Various CRLs | 47 Validated substrates | [84] | |
| High-throughput quantitative microscopy (HCA) | SCFGrr1 | 106 Putative substrates and 7 validated substrates | [85] |
| Differential expression (shotgun) proteomics | ASB2 | 2 Known substrates | [86] |
| MARCH9 | 12 Candidate substrates and 6 validated | [87] | |
| Affinity purification proteomics | SCF (Saf1) | 18 Putative substrates, 4 validated substrates and 17 known substrates | [88] |
| FBXL family | 88 Interacting proteins and 13 putative substrates | [89] | |
| FBXW11 | 96 Interacting proteins, 23 putative substrates and 1 validated | [90] | |
| BARD1 | 14 Putative substrates and 2 validated substrates | [91] | |
| βTrCP | 221 Putative substrates | [92] | |
| MuRF1 | 20 Putative substrates, 3 validated substrates and 3 known substrates | [93] | |
| Ubiquitin ligase trapping and proximity labelling | FBXL family | 17 Known substrates and 18 novel substrates | [94] |
| Rsp5, Itch, Psh1, RNF126, RNF168 | Numerous | [95] | |
| βTrCP1/2 | 50 Candidate substrates and 12 validated | [96] | |
| XIAP | 50 Putative substrates | [97] | |
| diGly remnant affinity proteomics | CRL | 253 Ubiquitylation sites, 59 known Ub-sites or CRL components | [65] |
| PARKIN | 1654 Putative substrates | [98] | |
| SPOP | 12 Ubiquitylation sites and 4 likely substrates fated for degradation | [99] | |
| FBXO21 | 36 Putative substrates, 2 validated substrates — one novel ubiquitylation site and 5 previously reported sites identified | [60] | |
| Integrated approaches | HRD1 | 9 High-confidence substrates and 1 validated substrate | [100] |
| HUWE1 | 1 Substrate and 1 interactor confirmed | [101] | |
| Trim32 | 19 High-confidence substrates and 1 validated substrate | [102] | |
| ASB2α | 2 Validated substrates | [103] |
Interacting proteins represent potential substrates, but require further validation by orthogonal methods.
Genetics and functional genomics
Genetic screens have been developed in various formats to facilitate the identification of E3 ligase protein–protein interaction (PPI) networks in eukaryotes and prokaryotes. These include cell-based [e.g. yeast two-hybrid (Y2H)] and in vitro methods using reconstituted ubiquitylation assays (e.g. phage display screens and protein microarrays). Each technique is scalable to screen large gene libraries from human, yeast and other species and can be paired with various reporters (e.g. β-galactosidase and luciferase) for rapid identification of interactors/substrates. A common general restraint across the various genetic approaches used is limitations on the prey libraries available. For example, cDNA libraries may be incomplete, biased towards shorter genes, or contain truncated genes. Phage display systems also place severe restrictions on the size of genes that can be expressed.
Yeast two-hybrid
Y2H screening identifies PPIs in live yeast cells via the reconstitution of a functional transcription factor (Gal4) and subsequent reporter gene expression following the interaction of bait and prey proteins fused to the N-terminal DNA-binding domain and the C-terminal (transcriptional) activation domain of Gal4, respectively. Commonly used reporter systems include a GAL1–lacZ fusion gene, encoding β-galactosidase enzyme that labels the yeast cell blue when cultured on indicator plates, or facilitates selection-based screens based on the ability of yeast cells to grow on galactose [104–107]. Y2H has been applied to identify putative substrates of many E3 ligases. For example, the PDZ protein interaction domains of the LNX family of E3s were used as baits to screen a random peptide library, identifying 64 putative interacting proteins [108]. Similarly, the pVHL substrate recognition subunit was used as a bait to screen a human testis cDNA library to identify substrates of the CBCVHL E3 ligase complex [62]. Eight novel interactors were identified, along with previously known substrates [71]. Y2H is a relatively quick and simple technique that can be adapted to high-throughput format. However, the technique appears to be particularly susceptible to high rates of false-positive and false-negative (24–51% false-positive rate and 45–96% false-negative rate) [109–111]. Furthermore, Y2H may not be compatible with the expression of non-yeast proteins, which may not fold correctly or are potentially toxic. Importantly, Y2H is limited to the number of genes in a library and to PPIs that occur in the nucleus.
In vitro ubiquitylation screens
Many techniques for screening ubiquitin substrates rely on reconstituted in vitro assays using recombinant components (Ub, E1, E2 and E3) applied to a library of potential substrates (e.g. phage display libraries, recombinant protein libraries or protein microarrays). While these assays can be adapted to high-throughput formats and provide a direct readout of substrate ubiquitlyation, their main drawback is that they cannot account for cellular context (e.g. sub-cellular localization, differential expression and co-regulation by DUBs). The requirement for active recombinant enzymes can also be problematic as these enzymes are not always readily available and their activity may be regulated by PTMs and/or cofactors. A further consideration with in vitro ubiquitylation assays is the issue of E2 specificity. While many E3 ligases exhibit activity with ‘promiscuous’ E2 enzymes, such as UBE2D, some E3 ligases only function effectively with a specific E2 enzyme [25]. Protein microarray sub-screens to identify optimal E2 partners for E3 ligases (discussed below) may be useful in this context, but the issue should be considered whether using in vitro assays in either screening or validation formats.
Phage display
An in vitro ubiquitylation assay coupled with a phage display library using His-tagged ubiquitin, a human brain cDNA library and recombinant GST-MDM2, has been used to identify MDM2 substrates. Following purification and amplification of ubiquitylated phages, individual clones were PCR-amplified and sequenced to identify 16 ubiquitylated MDM2 substrates [76]. This method is fast and simple but may suffer from limitations in the size of peptide that can be expressed by the phage display system, meaning that potential conformational determinants and allosteric regulatory regions of substrates may not be accommodated.
Luminescent in vitro ubiquitylation assay
Luminescent in vitro ubiquitylation assays use a collection of putative substrates as GST-tagged fusion proteins bound to anti-GST acceptor beads, while biotin–ubiquitin is bound to streptavidin donor beads [77]. Ubiquitylation of GST-tagged proteins with biotin–ubiquitin brings acceptor and donor beads (which also contain a photosensitizer) into close proximity. Upon excitation (680 nm), a series of luminescent energy transfers between chemiluminescent and fluorescent compounds in the acceptor beads are measured by a micro-plate reader [77,112]. This assay was used to screen for yeast Rsp5 E3 ligase substrates using 188 GST-tagged fusion proteins, identifying and validating 9 Rsp5 substrates [77]. A significant limitation of this approach is the need to generate tagged, recombinant proteins — which may fold incorrectly, lack essential PTMs or suffer interference by affinity tags.
Protein microarrays
Functional protein microarrays are gaining increasing popularity for high-throughput, systematic identification of E3 ligase substrates, in part because this assay is now offered as a commercial service. This method uses a library of putative substrates — in the form of recombinant proteins — arrayed to a carrier surface, such as glass slides [113,114], combined with a reconstituted in vitro ubiquitylation cascade (as above) and a sensitive method (radiological, fluorescence or chemiluminescence) for the detection of conjugated ubiquitin. This system has the advantage of also facilitating sub-screens to identify optimal E2 partners for E3 ligase activity using an arrayed panel of E2 enzymes, but still suffers from the usual restrictions of in vitro assays and limitations on the arrayed substrate library (see above). This method was first used with a 4000 protein yeast proteome microarray to identify 40 substrates of the HECT E3 ligase Rsp5, identifying 11 previously known substrates, with 6 further validated [78]. A similar technique to identify Rsp5 substrates with a different array containing 5800 proteins [79] included a dosage lethality/suppression screen that validated 28 substrates (out of 86 candidates). Interestingly, there was little overlap between the substrates identified in two screens for the same E3 ligase, highlighting the need for orthogonal approaches and validation.
