Abstract
Reactive oxygen species (ROS) formation by mitochondria is an incompletely understood eukaryotic process. I proposed a kinetic model [BioEssays (2011) 33, 88–94] in which the ratio between electrons entering the respiratory chain via FADH2 or NADH (the F/N ratio) is a crucial determinant of ROS formation. During glucose breakdown, the ratio is low, while during fatty acid breakdown, the ratio is high (the longer the fatty acid, the higher is the ratio), leading to higher ROS levels. Thus, breakdown of (very-long-chain) fatty acids should occur without generating extra FADH2 in mitochondria. This explains peroxisome evolution. A potential ROS increase could also explain the absence of fatty acid oxidation in long-lived cells (neurons) as well as other eukaryotic adaptations, such as dynamic supercomplex formation. Effective combinations of metabolic pathways from the host and the endosymbiont (mitochondrion) allowed larger varieties of substrates (with different F/N ratios) to be oxidized, but high F/N ratios increase ROS formation. This might have led to carnitine shuttles, uncoupling proteins, and multiple antioxidant mechanisms, especially linked to fatty acid oxidation [BioEssays (2014) 36, 634–643]. Recent data regarding peroxisome evolution and their relationships with mitochondria, ROS formation by Complex I during ischaemia/reperfusion injury, and supercomplex formation adjustment to F/N ratios strongly support the model. I will further discuss the model in the light of experimental findings regarding mitochondrial ROS formation.
Keywords: FADH2/NADH ratio, mitochondria, peroxisomes, reactive oxygen species, reverse electron transport, uncoupling
Nothing in Biology Makes Sense Except in the Light of Evolution — T. Dobzhansky
A kinetic theory in which FADH2/NADH ratios determine mitochondrial ROS formation
When primordial eukaryotes evolved from archaeal hosts that took in an alpha-proteobacterium, they gained the ability to combine catabolic pathways with highly efficient ATP generation using the respiratory chain and ATP synthase (i.e. oxidative phosphorylation [1]). The endosymbiont presumably used amino acids, glycerol, and fatty acids (FAs) as energy sources, breaking them down with pathways such as those catalysed by (part of) the tricarboxylic (citric) acid (TCA) cycle enzymes and β-oxidation enzymes. During these processes, energy-rich reduced intermediates such as FADH2 and NADH are generated, which in turn are oxidized by the respiratory chain [2–4]. The archaeon could, most probably, use glycolysis to generate ATP from sugars, but this process, ending in pyruvate (or lactate, accounting for NAD+ regeneration, in the absence of a respiratory chain), became linked to the TCA cycle and oxidative phosphorylation via pyruvate dehydrogenase (PDH) activity in the endosymbiont.
Clearly, this very efficient use of alternating energy sources (FAs, carbohydrates, and proteins) has contributed tremendously to the eukaryotes' evolutionary success. But for this merger to become successful, hurdles had to be overcome and a long-term prize had to be awarded. This is due to the inescapable, highly reactive, toxic by-products of mitochondrial (mt) respiration: reactive oxygen species (ROS). Two important aspects of the fallout of endogenous oxidation of many different substrates (becoming feasible upon merging two cells) have not been sufficiently highlighted. First of all, ROS formation is not a peripheral bacterial phenomenon anymore: it now occurs in the middle of the new organism. Secondly, alternatively using different catabolic substrates, as described above, gave rise to more ROS formation [5,6]. Why should this be so?
Relative amounts of the intermediates FADH2 and NADH (the aforementioned F/N ratios) generated during complete oxidative breakdown (to CO2 and H2O) vary for the different catabolic substrates listed [5]. We find a minimum for glucose, with a ratio of 0.2 (one FADH2 formed for five molecules of NADH) if ‘normal’ NADH import into mitochondria, using the aspartate/malate shuttle, occurs. Saturated long-chain FAs represent a maximum, approaching 0.5 (one molecule of FADH2 per two NADH molecules); all others falling somewhere in between [5]. When discussing F/N ratios, a possible point of confusion has to be addressed first. The energetic content of NADH, which is a freely moving compound, is ‘fixed’. However, the energetic content of the bound prosthetic group FAD(H2) depends on its specific attachment. PDH (and the closely related 2-oxoglutarate dehydrogenase) multienzyme complexes use FAD as the electron carrier of their third enzyme. These bound FADH2 groups have such low redox potentials that they can donate their electrons to NAD+, forming NADH. In F/N ratios, we, of course, are only considering FADH2 which donates its electrons to Q (i.e. belonging to isopotential group 2; see below).
To grasp why breakdown of high F/N substrates produces more ROS formation (all other things being equal; see below), we have to focus on Complex I (NADH:ubiquinone oxidoreductase) as saturated long-chain FAs are broken down. Large amounts of NADH have to be oxidized by Complex I, which is only capable of doing so when its acceptor (fully oxidized) ubiquinone (Q) is easily available. However, compared with glucose catabolism, FA catabolism results in extra competition for Q because (reduced) ubiquinol (QH2) increases disproportionally. Instead of only competing with Complex II (succinate coenzyme Q reductase; using FAD/FADH2 as an electron carrier) for Q, Q is now reduced by electron transfer flavoprotein:ubiquinone oxidoreductase (ETF:QO; oxidizing FADH2) as well [7] (compare Figure 1A and B). FADH2 used by Complex II is of course produced in the TCA cycle during the oxidation of acetyl-CoA: produced by PDH during the oxidation of glucose or as the recurring end product of the four-step FA breakdown by mt β-oxidation. Acetyl-CoA generates one FADH2 (produced and oxidized by succinate coenzyme Q reductase, Complex II, using Q as the acceptor) per three NADH molecules during breakdown in the TCA cycle [8]. Every β-oxidation cycle also generates one FADH2 and one NADH. Thus, especially when longer FAs are catabolized, Complex I could be faced with a lack of Q, its electron acceptor. I formulated a kinetic model in which this results in increased ROS formation, with especially Complex I as a major source of mt ROS [5]. Thus, increased electron fluxes using Complex I, II, and ETF directly lead to a general increase in both the QH2/Q and NADH/NAD+ ratios (see Figure 1B). This in turn would lead to radical formation when oxidation of QH2 cannot keep up [6]. When the oxidation rate of QH2 by Complex III (CoQH2–cytochrome c reductase) is sufficient to suppress ROS formation upstream during glucose catabolism, the same oxidation rate would not necessarily suffice during FA breakdown [5] (see also [9]). As we will see, Complex I has (a) vulnerable, ROS-producing site(s). Every process, for example oxidation of high F/N ratio metabolites, that increases the time that such downstream sites are occupied by electrons could enhance ROS formation. Let us look at some further aspects of such a kinetic theory of radical formation to clarify this.
Figure 1. Schematic depiction of ROS formation in Complex I and elsewhere due to high QH2 levels during β-oxidation.
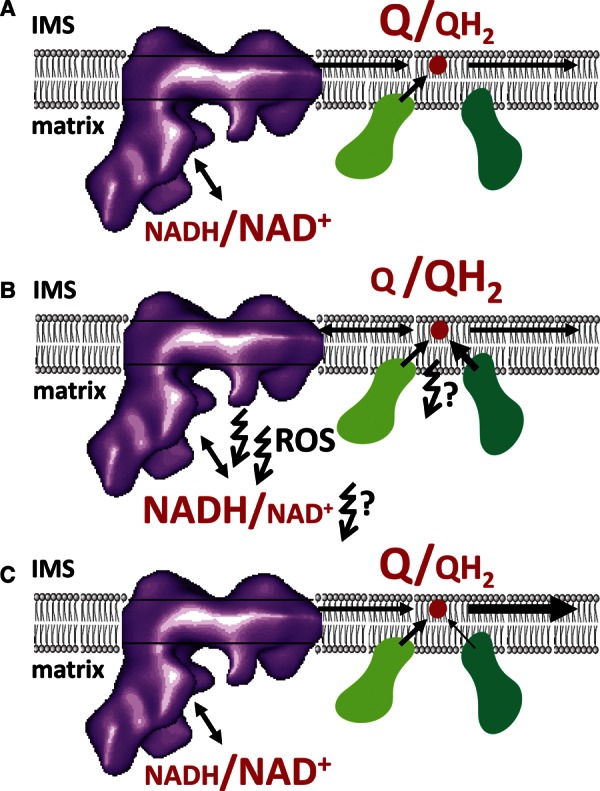
(A) Glucose oxidation (low F/N ratio) with adequate electron acceptor (Q) for Complex I. (B) Fatty acid oxidation (high F/N ratio) with insufficient electron acceptor (Q) for Complex I; ROS formation at Complex I via RET (or unknown other mechanisms? [74]), Complex II, and/or the matrix dehydrogenases (e.g. isocitrate dehydrogenase or α-ketoglutarate dehydrogenase, which depend on NAD+). (C) ROS reduction via either less FA oxidation or enhanced QH2 oxidation (e.g. more ATP use, uncoupling processes). IMS, intermembrane space; Complex I (not to scale; http://physioweb.uvm.edu/radermacher-lab/research-interests/complex/ [197]), purple; Complex II, light green; ETF complex, dark green; ubiquinone (Q), red. For details see the main text.
Further important aspects of the kinetic theory of radical formation
Some crucial aspects of the kinetic model should be highlighted first. To simplify, I will leave out many eukaryotic inventions that both suppress endogenous ROS formation and lessen their impact for now. A crucial aspect of ROS formation (highlighting why it is inescapable during mt respiration) is that it occurs in the context of an important mechanism of energy conservation: respiratory control. This mechanism has the availability of ADP (signalling the need for energy of the cell) control the ATP synthase activity, and with it the release of membrane potential (Mitchell's proton motive force, Δp [1]). This simple link makes respiration dependent on ADP concentrations. Tethering catabolism to energy need is of enormous importance for normal mt function and evolutionary competiveness. Whether bacteria also use it as such had been debated, but Burstein et al. [10] demonstrated its existence in Escherichia coli. This ‘anti-waste’ mechanism is an important evolutionary asset, especially for the first free-living single cellular eukaryotes such as the last eukaryotic common ancestor (LECA), which possibly had to confront quickly changing environments [11,12]. However, the respiratory chain can thus end up in a reduced state, when both substrate and final electron acceptor, molecular oxygen, are plentiful while insufficient ATP hydrolysis keeps them apart. This would mean that the pre-mitochondrion inside the archaeon might end up in a highly reactive state with a high membrane potential, in the presence of abundant O2. This also happens regularly in modern day mitochondria (see [13]).
When high F/N ratio substrates are used for energy generation, Complex I clearly is a likely candidate for ROS formation via reverse electron transport (RET) at the quinone reduction site (site IQ) and/or the flavin site (site IF). The relative contributions of each site are heavily debated (see [14–17]). However, the high QH2/Q ratio associated with high F/N ratios might increase radical formation by both FADH2-oxidizing complexes as well. How ROS formation from the unstable radical form semiquinone, and its production, are influenced under these conditions is not clear [14,18,19].
Important attributes of the kinetic theory of ROS formation stem from the fact that cells have two main mechanisms to lower the total mt radical burden (see Figure 1C). First, reducing the amount of high F/N ratio substrates oxidized by the mitochondrion. This solution is found in the breakdown of, especially (very-)long-chain, FAs at separate locations in the cell: the peroxisomes. Here, β-oxidation occurs without the generation of FADH2 to be oxidized in mitochondria; high-energy electrons stored in FADH2 are directly transferred to O2 forming hydrogen peroxide (H2O2), reflected in the organelles' name. This reaction is catalysed by the peroxisomal acyl-CoA oxidase. H2O2 is then split into water and O2 by the highly abundant peroxisomal enzyme catalase [5,20]. Thus, to suppress mt ROS, ATP is lost, because FADH2 oxidation does not contribute to Δp in mitochondria, while reactive H2O2 is formed. Eukaryotes and different tissues from complex multicellular eukaryotes, such as ourselves, differ enormously in the levels and kinds of β-oxidation relegated to peroxisomes (see below). Another example is found in neuronal metabolism, foregoing mt β-oxidation (neurons do not use FAs as an energy source, instead normally relying on glucose/lactate), despite large energy needs ([5,21]; see below).
The second way to lower high QH2/Q ratios, resulting from high F/N ratios, does not reduce QH2 formation. Instead of suppressing production, removal of QH2 by enhancing its oxidation will also lessen ROS formation (Figure 1C). However, this entails ‘overruling’ the principle of respiratory control. Thus, enhanced oxidation will also come at a price of reduced fuel efficiency (in this respect resembling the β-oxidation in peroxisomes). This is accomplished by uncoupling obligatory synthesis of ATP by ATP synthase from the return of protons to the mt matrix passing the inner membrane (the original bacterial membrane). Of course, this has to occur in a highly controlled manner, for example, using protein channels that (either as uniporters or symporters that co-transport metabolites; see [22]) constitute conduits for protons: uncoupling proteins. Of note, this would mean that higher oxygen consumption under these circumstances would lead to less ROS formation, instead of more as intuitively predicted by a simple mass action law. This important observation explains the lack of correlation between the amount of ROS produced and oxygen consumed, as well as the confusion in discussions regarding the mt free radical theory of ageing (for a balanced view, see [23]). Substrates with high F/N ratios, such as FAs, have somewhat lower P/O ratios — they require a bit more oxygen per amount of ATP formed than glucose — because they use relatively more FADH2 which contributes less to Δp per amount of O2 reduced by Complex IV, cytochrome c oxidase, than NADH [24]. Due to uncoupling, P/O ratios are lowered further. The optimal P/O ratio obtained with aerobic glucose catabolism (with the lowest F/N ratio, 0.2) could have driven the complete integration of cytoplasmic glycolysis, PDH, TCA cycle, and respiratory chain at the origin of the eukaryotes: most ATP at lowest amounts of oxygen consumption.
To distil the contours of the kinetic theory of mt ROS formation using isolated mitochondria is tricky as isolation of mitochondria plays havoc with the Δp and fully coupled isolated mitochondria are presumably impossible to obtain [14]. Apart from that, later eukaryotic and tissue-specific adaptations, such as the uncoupling proteins mentioned, make results hard to interpret. ROS detection is further hampered by the sheer reactivity of radical species while detection probes have to compete with a range of antioxidant mechanisms that eukaryotic cells evolved to deal with enhanced endogenous ROS formation, like enzymes and antioxidant molecules ([19,25,26]; see below).
Peroxisomes
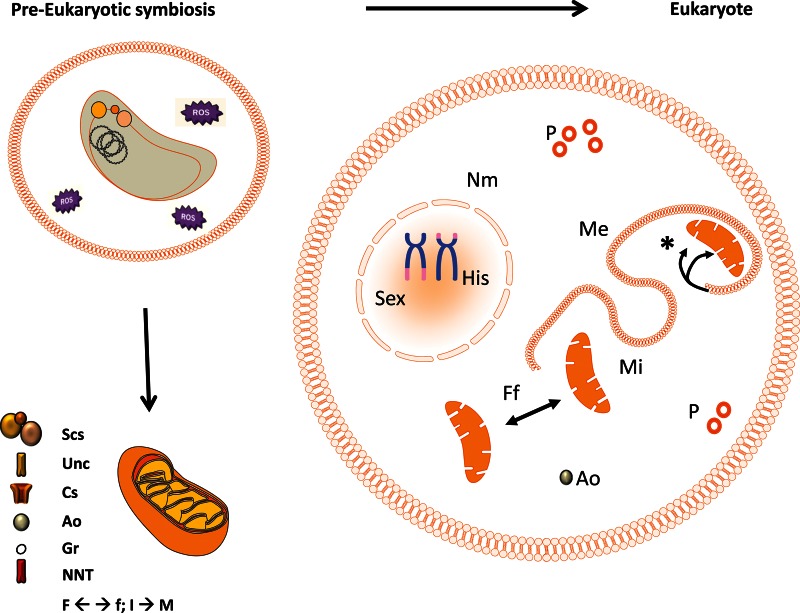
How strong is the evidence that peroxisomes evolved as a way to lower F/N ratios during FA breakdown in eukaryotes? Could the ‘first’ eukaryote easily evolve such an organelle (all eukaryotes known having peroxisomes/microbodies [27])? Recently, the closest relative yet to the archaeon host leading to all extant eukaryotes was reconstructed [28,29]. This ‘Lokiarchaeon’ encodes a cytoskeleton and some genes associated with endocytosis/internal membrane formation. A new exciting model posits that these genes plus some endosymbiont ones would allow LECA to develop internal control over secreted endosymbiont membranes and meet challenges faced during the evolution towards complete symbiosis, such as sequestering biochemical pathways, e.g. peroxisomal FA breakdown, and synchronizing cell divisions of the two organisms. This model also nicely explains why eukaryotic membrane lipids are bacterial (with ester linkages, glycerol 3-phosphate backbones, and fatty acids replacing archaeal ether linkages, glycerol 1-phosphate backbones, and isoprenoids) [30]. During the last decade, genome studies of free living eukaryotes representing different supergroups (as found in [31,32]) have opened up the possibility to reconstruct the LECA [33] (see also [11]). LECA had mitochondria, peroxisomes, and a highly adaptable and complex metabolic repertoire, of course topped by very efficient ATP generation using mt respiration [32]. The peroxisome, earlier on seen as an endosymbiotic remnant [34], was convincingly shown to be a eukaryotic invention [35,36], especially by extensive analysis of the targeting of peroxisomal membrane proteins [37–39]. The rapid evolution of peroxisomes makes sense in this merger starting out with multiple not efficiently integrated metabolic pathways and sources of energy. The alpha-proteobacterium would kick off the process with high ROS formation upon FA oxidation (owing to high F/N ratios). Internal radicals led to severe protein, lipid, and nucleic acid damage. Thus, the reduction of mt FADH2 formation would be strongly selected for. However, the complexity hypothesis [40] tells us that moving protein complexes to a different location in the cell is almost impossible (not to mention the effects on the efficiencies of ATP generation), but simple pathways can be relocated. β-Oxidation is a relatively simple repetitive four-step catabolic process, mostly ending with acetyl (2C atoms)-CoA moieties only. The recurring mt steps: FA (β-)oxidation by one of the acyl-CoA dehydrogenases (flavoprotein dehydrogenases), ending in double-bond formation between C-2(β) and C-3(γ) (placing electrons with FAD, forming FADH2), hydration of the double bond using enoyl-CoA hydratase, followed by oxidation by NAD+ forming NADH, giving l-β-hydroxyacyl-CoA (a step performed by l-β-hydroxyacyl-CoA dehydrogenase). The final step is thiolysis by CoA-SH, between C-2 and C-3, to end with acetyl-CoA and a shortened FA (catalysed by β-ketothiolase) [8]. Mt and peroxisomal β-oxidation pathways are highly similar, only the first step is different. As mentioned, initial double-bond formation is coupled to direct oxygen reduction. This bypasses the respiratory chain, leading to loss of ATP generation from FADH2 oxidation and H2O2 formation. Interestingly, it was recently found that all peroxisomal enzymes involved (two enzymes, as steps 2 and 3 are catalysed by a single bifunctional enzyme after gene fusion), of course apart from the peroxisomal acyl-CoA oxidase used in step 1, are derived from mitochondria [41]. Though ‘peroxisomes’ now have accreted many extra functions (differing per species and tissue), comparative analysis shows β-oxidation to be their most ancient pathway [27,41]. More complicated pathways are presumed to have been relocated in eukaryotes, maybe following positive selection of low levels of proteins ending up in aberrant cellular environments [42].
