Abstract
Osteosarcoma is the most common type of primary malignant bone tumor, and deregulated microRNAs (miRNAs or miRs) in osteosarcoma have attracted great attention. In the present study, through miRNA microarray analysis, it was identified that miR-148a expression was significantly increased in osteosarcoma tissues. Increased miR-148a expression was significantly correlated with tumor progression and prognosis. Furthermore, increased miR-148a expression could promote osteosarcoma growth in vitro and in vivo, and the tumor-promoting effect was due to enhanced activation of the phosphoinositide 3-kinase signaling pathway caused by miR-148a-mediated inhibition of phosphatase and tensin homolog expression. Together, the present results suggest a role for miR-148a in osteosarcoma development and its potential use in prognosis prediction and cancer therapy.
Keywords: microRNA, miR-148a, proliferation, PTEN, PI3K, osteosarcoma
Introduction
MicroRNAs (miRNAs or miRs) are small non-coding RNAs with 17–25 nucleotides that are present in eukaryotic organisms (1). miRNAs are important in diverse biological processes by post-transcriptionally regulating gene expression through base pairing to complementary sites in the 3′-untranslated region (UTR) of selected transcripts (1). Since their initial identification, ~1,000 miRNA sequences have been reported thus far (1), and their biological functions have attracted great attention, although further investigation into their mechanism of action is still required. Recently, miRNAs have been observed to participate in the regulation of diverse physiological and pathological processes, particularly in cancer development (2). Aberrant miRNA expression is associated with the pathogenesis of various types of cancer, and they are associated with clinical outcomes of cancer patients (3). However, identifying the deregulated miRNAs and their roles in carcinogenesis and cancer progression, particularly in osteosarcoma development, is an ongoing process (4).
Osteosarcoma is the most common type of human primary malignant bone tumor, and usually develops in children and young adults, with an incidence rate of 3 per million per year (5). It is characterized by an aggressive clinical course, with a 5-year survival rate of 60–70% in patients with no metastatic disease (5–7). The clinical outcomes are considerably worse in patients with metastatic disease (5–7). Currently, the mechanisms responsible for the oncogenic insults in the initiation and progression of osteosarcoma remain to be fully elucidated. To date, a number of miRNAs have been determined to be deregulated in osteosarcoma and to participate in osteosarcoma development, including miR-212 (8), miR-33b (9), miR-503 (10) and miR-451 (11). In addition, the present authors also reported that miR-143 and miR-133a are downregulated in osteosarcoma, and they function as anti-oncomiRs in osteosarcoma development (12,13). However, the miRNA expression profile and the roles of deregulated miRNAs in osteosarcoma carcinogenesis and progression require further investigation.
In order to identify the deregulated miRNAs involved in osteosarcoma, paired clinically resected osteosarcoma tissues and adjacent normal tissues were obtained from 92 primary osteosarcoma patients, and their miRNA expression profile was elucidated using miRNA microarray analysis in the present study. A set of deregulated miRNAs in osteosarcoma tissues were identified, including upregulated miR-31, miR-210 and miR-148a, and downregulated miR-206, miR-1 and miR-133a. Since the roles of miR-31, miR-210, miR-206, miR-1 and miR-133a in osteosarcoma progression have been previously reported (13–17), and the roles of miR-148a in osteosarcoma development are elusive, the present study focused on the expression and biological function of miR-148a in osteosarcoma carcinogenesis and progression.