Many studies have used protein microarrays to profile substrates of mammalian E3 ligases. For example, a microarray containing 8222 human proteins was used to identify ∼50 putative substrates each for the human Rsp5 orthologues hNedd4-1, hNedd4-2 and the rat orthologue rNedd4-1 [80]. A smaller human protein microarray (containing ∼2000 proteins) was used to identify substrates of the human E3 ligase Praja1 [82]. This study used a commercially available E2 profiling kit to determine that UBE2D3 was the optimal E2 for Praja1. An interesting aspect of this study was the use of duplicate microarrays — one probed with an anti-ubiquitin antibody that detects both mono- and poly-ubiquitylated proteins and the other probed with biotinylated tandem ubiquitin-binding entity (TUBE) specific for ubiquitin chains. Fourteen putative Praja1 substrates were identified in common to both detection methods. A major shortcoming of this technique is that currently available commercial protein microarrays do not have coverage of the entire proteome [115] and do not account for cellular context.
Global protein stability profiling
Significant advances in functional genomic tools and models have made possible the systematic and global identification of E3 ligase substrates in live mammalian cells, which is a key consideration in understanding UPS function in the context of cell signalling. These approaches can identify differential degradation of proteins following the modulation of E3 ligase activity or expression, using reporters such as GFP to permit observation of spatio-temporal context and rapid validation using traditional biochemical techniques such as co-IP. Global protein stability (GPS) profiling is a fluorescence-based, high-throughput system to monitor protein turnover (but not protein ubiquitylation directly) in mammalian cells using a retroviral reporter construct containing a single promoter driving the bi-cistronic translation of two fluorescent proteins from one mRNA transcript [116]. Discosoma sp. red fluorescent protein (DsRed) acts as an internal control, whereas enhanced GFP (eGFP) is expressed fused to a protein of interest. As they are translated from a single mRNA, both fluorescent proteins are produced at approximately the same rate and DsRed expression should remain relatively constant, controlling for background protein stability. Therefore, changes in the green/red fluorescence (eGFP/DsRed) ratio of a cell under different conditions are a direct readout of eGFP-labelled fusion protein degradation [116].
This reporter system was coupled with the hORFeome library, fluorescence-activated cell sorting and microarray deconvolution to identify differential degraded proteins, following genetic alteration of Skp Cullin F-box (SCF) activity (i.e. putative substrates) [83]. This study identified 73% of known SCF substrates and 359 putative substrates, of which 31 were validated [83]. This method was further extended by integration with proteomics to identify CRL substrates following the treatment of cells with a CRL inhibitor (described in detail below — Integrated Approaches).
GPS profiling provides significant advantages over other genetic screening methods, probably the most important being the ability to monitor dynamic changes in protein stability in live mammalian cells following pharmacological or genetic manipulation [83,116]. Even with limitations of the hORFeome library used (containing only ∼8000 of the estimated ∼20 000 proteins in the proteome), GPS profiling represents a significant expansion of screening coverage in mammalian cells. Potential disadvantages include the time-consuming cloning of each protein into the reporter construct. The EGFP tag may also interfere with protein folding, localization, PTMs and protein stability [116]. An important caveat of GPS profiling from the perspective of understanding UPS function is that the assay measures protein stability, and is not a direct measure of protein ubiquitylation. Hence, other factors influencing protein stability must be considered when interpreting results, and GPS profiling is not able to distinguish ubiquitylation events with signalling outcomes other than protein degradation.
High-throughput quantitative microscopy (high-content analysis)
High-throughput microscopy (also known as high-content analysis, HCA) was utilized to identify SCFGrr1 substrates by comparing isogenic wild-type GRR1 and GRR1 knockout (grr1Δ) yeast cells simultaneously. GRR1 was replaced with red fluorescent protein (RFP) in grr1Δ cells, and the grr1Δ::RFP strain was then crossed with a library of 4000 yeast strains that each express a GFP-tagged ORF and imaged in 96-well format using HCA [85]. GRR1 substrates were expected to have increased GFP intensity in grr1Δ (RFP-positive) cells compared with GRR1 cells (RFP-negative). Similar to GPS profiling, this technique does not provide a direct measure of protein ubiquitylation and similar caveats around limitations in library size and potential interference from GFP tag apply. However, the utility of this screening strategy is clear, identifying 106 putative GRR1 substrates, with 7 validated. Furthermore, the ability to screen in live cells provides clear advantages in understanding dynamic processes in relevant spatio-temporal context.
Proteomics
Proteomics has emerged as a powerful tool for identifying E3 ligase substrates, as continuing improvements in biochemical techniques and instrument sensitivity allow more precise qualitative and quantitative analyses of the E3 ligase-regulated proteome. Mass spectrometry (MS)-based proteomics has been coupled with a variety of model systems and enrichment strategies to identify E3 ligase substrates at both the protein and site level (>23 000 modified sites mapping to >4600 proteins) [65,66]. These strategies include defining PPI networks of E3 ligases and profiling differential changes in the global proteome — or ubiquitome specifically — following the modulation of E3 ligase expression or activity [86,98,101].
Shotgun proteomics
Label-free shotgun proteomics was used to identify ASB2 (ankyrin repeat-containing protein with a suppressor of cytokine signalling box 2) E3 ligase substrates. Proteins that had reduced abundance in ASB2 wild-type expressing cells, compared with ligase-inactive mutant-expressing cells, were likely to be ubiquitylated by ASB2 and degraded by the proteasome [86]. This technique identified two known ASB2 substrates [117]. Similarly, Hör et al. [87] used SILAC (stable isotope labelling by amino acids in cell culture)-MS to identify differentially expressed plasma membrane proteins in cells overexpressing GFP-tagged MARCH9 E3 ligase or GFP alone. Twelve down-regulated proteins were identified in GFP-MARCH9 overexpressing cells compared with GFP alone, and six were validated as substrates by flow cytometry. The major caveat of shotgun proteomics strategies (as with GPS profiling — above) is that they will identify Ub substrates fated for proteasomal degradation, whereas they will not detect substrates fated for other signalling events.
Affinity purification–tandem mass spectrometry
Various configurations of affinity purification coupled with mass spectrometry (AP–MS/MS) have been successfully applied to identify putative E3 ligase-interacting proteins and substrates. In their most basic format, these typically involve the expression of an E3 ligase or Ub bait fused to a small tag (such as HA) for enrichment, combined with various genetic or pharmacological manipulations. More sophisticated approaches to AP–MS/MS have recently been developed, which allow trapping and/or labelling of substrates, enrichment of specific poly-Ub chain types or site-specific identification of Ub modifications.
There are numerous examples of co-purifying interacting proteins of individual E3 ligase baits to identify putative substrates, but the emergence of systematic approaches is proving very powerful. For example, parallel adaptor capture (PAC) proteomics coupled with CompPASS (Comparative Proteomics Analysis Software Suite) was developed to systematically identify substrates of 19 F-box proteins (FBXLs) [89]. Interactors of HA-tagged FBXL proteins, overexpressed in HEK293 cells, were identified by parallel α-HA affinity purification, MS/MS and CompPASS analysis. This study identified 22 high confidence-interacting proteins (HCIPs), and 13 were found to have increased stability in the presence of the Nedd8-activating enzyme inhibitor MLN4924, which inactivates FBXL-containing cullin–RING E3 ligases [89]. This strategy was repeated in a colon cancer cell line, which identified 88 HCIPs, 44 of which were also identified in the HEK293 experiment [89]. If known, the isolated substrate-binding region of an E3 can also be used to purify interacting proteins. For example, WNKs were co-purified and identified using the CUL3 substrate adaptor KLHL3 as a bait [118]. While this is an effective and efficient method to identify putative E3 ligase substrates, not all interacting proteins identified are necessarily substrates, making the use of inhibitors or functional mutants an essential aspect of these experiments.
The great power of the PAC–CompPASS method is the ability to simultaneously interrogate substrates of multiple E3 ligases. However, the requirement for affinity purifications of multiple proteins performed in parallel and consequently high demand on instrument and analytical time may put this technique beyond the reach of many investigators. Furthermore, CompPASS can identify proteins based on a single peptide, increasing the potential for incorrect protein identifications or false-positives [89]. Importantly, PAC-based approaches are unlikely to enrich for monoubiquitylated substrates and substrates that are not fated to be degraded [90]. The PAC–CompPASS approach was recently applied to identify substrates of the FBXW11 ubiquitin ligase adaptor protein [90]. This study identified 96 interacting proteins, 23 of which are putative substrates, with 1 independently validated as a substrate [90]. Interestingly, a similar approach has also been applied to systematic identification of DUB substrates [119].