Thus, the stage for the ‘invention’ of peroxisomes is set. As catalase (a prokaryotic enzyme which can be secreted [43,44]) might easily wind up in membrane vesicles, only the arrival of the ‘new’ acyl-CoA oxidase was needed to have part of the β-oxidation occur outside of mitochondria. Longer FA substrate targeting, now performed using acyl-carnitine/CoA esters [45], was not needed because FAs, especially longer ones, associate with membrane vesicles. ROS formation being reduced by lowering F/N ratios, such as breakdown of FAs without mt FADH2 generation, would be beneficial. This, of course, would lead to peroxisomes, explaining their most ancient function and evolution [5,20]. There are strong evolutionary links between mitochondria and peroxisomes, consistent with the redistribution of FA oxidation as an important driving force behind the invention of peroxisomes. As mentioned, the oldest peroxisomal metabolic route is its β-oxidation pathway, with all responsible enzymes, save one, originating from the symbiont: a clear indication of coevolution of these organelles with FA oxidation redistribution as the driving force. Peroxisomes and mitochondria go together: Giardia lamblia, which instead of mitochondria has hydrogenosomes, also lost peroxisomes. Observations linking mitochondria and peroxisomes grow numerous. We find evidence for communication and co-operation [46]. Peroxisomes also turn out to contain export systems in which FAs are coupled to carnitine instead of CoA, targeting them for further processing in mitochondria [47]. Peroxisomes can use, as well as export, carnitine esters, the ‘mt’ FA form, and even oxidize the C-12 ester: using human fibroblasts manipulated to cripple mt C12-carnitine oxidation (interfering with carnitine/acyl-carnitine translocase and mt carnitine palmitoyltransferase II, CPT2), C12-carnitine oxidation was performed by peroxisomes [48]. This last feat is indicative of very close organellar complementation with regard to FA oxidation (see also the ‘Balancing ROS and ATP: mammalian metabolic tissue differentiation’ section). In HeLa cells, cargo-selected transport from mitochondria to peroxisomes was demonstrated [49]. In mitochondria, a degradation system for misfolded proteins has been found [50–52], which is evolutionarily related to the endoplasmic reticulum associated degradation (ERAD) system [53]. ERAD itself has been consistently linked to the peroxisomal protein import machinery, making rapid coevolution of the three systems in early eukaryotic evolution likely. In conclusion, ROS formation inside of the new cellular entity (the ‘eukaryote to be’) led to evolution of peroxisomes for partial β-oxidation of fatty acids.
An important point, diverse relative selective pressures in different eukaryotic organisms/tissue cell types with respect to:
optimal energy use (favouring the retention of as much β-oxidation in the mitochondria as possible),
suppression of internal radical generation (relegating FA oxidation to peroxisomes, especially useful in the case of the longest FAs, having the highest F/N ratios),
importance of different biosynthetic (anabolic) routes using, for example, FAs or acetyl-CoA, and
longevity demands (i.e. does the cell have apoptotic options or is it irreplaceable?),
would lead to large differences in amounts of β-oxidation found, as well as in the relative contributions of peroxisomes and mitochondria to FA breakdown.
The delicate balance between ROS and ATP generation
Looking at a few examples of the trade-offs described will be enlightening. As mentioned, large-scale organellar interplay between peroxisomes and mitochondria has often evolved [47,54]. In vertebrates, the longest FAs (very-long-chain FAs, VLCFAs, >18 C atoms) are exclusively broken down by peroxisomal β-oxidation. Consider cells using either glucose or FAs as an energy source, with F/N ratios ranging from 0.2 to about 0.5. If mitochondria would perform the breakdown of very-long-chain saturated FAs (e.g. cerotic acid, having 26 C atoms), this would give an F/N ratio (25/51) of ∼0.49. Cerotic acid breakdown via peroxisomes (with breakdown presumably proceeding to ∼C-8 before allowing mt β-oxidation; see also below) results in a reduced F/N ratio (16/51) of ∼0.31. This set-up thus lowers F/N ratios and buffers against large ratio variations over time, probably to efficiently suppress ROS formation. Both the creation of H2O2 and the loss in ATP generation underscore that getting rid of the FADH2 formed during VLCFA oxidation is crucial. Indeed, the so-called PPAR transcription factors strongly induce peroxisomes upon high-fat diets. Under such circumstances, peroxisomal FA breakdown without concomitant FADH2 generation in mitochondria can make a much larger contribution to overall fatty acid breakdown, lowering F/N ratios [55].
Yeasts evolved and diversified to take advantage of large amounts of biomolecules available in nutrient-rich environments resulting from ecological changes due to the rise of the eukaryotes, especially plants, upon secondary endosymbiosis of the cyanobacteria-forming chloroplasts [56]. This means that, for many yeasts, ATP generation was not as ‘limiting’ anymore and they could rewire their metabolic routes accordingly. Baker's yeast (Saccharomyces cerevisiae) is an excellent example (see also Figure 2). Leaving out anaerobic modes (such as fermentation in which ethanol is produced from glucose with severely reduced ATP generation per glucose molecule), it also has a reduced aerobic metabolism in which peroxisomes play a major part [57]. First of all, instead of the ‘normal’ Complex I, it has a highly size-reduced, single subunit type 2 NADH dehydrogenase encoded by the Ndi1 gene [58] that is not membrane-spanning and does not contribute to Δp and concomitant ATP production. Transgene experiments, placing it in mammalian backgrounds, show it to be less prone to ROS formation than Complex I [59]. Its expression in ‘laboratory’ Drosophila increased lifespan and countered age-associated decline in respiratory capacity [60]. This again stresses detrimental effects of ROS formation by Complex I. In a surprising parallel development, the dihydro-orotate dehydrogenase DHODH, involved in the biosynthesis of pyrimidine, yet another mt FAD-containing protein using Q as its final acceptor, has become cytosolic in S. cerevisiae [61]. The strongly conserved mt NAD(P) transhydrogenase (nicotinamide nucleotide-NAD(P) transhydrogenase, NNT; see Figure 2 and [62], and references therein), which normally exchanges NADH for NADPH [63] in the mt matrix (using Δp and NADH, thus both lowering ROS formation and generating ROS-scavenging equivalents in the form of NADPH, for example to be used by glutathione and thioredoxin), has also been lost [64], arguing for an important role of NNT in battling ROS formation by Complex I along the lines sketched above. Of note, NNT seems to be highly expressed in neurons of Caenorhabditis elegans and humans [65]. NNT deficiencies can lead to neurological problems, as also illustrated by the extra sensitivity to neurotoxic agents observed in mice in its absence ([62]; see below and [66]). In S. cerevisiae, not only Complex I and NNT have gone, but β-oxidation has been completely (!) relocated to the less energy-efficient peroxisomes as well. Clearly, in the ROS/ATP trade-off, S. cerevisiae has taken up a position different from that of humans and other animals. Not all fungi do this, as Ustilago maydis has all of the appropriate enzymes for co-operative peroxisomal and mt fatty acid β-oxidation, resembling animals. The U. maydis and S. cerevisiae peroxisomal systems are both inducible, as in mammals [67]. Why yeast species favour alcohol, glucose, or organic acids for energy production instead of FA oxidation is not known [66], but the concomitant reduction in mt ROS formation looks like an enticing explanation. The correlation between the suppression of ROS formation and alternative fermentation pathways could be telling as well. Figure 2 gives an overview of eukaryotic variations on, and extensions of, oxidative phosphorylation in the light of the ‘battle’ between ATP and ROS generation.
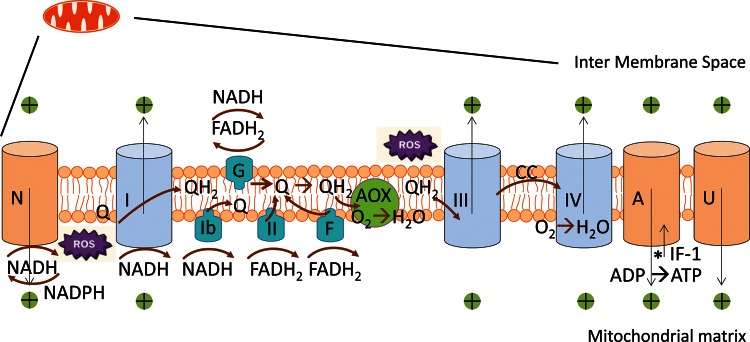
Figure 2. Mitochondrial oxidative phosphorylation in the light of the battle between ATP and ROS generation.
The ‘normal’ respiratory chain starts with Complex I (NADH dehydrogenase), which oxidizes NADH (forming NAD+) and reduces Q (forming QH2). QH2 is used by Complex III (cytochrome c reductase) to reduce cytochrome c (CC), this is re-oxidized by Complex IV, with molecular oxygen functioning as the final electron acceptor. All of these complexes (light blue) pump protons, are thus coupled to the membrane potential (Δp), and influenced in their tendencies to generate ROS by Δp. The Δp is used (light orange channels) by ATP synthase (A) to make ATP, by ‘uncoupling’ activities/proteins (U), that lower Δp (for instance when using it to import mitochondrial substrates) or by NNT (the transhydrogenase that exchanges NADH for NADPH, involved in ROS scavenging). ATP synthase can function as an ATPase to sustain Δp in the absence of sufficient respiratory chain activity for such import activities (*). ATP synthase activity can be inhibited by the IF-1 inhibitor protein. Not counting Complex I, Q can also be reduced by electrons coming from proteins with FAD/FADH2 as a prostethic cofactor: Complex II (succinate dehydrogenase, a TCA cycle enzyme), complex ‘F’ (the ETF complex involved in fatty acid oxidation), and ‘G’ (the glycerol 3-phosphate shuttle allowing cytoplasmic NADH to be oxidized in the mitochondrion, when aspartate/malate shuttling is insufficient). A yeast alternative dehydrogenase (Ndi1; Ib) also oxidizes NADH to reduce Q. Three other enzymes reducing Q are not shown: malate:quinone oxidoreductase (MQO; mostly in parasites), sulfide:quinone oxidoreductase (SQR) and dihydroorotate dehydrogenase (DHODH). All such complexes (turquoise) do not contribute to Δp. QH2 can be oxidized by an alternative non-vertebrate oxidase (AOX; green), bypassing both III and IV, and thus not contributing to Δp either. ROS can be formed by complexes I and III, especially at the IF and/or IQ binding site of I (the IQ site). Proton movement indicated by thin black arrows; electron transport by thick brown arrows.
Balancing ROS and ATP: mammalian metabolic tissue differentiation
Also enlightening is the mammalian metabolic tissue differentiation in FA oxidation, as reflected in relative mRNA amounts for transcripts involved in peroxisomal acylcarnitine (see also below) and free (non-esterified) FA (FFA) formation [47]. Looking at transcript levels for acylcarnitine transferases (involved in esterification with small —acetyl/propionyl — or medium FAs) and thioesterases (again either releasing small or medium FFAs) researchers found informative complementary expression patterns in different tissues. One can interpret the products of all of these peroxisomal enzymes along two different axes. FFA or acylcarnitine: only the second form is directly destined for burning in mitochondria. Small and medium FFAs both are probably destined for anabolic processes, but the first has already been ‘oxidized’. Comparing tissues, striking differences in mt FA oxidation were found. The following were used: mouse brain (neurons having no mt β-oxidation; see below), heart (full-fledged mt β-oxidation), brown and white adipose tissue (BAT and WAT; full-fledged, but uncoupled, and low mt β-oxidation, respectively), and liver and intestine (varying mt β-oxidation). Overall, tissues seemed to express either the transferase or the thioesterase (and this holds true for both small and medium FA substrates). The relevant specific features (using RNA levels as a proxy for activities) are as follows. BAT: only high amounts of acetylcarnitine (see Figure 2B) excreted from peroxisomes (peroxisomal and uncoupled mt FA oxidation both ‘just’ release heat, so having the FAs completely β-oxidized in the peroxisomes comes to exactly the same thing). WAT and brain: only (especially in brain) formation of medium-chain FFAs, no carnitine ester formation, implying the absence of further mt involvement. The medium-chain FFAs are most probably released for anabolic processes (such as storage as fat in WAT). Heart: relatively low contribution of the peroxisomal as opposed to the mt acylcarnitine transferases. Liver and intestinal peroxisomes would seem to produce either very small carboxylic acids (acetate/propionate) or medium-chain carnitine esters, demonstrating involvement of both organelles in ongoing β-oxidation. Especially the liver showed an interesting phenotype with respect to β-oxidation. This tissue had high expression of the medium-chain carnitine transferase and, in the case of the acetylcarnitine transferase, which is found in both mitochondria and peroxisomes (due to alternative splicing), it had the highest relative contribution of the peroxisomal variant (peroxisomes being most abundant in hepatocytes). One might conclude that liver cells have to perform β-oxidation, but a relatively large portion is taken care of in peroxisomes. As peroxisomes and mitochondria are highly abundant in mammalian (e.g. rat) liver, hepatocytes have been studied extensively in regard to these organelles. However, the liver with its highly complex metabolic role (taking care of energy homoeostasis for the rest of the body) probably has the most intricate, layered, repertoire of mt/peroxisomal adaptations of all cell types. The complex results obtained while studying peroxisomal and mt function in hepatocytes are discussed in the light of the kinetic theory of radical formation in the Supplementary Material: ‘The multi-layered response of hepatocytes in fatty acid metabolism’. For now, let us leave theoretical considerations aside and look at ROS formation by purified mitochondria burning FAs.
ROS formation in isolated mitochondria oxidizing FAs: is there systemic underestimation?