Another AP–MS/MS-based strategy to identifying E3 ligase substrates exploits the attachment of His-tagged Ub (and various mutants) to substrates following E3 ligase protein overexpression [91]. Using quantitative mass spectrometry analysis, this study identified 14 proteins with increased ubiquitylation following BRCA1/BARD1 overexpression [91]. A similar approach combined bioinformatics and AP–MS/MS to identify SCFβTrCP E3 ligase substrates. First, a consensus motif derived from reported βTrCP substrates was generated and used to search biological sequence databases for substrates [92]. Subsequently, using AP–MS/MS, βTrCP-interacting proteins were identified and the data were integrated with the bioinformatics prediction data [92]. This approach identified 221 βTrCP interactors that also harbour conserved sequences similar to the consensus motif of known substrates [92].
Alternative affinity purification strategies have been developed that not only identify substrates destined for proteasomal degradation but also identify substrates labelled with alternative ubiquitin chain topologies signalling various fates. For example, TUBEs, containing four tandem UBA domains and additional tags, such as 6xHis for detection, efficiently bind mono- and poly-ubiquitylated proteins [120]. This method does not rely on protein overexpression, tag fusion proteins, inhibitors or chemical and genetic manipulations and therefore is ideal for the detection of endogenous ubiquitylated proteins [121,122]. To identify substrates of the MuRF1 E3 ligase, TUBEs coupled with two-dimensional differential in-gel electrophoresis (2D-DIGE) and MS were used to compare cells overexpressing either MuRF1 or GFP [93]. This study identified 20 proteins with differential ubiquitylation following MuRF1 overexpression, 3 of which were previously known substrates and 3 that were validated as novel substrates. One limitation arises from the use of 2D-DIGE, as low-abundance protein bands may not be visible, very hydrophobic proteins are difficult to detect, and there may be sample loss during gel processing. More contemporary proteomic approaches have largely made the use of 2D-DIGE redundant and promise much more sensitive detection of proteins that underwent TUBE enrichment.
Ubiquitin ligase trapping and proximity labelling
Many techniques have recently been developed that use various formats of E3 ligase fusion proteins to either ‘trap’ putative ligase substrates or label proteins in close proximity to the E3 ligase active site for downstream identification using proteomics.
The ubiquitin ligase trapping method changes the kinetics of the ligase–substrate interaction (i.e. decreases the off-rate), essentially increasing the affinity of a ligase for its polyubiquitylated substrates and providing a means for purification and identification of the substrate. The first iteration of this technique used a synthetic fusion protein, consisting of an UBA domain fused to the N-terminus of an E3 via a linker composed of three tandem FLAG epitopes, coupled with mass spectrometry, [94], to identify substrates of 8 FBXLs in yeast [88]. This study identified 17 known FBXL substrates and 18 novel substrates, but very little overlap was observed in the substrates identified compared with the PAC–CompPASS method described above [89], possibly reflecting differences between yeast and human FBXLs. A possible caveat of this approach is the potential for the UBA domain fused to the FBXL N-terminus to disrupt substrate binding. Furthermore, UBA domains have preference for different ubiquitin chain topologies [94,123,124] and hence, the specific UBA domain used may not detect all possible substrates of a given ligase. A more recently developed trapping approach uses E3-ubiquitin fusion proteins (ubiquitin-activated interaction traps, UBAITs) to covalently capture proteins that interact with the E3 in an E1- and E2-dependent manner. This technique was successfully identified binding partners of both HECT (Rsp5 and Itch) and RING (Psh1, RNF126 and RNF168) E3 ligases [95].
Proximity-dependent biotin labelling (BioID) uses a fusion protein of an E3 coupled with a biotin-conjugating enzyme (BirA*). BirA* activates biotin, which then reacts with nearby amine groups on lysine residues in interacting proteins. This assay was coupled with semi-quantitative mass spectrometry on cells treated with MG132 to identify 50 new SCFβ-TrCP1/2 interacting proteins, with 12 being validated as E3 ligase substrates [96]. A more sophisticated version of the proximity labelling strategy uses an engineered chimeric protein called the NEDDylator, which was generated by removing the RING domain of XIAP and then fusing the E2 for NEDD8 (Ubc12) to the substrate-binding domain of the ligase via a flexible linker. NEDDylator–XIAP maintains specific substrate-binding capacity, but substitutes conjugation of the ubiquitin-like protein NEDD8 to the substrate instead of Ub [97]. Compared with Ub, NEDD8 is a relatively rare PTM in cells and is generally thought to regulate protein activity, not stability [97]. Jurkat cells were incubated with NEDDylator–XIAP and HB–NEDD8 followed by purification of HB–NEDD8-tagged substrates and mass spectrometry analysis. More than 50 putative XIAP substrates were identified [97]. An advantage of this strategy is that it enriches for proteins that are specifically ubiquitylated by the chimeric protein, not just interacting proteins in close proximity. However, generation of the recombinant chimeric protein may be difficult for some larger multidomain E3s. In addition, the utility of the NEDDylator may be restricted to RING domain ligases, as unlike HECT domain ligases, these do not directly catalyze the ubiquitylation of their substrates.
Gly–Gly (diGly) remnant affinity purification
An important breakthrough in understanding cellular ubiquitin signalling was the development of monoclonal antibodies that specifically recognize the characteristic Gly–Gly (diGly) remnant present on the ε-amine of lysine following trypsin digestion of ubiquitylated proteins. This modification is detected by MS analysis as a distinct mass shift of 114.04 Da [125]. Several laboratories have developed di-Gly antibodies [65,125–127], driving a significant increase in the available resolution and coverage of ubiquitylation detection. The potential of diGly antibodies is underscored by the widespread utility and analytical power provided by the development of phospho-specific antibodies in understanding kinase signalling [128] or methyl-/acetyl-specific antibodies in characterizing histone modifications [129,130].
One important limitation of diGly remnant affinity purification for identifying ubiquitylation sites is that an identical remnant is left following trypsin digestion of proteins conjugated to ubiquitin-like modifiers, such as ISG15 and NEDD8, making them indistinguishable by tandem MS [125,131,132]. One way to overcome this limitation is a two-step enrichment strategy where ubiquitylated proteins are first purified from cells expressing 6xHis-tagged ubiquitin followed by trypsin digestion and diGly immuno-purification proteomics (diGly proteomics). This strategy was used to identify 374 diGly-modified peptides on 236 proteins, but nullifies one of the most powerful aspects of the diGly technique, namely the ability to detect endogenous ubiquitylation without the possible confounding factors introduced by the use of tagged protein overexpression [125].
Initially, global analysis of lysine ubiquitylation using diGly antibodies under normal physiological conditions or following pharmacological treatment (e.g. with proteasome inhibitor, MG132) was deployed to identify between 1664–20 000 diGly-modified sites [65,66,127,133]. However, this strategy could not match the ubiquitylated site with the responsible E3 ligase, nor reveal ubiquitin chain topology. DiGly proteomics is increasingly being used in conjunction with chemical inhibition or genetic manipulation of E3 ligase activity or expression to couple ubiquitylation sites with the responsible E3 ligase. For example, this approach was utilized to identify CRL substrates, identifying 253 diGly sites with reduced abundance following CRL and proteasome inhibition — of which 59 sites were on known CRL substrates or components of CRLs [65]. Another technique combined 73 control and diGly proteomic experiments with PARKIN overexpression and/or mitochondrial depolarization by carbonyl cyanide m-chlorophenyl hydrazone treatment to identify 4772 dynamically regulated ubiquitylation sites in 1654 proteins [98]. Several of these were subsequently validated by western blot analysis using PARKIN siRNA. A similar approach was used to identify cullin–RING ubiquitin ligase adaptor protein speckle-type POZ protein (SPOP) ubiquitylation sites by performing diGly proteomics on cells overexpressing either wild-type SPOP (SPOP-WT), prostate cancer relevant mutants (SPOP-MT) or empty vector [99]. Twelve diGly sites identified were more abundant in SPOP-MT cells compared with SPOP-WT or empty vector, and four proteins had an inverse correlation with protein expression [99].
A newer approach, combining trypsin-resistant tandem ubiquitin-binding entity(ies) (TR-TUBE) technology with diGly proteomics, was recently deployed to identify substrates of the uncharacterized F-box protein FBXO21. TR-TUBE is based on a modification of the previously described TUBE technology [120] to protect ubiquitin chains from trypsin digestion [60]. Overexpression of FLAG-TR-TUBE and E3 ligase followed by enrichment with anti-FLAG antibody and diGly proteomics identified putative substrates with less background compared with enrichment by TR-TUBE alone [60]. In addition, substantially less starting material is required than using diGly proteomics only [60]. This approach was applied to identify several putative substrates of FBXO21, of which two were independently validated [60].
Integrated approaches
Integrating orthogonal approaches is potentially a rapid means to identify high-confidence E3 ligase interactors and substrates, substantially reducing the number of hits required to validate using more traditional biochemical techniques. Aiming to provide a more comprehensive survey, Lee et al. used an integrated strategy combining diGly proteomics and AP–MS. Mono- and poly-ubiquitylated proteins were purified from cells expressing 6xHis-Ub and HRD1 siRNA, followed by mass spectrometry analysis [100]. This approach identified 29 ubiquitylated proteins that had decreased abundance in HRD1 siRNA samples compared with control siRNA [100]. The second strategy utilized diGly proteomics following attenuated expression of HRD1 by siRNA [100]. This study identified 117 ubiquitylated peptides with reduced abundance in HRD1 siRNA sample compared with control. Nine high-confidence substrates were identified by both methods, highlighting the advantage of using a combined strategy.
To identify high-confidence E3 ligase substrates in a physiological setting, several studies have employed integrated approaches utilizing tissue from isogenic mouse models coupled with label-free quantitative (LFQ) mass spectrometry. This method was implemented in the search for substrates of Trim32 in skeletal muscle cells [102]. Comparing Trim32 knockout mouse (T32KO) with wild-type control, this study identified 19 proteins that had increased abundance in T32KO muscle samples [102]. Following independent validation of two putative substrates, NDRG2 was identified as a novel substrate of Trim32 [102]. Spinner et al. [103] utilized this strategy to identify substrates of the E3 ligase ASB2α in different dendritic cell (DC) subsets. This study identified FLNa and FLNb as the only proteins that significantly accumulated in ASB2α-deficient DCs compared with ASB2α-expressing DCs, indicating that FLNa and FLNb are highly specific targets of ASB2α in haematopoietic cells [103]. This approach highlights the power of isogenic mouse models coupled with LFQ mass spectrometry to identify E3 ligase substrates in a physiological setting, but validation using independent biochemical approaches is still required.
Validation methods
Even with advances in proteomics, genomics and in vitro assays that address many of the significant challenges outlined above, identifying E3 ligase substrates — or determining which E3 ligase targets a specific substrate — remains a significant challenge. Notably, these technological advances do not directly address a significant remaining bottleneck in the quest to identify genuine E3 ligase targets — validation of the often large number of putative substrates identified, particularly in a functional context. There are many ways to validate putative E3 ligase substrates and interacting proteins, including IP–western blot experiments, in vitro ubiquitylation assays and cell-based assays.
The decision about which validation method(s) to adopt is heavily dependent on the ultimate aim of the study. While many consider in vitro ubiquitlyation assays using recombinant components to be the gold standard, theses cannot account for cellular context, regulation by PTMs or feedback regulation by DUBs. Confirming PPIs in the cellular context using Co-IP, IF or BiFC is an important consideration. Similarly, assays combining site-directed mutagenesis, si/shRNA or CRISPR with assays of cellular function (e.g. viability and proliferation) or more specific readouts of signal transduction pathway output may be desirable to confirm functional roles. Animal models of disease and/or gene function may also be used in this context.
However, these are not generally suitable to validate large numbers of hits from high-throughput screening platforms. Instead, novel strategies are required to rapidly and efficiently validate a large number of putative E3 substrates, or at least render the large data set down to a more manageable list of high-confidence hits. Currently, integrating orthogonal approaches, such as diGly proteomics and AP–MS with genetic/chemical inhibition of E3 ligase activity or protein levels, seem to be the most effective strategy. DiGly proteomics requires putative sites to be independently validated, but western blot analysis is unable to detect differences in ubiquitylation at the site level [65,66]. While ORFeome libraries and high-content imaging technologies enable high-throughput validation of PPIs, validation of individual ubiquitin-modified sites requires time-consuming site-directed mutagenesis studies. Ideally, functional validation of substrates and ubiquitylation sites using knockout animal models and/or isogenic cell lines is necessary. For example, the identification of putative Nedd4-1 and Nedd4-2 substrates using human protein microarrays was functionally validated by showing that Nedd4-1 negatively regulates signalling via the fibroblast growth factor receptor 1 (FGFR1) [80]. Following on from this study, the Rotin laboratory identified the sequence motif necessary for Nedd4-1 to bind to and ubiquitylate FGFR1 [134]. Deletion of the sequence motif (FGFR1-Δ6) stimulated neuronal stimulation in human embryonic neuronal stem cells [134]. Importantly, FGFR1-Δ6 expression in Zebrafish embryos caused defective anterior neuronal patterning, indicative of excessive FGFR1 signalling [134]. This study exemplifies the utility of an integrated approach to systematically identify an E3 ligase substrate, from high-throughput screen (i.e. protein array) to in vitro and in vivo functional validation.
Conclusions
The recent development of highly innovative methods to identify substrates of the UPS has gone a long way to address a significant challenge in the field. These systematic approaches are underpinning rapid advances in our understanding of the biochemistry and biology of the UPS and are already having significant impact on our understanding of a diverse range of disease processes. Investigators need to consider several factors in choosing the most appropriate approach to identifying E3 ligase substrates in the particular context of interest. 1. Access to analytical platforms and expertise. 2. Speed and cost. 3. Availability and suitability of model systems for both discovery and validation (i.e. cell lines and animals). 4. Availability of reagents (e.g. recombinant proteins, inhibitors and expression libraries). Integrated approaches — combining genomics or proteomics with isogenic models or chemical inhibition — are emerging as the optimal strategy to identify E3 ligase substrates and should be favoured whenever possible. Spatial and temporal cellular context is also an important consideration. E3 ligase substrates identified using reconstituted in vitro assays may not necessarily be relevant in vivo if the ligase–substrate are not expressed in the same cellular compartment at the same time. Conversely, demonstrating in vitro ligase activity towards a particular substrate is still considered the gold standard by many.
Functional validation of ubiquitylation sites and putative substrates in cellular and in vivo contexts still presents a major bottleneck, due to both the sheer number of putative substrates identified and a shortage of suitable high-throughput biochemical methods. The future development of improved model systems and research tools will be key to driving the gaining momentum in the field. In particular, many researchers would welcome the development of selective and potent pharmacological inhibitors across all classes of E3 ligases. The recent development of selective Ub variant probe libraries [44], targeting a panel of HECT ligases, is a significant advance in this respect and should provide a powerful toolkit for the discovery and functional validation of E3 ligase substrates. Rapid improvements in both the speed and precision of gene targeting approaches (e.g. using CRISPR) to generate isogenic knockout/knockin mouse and cell line models for in vivo discovery and validation of E3 ligase function and substrates will also be important. One significant outstanding hurdle to enabling a systematic and comprehensive characterization of UPS substrates is the lack of a central, co-ordinated database of ligase–substrate pairs. A practical and reliable knowledge base would provide a key resource — not only for researchers studying the UPS, but also more widely in pathophysiology and drug development in a range of disease contexts.