Before looking at experimental results with isolated mitochondria, two points should be made: (i) Complexes I and III are the major superoxide producers in the respiratory chain [14], specific mechanisms possibly involved being studied in both [68,69], and (ii) the ‘isopotential group’ concept has to be introduced. The three groups of redox centres of the respiratory chain operate around ever higher redox potentials (Ehs): NADH/NAD+ at Eh ∼ −280 mV, QH2/Q at Eh ∼ +20 mV, and cytochrome c at Eh ∼ +320 mV [70,71]. This entails that RET from isopotential group 2 to group 1 is thermodynamically unfavourable and will be sensitive to loss of Δp, making isolated mitochondria not ideal to study RET. Indeed, experimental results with regard to ROS formation by isolated mitochondria performing FA oxidation have always shown rather mixed and unexpected effects, though enhanced radical formation with FAs as substrate can quite often be observed [72–74]. In rat skeletal muscle mitochondria, Quinlan et al. [16] found very high ROS at site IQ when using succinate, but none at this site when using palmitoylcarnitine and carnitine. Skeletal muscle is of course adapted to fatty acid oxidation, so this might reflect a preadaptation such as the presence of specific supercomplexes; see point (iii) below. Thus, on the one hand, biological evidence for very strong ROS formation in whole organisms, tissues, and cells burning fat abounds (see [75]); on the other hand, experiments performed with FA carnitine (with or without extra carnitine) esters in mitochondria often show very low ROS formation (comparable with the amounts observed with completely uncoupled mitochondria, using, for example, trifluorocarbonylcyanide phenylhydrazone). This discrepancy is often explained by claiming that: (i) non-mt xanthine and NADPH oxidase-dependent enzymes are responsible for the majority of ROS generation in the living organism, as they are activated by high FFA levels [76,77], (ii) ROS levels are influenced by the uncoupling effect of FFAs, (iii) separate ubiquinone pools exist: the one used by succinate dehydrogenase (Complex II) capable of RET to Complex I and the one used by ETF:QO (involved in oxidizing FAs) not being able to do so. This last conjecture has been explained by positing that Complex II might be present in a supercomplex with Complex I allowing RET, while such a complex is not formed between Complex I and ETF [78]. However, Complex II seems to be absent from Complex I containing supercomplexes in the large majority of studies published thus far (another caveat here concerns tissue differences in as far as supercomplex formation is concerned) [79,80]. An interesting explanation for the overall low-level ROS abundancies observed with FAs could lie in the fact that the experiments have to be performed using carnityl esters. FFAs or CoA esters cannot be used because of uncoupling effects and because FAs have to be imported as activated carnitine esters. However, as discussed below, there are quite a few indications that carnitine (see Figure 3A) functions as an effective antioxidant that aggregates in the mt matrix upon import and will efficiently compete with the ROS detection systems used [6,81,82].
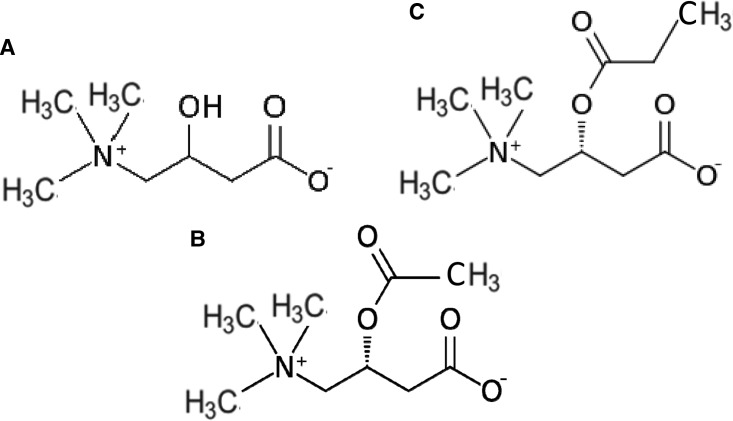
Figure 3. Structural formulas of carnitine and simple acylcarnitine esters.
(A) Carnitine, (B) l-acetylcarnitine, and (C) l-propionylcarnitine as intermediates of fatty acid metabolism and related ROS scavengers. The fatty acid is esterified to the hydroxy group of carnitine for import into the mitochondrion. See the main text.
Isolated mitochondria: does RET from ETF:QO lead to ROS formation by Complex I?
Performing experiments with isolated mitochondria, one has the intuitive tendency (because of the reduction in complexity upon purification of a single ‘organelle’) to greatly underestimate the complexity of the mt preparation, when looking at readouts all (in)directly reflecting oxidative phosphorylation. Different tissues demanding different purification protocols end up with differing contaminations. Even more importantly, isolated mitochondria from different tissues (or even the same tissues in different metabolic states [74,83]) show myriad and striking differences in respiration and ROS formation [72,84], illustrating how complex these systems are. The observed values not only stem from differences in Complexes I–V, ETF:QO oxidoreductase, glycerol-3-phosphate dehydrogenase, and DHODH themselves or their involvement in supercomplexes, but also from the relative amounts of uncoupling (proteins), the (iso)forms of the mt importers, and the presence/absence of matrix enzymes with differences in Km and Vmax (see [84]). As an example of ‘conflicting’ results, let us consider measurements of ROS formation (using the Amplex Red/horseradish peroxidase system) in rat kidney mitochondria (either obtained from the whole organ [72] or from purified cortical tubule [74]). If we only consider succinate and palmitoylcarnitine incubations, in both preparations succinate gives the higher H2O2 readout, which drops significantly upon rotenone administration, but is restored with antimycin A. However, palmitoylcarnitine oxidation induced H2O2 formation is not lowered by rotenone in the whole organ preparation (which, however, uses state 4 measurements), while a significant decrease is observed with the cortical tubule mitochondria (state 3). Of note, in [72], palmitoylcarnitine is not supplemented with malate. Thus, extensive ‘benchmark’ studies with isolated mitochondria from a range of tissues such as those presented in [72] are important.
Crucially, the standard incubations chosen with regard to substrate try to single out specific complex activity. For Complex I, a combination of glutamate/malate is mostly used, effectively leading to ongoing high use of Complex I ‘only’ in (liver) mitochondria, whereas Complex II is tested with succinate only. However, oxaloacetate formed further downstream in the TCA cycle will strongly inhibit Complex II, explaining why inhibiting Complex I with rotenone, essential for oxaloacetate formation, can actually enhance oxygen consumption using succinate, under such conditions. Last, but not least, ETF:QO cannot be tested on its own and is probed with FA-carnityl esters, with or without malate as a C4 source to allow the acetyl-CoA resulting from FA oxidation to donate C2 to oxaloacetate for the synthesis of citrate, in the condensation step of the TCA cycle. During FA oxidation NADH, FADH2, and acetyl-CoA are formed, so Complex I cannot be inhibited without severe effects on β-oxidation [84].
Considering all of this, can we demonstrate RET from ETF:QO during FA oxidation leading to ROS at Complex I in isolated mitochondria? Electrons from ubiquinone stemming from FADH2, when functioning as the cofactor of Complex II, have been shown again and again to lead to ROS formation by Complex I upon RET [14]. As there is no principal difference in QH2 coming from either Complex II or ETF:QO, RET is perfectly feasible during FA oxidation by isolated mitochondria. However, we get no clear indications for this with most mt incubations (see [72,78]). Exceptionally, the experiments in kidney cortical tubule mitochondria [74] do seem to give the classical significant reduction in the H2O2 readout upon rotenone administration indicative of RET. However, the authors cannot interpret it thus, because, somewhat surprisingly, they also observe a similar reduction following rotenone addition using the Complex I substrate pyruvate/malate. The experiments do show large mt ROS formation associated with β-oxidation, which is even very strongly enhanced in rats with the onset of diabetes. For those who do not believe that Δp ever reaches levels high enough for RET in real life, this is a first glimpse of how high QH2 resulting from FA oxidation might lead to abundant ROS formation ([74]; see also below). Mammalian cells, as complicated eukaryotic descendants, have developed adaptations (different Q pools, supercomplex formation, and inherent uncoupling) making detection of FA oxidation-associated RET in isolated mitochondria next to impossible. Schönfeld et al. [78] showed that RET-associated ROS formation does not seem to occur in isolated heart mitochondria upon FA oxidation. However, there is some discussion whether the membrane potential in these experiments was still high enough to allow RET [85].
The relevance of such comparisons is also not quite clear. In the case of succinate incubations (also used in [78] for the purpose of comparison), RET-associated ROS formation is shown by the familiar drop in ROS formation upon the addition of rotenone (taking out the contribution to ROS formation at Complex I; see [86]). But the effect of rotenone addition is completely different when comparing succinate and FA oxidation. As explained above, succinate oxidation can actually be enhanced by rotenone. FA oxidation (also in the presence of malate, as performed by Schönfeld et al. [78]), however, is dependent on Complex I activity and will in principle be inhibited by rotenone. Thus, this comparison cannot tell us whether RET-associated ROS formation can occur during FA oxidation in the heart, in vivo.
Isolated mitochondria: experimental findings consistent with the kinetic model
As we have seen, studying and interpreting radical formation with isolated mitochondria is quite a challenging undertaking. ROS formation is strongly determined by the coupling between the respiratory chain and the proton gradient, easily lost upon isolation. On top of this, later evolutionary inventions such as uncoupling (proteins), tissue-specific adjustments, the arrival of carnitine, supercomplex formation, and diverse antioxidant mechanisms severely complicate findings obtained with isolated mitochondria when compared with the ‘simple’ ancestral state envisaged. Thus, differences in tissue and isolation procedures, plus the sheer complexity of the system, might explain the conflicting results and models.
However, certain conclusions seem to be solid. Abundant radical formation via RET at Complex I is found to be associated with electrons ending up in ubiquinone from succinate, fatty acid oxidation, or glycerol 3-phosphate. Paraphrasing from [14]: extensive RET-induced radical formation occurs at Complex I in isolated mitochondria (this occurs at high Δp and with the electron supply to Q seeming to come from succinate, glycerol 3-phosphate, or fatty acid oxidation). ROS production from electrons entering Complex I through Q-binding sites is controlled by Δp and seems to disappear upon minor decreases in Δp. It clearly is most strongly influenced by the ΔpH (hence not the Δψ part) of Δp. All of this is in agreement with the kinetic model. Treberg et al. [17] characterized the high maximum (!) superoxide formation site in Complex I: it depends on both high OH2/Q (reduced) state and high Δp. As superoxide formation can be inhibited by rotenone, the IQ site is a possible candidate [17], which is not totally unexpected, as it is located in the membrane periphery (see below). However, the matrix-located IF site clearly cannot be excluded.
Let us look at some further findings in support of the model: strong radical formation can occasionally be observed with FAs as a substrate [72–74]. When NADH is oxidized by Complex I but the QH2/Q ratio is concomitantly kept high artificially by cytochrome c removal, enhanced radical formation is observed [87]. Very strong support, as we have seen, comes from succinate incubations of isolated mitochondria (see [88]). Interestingly, if mitochondria burning NADH are incubated with succinate (a direct source of Q reducing FADH2), or the other way around, one observes induction of radical formation by Complex I, stressing the importance of the concepts of F/N ratios and competition for Q [89]. Complex II does not seem to be an important source of radical formation (as possibly is the case for glycerol-3-phosphate dehydrogenase and ETF:CoQ reductase as well [14,19]; but see [74]); however, it can ‘overload’ Complex I [14,18]. Surprisingly, succinate activation of radical formation was found to be suppressed by long-chain FA acyl-CoAs in [90], though FADH2 levels could, in theory, rise further because of added FA oxidation. However, it is found that FAs are not oxidized under these conditions and that FA acyl-CoAs are able to uncouple isolated mitochondria due to their hydrophobicity. In practically every incubation of mt preparations, radical formation is highest with succinate, operating at a rather extreme F/N ratio (P/O measurements indicating that Complex I does not seem to contribute very significantly to the Δp in most cases). The observation that the so-called respiratory control rates (ratio of oxygen consumption with or without ADP present, a measure of coupling/membrane integrity) of these preparations are always highest with ‘strict’ NADH donors (e.g. glutamate/malate [84]) seems to indicate that strict FADH2 donors quickly compromise membrane integrity by excessive radical formation. As mentioned above, experimental evidence for FA oxidation activating ROS formation by Complex I involving RET from ETF:CoQ reductase in isolated mitochondria is debated [73,78,85,91], while evidence for RET from succinate is incontrovertible. The discrepancy is due to differences in coupling, supercomplex formation [92], and/or to the role of carnitine (see below and [82]).
In vivo: RET from Complex II leads to ROS formation by Complex I
RET only occurs at high Δp. A cell always uses energy and thus dissipates the mt membrane potential. Even in neurons, basal respiration only seems to constitute 50% of maximal respiration [93]. Thus, is RET not an artefact of the mt incubation with high [succinate]? Do state 4, resting, mitochondria ever occur in vivo? To put it another way: is ATP synthase ever severely limited by lack of ADP and does this have important physiological consequences? A host of recent papers shows RET-induced ROS formation at Complex I to be absolutely crucial.
Recently, Chouchani et al. [94] completely elucidated the source of mt ROS in ischaemia/reperfusion (IR) injury. In a high-quality study, they perfectly illustrate certain aspects of the kinetic model and show:
selective accumulation of succinate (via reversal of succinate dehydrogenase, Complex II, under ischaemia),
rapid re-oxidation of the accumulated succinate upon reperfusion,
rapid hyperpolarization upon reperfusion (first minute ‘delay’ of ATP synthase), and
mt Complex I ROS generation upon RET from Complex II via QH2.
The outcome is exactly as the kinetic model predicts, but though this occurs in vivo, one can hardly call conditions of IR injury ‘natural’. The ischaemia period leads to a change in the AMP/ADP ratio, such that the ‘low ADP delay’ at ATP synthase can lead to hyperpolarization. Is mt hyperpolarization found under more normal circumstances? It occurs in T-cell activation [95,96], is supposed to be a factor in disease complications upon high blood glucose in diabetes [97], has been observed over longer periods of stress in astrocytes [98], is a characteristic of arterial smooth muscle cells in pulmonary hypertension [99], and is a ‘sensitizing feature’ in apoptotic potential of Caco-2 intestinal cells [100]. Neurons tolerant of transient glutamate excitation show significant mt hyperpolarization in response [101], and it is quite common in many types of cancer cells [102,103]. Thus, one might be tempted to conclude that hyperpolarization mostly occurs under (transient) extreme conditions, leading to detrimental local ADP deficits (like the delay under IR injury mentioned above or possible temporary ADP/ATP translocase limits). But even if hyperpolarization in mitochondria were indeed rare, RET-induced ROS formation by Complex I during β-oxidation does not seem to be.
In vivo: RET from ETF:QO leads to crucial ROS formation by Complex I
A very recent extensive study settles the question regarding the relevance of RET and specific ROS formation at Complex I once and for all [104]: using mice fibroblast cell lines, Enriquez and colleagues demonstrate that saturation of CoQ oxidation capacity upon FA oxidation indeed induces RET from reduced CoQ to Complex I. With the aid of mass spectrometric analysis, they prove that the resulting local generation of ROS oxidizes specific Complex I proteins, triggering their degradation and the disintegration of Complex I. To prevent further ROS formation, the respiratory chain superstructure (i.e. the configuration of supercomplexes) is reconfigured by this controlled degradation of Complex I, liberating associated Complex III. This, in turn, increases electron flux via FADH2 at the expense of NADH, adjusting the electron transport chain to the high F/N ratio of β-oxidation. Thus, CoQ redox status can act as a metabolic sensor adjusting the respiratory chain upon changes in the overall catabolic substrate availability. In related research, using macrophages, respiratory chain adaptations were induced by ROS coming from a phagosomal NADPH oxidase triggered by uptake of bacteria. This ROS signal activates ROS-dependent tyrosine kinase Fgr, which activates Complex II by phosphorylation, while Complex I-containing supercomplexes also decrease in concentration. Overall Complex I involvement goes down, while the glycerol 3-phosphate shuttle, exchanging cytoplasmic NADH for mt FADH2, is activated [105]. These processes were crucial for cell activation and antibacterial host defence. It is well known that metabolic adaptations, reflecting changes in carbon source preference, are essential for macrophage activation (as well as for pluripotency and cancer phenotypes) [106]. This research shows ROS-triggered electron chain rearrangement to be crucially involved in this process. In earlier work, using a.o. mice fibroblast cells, direct Fgr activation by mt ROS also allowed phosphorylation of Complex II and subsequent adaptation to different fuel use [107]. In a surprising study, monitoring Drosophila brains, RET was shown to lead to ROS formation in Complex I which, in this instance, delays ageing [108]! This might seem to conflict with the fact that expressing yeast NADH dehydrogenase Ndi1 (lowering ROS formation by bypassing Complex I; see Figure 2) in Drosophila confers increased lifespan [60]. However, it is just an important reminder of the highly integrated role of mt ROS in cellular metabolism, as reflected in the concept of mitohormesis (see the ‘Conclusions’ section), which states that the cellular outcome of ROS generation critically depends on levels and sites of origin [109].
If RET is commonplace in tightly regulated complex organisms, how about many present-day single-celled eukaryotes (as the original free-living LECA), which are directly confronted with extremes in the form of strong competition for energy sources as well as with possible fluctuating [O2]? Energy-conserving respiratory control will block ATPase in the absence of ADP and high Δp should allow RET. Respiratory control is a major factor in mt functioning and under conditions such as high energy input of highly reduced substances (whether carbohydrates or FAs), high mt membrane potentials can be observed. ROS formation will thus always be a problem, especially in the light of positive feedback loops (oxidative damage leading to more ROS). Cells might actively minimize extreme fluctuations in energy intake, as well as in F/N ratios of said intake, in as far as the environment and (in multicellular organisms) function allow. This could help explain the ‘Randle cycle’: the mutual exclusion of glucose and fatty acids in catabolism in mammalian tissues [110]. One can predict that in multicellular organisms, ‘irreplaceable’ cells such as neurons would tend to use energy sources with the lowest F/N ratios with as little fluctuation as possible. Let us look at neuronal metabolism.