Abbreviations
- 2D-DIGE
two-dimensional differential in-gel electrophoresis
- APC/C
anaphase-promoting complex/cyclosome
- AP–MS/MS
affinity purification coupled with mass spectrometry
- ASB2
ankyrin repeat-containing protein with a suppressor of cytokine signalling box 2
- BioID
biotin labelling
- CompPASS
Comparative Proteomics Analysis Software Suite
- CRBN
cereblon
- CRL
Cullin-RING ubiquitin ligase
- DC
dendritic cell
- DsRed
Discosoma sp. red fluorescent protein
- DUBs
deubiquitinases
- eGFP
enhanced GFP
- FGFR1
fibroblast growth factor receptor 1
- GPS
global protein stability
- HCA
high-content analysis
- HECT
homology to E6-Ap carboxyl terminus
- IAP
inhibitor of apoptosis
- IMiDs
immunomodulatory drugs
- IP
immunoprecipitation
- LFQ
label-free quantitative
- MS
mass spectrometry
- ORF
open reading frame
- PAC
parallel adaptor capture
- PPI
protein–protein interaction
- PTM
post-translational modification
- RBR
RING-in-between-RING domains
- RFP
red fluorescent protein
- RING
really interesting new gene
- SCF
Skp Cullin F-box
- SPOP
speckle-type POZ protein
- SPOP-MT
SPOP relevant mutants
- SPOP-WT
wild-type SPOP
- T32KO
Trim32 knockout mouse
- Ub
ubiquitin
- UBAITs
ubiquitin-activated interaction traps
- UBv
Ub variant
- UPS
ubiquitin proteasome system
- USPs
ubiquitin-specific proteases
- Y2H
yeast two-hybrid.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Chau V., Tobias J.W., Bachmair A., Marriott D., Ecker D.J., Gonda D.K. et al. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 PMID: [DOI] [PubMed] [Google Scholar]
- 2.Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P.P., Dikic I. et al. (2003) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell. Biol. 5, 461–466 doi: 10.1038/ncb983 [DOI] [PubMed] [Google Scholar]
- 3.Hicke L. and Riezman H. (1996) Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84, 277–287 doi: 10.1016/S0092-8674(00)80982-4 [DOI] [PubMed] [Google Scholar]
- 4.Staub O., Gautschi I., Ishikawa T., Breitschopf K., Ciechanover A., Schild L. et al. (1997) Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 16, 6325–6336 doi: 10.1093/emboj/16.21.6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koepp D.M., Harper J.W. and Elledge S.J. (1999) How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97, 431–434 doi:http://dx.doi.org/10.1016/S0092-8674(00)80753-9 [DOI] [PubMed] [Google Scholar]
- 6.Jin L., Williamson A., Banerjee S., Philipp I. and Rape M. (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665 doi: 10.1016/j.cell.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spence J., Sadis S., Haas A.L. and Finley D. (1995) A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15, 1265–1273 doi: 10.1128/MCB.15.3.1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies A.A., Huttner D., Daigaku Y., Chen S. and Ulrich H.D. (2008) Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell 29, 625–636 doi: 10.1016/j.molcel.2007.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L., Li L., Zhou B., Qin Z. and Dou Y. (2014) H2b ubiquitylation promotes RNA Pol II processivity via PAF1 and pTEFb. Mol. Cell 54, 920–931 doi: 10.1016/j.molcel.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grøfte M. et al. (2012) TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 150, 697–709 doi: 10.1016/j.cell.2012.06.039 [DOI] [PubMed] [Google Scholar]
- 11.Yoshida M., Yoshida K., Kozlov G., Lim N.S., De Crescenzo G., Pang Z. et al. (2006) Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 25, 1934–1944 doi: 10.1038/sj.emboj.7601079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleynen I., Vazeille E., Artieda M., Verspaget H.W., Szczypiorska M., Bringer M.-A. et al. (2014) Genetic and microbial factors modulating the ubiquitin proteasome system in inflammatory bowel disease. Gut 63, 1265–1274 doi: 10.1136/gutjnl-2012-303205 [DOI] [PubMed] [Google Scholar]
- 13.Komander D. and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 doi: 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- 14.Jung C.-R., Hwang K.-S., Yoo J., Cho W.-K., Kim J.-M., Kim W.H. et al. (2006) E2-EPF UCP targets pVHL for degradation and associates with tumor growth and metastasis. Nat. Med. 12, 809–816 doi: 10.1038/nm1440 [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q., Wang H., Schwartz D.M., Stoffels M., Park Y.H., Zhang Y. et al. (2015) Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat. Genet. 48, 67–73 doi: 10.1038/ng.3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan Z., Sun X., Hou F.-S., Oh H.-W., Hilgenberg L.G.W., Hol E.M. et al. (2007) Mutant ubiquitin found in Alzheimer's disease causes neuritic beading of mitochondria in association with neuronal degeneration. Cell Death Differ. 14, 1721–1732 doi: 10.1038/sj.cdd.4402180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chesarino N.M., McMichael T.M. and Yount J.S. (2015) E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog. 11, e1005095 doi: 10.1371/journal.ppat.1005095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershko A. and Ciechanover A. (1992) The ubiquitin system for protein degradation. Annu. Rev. Biochem. 61, 761–807 doi: 10.1146/annurev.bi.61.070192.003553 [DOI] [PubMed] [Google Scholar]
- 19.Bernassola F., Karin M., Ciechanover A. and Melino G. (2008) The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21 doi: 10.1016/j.ccr.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 20.Rotin D. and Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 10, 398–409 doi: 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- 21.Dove K.K., Stieglitz B., Duncan E.D., Rittinger K. and Klevit R.E. (2016) Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep. 17, 1221–1235 doi: 10.15252/embr.201642641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu J., Sheedlo M.J., Yu K., Tan Y., Nakayasu E.S., Das C. et al. (2016) Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533, 120–124 doi: 10.1038/nature17657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siepmann T.J., Bohnsack R.N., Tokgoz Z., Baboshina O.V. and Haas A.L. (2003) Protein interactions within the N-end rule ubiquitin ligation pathway. J. Biol. Chem. 278, 9448–9457 doi: 10.1074/jbc.M211240200 [DOI] [PubMed] [Google Scholar]
- 24.Lee I. and Schindelin H. (2008) Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell 134, 268–278 doi: 10.1016/j.cell.2008.05.046 [DOI] [PubMed] [Google Scholar]
- 25.Christensen D.E. and Klevit R.E. (2009) Dynamic interactions of proteins in complex networks: identifying the complete set of interacting E2s for functional investigation of E3-dependent protein ubiquitination. FEBS J. 276, 5381–5389 doi: 10.1111/j.1742-4658.2009.07249.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nijman S.M.B., Luna-Vargas M.P.A., Velds A., Brummelkamp T.R., Dirac A.M.G., Sixma T.K. et al. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 doi: 10.1016/j.cell.2005.11.007 [DOI] [PubMed] [Google Scholar]
- 27.Metzger M.B., Hristova V.A. and Weissman A.M. (2012) HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell. Sci. 125, 531–537 doi: 10.1242/jcs.091777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nalepa G., Rolfe M. and Harper J.W. (2006) Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 5, 596–613 doi: 10.1038/nrd2056 [DOI] [PubMed] [Google Scholar]
- 29.Kee Y., Lyon N. and Huibregtse J.M. (2005) The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 24, 2414–2424 doi: 10.1038/sj.emboj.7600710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutz S., Kayagaki N., Phung Q.T., Eidenschenk C., Noubade R., Wang X. et al. (2014) Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature 518, 417–421 doi: 10.1038/nature13979 [DOI] [PubMed] [Google Scholar]
- 31.Xie Y., Avello M., Schirle M., McWhinnie E., Feng Y., Bric-Furlong E. et al. (2013) Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J. Biol. Chem. 288, 2976–2985 doi: 10.1074/jbc.M112.430066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D., Frezza M., Schmitt S., Kanwar J. and Dou Q.P. (2011) Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr. Cancer Drug Targets 11, 239–253 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoeller D. and Dikic I. (2009) Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 doi: 10.1038/nature07960 [DOI] [PubMed] [Google Scholar]