Aspects of neuronal metabolism reflecting the kinetic theory of ROS formation
Cells will adapt to changes in food supply and tolerate some variation in oxygen supply. This, however, will lead to fluctuations in ROS formation and possible mt damage in spite of defence by (oxidative) stress proteins [111], backed up by increases in mitophagy [112]. In multicellular organisms, an option is available to sacrifice cells upon excess ROS formation: apoptosis [111]. However, not all cell types are so easily replaced. Especially neurons in the brain contain vital and irreplaceable information, encoded in the vast network of neuronal contacts. Neurons constitute some of the most energy-demanding cells, but do not seem to use our most energy-rich substrate, fatty acids, instead preferring glucose (though FA contains 2.2-fold the amount of energy per gram and is stored much more efficiently and abundantly in the form of fat). This fact could be invoked to explain the inverse correlation between brain size and fat deposits in mammals [113,114]. The best explanation for the fact that neurons normally use only high catabolic oxidative glucose/lactate rates to sustain and regenerate large ion potentials over their membranes is their need to function at low ‘constant’ F/N ratios. Completely excluding mt β-oxidation allows them to do so. The lactate consumed comes from oligodendrocytes and astrocytes (relying more on glycolysis than on oxidative phosphorylation), which themselves use glucose and glycogen, in a process known as the astrocyte–neuron lactate shuttle [115,116]. Of course, lactate and glucose have the same F/N ratio, but using lactate possibly allows the down-regulation of glycolysis, freeing up substrate for the pentose phosphate pathway (PPP) instead. Thus, NADPH, a cofactor in ROS defence, can be produced. Findings of inverse regulation of glycolysis and PPP in stem-like glioblastoma cells fit in with this picture [117]. Under hypoxia, these cells switch to glycolysis for energy production, lowering PPP, while elevating PPP activity during normal oxidative growth, possibly to protect against ROS-related damage.
Lactate seems the optimal energy source for neurons, which, using the malate/aspartate shuttle to import NADH, produced by lactate dehydrogenase, into mitochondria, comes in at an FADH2/NADH ratio of 0.2. Thus, neurons are able to produce the necessary large amounts of ATP, but with minimal free radical production. However, there is occasional ‘trouble in paradise’. During starvation, the liver has to supplement glucose by ketone bodies [8]. How does this reflect the kinetic theory?
Breakdown of ketone bodies in neurons perfectly illustrates the kinetic model
As discussed, neurons are obliged to combine high ATP and low ROS generation. They invariably use lactate at the lowest F/N ratio. Presumably, lactate/glucose is the ‘best’ catabolic substrate because it also needs the smallest quantity of molecular oxygen per quantity of ATP produced, and it has the highest P/O ratio. But, when the liver can no longer sustain normal glucose levels, it is supplemented with ketone bodies. Two different ketone bodies, acetoacetate and β-hydroxybutyrate, are supplied. So what happens in starvation? In an interesting study, Cahill Jr [118] got three obese volunteers to fast for several weeks. He describes the switch in brain substrate utilization as follows. From 100% glucose, only 33% of the metabolic needs of brain (i.e. neurons, see also below) can still be met by glucose. Of the two ketone bodies, acetoacetate turns out to be the minor component, supplying 11%, and β-hydroxybutyrate the major component, supplying more than half (56%) of the brain's energy needs. Brains use mostly β-hydroxybutyrate. This makes sense: ketone bodies do not have the same F/N ratios. Acetoacetate (yielding two acetyl-CoA moieties for use by the TCA cycle) is formed from β-hydroxybutyrate by mt d-β-hydroxybutyrate dehydrogenase, which generates one molecule of NADH. Acetoacetate has an F/N ratio of 0.333 (2/6), while β-hydroxybutyrate thus ends up at 0.286 (2/7). Again, neurons seem to prefer lower F/N ratios. Calculating the overall change from fully fed to starved, with respect to the energy supply of neurons, we go from 0.2 to 0.263, and after several weeks of starvation, neurons only have to adjust to an ∼30% increase. Thus, the relative amounts of different forms of ketones found in ‘brain diets’ upon starvation are telling, underscoring the imperative of relatively stable supplies of catabolic substrates with low F/N ratios for neurons.
Is neuronal catabolism really ‘fat free’?
There seems to be relatively little discussion regarding the metabolic needs of the brain, as reflected by a review entitled ‘Why does brain metabolism not favor burning of fatty acids to provide energy? Reflections on disadvantages of the use of free fatty acids as fuel for brain’ [119]. But some healthy scepticism [85] is also expressed. However, especially the discussion with regard to neurons seems to be over, in the light of the following:
Import of FAs into the brain does not automatically imply β-oxidation (for which very little evidence indeed exists; see [85,119], and references therein).
‘Brain’ cells are actually neurons, astrocytes, and oligodendrocytes; all of them have different overall metabolism, with not many indications for β-oxidation, but especially neurons cannot be linked to any FA oxidation in vivo.
Even if one would have some indications for FA oxidation in vitro, this does not make it relevant for the in vivo situation, especially if all indications are that neurons do not use the process in real life.
The specific severity of PDH impairment in neurons [120] shows them to be highly dependent on pyruvate.
Considering the overall response of the liver in fasting described above, and the fact that FAs are easily taken up by the brain, the conclusion that brain cells (especially neurons) do not use FAs as fuel seems to be a safe bet.
Though cell culture studies are dangerous (dedifferentiation can occur), the study by Edmond et al. [121] is interesting in this respect. These researchers established neuron, astrocyte, and oligodendrocyte cultures from developing rat brain and were only able to detect some FA oxidation in cultured astrocytes, using highly sensitive 14C label detection. In contrast, Panov et al. [85] state that they found indications for rat ‘brain mitochondria’ oxidizing FAs (albeit at low efficiency). This was also found in [72], with mitochondria of synaptic and non-synaptic origin. Some points have to be raised. Panov et al. [85] prefer mt incubations with mixtures of substrates (stating that these reflect the in vivo situation), which makes their experiments all the more complicated to interpret. They also cannot be sure that ‘astroglia’ mitochondria are not present in their mt preparation. All in all, we can conclude that neurons, as long-lived high energy-consuming cells, use a ‘fat-free diet’ to protect against internal ROS formation upon mt respiration of high F/N substrates.
The special position of the brain and Complex I
The special position of brains regarding ROS sensitivity is illustrated by evidence that disrupted mt function, mostly at Complexes I and III, enhances ROS production in many neurodegenerative disorders. For a discussion of the issues in the light of the kinetic theory, see [66]. Despite not using β-oxidation, neuronal radical formation by Complex I still seems to be involved in brain pathogenesis over the long human lifespan. A possible high maximum superoxide-forming site in Complex I, the IQ (quinone-binding) site, is probably located in a hinge-like space at the interface of the hydrophobic membrane-embedded part and the hydrophilic domain, abutting the mt matrix [122]. Complex I has had the tendency to grow during its evolution, picking up so-called supernumerary subunits. It started out with 14 bacterial subunits, but has 30 subunits in algae and plants, 37 in fungi and a minimum of 44 in Homo sapiens [123]. A doubling of the amount of bacterial subunits had most probably already occurred in the LECA [124,125], and some extra subunits hint at links with FA oxidation [126]. One might hypothesize that some supernumerary subunits function in minimizing the rate of ROS formation or combat their effects, especially since the IQ site can be near membrane-linked mt DNA [127]. They would then be concentrated around the Q-binding site and might be brain-specific. Brain specificity also occurs in the case of the uncoupling proteins (UCPs). Humans encode five different UCPs, and UCP-4 and -5 are brain-specific [128]. Which brings us to uncoupling.
Uncoupling (proteins)
A combination of high Δp, high NADH, and high QH2/Q, the result of FA breakdown, induces radical formation, especially with longer FAs [14], but uncoupling (e.g. using protein channels; the so-called UCPs) could reduce Δp. These proteins are ancient eukaryotic inventions, which can be activated by FAs [129]. UCP-1 is the well-known uncoupler in BAT, using ATP to generate heat. For a single-celled eukaryote, such a UCP would entail suicidal ATP loss in the form of heat. However, UCPs evolved as transport proteins that were also selected for their uncoupling effects as H+ (co-)transporters (see below). Even mild uncoupling, as we have seen, is effective. QH2/Q ratios are then lowered by increasing the amount of QH2 oxidation and low local [O2] is lowered even further by enhanced electron flow. Of note, isolated mitochondria mostly show linear depletion of O2 by cytochrome c oxidase (Complex IV; Kd for O2 <1 µM [130]). Ergo, reducing Δp, for example by expressing UCPs, will efficiently reduce mt ROS formation. Thus, eukaryotes had to evolve new ways to allow minor QH2 oxidation short-circuiting ATP synthase. ‘Minor’, as the principle of respiratory control remains paramount (burn food only when ADP levels indicate the need), and (as seen in mt respiration experiments) as this is already sufficient to stop RET to Complex I. Though uncoupling can use many different mechanisms (e.g. an ATP-sensitive K+ channel [77]; see also Supplementary Material), I will concentrate on the UCP family of mt membrane transporters, as they specifically illuminate the kinetic theory.
UCPs are mt inner membrane carriers a.o. involved in protection against oxidative damage and thermoregulation due to uncoupling activity [131–134]. The members of this carrier family can transport amino acids, keto acids, inorganic ions (e.g. phosphate, H+, and K+), nucleotides, and cofactors [135]. The precise mechanism of uncoupling of the canonical UCP-1 was studied extensively. UCP-1 was demonstrated to be a long-chain fatty acid (LCFA) anion/H+ symporter, showing an intrinsic association with FA catabolism [136]. The LCFA anions seem unable to dissociate from UCP-1 efficiently, so it operates as an LCFA-activated proton channel. Other UCPs use other mechanisms. By using a set of experimentally challenging assays (i.e. reconstituting lipid vesicles with active transporter), UCP-2 could be shown to be a highly specific (matrix) carbon-4/H+, phosphate antiporter [22]. Thus, the protein can play an essential role in metabolic rewiring of cells (by export of, for example, TCA cycle intermediates for biosynthesis), overall lowering of the mt membrane potential, and reducing ROS formation. Strong indications that UCPs evolved in association with pressure to reduce ROS formation during β-oxidation can be found in regulation of expression and activity. The first important experiments were performed by the Brand group using yeast mitochondria expressing mammalian UCP-1 [137]. Here, palmitate (saturated C-16) and an oxidized (!) FA (4-hydroxy-2-nonenal) enhanced UCP proton transport. Synergistic effects were observed using both [137]. Oxidized FAs like 4-hydroxy-2-nonenal signal encountering ROS. The uncoupling predicted by the kinetic model indeed should be enhanced during β-oxidation, and even more so upon ROS production, as monitored by oxidized FAs. This completely fits with the observations [9,138]. Recently, even direct ROS involvement in in vivo thermogenesis could be demonstrated. β-Oxidation was specifically interfered with in the adipose tissue of a mice model, which lessened ROS formation on high-fat diets, but abolished cold-induced thermogenesis [75]. Studying UCP-1 specifically, ROS-dependent sulfenylation of UCP-1 Cys253 was shown to drive both increased respiration and uncoupling in BAT [139].
UCP-2 has been intensively studied at the level of transcription, (post-)translation, and even at the level of activity. An extensive overview can be found in [134]. Results are summarized in Table 1 (see also references therein). At physiologically relevant levels, UCP-2 activity can be strongly inhibited by purines (such as ATP and GTP). One could envisage the ratio between tri- and di-phosphates, reflecting the cell's energy state, being important here. Low ADP lessens ATP synthase activity (respiratory control): saving energy, but increasing ROS formation. Should ATP inhibit UCPs less than ADP, low-level UCP activity would reduce ROS formation as ADP is replaced by ATP. Only UCP-3 is inhibited more strongly by ADP [140], so this elegant mechanism cannot play a role in UCP-2 regulation. Purine nucleotides inhibit UCP activity, but are strongly overruled by ROS. Long-term regulation is exemplified by the transcription factor forkhead box A1 (Foxa1), which inhibits, and the adrenal hormone tri-iodothyronine (T3), which enhances UCP-2 expression. Foxa1 needs NAD+-dependent SIRT1 for its inhibitory activity. This makes sense, as a high NAD+/NADH ratio (reflecting low energy levels) should down-regulate energy-dissipating UCP-2 when abundant substrate is lacking.
Table 1. Some regulatory molecules and environmental stimuli influencing UCP-2 activity (adapted and extended from [6]).
| Signal | +/− | Mechanism | Relevance to the kinetic model of ROS generation induced by high F/N ratios | Reference(s) |
|---|---|---|---|---|
| ANP/GNP | −* | A | Mediators of respiratory control; see text | [140,147] |
| (u)FAs | +# | A | Lowering QH2/Q by dissipating PMF (direct activation by FA) | [148] |
| Foxa1 | − | T | Possibly signals NADH shortage (via NAD+-dependent SIRT1 interaction) | [149] |
| PPARs | + | T | Lowering QH2/Q by dissipating PMF (indirect effect of FA) | [150,151] |
| QH2 | + | A | Lowering QH2/Q by dissipating PMF (direct activation by overriding inhibitory purine nucleotides; see text) | [152–154] |
| ROS | + | T, Tl, Ptl, A | Lowering QH2/Q by dissipating PMF | [155–158] |
| Cold | + | T | UCP-2 functioning as UCP-1 | [143] |
| Fasting | + | T | Only in skeletal muscle; see text | [143] |
| FABP-FFA equilibrium | −/+ | T | Amount of cytoplasmic FFAs controls UCP-2 expression (via PPAR) | [159] |
| Adrenal T3 | + | T | Enhances the basic metabolic rate | [160] |
Triphosphates, on the whole, clearly inhibit more potently than di/mono-phosphates.
#Activation of polyunsaturated FA > unsaturated > saturated.
Response time decreases from T (hours to days) to A (seconds). Abbreviations: ANP/GNP, purine nucleotides (ATP, ADP, AMP, GTP, GDP, and GMP); (u/F)FAs, unsaturated/free fatty acids; ROS, reactive oxygen species; +, enhances activity, −, inhibits activity; T, transcriptional; Tl, translational; Ptl, post-translational; A, direct impact on UCP-2 activity.
Let us concentrate on aspects of UCP-2 regulation that strongly support the kinetic model. FAs (especially unsaturated ones) enhance UCP function directly, reflecting that β-oxidation, without suppression mechanisms, increases ROS. Fatty acid sensing PPAR transcription factors up-regulate UCP-2 protein expression in the presence of this mt substrate. The most telling correlation is that ROS strongly induces UCP-2 at all possible levels (Table 1). One might say that the regulation by FAs and ROS represents strong, but circumstantial, evidence for the kinetic model. There is also a direct connection. As explained, high F/N ratios translate into high QH2 levels, competition for Q, RET to vulnerable sites of Complex I, and increased ROS formation. Crucial experiments by Sluse et al. (reviewed in [141]) show the inhibitory effects of purine nucleotides to be completely overruled by QH2, and even a direct correlation: the higher the QH2/Q ratio, the stronger is the UCP-2 activation. A specific (indirect) link between UCP2 expression and suppression of RET to Complex I has also been found. The antidiabetic drug metformin induces UCP-2 [142]. Batandier et al. [86] showed that precisely the RET-induced ROS formation at Complex I in rat liver mitochondria could be reduced by pretreating the rats with metformin.
With the identification of the UCP-2 mechanism, there has been a tendency to state that the ‘real’ function is metabolic rewiring, and the uncoupling has been downplayed as ‘mild’ [22]. However, metabolic rewiring can only occur in the context of ROS formation and mt membrane potential, and should not be separately discussed. For example, when stating that the observed induction of UPC2 in rat kept under cold conditions is acceptable for an uncoupling protein, while the same under fasting is not (as ATP is dissipated), we are guilty of oversimplification. In the experiments referred to, cold exposure led to increased UCP2 mRNA in BAT, heart, and skeletal muscle, while fasting only gave rise to an increase in skeletal muscle [143]. Skeletal muscle normally uses both glucose and FAs as substrates for energy generation. However, upon starvation, the muscle is broken down and carbon in the form of alanine is exported for gluconeogenesis by the liver. Thus, all energy in the muscle has to come from FA oxidation as continued glucose utilization would reduce alanine output. So, the C4 export as well as the uncoupling function of UCP-2 could be relevant here. Skeletal muscle also has inducible UCP-3, elevated whenever FA oxidation is intensified under exercise and robustly reducing radical formation due to FA use [144]. UCP-3 is up-regulated, and activated, by oxidative stress via the transcription factor Nrf2 [145], which signals to uncouple mitochondria when FAs are detected [146]. The brain-specific UCP-4 and -5 are implicated in protection against mt ROS and dysfunction as well [128]. In conclusion, all UCP proteins support roles in dealing with mt ROS formation (by far most of them with links to FA oxidation).
Evolution of the UCP family
As mt carriers are crucial components of the organellar interface, they presumably arose as early adaptations of the developing eukaryote [40]. UCPs have been detected in present-day animals, plants, and unicellular eukaryotes. In mammals, wildly different patterns of tissue distribution are observed [131]. Branching out, UCP evolution has a history of multiple duplications in vertebrates [161]. Vertebrates now have five UCPs [129]. As described, the precise mechanisms and roles of UCPs are coming into focus. All function in specific transport processes that also reduce ROS by lowering mt membrane potential. The role of mammalian UCP-1 is related, but a bit different: the protein generates heat for hibernating and newborn animals using BAT energy stores. Surprisingly, BAT is also very important in adults, its absence correlating with both the metabolic syndrome and obesity [162]. Of course, this UCP-1 adaptation is a later specific function of homeothermic mammals. UCP-1, UCP-2, and UCP-3 in other species are all involved in ROS reduction. UCP-4 and -5, mainly expressed in the brain, have unfortunately not been studied extensively [128]. Their further study should be enlightening: are they involved in neuroprotective ROS suppression?
Did carnitine evolve as an efficient ROS scavenger in the context of β-oxidation?
I will try to show that the eukaryotic invention of carnitine shuttles can be explained by the kinetic theory. It limits the overall β-oxidation flux and thus reduces ROS generation when appropriate. High local mt matrix concentrations of carnitine (shown to have antioxidant capacities [81,82]) will mitigate adverse effects of ROS formation. We will return to the question whether zwitterionic carnitine (see Figure 3A) really is an effective antioxidant later. The use of carnitine as a carrier for transport of FAs over mt (and peroxisomal) membranes clearly is an early eukaryotic invention [163]. It can, for example, be synthesized upon protein breakdown, which liberates trimethyl-lysine. Synthesis mostly occurs in the cytoplasm. Only the first of the four sequential steps is catalysed by a mt trimethyl-lysine dioxygenase, which uses O2 [163]. This could allow single cells to probe local mt [O2] before starting import of FAs. Humans take up 75% of total carnitine directly from food, especially meat.
Getting LCFAs into mitochondria for β-oxidation has become a complicated process [45]. The mt outer membrane CPT1 transfers activated FA from CoA to carnitine. Next, this acylcarnitine is exchanged for matrix carnitine by inner membrane-located carnitine/acylcarnitine translocase (CACT). Another inner membrane protein, CPT2, reacting with CoA, gives acyl-CoA and releases carnitine. This is quite an elaborate and complex scheme, but it has real evolutionary advantages which can be deduced from the following [6]. CACT does not only exchange acylcarnitines for carnitines by an antiport function, but it also exhibits low-rate uniport carnitine transport [164]. This gives high equilibrated concentrations of approximately 2–5 mM carnitine in both cytoplasm and mt matrix upon β-oxidation. Interestingly, the affinity of CACT for carnitine in the matrix is low (10 mM [165]), so export is far below maximal rates. This in turn limits coupled acylcarnitine import, and, thus, the overall β-oxidation rate. In this way, differences in translocase (acyl)carnitine-binding parameters on respective sides of the inner membrane ensure that FA oxidation will only occur with high matrix concentrations of carnitine [45]. This gives control over the overall rate, and positions the antioxidant (acyl)carnitine [81,82] at high concentrations at the most optimal position, considering the ROS generating part of Complex I. The most extremely damaging ROS effects are probably lipid peroxidation of the mt inner membrane and oxidative damage to the mt DNA nearby [166–168]. As an example, l-propionyl carnitine (Figure 3B) was shown to reduce hydroxyl radical production because it chelates iron. It also demonstrates dose-dependent direct free radical scavenging. The carnitine shuttle thus sustains antioxidant saturation where and when it really matters. Its ability to limit overall rates of β-oxidation, the extremely high mt carnitine concentrations, as well as its antioxidant capacities, all attest to the evolutionary forces (elucidated by the kinetic theory) probably associated with its emergence. I just stated confidently that carnitine is an antioxidant, but why is it efficient as such? Looking at carnitine (Figure 3A), it is not immediately apparent what the chemical basis for the antioxidant property would be. The molecule itself is relatively small and there are no obvious (conjugated) double-bond structures or sulphur atoms, all molecular aspects we associate with radical scavenging. However, carnitine is a quaternary ammonium compound. Such compounds are known to be relatively stable molecules which might enhance and target ROS scavenging under these conditions. We can envisage ROS (such as the superoxide anion) to be drawn to the positively charged nitrogen. As this atom is surrounded by relatively reduced carbon atoms, these molecules might preferentially react with ROS. Finally, it has indeed been shown that the presence of quaternary nitrogen in compounds increases the strength of scavenging effects (see [169]). Further chemical analysis is needed to elucidate the mechanism(s) involved.
Separate quinone pools and supercomplexes
I already mentioned the role of dynamic supercomplex formation in F/N ratio adjustment. Extensive and flexible formation of supercomplexes (which seem to be already present in respiring bacteria [79,170]) represents a further eukaryotic adaptation. Supercomplex analysis is performed by the powerful blue native gel electrophoresis technique, pioneered by Schägger and von Jagow [171]. Formation is dependent on the specific bacterial/mt inner membrane lipid cardiolipin [172]. Most supercomplexes do not seem to involve Complex II ([173], but see also [78]). As an example, Complexes I, III, and IV are found to be associated in quaternary structures, without (FAD-containing) Complex II [174]. Thus, supercomplexes could allow Complex I access to a ‘private’ ubiquinone pool, suppressing RET from QH2 reduced by Complex II. How this influences Q membrane kinetics is not completely clear [92]. Presumably, supercomplex formation smoothens out electron flow, e.g. reducing the competition for Q between Complexes I, II, and ETF, as described. Loss of the ability to form supercomplexes indeed gives rise to ROS increase and a mt ageing phenotype [175]. Thus, supercomplexes reduce radical formation [176,177] and allow the respiratory chain to dynamically deal with varying carbon sources [104,107,178]: the ‘plasticity’ model [80]. Earlier results indicate that I/III/IV supercomplexes decrease upon FA oxidation with increased FADH2 oxidation via III/IV supercomplexes and free Complex III and Complex IV [179] relied on experiments with so-called SCAF1+/− mice. However, the phenotypes of these mice were heavily debated [180]. Using a completely different approach, Maranzana et al. [181] showed that supercomplex formation indeed limits ROS production from Complex I. Even DHODH (see above), using Q as its electron acceptor, seems to form supercomplexes with Complexes II and III [182]. All discussion regarding the crucial role of dynamic supercomplex formation in long-term ROS reduction and adjustment to catabolic substrate ended with the string of recent studies described above (see the ‘In vivo: RET from ETF:QO leads to crucial ROS formation by Complex I’ section).
The glycerol 3-phosphate shuttle
I described how, in macrophages, respiratory chain adaptations are induced by ROS triggered following the uptake of bacteria. This ROS signal activates Complex II, whereas overall Complex I involvement is down-regulated. Under these conditions, cytoplasmic NADH is actively exchanged for mt FADH2 by the glycerol 3-phosphate shuttle [105]. Such extensive metabolic adaptations are known from a.o. macrophages, T-cells, and cancers [106]. The glycerol 3-phosphate shuttle normally regenerates cytoplasmic NAD+, for example to sustain ongoing glycolysis, from NADH. Electrons are shuttled via the inner membrane-bound mt glycerol-3-phosphate dehydrogenase to Q, resulting in ATP loss and artificial increase in the F/N ratio, by exchanging NADH for FADH2. This constitutes an alternative for the ‘normal’ malate/aspartate shuttle used for export of NADH to mitochondria, which is driven by differences in [NADH]c and [NADH]m as well as by Δp. Sometimes, these two factors cannot sustain full rapid glycolysis, for example, in muscle cells with sufficient oxygen. As one cytoplasmic NADH is exchanged for a mt FADH2, the Gibbs-free energy difference represents a strong driving force. The following are a few points of interest:
The overall contribution of this shuttle in muscle, only used during full oxidative glycolysis (the process with the lowest F/N ratio), is clearly much lower than that of the default malate/aspartate shuttle.
In the example of macrophages, we found it involved upon active down-regulation of Complex I.
Glycerol-3-phosphate dehydrogenase activity in neurons is disputed [183,184].
When isolated brain mitochondria of ‘synaptic, as well as non-synaptic’ origin (i.e. also containing non-neuronal cells), are incubated with glycerol 3-phosphate, they give rise to ROS via Complex I, though at lower levels than obtained with succinate [185].
Evolutionary speculations: enhanced endogenous ROS formation as a major driving force in many eukaryotic inventions
The emerging eukaryote was not only confronted with the sudden appearance of an endogenous ROS source, but upon integrating the diverse pathways the two different genomes encoded, amounts of ROS formation increased as well, as described here. I will give a partially speculative extended list of eukaryotic ‘inventions’ (for both archaeon and alpha proteobacterium) possibly shaped by endogenous ROS formation as a driving force (see Figure 4).
Figure 4. Eukaryotic inventions possibly influenced by enhanced internal ROS formation.
Adaptations of the mitochondrion (Mi): dynamic supercomplexes (Scs) formation, uncoupling proteins (Unc), carnitine shuttles (Cs), antioxidant mechanisms (Ao), extreme mitochondrial genome reduction (Gr), NNT shuttling electrons from NADH to antioxidant defence, fusion fission cycles (Ff), and replacing isoleucine by the antioxidant methionine via AUA codon reassignment (I → M; [191]). Adaptations of the cell: peroxisome formation (P), antioxidant mechanisms (Ao), internal membrane (Me) formation (autophagy/mitophagy *), nuclear membrane (Nm) formation, size increase, meiotic sex (Sex), DNA protection by histone coverage/chromatin structure (His), and superior DNA repair? Adapted from [11].
The adaptations of the archaeon (cell) can be listed as follows. Clearly, extending and inventing an array of antioxidant mechanisms was the first order of business. The invention of peroxisomes along the lines I sketched [5] to reduce FA-associated ROS formation was fully accomplished in LECA. Internal membrane formation (an archaeon capability [28] or, more probably, coming from the endosymbiont [30]) would have led to nuclear/peroxisomal membrane formation and autophagy. This would shield cell compartments from ROS, lessen ROS production, and remove ROS damage. The mutational power of ROS could further have led to DNA protection by histone coverage/overall chromatin structure, meiotic sex [11], superior DNA repair, and maybe even cellular size increase.
Next, the possible adaptations of the alpha proteobacterium (mitochondrion). Antioxidant mechanisms go without saying. Uncoupling proteins do not only transport metabolites at the interphase of the rest of the cell and the mitochondrion, but they also evolved in a context where ROS suppression was a major driving force. The extreme mt genome reduction can best be understood by the fact that the bacterial genome was close to major sites of ROS formation located in Complex I [186]. Carnitine shuttles and flexible supercomplex formation can also be seen as adaptations to lessen both the amounts of ROS formation and their effects. Especially supercomplexes seem to have evolved under constraints to minimize competition for Q. The mt fusion fission cycles, linked to mitophagy, seem involved in quality control necessitated by ROS [187,188] and in cellular adaptations to lessen ROS formation with specific carbon sources [189]. Mt morphology is thus dynamically and complexly regulated by the redox homoeostasis of different cell types, but seems to have a constant feature: ROS leading to fragmentation and a ‘mitophagy permissive state’ [188]. Indeed, enhanced mt fusion in cultured adipocytes upon knockdown of the mt dynamics regulator BNIP3 results in increases in Δp and FA-induced ROS damage [190]. Last but not least, the AUA codon reassignment (replacing isoleucine by its antioxidant ‘mimic’ methionine), which occurred in most mitochondria, can be seen as an example of antioxidant pressures [191]. A more detailed description of the crucial and multifaceted role of endogenous ROS formation in eukaryotic evolution can be found in [192].
Conclusion
Recently, in an essay about mitochondria as signalling organelles, it was stated that “mitochondria have primarily been viewed as bioenergetic and biosynthetic organelles that autonomously coexist within the cell” [193]. It is hardly likely that anybody who gave the question some thought would agree. The eukaryote is so clearly a tightly linked new entity in which the metabolic contribution of the mitochondria is paramount that to think of it as a ‘signalling organelle’ would need neither explanation nor justification. The most important signalling molecules have to be ROS, because of their sheer omnipresence and informational value. ‘Cell, if there is no ROS: something is wrong; if ROS increases: take antioxidant measures and rewire metabolism; if there is too much ROS: prepare to die (but do it apoptotically, if you can).’ Such a description of low levels of ROS being ‘healthy’, but larger amounts being toxic due to indiscriminate reactivity, constitutes ‘mitohormesis’ [109]. ROS signalling research is coming up with more and more rather precise regulated examples, (discussed in [194,195]), while differences are seen between oxygen radicals and H2O2. The latter molecule can signal, for example, via specific interactions using peroxiredoxins [196].
Here I have tried to reconstruct how some of the pathways involved came about and how they function: implications of the kinetic theory of mt respiratory chain ROS formation. Those studying mitochondria are faced with challenges; not only the complexity of isolated ‘mitochondria’, but also many layers of adaptations that mitochondria, cells, and tissues harbour to cope with different substrates and concomitant ROS formation. I would argue, however, that the evolutionary evidence, as illustrated by the invention of peroxisomes, the absence of neuronal β-oxidation, the invention of carnitine and regulated uncoupling, the RET-related ROS formation by Complex I, dynamic supercomplex formation, as well as the many links between FA oxidation and a myriad of ROS-related mechanisms and adaptations make the kinetic model enticing for further study.
Acknowledgements
I thank Michael M. Murphy and Alex Andreyev for their kind critical email feedback regarding earlier publications concerning the kinetic theory of mitochondrial ROS formation.
Abbreviations
- Δp
proton motive force
- BAT
brown adipose tissue
- CACT
carnitine/acylcarnitine translocase
- CPT1/2
carnitine palmitoyltransferase I/II
- DHODH
dihydro-orotate dehydrogenase
- Ehs
ever higher redox potentials
- Eh(s)
redox potential(s)
- ERAD
endoplasmic reticulum-associated degradation
- ETF
electron transfer flavoprotein
- ETF:QO
electron transfer flavoprotein:ubiquinone oxidoreductase
- F/N
FADH2/NADH
- FA
fatty acid
- FABP
fatty acid binding protein
- FFA
free FA
- Foxa1
forkhead box A1
- IR
ischaemia/;reperfusion
- LCFA
long-chain fatty acid
- LECA
last eukaryotic common ancestor
- mt
mitochondrial
- NNT
nicotinamide nucleotide-NAD(P) transhydrogenase
- PPAR
peroxisome proliferator-activated receptor
- PDH
pyruvate dehydrogenase
- PMF
proton motive force
- PPP
pentose phosphate pathway
- Q
ubiquinone
- QH2
ubiquinol
- RET
reverse electron transport
- ROS
reactive oxygen species
- Sc(s)
supercomplex(es)
- SIRT
sirtuin
- TCA
tricarboxylic acid
- T3
tri-iodothyronine
- UCP
uncoupling protein
- VLCFAs
very-long-chain FAs
- WAT
white adipose tissue.
Competing Interests
The Author declares that there are no competing interests associated with this manuscript.