- 34.Mattern M.R., Wu J. and Nicholson B. (2012) Ubiquitin-based anticancer therapy: carpet bombing with proteasome inhibitors vs surgical strikes with E1, E2, E3, or DUB inhibitors. Biochim. Biophys. Acta, Mol. Cell Res. 1823, 2014–2021 doi: 10.1016/j.bbamcr.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pal A., Young M.A. and Donato N.J. (2014) Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res. 74, 4955–4966 doi: 10.1158/0008-5472.CAN-14-1211 [DOI] [PubMed] [Google Scholar]
- 36.Adams J., Palombella V.J., Sausville E.A., Johnson J., Destree A., Lazarus D.D. et al. (1999) Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 59, 2615–2622 PMID: [PubMed] [Google Scholar]
- 37.Goldberg A.L. (2012) Development of proteasome inhibitors as research tools and cancer drugs. J. Cell. Biol. 199, 583–588 doi: 10.1083/jcb.201210077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orlowski R.Z., Stinchcombe T.E., Mitchell B.S., Shea T.C., Baldwin A.S., Stahl S. (2002) Phase I trial of the proteasome inhibitor PS-341 in patients with refractory hematologic malignancies. J. Clin. Oncol. 20, 4420–4427 doi: 10.1200/JCO.2002.01.133 [DOI] [PubMed] [Google Scholar]
- 39.Ocio E.M., Richardson P.G., Rajkumar S.V., Palumbo A., Mateos M.V., Orlowski R. et al. (2014) New drugs and novel mechanisms of action in multiple myeloma in 2013: a report from the International Myeloma Working Group (IMWG). Leukemia 28, 525–542 doi: 10.1038/leu.2013.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y. et al. (2010) Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 doi: 10.1126/science.1177319 [DOI] [PubMed] [Google Scholar]
- 41.Zhu Y.X., Braggio E., Shi C.-X., Bruins L.A., Schmidt J.E., Van Wier S. et al. (2011) Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood 118, 4771–4779 doi: 10.1182/blood-2011-05-356063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sackton K.L., Dimova N., Zeng X., Tian W., Zhang M., Sackton T.B. et al. (2014) Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature 514, 646–649 doi: 10.1038/nature13660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang Q., Dexheimer T.S., Zhang P., Rosenthal A.S., Villamil M.A., You C. et al. (2014) A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 10, 298–304 doi: 10.1038/nchembio.1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W., Wu K.-P., Sartori M.A., Kamadurai H.B., Ordureau A., Jiang C. et al. (2016) System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes. Mol. Cell 62, 121–136 doi: 10.1016/j.molcel.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ernst A., Avvakumov G., Tong J., Fan Y., Zhao Y., Alberts P. et al. (2013) A strategy for modulation of enzymes in the ubiquitin system. Science 339, 590–595 doi: 10.1126/science.1230161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Obrist F., Manic G., Kroemer G., Vitale I. and Galluzzi L. (2015) Trial watch: proteasomal inhibitors for anticancer therapy. Mol. Cell. Oncol. 2, e974463 doi: 10.4161/23723556.2014.974463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W. and Sidhu S.S. (2014) Development of inhibitors in the ubiquitination cascade. FEBS Lett. 588, 356–367 doi: 10.1016/j.febslet.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ofir-Rosenfeld Y., Boggs K., Michael D., Kastan M.B. and Oren M. (2008) Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell 32, 180–189 doi: 10.1016/j.molcel.2008.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliner J.D., Pietenpol J.A., Thiagalingam S., Gyuris J., Kinzler K.W., Vogelstein B. et al. (1993) Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362, 857–860 doi: 10.1038/362857a0 [DOI] [PubMed] [Google Scholar]
- 50.Roth J., Dobbelstein M., Freedman D.A., Shenk T. and Levine A.J. (1998) Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17, 554–564 doi: 10.1093/emboj/17.2.554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vassilev L.T., Vu B.T., Graves B., Carvajal D., Podlaski F., Filipovic Z. et al. (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 doi: 10.1126/science.1092472 [DOI] [PubMed] [Google Scholar]
- 52.Hird A.W., Aquila B.M., Hennessy E.J., Vasbinder M.M. and Yang B. (2015) Small molecule inhibitor of apoptosis proteins antagonists: a patent review. Expert Opin. Ther. Pat. 25, 755–774 doi: 10.1517/13543776.2015.1041922 [DOI] [PubMed] [Google Scholar]
- 53.Tasaki T., Mulder L.C.F., Iwamatsu A., Lee M.J., Davydov I.V., Varshavsky A. et al. (2005) A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 25, 7120–7136 doi: 10.1128/MCB.25.16.7120-7136.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jadhav T., Geetha T., Jiang J. and Wooten M.W. (2008) Identification of a consensus site for TRAF6/p62 polyubiquitination. Biochem. Biophys. Res. Commun. 371, 521–524 doi: 10.1016/j.bbrc.2008.04.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen H.I. and Sudol M. (1995) The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl Acad. Sci. USA 92, 7819–7823 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deckert M., Elly C., Altman A. and Liu Y.-C. (1998) Coordinated regulation of the tyrosine phosphorylation of Cbl by Fyn and Syk tyrosine kinases. J. Biol. Chem. 273, 8867–8874 doi: 10.1074/jbc.273.15.8867 [DOI] [PubMed] [Google Scholar]
- 57.Jiang W., Wang S., Xiao M., Lin Y., Zhou L., Lei Q. et al. (2011) Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell 43, 33–44 doi: 10.1016/j.molcel.2011.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komander D., Reyes-Turcu F., Licchesi J.D.F., Odenwaelder P., Wilkinson K.D., Barford D. et al. (2009) Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 10, 466–473 doi: 10.1038/embor.2009.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pierce N.W., Kleiger G., Shan S.-o. and Deshaies R.J. (2009) Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462, 615–619 doi: 10.1038/nature08595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida Y., Saeki Y., Murakami A., Kawawaki J., Tsuchiya H., Yoshihara H. et al. (2015) A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc. Natl Acad. Sci. USA 112, 4630–4635 doi: 10.1073/pnas.1422313112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jain A.K. and Barton M.C. (2009) Regulation of p53: TRIM24 enters the RING. Cell Cycle 8, 3668–3674 doi: 10.4161/cc.8.22.9979 [DOI] [PubMed] [Google Scholar]
- 62.Harper J.W. and Tan M.-K.M. (2012) Understanding cullin-RING E3 biology through proteomics-based substrate identification. Mol. Cell. Proteomics 11, 1541–1550 doi: 10.1074/mcp.R112.021154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swatek K.N. and Komander D. (2016) Ubiquitin modifications. Cell. Res. 26, 399–422 doi: 10.1038/cr.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buetow L. and Huang D.T. (2016) Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. doi: 10.1038/nrm.2016.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A. et al. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 doi: 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wagner S.A., Beli P., Weinert B.T., Nielsen M.L., Cox J., Mann M. et al. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284 doi: 10.1074/mcp.M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Komander D. (2009) The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 37, 937–953 doi: 10.1042/BST0370937 [DOI] [PubMed] [Google Scholar]
- 68.Ordureau A., Munch C. and Harper J.W. (2015) Quantifying ubiquitin signaling. Mol. Cell 58, 660–676 doi: 10.1016/j.molcel.2015.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sylvestersen K.B., Young C. and Nielsen M.L. (2013) Advances in characterizing ubiquitylation sites by mass spectrometry. Curr. Opin. Chem. Biol. 17, 49–58 doi: 10.1016/j.cbpa.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 70.Guo Z.G., Song E., Ma S., Wang X., Gao S., Shao C. et al. (2012) Proteomics strategy to identify substrates of LNX, a PDZ domain-containing E3 ubiquitin ligase. J. Proteome Res. 11, 4847–4862 doi: 10.1021/Pr300674c [DOI] [PubMed] [Google Scholar]
- 71.Bex C., Knauth K., Dambacher S. and Buchberger A. (2007) A yeast two-hybrid system reconstituting substrate recognition of the von Hippel-Lindau tumor suppressor protein. Nucleic Acids Res. 35, e142 doi: 10.1093/nar/gkm932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murillas R., Simms K.S., Hatakeyama S., Weissman A.M. and Kuehn M.R. (2002) Identification of developmentally expressed proteins that functionally interact with Nedd4 ubiquitin ligase. J. Biol. Chem. 277, 2897–2907 doi: 10.1074/jbc.M110047200 [DOI] [PubMed] [Google Scholar]
- 73.Kishi T., Ikeda A., Koyama N., Fukada J. and Nagao R. (2008) A refined two-hybrid system reveals that SCFCdc4-dependent degradation of Swi5 contributes to the regulatory mechanism of S-phase entry. Proc. Natl Acad. Sci. USA 105, 14497–14502 doi: 10.1073/pnas.0806253105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fortier J.M. and Kornbluth J. (2006) NK lytic-associated molecule, involved in NK cytotoxic function, is an E3 ligase. J. Immunol. 176, 6454–6463 doi: 10.4049/jimmunol.176.11.6454 [DOI] [PubMed] [Google Scholar]
- 75.Locke M., Tinsley C.L., Benson M.A. and Blake D.J. (2009) TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum. Mol. Genet. 18, 2344–2358 doi: 10.1093/hmg/ddp167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo Z., Wang X., Li H. and Gao Y. (2013) Screening E3 substrates using a live phage display library. PLoS ONE 8, e76622 doi: 10.1371/journal.pone.0076622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kus B., Gajadhar A., Stanger K., Cho R., Sun W., Rouleau N. et al. (2005) A high throughput screen to identify substrates for the ubiquitin ligase Rsp5. J. Biol. Chem. 280, 29470–29478 doi: 10.1074/jbc.M502197200 [DOI] [PubMed] [Google Scholar]
- 78.Gupta R., Kus B., Fladd C., Wasmuth J., Tonikian R., Sidhu S. et al. (2007) Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol. Syst. Biol. 3, 116 doi: 10.1038/msb4100159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu J.-y., Lin Y.-y., Qian J., Tao S.-c., Zhu J., Pickart C. et al. (2007) Functional dissection of a HECT ubiquitin E3 ligase. Mol. Cell. Proteomics 7, 35–45 doi: 10.1074/mcp.M700353-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Persaud A., Alberts P., Amsen E.M., Xiong X., Wasmuth J., Saadon Z. et al. (2009) Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4-2 using proteome arrays. Mol. Syst. Biol. 5, 333 doi: 10.1038/msb.2009.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Andrews P.S., Schneider S., Yang E., Michaels M., Chen H., Tang J. et al. (2010) Identification of substrates of SMURF1 ubiquitin ligase activity utilizing protein microarrays. Assay Drug Dev. Technol. 8, 471–487 doi: 10.1089/adt.2009.0264 [DOI] [PubMed] [Google Scholar]
- 82.Loch C.M., Eddins M.J. and Strickler J.E. (2011) Protein microarrays for the identification of Praja1 E3 ubiquitin ligase substrates. Cell Biochem. Biophys. 60, 127–135 doi: 10.1007/s12013-011-9180-x [DOI] [PubMed] [Google Scholar]
- 83.Yen H.-C.S. and Elledge S.J. (2008) Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322, 923–929 doi: 10.1126/science.1160462 [DOI] [PubMed] [Google Scholar]
- 84.Emanuele M.J., Elia A.E.H., Xu Q., Thoma C.R., Izhar L., Leng Y. et al. (2011) Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 doi: 10.1016/j.cell.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Benanti J.A., Cheung S.K., Brady M.C. and Toczyski D.P. (2007) A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat. Cell. Biol. 9, 1184–1191 doi: 10.1038/ncb1639 [DOI] [PubMed] [Google Scholar]
- 86.Burande C.F., Heuze M.L., Lamsoul I., Monsarrat B., Uttenweiler-Joseph S., Lutz P. G. et al. (2009) A label-free quantitative proteomics strategy to identify E3 ubiquitin ligase substrates targeted to proteasome degradation. Mol. Cell. Proteomics 8, 1719–1727 doi: 10.1074/mcp.M800410-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hör S., Ziv T., Admon A. and Lehner P.J. (2009) Stable isotope labeling by amino acids in cell culture and differential plasma membrane proteome quantitation identify new substrates for the MARCH9 transmembrane E3 ligase. Mol. Cell. Proteomics 8, 1959–1971 doi: 10.1074/mcp.M900174-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mark K.G., Simonetta M., Maiolica A., Seller C.A. and Toczyski D.P. (2014) Ubiquitin ligase trapping identifies an SCFSaf1 pathway targeting unprocessed vacuolar/lysosomal proteins. Mol. Cell 53, 148–161 doi: 10.1016/j.molcel.2013.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tan M.-K.M., Lim H.-J., Bennett E.J., Shi Y. and Harper J.W. (2013) Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol. Cell 52, 9–24 doi: 10.1016/j.molcel.2013.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim T.Y., Siesser P.F., Rossman K.L., Goldfarb D., Mackinnon K., Yan F. et al. (2015) Substrate trapping proteomics reveals targets of the βTrCP2/FBXW11 ubiquitin ligase. Mol. Cell. Biol. 35, 167–181 doi: 10.1128/MCB.00857-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song M., Hakala K., Weintraub S.T. and Shiio Y. (2011) Quantitative proteomic identification of the BRCA1 ubiquitination substrates. J. Proteome Res. 10, 5191–5198 doi: 10.1021/pr200662b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Low T.Y., Peng M., Magliozzi R., Mohammed S., Guardavaccaro D., Heck A.J.R. et al. (2014) A systems-wide screen identifies substrates of the SCF TrCP ubiquitin ligase. Sci. Signal. 7, rs8 doi: 10.1126/scisignal.2005882 [DOI] [PubMed] [Google Scholar]
- 93.Rubel C.E., Schisler J.C., Hamlett E.D., DeKroon R.M., Gautel M., Alzate O. et al. (2013) Diggin′ on U(biquitin): a novel method for the identification of physiological E3 ubiquitin ligase substrates. Cell. Biochem. Biophys. 67, 127–138 doi: 10.1007/s12013-013-9624-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mark K.G., Loveless T.B. and Toczyski D.P. (2016) Isolation of ubiquitinated substrates by tandem affinity purification of E3 ligase-polyubiquitin-binding domain fusions (ligase traps). Nat. Protoc. 11, 291–301 doi: 10.1038/nprot.2016.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O'Connor H.F., Lyon N., Leung J.W., Agarwal P., Swaim C.D., Miller K.M. et al. (2015) Ubiquitin-activated interaction traps (UBAITs) identify E3 ligase binding partners. EMBO Rep. 16, 1699–1712 doi: 10.15252/embr.201540620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coyaud E., Mis M., Laurent E.M.N., Dunham W.H., Couzens A.L., Robitaille M. et al. (2015) BioID-based identification of Skp Cullin F-box (SCF)β-TrCP1/2 E3 ligase substrates. Mol. Cell. Proteomics 14, 1781–1795 doi: 10.1074/mcp.M114.045658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhuang M., Guan S., Wang H., Burlingame A.L. and Wells J.A. (2013) Substrates of IAP ubiquitin ligases identified with a designed orthogonal E3 ligase, the NEDDylator. Mol. Cell 49, 273–282 doi: 10.1016/j.molcel.2012.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sarraf S.A.., Raman M., Guarani-Pereira V., Sowa M.E., Huttlin E.L., Gygi S.P. et al. (2013) Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 doi: 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Theurillat J.-P.P., Udeshi N.D., Errington W.J., Svinkina T., Baca S.C., Pop M. et al. (2014) Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science 346, 85–89 doi: 10.1126/science.1250255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee K.A., Hammerle L.P., Andrews P.S., Stokes M.P., Mustelin T., Silva J.C. et al. (2011) Ubiquitin ligase substrate identification through quantitative proteomics at both the protein and peptide levels. J. Biol. Chem. 286, 41530–41538 doi: 10.1074/jbc.M111.248856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thompson J.W., Nagel J., Hoving S., Gerrits B., Bauer A., Thomas J.R. et al. (2014) Quantitative Lys-ɛ-Gly-Gly (diGly) proteomics coupled with inducible RNAi reveals ubiquitin-mediated proteolysis of DNA damage-inducible transcript 4 (DDIT4) by the E3 ligase HUWE1. J. Biol. Chem. 289, 28942–28955 doi: 10.1074/jbc.M114.573352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mokhonova E.I., Avliyakulov N.K., Kramerova I., Kudryashova E., Haykinson M.J., Spencer M.J. et al. (2015) The E3 ubiquitin ligase TRIM32 regulates myoblast proliferation by controlling turnover of NDRG2. Hum. Mol. Genet. 24, 2873–2883 doi: 10.1093/hmg/ddv049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spinner C.A., Uttenweiler-Joseph S., Metais A., Stella A., Burlet-Schiltz O., Moog-Lutz C. et al. (2015) Substrates of the ASB2α E3 ubiquitin ligase in dendritic cells. Sci. Rep. 5, 16269 doi: 10.1038/srep16269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fields S. and Song O.-k. (1989) A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 doi: 10.1038/340245a0 [DOI] [PubMed] [Google Scholar]