References
- 1.Mitchell P. (1961) Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191, 144–148 doi: 10.1038/191144a0 [DOI] [PubMed] [Google Scholar]
- 2.Hackstein J.H.P., Tjaden J. and Huynen M. (2006) Mitochondria, hydrogenosomes and mitosomes: products of evolutionary tinkering! Curr. Genet. 50, 225–245 doi: 10.1007/s00294-006-0088-8 [DOI] [PubMed] [Google Scholar]
- 3.Gabaldon T. and Huynen M.A. (2003) Reconstruction of the proto-mitochondrial metabolism. Science 301, 609 doi: 10.1126/science.1085463 [DOI] [PubMed] [Google Scholar]
- 4.Gabaldon T. and Huynen M.A. (2004) Shaping the mitochondrial proteome. Biochim. Biophys. Acta 1659, 212–220 doi: 10.1016/j.bbabio.2004.07.011 [DOI] [PubMed] [Google Scholar]
- 5.Speijer D. (2011) Oxygen radicals shaping evolution: Why fatty acid catabolism leads to peroxisomes while neurons do without it. BioEssays 33, 88–94 doi: 10.1002/bies.201000097 [DOI] [PubMed] [Google Scholar]
- 6.Speijer D. (2014) How the mitochondrion was shaped by radical differences in substrates. BioEssays 36, 634–643 doi: 10.1002/bies.201400033 [DOI] [PubMed] [Google Scholar]
- 7.Watmough N.J. and Frerman F.E. (2010) The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim. Biophys. Acta, Bioenergetics 1797, 1910–1916 doi: 10.1016/j.bbabio.2010.10.007 [DOI] [PubMed] [Google Scholar]
- 8.Berg J.M., Tymoczko J.L. and Stryer L. (2006) Biochemistry, 6th edn, W. H. Freeman, New York [Google Scholar]
- 9.Brand M.D., Buckingham J.A., Esteves T.C., Green K., Lambert A.J., Miwa S. et al. (2004) Mitochondrial superoxide and aging: uncoupling-protein activity and superoxide production. Biochem. Soc. Symp. 71, 203–213 doi: 10.1042/bss0710203 [DOI] [PubMed] [Google Scholar]
- 10.Burstein C., Tiankova L. and Kepes A. (1979) Respiratory control in Escherichia coli K12. Eur. J. Biochem. 94, 387–392 doi: 10.1111/j.1432-1033.1979.tb12905.x [DOI] [PubMed] [Google Scholar]
- 11.Speijer D., Lukeš J. and Eliáš M. (2015) Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc. Natl Acad. Sci. USA 112, 8827–8834 doi: 10.1073/pnas.1501725112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behnke A., Bunge J., Barger K., Breiner H.-W., Alla V. and Stoeck T. (2006) Microeukaryote community patterns along an O2/H2S gradient in a supersulfidic anoxic fjord (Framvaren, Norway). Appl. Environ. Microbiol. 72, 3626–3636 doi: 10.1128/AEM.72.5.3626-3636.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kikusato M. and Toyomizu M. (2013) Crucial role of membrane potential in heat stress-induced overproduction of reactive oxygen species in avian skeletal muscle mitochondria. PLoS ONE 8, e64412 doi: 10.1371/journal.pone.0064412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy M.P. (2009) How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13 doi: 10.1042/BJ20081386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pryde K.R. and Hirst J. (2011) Superoxide is produced by the reduced flavin in mitochondrial complex I: a single, unified mechanism that applies during both forward and reverse electron transfer. J. Biol. Chem. 286, 18056–18065 doi: 10.1074/jbc.M110.186841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinlan C.L., Perevoshchikova I.V., Hey-Mogensen M., Orr A.L. and Brand M.D. (2013) Sites of reactive oxygen species generation by mitochondria oxidizing different substrates. Redox. Biol. 1, 304–312 doi: 10.1016/j.redox.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treberg J.R., Quinlan C.L. and Brand M.D. (2011) Evidence for two sites of superoxide production by mitochondrial NADH-ubiquinone oxidoreductase (complex I). J. Biol. Chem. 286, 27103–27110 doi: 10.1074/jbc.M111.252502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brand M.D. (2010) The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 45, 466–472 doi: 10.1016/j.exger.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreyev A.Y., Kushnareva Y.E., Murphy A.N. and Starkov A.A. (2015) Mitochondrial ROS metabolism: 10 years later. Biochemistry (Moscow) 80, 517–531 doi: 10.1134/S0006297915050028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speijer D. (2014) Reconsidering ideas regarding the evolution of peroxisomes: the case for a mitochondrial connection. Cell. Mol. Life Sci. 71, 2377–2378 doi: 10.1007/s00018-013-1507-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S.Y., He X.Y. and Schulz H. (1987) Fatty acid oxidation in rat brain is limited by the low activity of 3-ketoacyl-coenzyme A thiolase. J. Biol. Chem. 262, 13027–13032 PMID: [PubMed] [Google Scholar]
- 22.Vozza A., Parisi G., De Leonardis F., Lasorsa F.M., Castegna A., Amorese D. et al. (2014) UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc. Natl Acad. Sci. USA 111, 960–965 doi: 10.1073/pnas.1317400111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barja G. (2013) Updating the mitochondrial free radical theory of aging: an integrated view, key aspects, and confounding concepts. Antioxid. Redox Signal. 19, 1420–1445 doi: 10.1089/ars.2012.5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinkle P.C. (2005) P/O ratios of mitochondrial oxidative phosphorylation. Biochim. Biophys. Acta, Bioenergetics 1706, 1–11 doi: 10.1016/j.bbabio.2004.09.004 [DOI] [PubMed] [Google Scholar]
- 25.Andreyev A.Y., Kushnareva Y.E. and Starkov A.A. (2005) Mitochondrial metabolism of reactive oxygen species. Biochemistry (Moscow) 70, 200–214 doi: 10.1007/s10541-005-0102-7 [DOI] [PubMed] [Google Scholar]
- 26.Rizzo A.M., Berselli P., Zava S., Montorfano G., Negroni M., Corsetto P. et al. (2010) Endogenous antioxidants and radical scavengers. Adv. Exp. Med. Biol. 698, 52–67 doi: 10.1007/978-1-4419-7347-4_5 [DOI] [PubMed] [Google Scholar]
- 27.Gabaldon T. (2010) Peroxisome diversity and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 765–773 doi: 10.1098/rstb.2009.0240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spang A., Saw J.H., Jorgensen S.L., Zaremba-Niedzwiedzka K., Martijn J., Lind A.E. et al. (2015) Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 doi: 10.1038/nature14447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Embley T.M. and Williams T.A. (2015) Evolution: steps on the road to eukaryotes. Nature 521, 169–170 doi: 10.1038/nature14522 [DOI] [PubMed] [Google Scholar]
- 30.Gould S.B., Garg S.G. and Martin W.F. (2016) Bacterial vesicle secretion and the evolutionary origin of the eukaryotic endomembrane system. Trends Microbiol. 24, 525–534 doi: 10.1016/j.tim.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 31.Fritz-Laylin L.K., Prochnik S.E., Ginger M.L., Dacks J.B., Carpenter M.L., Field M.C. et al. (2010) The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140, 631–642 doi: 10.1016/j.cell.2010.01.032 [DOI] [PubMed] [Google Scholar]
- 32.Koonin E.V. (2010) The incredible expanding ancestor of eukaryotes. Cell 140, 606–608 doi: 10.1016/j.cell.2010.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koonin E.V. (2010) The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 11, 209 doi: 10.1186/gb-2010-11-5-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Duve C. (2007) The origin of eukaryotes: a reappraisal. Nat. Rev. Genet. 8, 395–403 doi: 10.1038/nrg2071 [DOI] [PubMed] [Google Scholar]
- 35.Geuze H.J., Murk J.L., Stroobants A.K., Griffith J.M., Kleijmeer M.J., Koster A.J. et al. (2003) Involvement of the endoplasmic reticulum in peroxisome formation. Mol. Biol. Cell. 14, 2900–2907 doi: 10.1091/mbc.E02-11-0734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoepfner D., Schildknegt D., Braakman I., Philippsen P. and Tabak H.F. (2005) Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122, 85–95 doi: 10.1016/j.cell.2005.04.025 [DOI] [PubMed] [Google Scholar]
- 37.Tabak H.F., Hoepfner D., Zand A.v.d., Geuze H.J., Braakman I. and Huynen M.A. (2006) Formation of peroxisomes: present and past. Biochim. Biophys. Acta, Mol. Cell Res. 1763, 1647–1654 doi: 10.1016/j.bbamcr.2006.08.045 [DOI] [PubMed] [Google Scholar]
- 38.Tabak H.F., van der Zand A. and Braakman I. (2008) Peroxisomes: minted by the ER. Curr. Opin. Cell. Biol. 20, 393–400 doi: 10.1016/j.ceb.2008.05.008 [DOI] [PubMed] [Google Scholar]
- 39.van der Zand A., Braakman I. and Tabak H.F. (2010) Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol. Biol. Cell. 21, 2057–2065 doi: 10.1091/mbc.E10-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szklarczyk R. and Huynen M.A. (2010) Mosaic origin of the mitochondrial proteome. Proteomics 10, 4012–4024 doi: 10.1002/pmic.201000329 [DOI] [PubMed] [Google Scholar]
- 41.Bolte K., Rensing S.A. and Maier U.-G. (2015) The evolution of eukaryotic cells from the perspective of peroxisomes. BioEssays 37, 195–203 doi: 10.1002/bies.201400151 [DOI] [PubMed] [Google Scholar]
- 42.Martin W. (2010) Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 847–855 doi: 10.1098/rstb.2009.0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naclerio G., Baccigalupi L., Caruso C., De F.M. and Ricca E. (1995) Bacillus subtilis vegetative catalase is an extracellular enzyme. Appl. Environ. Microbiol. 61, 4471–4473 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin D.-H., Choi Y.-S. and Cho Y.-H. (2008) Unusual properties of catalase A (KatA) of Pseudomonas aeruginosa PA14 are associated with its biofilm peroxide resistance. J. Bacteriol. 190, 2663–2670 doi: 10.1128/JB.01580-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Indiveri C., Iacobazzi V., Tonazzi A., Giangregorio N., Infantino V., Convertini P. et al. (2011) The mitochondrial carnitine/acylcarnitine carrier: function, structure and physiopathology. Mol. Aspects Med. 32, 223–233 doi: 10.1016/j.mam.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 46.Schrader M. and Yoon Y. (2007) Mitochondria and peroxisomes: are the ‘big brother’ and the ‘little sister’ closer than assumed? BioEssays 29, 1105–1114 doi: 10.1002/bies.20659 [DOI] [PubMed] [Google Scholar]
- 47.Westin M.A.K., Hunt M.C. and Alexson S.E.H. (2008) Short- and medium-chain carnitine acyltransferases and acyl-CoA thioesterases in mouse provide complementary systems for transport of β-oxidation products out of peroxisomes. Cell. Mol. Life Sci. 65, 982–990 doi: 10.1007/s00018-008-7576-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Violante S., IJlst L., te Brinke H., Koster J., Tavares de Almeida I., Wanders R.J. et al. (2013) Peroxisomes contribute to the acylcarnitine production when the carnitine shuttle is deficient. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1831, 1467–1474 doi: 10.1016/j.bbalip.2013.06.007 [DOI] [PubMed] [Google Scholar]
- 49.Neuspiel M., Schauss A.C., Braschi E., Zunino R., Rippstein P., Rachubinski R.A. et al. (2008) Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr. Biol. 18, 102–108 doi: 10.1016/j.cub.2007.12.038 [DOI] [PubMed] [Google Scholar]
- 50.Margineantu D.H., Emerson C.B., Diaz D. and Hockenbery D.M. (2007) Hsp90 inhibition decreases mitochondrial protein turnover. PLoS ONE 2, e1066 doi: 10.1371/journal.pone.0001066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heo J.-M., Livnat-Levanon N., Taylor E.B., Jones K.T., Dephoure N., Ring J. et al. (2010) A stress-responsive system for mitochondrial protein degradation. Mol. Cell. 40, 465–480 doi: 10.1016/j.molcel.2010.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor E.B. and Rutter J. (2011) Mitochondrial quality control by the ubiquitin–proteasome system: Figure 1. Biochem. Soc. Trans. 39, 1509–1513 doi: 10.1042/BST0391509 [DOI] [PubMed] [Google Scholar]
- 53.Gabaldon T., Snel B., van Z.F., Hemrika W., Tabak H. and Huynen M.A. (2006) Origin and evolution of the peroxisomal proteome. Biol. Direct 1, 8 doi: 10.1186/1745-6150-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thoms S., Gronborg S. and Gartner J. (2009) Organelle interplay in peroxisomal disorders. Trends Mol. Med. 15, 293–302 doi: 10.1016/j.molmed.2009.05.002 [DOI] [PubMed] [Google Scholar]
- 55.Lalloyer F. and Staels B. (2010) Fibrates, glitazones, and peroxisome proliferator-activated receptors. Arterioscler. Thromb. Vasc. Biol. 30, 894–899 doi: 10.1161/ATVBAHA.108.179689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dyall S.D., Brown M.T. and Johnson P.J. (2004) Ancient invasions: from endosymbionts to organelles. Science 304, 253–257 doi: 10.1126/science.1094884 [DOI] [PubMed] [Google Scholar]
- 57.Hiltunen J.K., Mursula A.M., Rottensteiner H., Wierenga R.K., Kastaniotis A.J. and Gurvitz A. (2003) The biochemistry of peroxisomal β-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 27, 35–64 doi: 10.1016/S0168-6445(03)00017-2 [DOI] [PubMed] [Google Scholar]
- 58.De Vries S., Van Witzenburg R., Grivell L.A. and Marres C.A. (1992) Primary structure and import pathway of the rotenone-insensitive NADH-ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur. J. Biochem. 203, 587–592 doi: 10.1111/j.1432-1033.1992.tb16587.x [DOI] [PubMed] [Google Scholar]
- 59.Yagi T., Seo B.B., Nakamaru-Ogiso E., Marella M., Barber-Singh J., Yamashita T. et al. (2006) Can a single subunit yeast NADH dehydrogenase (Ndi1) remedy diseases caused by respiratory complex I defects? Rejuvenation Res. 9, 191–197 doi: 10.1089/rej.2006.9.191 [DOI] [PubMed] [Google Scholar]
- 60.Sanz A., Soikkeli M., Portero-Otin M., Wilson A., Kemppainen E., McIlroy G. et al. (2010) Expression of the yeast NADH dehydrogenase Ndi1 in Drosophila confers increased lifespan independently of dietary restriction. Proc. Natl Acad. Sci. USA 107, 9105–9110 doi: 10.1073/pnas.0911539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagy M., Lacroute F. and Thomas D. (1992) Divergent evolution of pyrimidine biosynthesis between anaerobic and aerobic yeasts. Proc. Natl Acad. Sci. USA 89, 8966–8970 doi: 10.1073/pnas.89.19.8966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rydström J. (2006) Mitochondrial NADPH, transhydrogenase and disease. Biochim. Biophys. Acta, Bioenergetics 1757, 721–726 doi: 10.1016/j.bbabio.2006.03.010 [DOI] [PubMed] [Google Scholar]
- 63.Murphy M.P. (2015) Redox modulation by reversal of the mitochondrial nicotinamide nucleotide transhydrogenase. Cell Metab. 22, 363–365 doi: 10.1016/j.cmet.2015.08.012 [DOI] [PubMed] [Google Scholar]
- 64.Nissen T.L., Anderlund M., Nielsen J., Villadsen J. and Kielland-Brandt M.C. (2001) Expression of a cytoplasmic transhydrogenase in Saccharomyces cerevisiae results in formation of 2-oxoglutarate due to depletion of the NADPH pool. Yeast 18, 19–32 doi: [DOI] [PubMed] [Google Scholar]
- 65.Arkblad E.L., Egorov M., Shakhparonov M., Romanova L., Polzikov M. and Rydstrom J. (2002) Expression of proton-pumping nicotinamide nucleotide transhydrogenase in mouse, human brain and C. elegans. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 133, 13–21 doi: 10.1016/S1096-4959(02)00107-0 [DOI] [PubMed] [Google Scholar]
- 66.Speijer D., Manjeri G.R. and Szklarczyk R. (2014) How to deal with oxygen radicals stemming from mitochondrial fatty acid oxidation. Philos. Trans. R Soc. Lond. B Biol. Sci. 369, 20130446 doi: 10.1098/rstb.2013.0446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Camões F., Islinger M., Guimarães S.C., Kilaru S., Schuster M., Godinho L.F. et al. (2015) New insights into the peroxisomal protein inventory: acyl-CoA oxidases and -dehydrogenases are an ancient feature of peroxisomes. Biochim. Biophys. Acta, Mol. Cell Res. 1853, 111–125 doi: 10.1016/j.bbamcr.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 68.Kussmaul L. and Hirst J. (2006) The mechanism of superoxide production by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc. Natl Acad. Sci. USA 103, 7607–7612 doi: 10.1073/pnas.0510977103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drose S. and Brandt U. (2008) The mechanism of mitochondrial superoxide production by the cytochrome bc1 complex. J. Biol. Chem. 283, 21649–21654 doi: 10.1074/jbc.M803236200 [DOI] [PubMed] [Google Scholar]
- 70.Nicholls D.G. and Ferguson S.J. (2013) Bioenergetics 4, Academic Press, London [Google Scholar]
- 71.Wilson D.F., Dutton P.L., Erecinska M., Lindsay J.G. and Sato N. (1972) Mitochondrial electron transport and energy conservation. Acc. Chem. Res. 5, 234–241 doi: 10.1021/ar50055a002 [DOI] [Google Scholar]