- 105.Formstecher E., Aresta S., Collura V., Hamburger A., Meil A., Trehin A. et al. (2005) Protein interaction mapping: a Drosophila case study. Genome Res. 15, 376–384 doi: 10.1101/gr.2659105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rual J.-F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N. et al. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 doi: 10.1038/nature04209 [DOI] [PubMed] [Google Scholar]
- 107.Uetz P., Fields S., Giot L., Cagney G., Mansfield T.A., Judson R.S. et al. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 doi: 10.1038/35001009 [DOI] [PubMed] [Google Scholar]
- 108.Guo Z., Song E., Ma S., Wang X., Gao S., Shao C. et al. (2012) Proteomics strategy to identify substrates of LNX, a PDZ domain-containing E3 ubiquitin ligase. J. Proteome Res. 11, 4847–4862 doi: 10.1021/pr300674c [DOI] [PubMed] [Google Scholar]
- 109.Deane C.M., Salwinski L., Xenarios I. and Eisenberg D. (2002) Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell Proteomics 1, 349–356 PMID: [DOI] [PubMed] [Google Scholar]
- 110.Edwards A.M., Kus B., Jansen R., Greenbaum D., Greenblatt J., Gerstein M. et al. (2002) Bridging structural biology and genomics: assessing protein interaction data with known complexes. Trends Genet. 18, 529–536 doi: 10.1016/S0168-9525(02)02763-4 [DOI] [PubMed] [Google Scholar]
- 111.Huang H., Jedynak B.M. and Bader J.S. (2007) Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput. Biol. 3, e214 doi: 10.1371/journal.pcbi.0030214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rouleau N., Turcotte S., Mondou M.H., Roby P. and Bosse R. (2003) Development of a versatile platform for nuclear receptor screening using AlphaScreen™. J. Biomol. Screen 8, 191–197 doi: 10.1177/1087057103252605 [DOI] [PubMed] [Google Scholar]
- 113.Afanassiev V., Hanemann V. and Wolfl S. (2000) Preparation of DNA and protein micro arrays on glass slides coated with an agarose film. Nucleic Acids Res. 28, E66 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu H., Snyder M., Klemic J.F., Chang S., Bertone P., Casamayor A. et al. (2000) Analysis of yeast protein kinases using protein chips. Nat. Genet. 26, 283–289 doi: 10.1038/81576 [DOI] [PubMed] [Google Scholar]
- 115.Sutandy F.X., Qian J., Chen C.S. and Zhu H. (2013) Overview of protein microarrays Curr. Protoc. Protein Sci. 72:27.1:27.1.1–27.1.16 doi:10.1002/0471140864.ps2701s72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yen H.C.-S., Xu Q., Chou D.M., Zhao Z. and Elledge S.J. (2008) Global protein stability profiling in mammalian cells. Science 322, 918–923 doi: 10.1126/science.1160489 [DOI] [PubMed] [Google Scholar]
- 117.Heuze M.L., Lamsoul I., Baldassarre M., Lad Y., Leveque S., Razinia Z. et al. (2008) ASB2 targets filamins A and B to proteasomal degradation. Blood 112, 5130–5140 doi: 10.1182/blood-2007-12-128744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ohta A., Schumacher F.-R., Mehellou Y., Johnson C., Knebel A., Macartney T.J. et al. (2013) The CUL3-KLHL3 E3 ligase complex mutated in Gordon's hypertension syndrome interacts with and ubiquitylates WNK isoforms: disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem. J. 451, 111–122 doi: 10.1042/BJ20121903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sowa M.E., Bennett E.J., Gygi S.P. and Harper J.W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 doi: 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hjerpe R., Aillet F., Lopitz-Otsoa F., Lang V., England P., Rodriguez M.S. et al. (2009) Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 doi: 10.1038/embor.2009.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lopitz-Otsoa F., Rodriguez-Suarez E., Aillet F., Casado-Vela J., Lang V., Matthiesen R. et al. (2012) Integrative analysis of the ubiquitin proteome isolated using tandem ubiquitin binding entities (TUBEs). J. Proteomics 75, 2998–3014 doi: 10.1016/j.jprot.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 122.Shi Y., Chan D.W., Jung S.Y., Malovannaya A., Wang Y., Qin J. et al. (2011) A data set of human endogenous protein ubiquitination sites. Mol. Cell. Proteomics 10, M110.002089 doi: 10.1074/mcp.M110.002089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Raasi S., Varadan R., Fushman D. and Pickart C.M. (2005) Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct., Mol. Biol. 12, 708–714 doi: 10.1038/nsmb962 [DOI] [PubMed] [Google Scholar]
- 124.Sims J.J., Haririnia A., Dickinson B.C., Fushman D. and Cohen R.E. (2009) Avid interactions underlie the Lys63-linked polyubiquitin binding specificities observed for UBA domains. Nat. Struct., Mol. Biol. 16, 883–889 doi: 10.1038/nsmb.1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu G., Paige J.S. and Jaffrey S.R. (2010) Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28, 868–873 doi: 10.1038/nbt.1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Udeshi N.D., Mani D.R., Eisenhaure T., Mertins P., Jaffe J.D., Clauser K.R. et al. (2012) Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol. Cell. Proteomics 11, 148–159 doi: 10.1074/mcp.M111.016857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Udeshi N.D., Svinkina T., Mertins P., Kuhn E., Mani D.R., Qiao J.W. et al. (2013) Refined preparation and use of anti-diglycine remnant (K- -GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12, 825–831 doi: 10.1074/mcp.O112.027094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rogne M. and Tasken K. (2013) Cell signalling analyses in the functional genomics era. New Biotechnol. 30, 333–338 doi: 10.1016/j.nbt.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 129.Carlson S.M. and Gozani O. (2014) Emerging technologies to map the protein methylome. J. Mol. Biol. 426, 3350–3362 doi: 10.1016/j.jmb.2014.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang K., Tian S. and Fan E. (2013) Protein lysine acetylation analysis: current MS-based proteomic technologies. Analyst 138, 1628–1636 doi: 10.1039/c3an36837h [DOI] [PubMed] [Google Scholar]
- 131.Zhang D. and Zhang D.E. (2011) Interferon-stimulated gene 15 and the protein ISGylation system. J. Interferon Cytokine Res. 31, 119–130 doi: 10.1089/jir.2010.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rabut G. and Peter M. (2008) Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 9, 969–976 doi: 10.1038/embor.2008.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Iwabuchi M., Sheng H., Thompson J.W., Wang L., Dubois L.G., Gooden D. et al. (2014) Characterization of the ubiquitin-modified proteome regulated by transient forebrain ischemia. J. Cereb. Blood Flow Metab. 34, 425–432 doi: 10.1038/jcbfm.2013.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Persaud A., Alberts P., Hayes M., Guettler S., Clarke I., Sicheri F. et al. (2011) Nedd4-1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. EMBO J. 30, 3259–3273 doi: 10.1038/emboj.2011.234 [DOI] [PMC free article] [PubMed] [Google Scholar]