- 72.Tahara E.B., Navarete F.D.T. and Kowaltowski A.J. (2009) Tissue-, substrate-, and site-specific characteristics of mitochondrial reactive oxygen species generation. Free Radic. Biol. Med. 46, 1283–1297 doi: 10.1016/j.freeradbiomed.2009.02.008 [DOI] [PubMed] [Google Scholar]
- 73.Seifert E.L., Estey C., Xuan J.Y. and Harper M.-E. (2010) Electron transport chain-dependent and -independent mechanisms of mitochondrial H2O2 emission during long-chain fatty acid oxidation. J. Biol. Chem. 285, 5748–5758 doi: 10.1074/jbc.M109.026203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rosca M.G., Vazquez E.J., Chen Q., Kerner J., Kern T.S. and Hoppel C.L. (2012) Oxidation of fatty acids is the source of increased mitochondrial reactive oxygen species production in kidney cortical tubules in early diabetes. Diabetes 61, 2074–2083 doi: 10.2337/db11-1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee J., Ellis J.M. and Wolfgang M.J. (2015) Adipose fatty acid oxidation is required for thermogenesis and potentiates oxidative stress-induced inflammation. Cell. Rep. 10, 266–279 doi: 10.1016/j.celrep.2014.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leclercq I.A., Farrell G.C., Field J., Bell D.R., Gonzalez F.J. and Robertson G.R. (2000) CYP2E1 and CYP4A as microsomal catalysts of lipid peroxides in murine nonalcoholic steatohepatitis. J. Clin. Invest. 105, 1067–1075 doi: 10.1172/JCI8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alberici L.C., Oliveira H.C., Paim B.A., Mantello C.C., Augusto A.C., Zecchin K.G. et al. (2009) Mitochondrial ATP-sensitive K+ channels as redox signals to liver mitochondria in response to hypertriglyceridemia. Free Radic. Biol. Med. 47, 1432–1439 doi: 10.1016/j.freeradbiomed.2009.08.013 [DOI] [PubMed] [Google Scholar]
- 78.Schönfeld P., Więckowski M.R., Lebiedzińska M. and Wojtczak L. (2010) Mitochondrial fatty acid oxidation and oxidative stress: lack of reverse electron transfer-associated production of reactive oxygen species. Biochim. Biophys. Acta, Bioenergetics 1797, 929–938 doi: 10.1016/j.bbabio.2010.01.010 [DOI] [PubMed] [Google Scholar]
- 79.Schagger H. and Pfeiffer K. (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 doi: 10.1093/emboj/19.8.1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Acin-Perez R. and Enriquez J.A. (2014) The function of the respiratory supercomplexes: the plasticity model. Biochim. Biophys. Acta, Bioenergetics 1837, 444–450 doi: 10.1016/j.bbabio.2013.12.009 [DOI] [PubMed] [Google Scholar]
- 81.Vanella A., Russo A., Acquaviva R., Campisi A., Di Giacomo C., Sorrenti V. et al. (2000) l-propionyl-carnitine as superoxide scavenger, antioxidant, and DNA cleavage protector. Cell. Biol. Toxicol. 16, 99–104 doi: 10.1023/A:1007638025856 [DOI] [PubMed] [Google Scholar]
- 82.Gülçin I. (2006) Antioxidant and antiradical activities of l-carnitine. Life Sci. 78, 803–811 doi: 10.1016/j.lfs.2005.05.103 [DOI] [PubMed] [Google Scholar]
- 83.Brown J.C.L., Chung D.J., Cooper A.N. and Staples J.F. (2013) Regulation of succinate-fuelled mitochondrial respiration in liver and skeletal muscle of hibernating thirteen-lined ground squirrels. J. Exp. Biol. 216(Pt 9), 1736–1743 doi: 10.1242/jeb.078519 [DOI] [PubMed] [Google Scholar]
- 84.Gnaiger E. (2014) Mitochondrial Pathways and Respiratory Control. An Introduction to OXPHOS Analysis, Oroboros, Innsbruck [Google Scholar]
- 85.Panov A., Orynbayeva Z., Vavilin V. and Lyakhovich V. (2014) Fatty acids in energy metabolism of the central nervous system. BioMed. Res. Int. 2014, 22 doi: 10.1155/2014/472459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Batandier C., Guigas B., Detaille D., El-Mir M.Y., Fontaine E., Rigoulet M. et al. (2006) The ROS production induced by a reverse-electron flux at respiratory-chain complex 1 is hampered by metformin. J. Bioenerg. Biomembr. 38, 33–42 doi: 10.1007/s10863-006-9003-8 [DOI] [PubMed] [Google Scholar]
- 87.Kushnareva Y., Murphy A.N. and Andreyev A. (2002) Complex I-mediated reactive oxygen species generation: modulation by cytochrome c and NAD(P)+ oxidation-reduction state. Biochem. J. 368(Pt 2), 545–553 doi: 10.1042/bj20021121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zoccarato F., Cavallini L. and Alexandre A. (2009) Succinate is the controller of O2−/H2O2 release at mitochondrial complex I: negative modulation by malate, positive by cyanide. J. Bioenerg. Biomembr. 41, 387–393 doi: 10.1007/s10863-009-9238-2 [DOI] [PubMed] [Google Scholar]
- 89.Muller F.L., Liu Y., Abdul-Ghani M.A., Lustgarten M.S., Bhattacharya A., Jang Y.C. et al. (2008) High rates of superoxide production in skeletal-muscle mitochondria respiring on both complex I- and complex II-linked substrates. Biochem. J. 409, 491–499 doi: 10.1042/BJ20071162 [DOI] [PubMed] [Google Scholar]
- 90.Bortolami S., Comelato E., Zoccarato F., Alexandre A. and Cavallini L. (2008) Long chain fatty acyl-CoA modulation of H2O2 release at mitochondrial complex I. J. Bioenerg. Biomembr. 40, 9–18 doi: 10.1007/s10863-008-9126-1 [DOI] [PubMed] [Google Scholar]
- 91.St-Pierre J., Buckingham J.A., Roebuck S.J. and Brand M.D. (2002) Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 277, 44784–44790 doi: 10.1074/jbc.M207217200 [DOI] [PubMed] [Google Scholar]
- 92.Lenaz G. and Genova M.L. (2009) Mobility and function of coenzyme Q (ubiquinone) in the mitochondrial respiratory chain. Biochim. Biophys. Acta, Bioenergetics 1787, 563–573 doi: 10.1016/j.bbabio.2009.02.019 [DOI] [PubMed] [Google Scholar]
- 93.Jekabsons M.B. and Nicholls D.G. (2004) In situ respiration and bioenergetic status of mitochondria in primary cerebellar granule neuronal cultures exposed continuously to glutamate. J. Biol. Chem. 279, 32989–33000 doi: 10.1074/jbc.M401540200 [DOI] [PubMed] [Google Scholar]
- 94.Chouchani E.T., Pell V.R., Gaude E., Aksentijevic D., Sundier S.Y., Robb E.L. et al. (2014) Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515, 431–435 doi: 10.1038/nature13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nagy G., Koncz A. and Perl A. (2003) T cell activation-induced mitochondrial hyperpolarization is mediated by Ca2+- and redox-dependent production of nitric oxide. J. Immunol. 171, 5188–5197 doi: 10.4049/jimmunol.171.10.5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Perl A., Gergely P., Jr, Nagy G., Koncz A. and Banki K. (2004) Mitochondrial hyperpolarization: a checkpoint of T-cell life, death and autoimmunity. Trends Immunol. 25, 360–367 doi: 10.1016/j.it.2004.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brownlee M. (2001) Biochemistry and molecular cell biology of diabetic complications. Nature 414, 813–820 doi: 10.1038/414813a [DOI] [PubMed] [Google Scholar]
- 98.Korenić A., Boltze J., Deten A., Peters M., Andjus P. and Radenovic L. (2014) Astrocytic mitochondrial membrane hyperpolarization following extended oxygen and glucose deprivation. PLoS ONE 9, e90697 doi: 10.1371/journal.pone.0090697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pak O., Sommer N., Hoeres T., Bakr A., Waisbrod S., Sydykov A. et al. (2013) Mitochondrial hyperpolarization in pulmonary vascular remodeling. Mitochondrial uncoupling protein deficiency as disease model. Am. J. Respir. Cell Mol. Biol. 49, 358–367 doi: 10.1165/rcmb.2012-0361OC [DOI] [PubMed] [Google Scholar]
- 100.Giovannini C., Matarrese P., Scazzocchio B., Sanchez M., Masella R. and Malorni W. (2002) Mitochondria hyperpolarization is an early event in oxidized low-density lipoprotein-induced apoptosis in Caco-2 intestinal cells. FEBS Lett. 523, 200–206 doi: 10.1016/S0014-5793(02)02972-1 [DOI] [PubMed] [Google Scholar]
- 101.Ward M.W., Huber H.J., Weisova P., Dussmann H., Nicholls D.G. and Prehn J.H. (2007) Mitochondrial and plasma membrane potential of cultured cerebellar neurons during glutamate-induced necrosis, apoptosis, and tolerance. J. Neurosci. 27, 8238–8249 doi: 10.1523/JNEUROSCI.1984-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen L.B. (1988) Mitochondrial membrane potential in living cells. Annu. Rev. Cell. Biol. 4, 155–181 doi: 10.1146/annurev.cb.04.110188.001103 [DOI] [PubMed] [Google Scholar]
- 103.Bonnet S., Archer S.L., Allalunis-Turner J., Haromy A., Beaulieu C., Thompson R. et al. (2007) A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell 11, 37–51 doi: 10.1016/j.ccr.2006.10.020 [DOI] [PubMed] [Google Scholar]
- 104.Guarás A., Perales-Clemente E., Calvo E., Acín-Pérez R., Loureiro-Lopez M., Pujol C. et al. (2016) The CoQH2/CoQ ratio serves as a sensor of respiratory chain efficiency. Cell. Rep. 15, 197–209 doi: 10.1016/j.celrep.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 105.Garaude J., Acín-Pérez R., Martínez-Cano S., Enamorado M., Ugolini M., Nistal-Villan E. et al. (2016) Mitochondrial respiratory-chain adaptations in macrophages contribute to antibacterial host defense. Nat. Immunol. 17, 1037–1045 doi: 10.1038/ni.3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stanley I.A., Ribeiro S.M., Giménez-Cassina A., Norberg E. and Danial N.N. (2014) Changing appetites: the adaptive advantages of fuel choice. Trends Cell. Biol. 24, 118–127 doi: 10.1016/j.tcb.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Acin-Perez R., Carrascoso I., Baixauli F., Roche-Molina M., Latorre-Pellicer A., Fernandez-Silva P. et al. (2014) ROS-triggered phosphorylation of complex II by Fgr kinase regulates cellular adaptation to fuel use. Cell Metab. 19, 1020–1033 doi: 10.1016/j.cmet.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scialo F., Sriram A., Fernandez-Ayala D., Gubina N., Lohmus M., Nelson G. et al. (2016) Mitochondrial ROS produced via reverse electron transport extend animal lifespan. Cell Metab. 23, 725–734 doi: 10.1016/j.cmet.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Held N.M. and Houtkooper R.H. (2015) Mitochondrial quality control pathways as determinants of metabolic health. BioEssays 37, 867–876 doi: 10.1002/bies.201500013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hue L. and Taegtmeyer H. (2009) The Randle cycle revisited: a new head for an old hat. Am. J. Physiol. Endocrinol. Metab. 297, E578–E591 doi: 10.1152/ajpendo.00093.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Orrenius S., Gogvadze V. and Zhivotovsky B. (2007) Mitochondrial oxidative stress: implications for cell death. Annu. Rev. Pharmacol. Toxicol. 47, 143–183 doi: 10.1146/annurev.pharmtox.47.120505.105122 [DOI] [PubMed] [Google Scholar]
- 112.Chen Y., Azad M.B. and Gibson S.B. (2009) Superoxide is the major reactive oxygen species regulating autophagy. Cell. Death Differ. 16, 1040–1052 doi: 10.1038/cdd.2009.49 [DOI] [PubMed] [Google Scholar]
- 113.Navarrete A., van Schaik C.P. and Isler K. (2011) Energetics and the evolution of human brain size. Nature 480, 91–93 doi: 10.1038/nature10629 [DOI] [PubMed] [Google Scholar]
- 114.Speijer D. (2012) Brains have a gut feeling about fat storage. BioEssays 34, 275–276 doi: 10.1002/bies.201200002 [DOI] [PubMed] [Google Scholar]
- 115.Rinholm J.E. and Bergersen L.H. (2012) Neuroscience: the wrap that feeds neurons. Nature 487, 435–436 doi: 10.1038/487435a [DOI] [PubMed] [Google Scholar]
- 116.Pellerin L. and Magistretti P.J. (2012) Sweet sixteen for ANLS. J. Cereb. Blood Flow Metab. 32, 1152–1166 doi: 10.1038/jcbfm.2011.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kathagen A., Schulte A., Balcke G., Phillips H.S., Martens T., Matschke J. et al. (2013) Hypoxia and oxygenation induce a metabolic switch between pentose phosphate pathway and glycolysis in glioma stem-like cells. Acta Neuropathol. 126, 763–780 doi: 10.1007/s00401-013-1173-y [DOI] [PubMed] [Google Scholar]
- 118.Cahill G.F., Jr (2006) Fuel metabolism in starvation. Annu. Rev. Nutr. 26, 1–22 doi: 10.1146/annurev.nutr.26.061505.111258 [DOI] [PubMed] [Google Scholar]
- 119.Schönfeld P. and Reiser G. (2013) Why does brain metabolism not favor burning of fatty acids to provide energy? Reflections on disadvantages of the use of free fatty acids as fuel for brain. J. Cereb. Blood Flow Metab. 33, 1493–1499 doi: 10.1038/jcbfm.2013.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Barnerias C., Saudubray J.-M., Touati G., De Lonlay P., Dulac O., Ponsot G. et al. (2010) Pyruvate dehydrogenase complex deficiency: four neurological phenotypes with differing pathogenesis. Dev. Med. Child Neurol. 52, e1–e9 doi: 10.1111/j.1469-8749.2009.03541.x [DOI] [PubMed] [Google Scholar]
- 121.Edmond J., Robbins R.A., Bergstrom J.D., Cole R.A. and de Vellis J. (1987) Capacity for substrate utilization in oxidative metabolism by neurons, astrocytes, and oligodendrocytes from developing brain in primary culture. J. Neurosci. Res. 18, 551–561 doi: 10.1002/jnr.490180407 [DOI] [PubMed] [Google Scholar]
- 122.Efremov R.G., Baradaran R. and Sazanov L.A. (2010) The architecture of respiratory complex I. Nature 465, 441–445 doi: 10.1038/nature09066 [DOI] [PubMed] [Google Scholar]
- 123.Gabaldon T. and Huynen M.A. (2005) Lineage-specific gene loss following mitochondrial endosymbiosis and its potential for function prediction in eukaryotes. Bioinformatics 21(Suppl 2), ii144–ii150 doi: 10.1093/bioinformatics/bti1124 [DOI] [PubMed] [Google Scholar]
- 124.Huynen M.A., de Hollander M. and Szklarczyk R. (2009) Mitochondrial proteome evolution and genetic disease. Biochim. Biophys. Acta, Mol. Basis Dis. 1792, 1122–1129 doi: 10.1016/j.bbadis.2009.03.005 [DOI] [PubMed] [Google Scholar]
- 125.Cardol P. (2011) Mitochondrial NADH:ubiquinone oxidoreductase (complex I) in eukaryotes: a highly conserved subunit composition highlighted by mining of protein databases. Biochim. Biophys. Acta, Bioenergetics 1807, 1390–1397 doi: 10.1016/j.bbabio.2011.06.015 [DOI] [PubMed] [Google Scholar]
- 126.Nouws J., Nijtmans L., Houten S.M., van den Brand M., Huynen M., Venselaar H. et al. (2010) Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab. 12, 283–294 doi: 10.1016/j.cmet.2010.08.002 [DOI] [PubMed] [Google Scholar]
- 127.Wang Y. and Bogenhagen D.F. (2006) Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 281, 25791–25802 doi: 10.1074/jbc.M604501200 [DOI] [PubMed] [Google Scholar]
- 128.Ramsden D.B., Ho P.W.-L., Ho J.W.-M., Liu H.-F., So D.H.-F., Tse H.-M. et al. (2012) Human neuronal uncoupling proteins 4 and 5 (UCP4 and UCP5): structural properties, regulation, and physiological role in protection against oxidative stress and mitochondrial dysfunction. Brain Behav. 2, 468–478 doi: 10.1002/brb3.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ricquier D. and Bouillaud F. (2000) The uncoupling protein homologues: UCP1, UCP2, UCP3, StUCP and AtUCP. Biochem. J. 345(Pt 2), 161–179 doi: 10.1042/bj3450161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wikstrom M., Krab K. and Saraste M. (1981) Cytochrome Oxidase: A Synthesis, Academic Press, London [Google Scholar]
- 131.Jastroch M., Divakaruni A.S., Mookerjee S., Treberg J.R. and Brand M.D. (2010) Mitochondrial proton and electron leaks. Essays Biochem. 47, 53–67 doi: 10.1042/bse0470053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Azzu V. and Brand M.D. (2010) The on-off switches of the mitochondrial uncoupling proteins. Trends Biochem. Sci. 35, 298–307 doi: 10.1016/j.tibs.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nicholls D.G., Bernson V.S. and Heaton G.M. (1978) The identification of the component in the inner membrane of brown adipose tissue mitochondria responsible for regulating energy dissipation. Experientia Suppl. 32, 89–93 doi: 10.1007/978-3-0348-5559-4_9 [DOI] [PubMed] [Google Scholar]
- 134.Donadelli M., Dando I., Fiorini C. and Palmieri M. (2014) UCP2, a mitochondrial protein regulated at multiple levels . Cell Mol. Life Sci. 71, 1171–1190 doi: 10.1007/s00018-013-1407-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kunji E.R. and Robinson A.J. (2010) Coupling of proton and substrate translocation in the transport cycle of mitochondrial carriers. Curr. Opin. Struct. Biol. 20, 440–447 doi: 10.1016/j.sbi.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 136.Fedorenko A., Lishko P.V. and Kirichok Y. (2012) Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 151, 400–413 doi: 10.1016/j.cell.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Esteves T.C., Parker N. and Brand M.D. (2006) Synergy of fatty acid and reactive alkenal activation of proton conductance through uncoupling protein 1 in mitochondria. Biochem. J. 395, 619–628 doi: 10.1042/BJ20052004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brand M.D., Affourtit C., Esteves T.C., Green K., Lambert A.J., Miwa S. et al. (2004) Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins . Free Radic. Biol. Med. 37, 755–767 doi: 10.1016/j.freeradbiomed.2004.05.034 [DOI] [PubMed] [Google Scholar]
- 139.Chouchani E.T., Kazak L., Jedrychowski M.P., Lu G.Z., Erickson B.K., Szpyt J. et al. (2016) Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature 532, 112–116 doi: 10.1038/nature17399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Echtay K.S., Winkler E., Frischmuth K. and Klingenberg M. (2001) Uncoupling proteins 2 and 3 are highly active H+ transporters and highly nucleotide sensitive when activated by coenzyme Q (ubiquinone). Proc. Natl Acad. Sci. USA 98, 1416–1421 doi: 10.1073/pnas.98.4.1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sluse F.E., Jarmuszkiewicz W., Navet R., Douette P., Mathy G. and Sluse-Goffart C.M. (2006) Mitochondrial UCPs: new insights into regulation and impact. Biochim. Biophys. Acta, Bioenergetics 1757, 480–485 doi: 10.1016/j.bbabio.2006.02.004 [DOI] [PubMed] [Google Scholar]
- 142.Anedda A., Rial E. and Gonzalez-Barroso M.M. (2008) Metformin induces oxidative stress in white adipocytes and raises uncoupling protein 2 levels. J. Endocrinol. 199, 33–40 doi: 10.1677/JOE-08-0278 [DOI] [PubMed] [Google Scholar]
- 143.Boss O., Samec S., Dulloo A., Seydoux J., Muzzin P. and Giacobino J.-P. (1997) Tissue-dependent upregulation of rat uncoupling protein-2 expression in response to fasting or cold. FEBS Lett. 412, 111–114 doi: 10.1016/S0014-5793(97)00755-2 [DOI] [PubMed] [Google Scholar]
- 144.Anderson E.J., Yamazaki H. and Neufer P.D. (2007) Induction of endogenous uncoupling protein 3 suppresses mitochondrial oxidant emission during fatty acid-supported respiration. J. Biol. Chem. 282, 31257–31266 doi: 10.1074/jbc.M706129200 [DOI] [PubMed] [Google Scholar]
- 145.Anedda A., Lopez-Bernardo E., Acosta-Iborra B., Saadeh Suleiman M., Landázuri M.O. and Cadenas S. (2013) The transcription factor Nrf2 promotes survival by enhancing the expression of uncoupling protein 3 under conditions of oxidative stress. Free Radic. Biol. Med. 61, 395–407 doi: 10.1016/j.freeradbiomed.2013.04.007 [DOI] [PubMed] [Google Scholar]
- 146.Jiang N., Zhang G., Bo H., Qu J., Ma G., Cao D. et al. (2009) Upregulation of uncoupling protein-3 in skeletal muscle during exercise: a potential antioxidant function. Free Radic. Biol. Med. 46, 138–145 doi: 10.1016/j.freeradbiomed.2008.09.026 [DOI] [PubMed] [Google Scholar]
- 147.Žáčková M., Škobisová E., Urbánková E. and Ježek P. (2003) Activating ω-6 polyunsaturated fatty acids and inhibitory purine nucleotides are high affinity ligands for novel mitochondrial uncoupling proteins UCP2 and UCP3. J. Biol. Chem. 278, 20761–20769 doi: 10.1074/jbc.M212850200 [DOI] [PubMed] [Google Scholar]
- 148.Beck V., Jaburek M., Demina T., Rupprecht A., Porter R.K., Jezek P. et al. (2007) Polyunsaturated fatty acids activate human uncoupling proteins 1 and 2 in planar lipid bilayers. FASEB J. 21, 1137–1144 doi: 10.1096/fj.06-7489com [DOI] [PubMed] [Google Scholar]
- 149.Bordone L., Motta M.C., Picard F., Robinson A., Jhala U.S., Apfeld J. et al. (2005) Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic β cells. PLoS Biol. 4, e31 doi: 10.1371/journal.pbio.0040031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nakatani T., Tsuboyama-Kasaoka N., Takahashi M., Miura S. and Ezaki O. (2002) Mechanism for peroxisome proliferator-activated receptor- activator-induced up-regulation of UCP2 mRNA in rodent hepatocytes. J. Biol. Chem. 277, 9562–9569 doi: 10.1074/jbc.M110132200 [DOI] [PubMed] [Google Scholar]
- 151.Patterson A.D., Shah Y.M., Matsubara T., Krausz K.W. and Gonzalez F.J. (2012) Peroxisome proliferator-activated receptor alpha induction of uncoupling protein 2 protects against acetaminophen-induced liver toxicity. Hepatology 56, 281–290 doi: 10.1002/hep.25645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Woyda-Ploszczyca A. and Jarmuszkiewicz W. (2011) Ubiquinol (QH2) functions as a negative regulator of purine nucleotide inhibition of Acanthamoeba castellanii mitochondrial uncoupling protein. Biochim. Biophys. Acta, Bioenergetics 1807, 42–52 doi: 10.1016/j.bbabio.2010.08.012 [DOI] [PubMed] [Google Scholar]
- 153.Jarmuszkiewicz W., Navet R., Alberici L.C., Douette P., Sluse-Goffart C.M., Sluse F.E. et al. (2004) Redox state of endogenous coenzyme q modulates the inhibition of linoleic acid-induced uncoupling by guanosine triphosphate in isolated skeletal muscle mitochondria. J. Bioenerg. Biomembr. 36, 493–502 doi: 10.1023/B:JOBB.0000047331.25248.7a [DOI] [PubMed] [Google Scholar]
- 154.Jarmuszkiewicz W., Swida A., Czarna M., Antos N., Sluse-Goffart C.M. and Sluse F.E. (2005) In phosphorylating Acanthamoeba castellanii mitochondria the sensitivity of uncoupling protein activity to GTP depends on the redox state of quinone. J. Bioenerg. Biomembr. 37, 97–107 doi: 10.1007/s10863-005-4133-y [DOI] [PubMed] [Google Scholar]
- 155.Chan S.H.H., Wu C.-A., Wu K.L.H., Ho Y.-H., Chang A.Y. and Chan J.Y.H. (2009) Transcriptional upregulation of mitochondrial uncoupling protein 2 protects against oxidative stress-associated neurogenic hypertension. Circ. Res. 105, 886–896 doi: 10.1161/CIRCRESAHA.109.199018 [DOI] [PubMed] [Google Scholar]
- 156.Giardina T.M., Steer J.H., Lo S.Z. and Joyce D.A. (2008) Uncoupling protein-2 accumulates rapidly in the inner mitochondrial membrane during mitochondrial reactive oxygen stress in macrophages. Biochim. Biophys. Acta, Bioenergetics 1777, 118–129 doi: 10.1016/j.bbabio.2007.11.006 [DOI] [PubMed] [Google Scholar]
- 157.Mailloux R.J. and Harper M.-E. (2011) Uncoupling proteins and the control of mitochondrial reactive oxygen species production. Free Radic. Biol. Med. 51, 1106–1115 doi: 10.1016/j.freeradbiomed.2011.06.022 [DOI] [PubMed] [Google Scholar]
- 158.Mailloux R.J., Seifert E.L., Bouillaud F., Aguer C., Collins S. and Harper M.-E. (2011) Glutathionylation acts as a control switch for uncoupling proteins UCP2 and UCP3. J. Biol. Chem. 286, 21865–21875 doi: 10.1074/jbc.M111.240242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xu H., Hertzel A.V., Steen K.A., Wang Q., Suttles J. and Bernlohr D.A. (2015) Uncoupling lipid metabolism from inflammation through fatty acid binding protein-dependent expression of UCP2. Mol. Cell. Biol. 35, 1055–1065 doi: 10.1128/MCB.01122-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Barbe P., Larrouy D., Boulanger C., Chevillotte E., Viguerie N., Thalamas C. et al. (2001) Triiodothyronine-mediated up-regulation of UCP2 and UCP3 mRNA expression in human skeletal muscle without coordinated induction of mitochondrial respiratory chain genes. FASEB J. 15, 13–15 doi: 10.1096/fj.00-0502fje [DOI] [PubMed] [Google Scholar]
- 161.Hughes J. and Criscuolo F. (2008) Evolutionary history of the UCP gene family: gene duplication and selection. BMC Evol. Biol. 8, 306 doi: 10.1186/1471-2148-8-306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lee P., Swarbrick M.M. and Ho K.K. (2013) Brown adipose tissue in adult humans: a metabolic renaissance. Endocr. Rev. 34, 413–438 doi: 10.1210/er.2012-1081 [DOI] [PubMed] [Google Scholar]
- 163.Strijbis K., Vaz F.M. and Distel B. (2010) Enzymology of the carnitine biosynthesis pathway. IUBMB Life 62, 357–362 doi: 10.1002/iub.323 [DOI] [PubMed] [Google Scholar]
- 164.Indiveri C., Tonazzi A. and Palmieri F. (1991) Characterization of the unidirectional transport of carnitine catalyzed by the reconstituted carnitine carrier from rat liver mitochondria. Biochim. Biophys. Acta, Biomembranes 1069, 110–116 doi: 10.1016/0005-2736(91)90110-T [DOI] [PubMed] [Google Scholar]
- 165.Indiveri C., Tonazzi A. and Palmieri F. (1994) The reconstituted carnitine carrier from rat liver mitochondria: evidence for a transport mechanism different from that of the other mitochondrial translocators. Biochim. Biophys. Acta, Biomembranes 1189, 65–73 doi: 10.1016/0005-2736(94)90281-X [DOI] [PubMed] [Google Scholar]
- 166.He J., Mao C.-C., Reyes A., Sembongi H., Di Re M., Granycome C. et al. (2007) The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell. Biol. 176, 141–146 doi: 10.1083/jcb.200609158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rajala N., Gerhold J.M., Martinsson P., Klymov A. and Spelbrink J.N. (2014) Replication factors transiently associate with mtDNA at the mitochondrial inner membrane to facilitate replication. Nucleic Acids Res. 42, 952–967 doi: 10.1093/nar/gkt988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kirkwood T.B.L. and Kowald A. (2012) The free-radical theory of ageing — older, wiser and still alive. BioEssays 34, 692–700 doi: 10.1002/bies.201200014 [DOI] [PubMed] [Google Scholar]
- 169.Traykova M., Traykov T., Hadjimitova V., Krikorian K. and Bojadgieva N. (2003) Antioxidant properties of galantamine hydrobromide. Z. Naturforsch C. 58, 361–365 doi: 10.1515/znc-2003-5-613 [DOI] [PubMed] [Google Scholar]
- 170.Berry E.A. and Trumpower B.L. (1985) Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. J. Biol. Chem. 260, 2458–2467 PMID: [PubMed] [Google Scholar]
- 171.Schägger H. and von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 doi: 10.1016/0003-2697(91)90094-A [DOI] [PubMed] [Google Scholar]
- 172.Paradies G., Paradies V., De Benedictis V., Ruggiero F.M. and Petrosillo G. (2014) Functional role of cardiolipin in mitochondrial bioenergetics. Biochim. Biophys. Acta, Bioenergetics 1837, 408–417 doi: 10.1016/j.bbabio.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 173.Dudkina N.V., Sunderhaus S., Boekema E.J. and Braun H.-P. (2008) The higher level of organization of the oxidative phosphorylation system: mitochondrial supercomplexes. J. Bioenerg. Biomembr. 40, 419–424 doi: 10.1007/s10863-008-9167-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Moreno-Lastres D., Fontanesi F., Garcia-Consuegra I., Martin M.A., Arenas J., Barrientos A. et al. (2012) Mitochondrial complex I plays an essential role in human respirasome assembly. Cell. Metab. 15, 324–335 doi: 10.1016/j.cmet.2012.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gómez L.A. and Hagen T.M. (2012) Age-related decline in mitochondrial bioenergetics: does supercomplex destabilization determine lower oxidative capacity and higher superoxide production? Semin. Cell. Dev. Biol. 23, 758–767 doi: 10.1016/j.semcdb.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lenaz G., Baracca A., Barbero G., Bergamini C., Dalmonte M.E., Del Sole M. et al. (2010) Mitochondrial respiratory chain super-complex I-III in physiology and pathology. Biochim. Biophys. Acta, Bioenergetics 1797, 633–640 doi: 10.1016/j.bbabio.2010.01.025 [DOI] [PubMed] [Google Scholar]
- 177.Winge D.R. (2012) Sealing the mitochondrial respirasome. Mol. Cell. Biol. 32, 2647–2652 doi: 10.1128/MCB.00573-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Porras C.A. and Bai Y. (2015) Respiratory supercomplexes plasticity and implications. Front. Biosci. (Landmark Ed). 20, 621–634 doi: 10.2741/4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lapuente-Brun E., Moreno-Loshuertos R., Acin-Perez R., Latorre-Pellicer A., Colas C., Balsa E. et al. (2013) Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570 doi: 10.1126/science.1230381 [DOI] [PubMed] [Google Scholar]
- 180.Mourier A., Matic S., Ruzzenente B., Larsson N.-G. and Milenkovic D. (2014) The respiratory chain supercomplex organization is independent of COX7a2l isoforms. Cell Metab. 20, 1069–1075 doi: 10.1016/j.cmet.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Maranzana E., Barbero G., Falasca A.I., Lenaz G. and Genova M.L. (2013) Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal. 19, 1469–1480 doi: 10.1089/ars.2012.4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fang J., Uchiumi T., Yagi M., Matsumoto S., Amamoto R., Takazaki S. et al. (2013) Dihydro-orotate dehydrogenase is physically associated with the respiratory complex and its loss leads to mitochondrial dysfunction. Biosci. Rep. 33, e00021 doi: 10.1042/BSR20120097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nguyen N.H., Bråthe A. and Hassel B. (2003) Neuronal uptake and metabolism of glycerol and the neuronal expression of mitochondrial glycerol-3-phosphate dehydrogenase. J. Neurochem. 85, 831–842 doi: 10.1046/j.1471-4159.2003.01762.x [DOI] [PubMed] [Google Scholar]
- 184.McKenna M.C., Waagepetersen H.S., Schousboe A. and Sonnewald U. (2006) Neuronal and astrocytic shuttle mechanisms for cytosolic-mitochondrial transfer of reducing equivalents: current evidence and pharmacological tools. Biochem. Pharmacol. 71, 399–407 doi: 10.1016/j.bcp.2005.10.011 [DOI] [PubMed] [Google Scholar]
- 185.Tretter L., Takacs K., Hegedus V. and Adam-Vizi V. (2007) Characteristics of α-glycerophosphate-evoked H2O2 generation in brain mitochondria. J. Neurochem. 100, 650–663 doi: 10.1111/j.1471-4159.2006.04223.x [DOI] [PubMed] [Google Scholar]
- 186.Alexeyev M., Shokolenko I., Wilson G. and LeDoux S. (2013) The maintenance of mitochondrial DNA integrity — critical analysis and update. Cold Spring Harb. Perspect. Biol. 5, a012641 doi: 10.1101/cshperspect.a012641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kowald A. and Kirkwood T.B.L. (2011) Evolution of the mitochondrial fusion-fission cycle and its role in aging. Proc. Natl Acad. Sci. USA 108, 10237–10242 doi: 10.1073/pnas.1101604108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Willems P.H.G.M., Rossignol R., Dieteren C.E.J., Murphy M.P. and Koopman W.J.H. (2015) Redox homeostasis and mitochondrial dynamics. Cell Metab. 22, 207–218 doi: 10.1016/j.cmet.2015.06.006 [DOI] [PubMed] [Google Scholar]
- 189.Liesa M. and Shirihai O.S. (2013) Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 17, 491–506 doi: 10.1016/j.cmet.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tol M.J., Ottenhoff R., van Eijk M., Zelcer N., Aten J., Houten S.M. et al. (2016) A PPARγ-Bnip3 axis couples adipose mitochondrial fusion-fission balance to systemic insulin sensitivity. Diabetes dx.doi.org/10.2337/db16-0243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bender A., Hajieva P. and Moosmann B. (2008) Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc. Natl Acad. Sci. USA 105, 16496–16501 doi: 10.1073/pnas.0802779105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Speijer D. (2015) Birth of the eukaryotes by a set of reactive innovations: new insights force us to relinquish gradual models. BioEssays 37, 1268–1276 doi: 10.1002/bies.201500107 [DOI] [PubMed] [Google Scholar]
- 193.Chandel N.S. (2015) Evolution of mitochondria as signaling organelles. Cell Metab. 22, 204–206 doi: 10.1016/j.cmet.2015.05.013 [DOI] [PubMed] [Google Scholar]
- 194.Yun J. and Finkel T. (2014) Mitohormesis. Cell Metab. 19, 757–766 doi: 10.1016/j.cmet.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Holmström K.M. and Finkel T. (2014) Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 15, 411–421 doi: 10.1038/nrm3801 [DOI] [PubMed] [Google Scholar]
- 196.Sies H. (2014) Role of metabolic H2O2 generation: redox signaling and oxidative stress. J. Biol. Chem. 289, 8735–8741 doi: 10.1074/jbc.R113.544635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Peng G., Fritzsch G., Zickermann V., Schägger H., Mentele R., Lottspeich F. et al. (2003) Isolation, characterization and electron microscopic single particle analysis of the NADH:Ubiquinone oxidoreductase (complex I) from the hyperthermophilic Eubacterium Aquifex aeolicus. Biochemistry 42, 3032–3039 doi: 10.1021/bi026876v [DOI] [PubMed] [Google Scholar]