Materials and methods
Patients samples
Human osteosarcoma tumor tissues and paired adjacent normal tissues were obtained from 92 primary osteosarcoma patients during surgery. The surgeries of these patients were performed between April 2006 and December 2009 at Changhai Hospital (Shanghai, China). The clinical information of these patients was previously published (13). The tissues were rapidly frozen in liquid nitrogen following surgical resection. All specimens were collected upon obtaining informed consent from all patients. The experiments in the present study were approved by the ethics committee of The Second Military Medical University (Shanghai, China).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA, including miRNA, was extracted as previously reported (12,13). For evaluating the expression of miR-148a, RT-qPCR analysis was performed using stem-loop RT primers, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAAAG-3′, and qPCR primers 5′-TCAGTGCACTACAGAA-3′ (forward) and 5′-GTGCAGGGTCCGAGGT-3′ (reverse). The primers were synthesized by Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The RT-qPCR assay was performed and analyzed as previously reported (12,13).
Cell culture and transfection
The human osteoblastic cell line hFOB 1.19 and the osteosarcoma cell lines MG63 and U2OS (American Type Culture Collection, Manassas, VA, USA) were cultured, seeded and transfected as previously described (12,13). miR-148a mimics, miR-148a inhibitor and their respective controls were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). miRNA mimics or inhibitor were transfected at a final concentration of 20 or 50 nM, respectively, using INTERFERin® transfection reagent (Polyplus-transfection SA, Illkirch, France), following the manufacturer's protocol.
Cell proliferation analysis
MG63 and U2OS cells were subjected to in vitro cell proliferation assay as previously reported (12,13). In brief, cells were seeded into 96-well plates, transfected and analyzed using the (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay at the indicated time points. Used cell medium was replaced with 200 µl fresh medium containing MTT 0.5 mg/ml. Cell cultural plates were incubated at 37°C for 4 h, then 100 µl dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO, USA) was added and the plates were shaken for 10 min. The absorbance was measured at 570 nm.
Tumor growth analysis in vivo
All animal experiments were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the ethics committee of Second Military Medical University, Shanghai, China. The nude mice used in the present study were obtained from Shanghai Laboratory Animal Research Center (Shanghai, China), and kept in pathogen-free conditions with normal chow, access to water and regular light/dark cycles. MG63 cells (1×106) suspended in 0.1 ml phosphate-buffered saline were injected subcutaneously into the posterior flank of BALB/c athymic nude mice. Two weeks later, 10 nmol cholesterol-conjugated small RNA agonist (ago)-miR or antagonist (antago)-miR of miR-148a (Guangzhou RiboBio Co., Ltd., Guangzhou, China) in 0.1 ml saline buffer was locally injected into the tumor mass (once every 3 days for 2 weeks). Tumor growth was measured as previously described (13).
3′-UTR luciferase reporter analysis
Human phosphatase and tensin homolog (PTEN) 3′-UTR luciferase reporter constructs and constructs with miR-148a target site deletion were constructed as previously reported (12,13), and verified by sequencing. Sequencing was performed by service provider Jie Li Biology (Shanghai, China). Cells were transfected and luciferase activity was analyzed as previously described (12,13).
Western blotting
Cells or grinded human tissues were lysed, and their total protein content was extracted and subjected to electrophoresis and western blot analysis as previously reported (12,13). Antibodies specific for PTEN (rabbit monoclonal; catalog no., 9188; 1:1,000) and β-actin (mouse monoclonal; catalog no., 3700; 1:2,000), and horseradish peroxidase (HRP)-coupled secondary antibodies against rabbit [anti-rabbit immunoglobulin G (IgG), HRP-linked; catalog no., 7074; 1:2,000] and mouse (anti-mouse IgG, HRP-linked; catalog no., 7076; 1:2,000) were obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA) and used following the standard protocols. Densitometric analysis was performed using LabWorks Image Acquisition and Analysis Software (UVP, Inc., Upland, CA, USA) as previously described (13).
Statistical analysis
Data are presented as the mean ± standard deviation. Student's t-test was used to analyze statistical comparisons between groups, and two-tailed P<0.05 was considered to indicate a statistically significant difference. Spearman's rank correlation coefficient in SPSS version 17.0 software (SPSS, Inc., Chicago, IL, USA) was used to analyze the correlation between miR-148a expression and clinical osteosarcoma stages, while Kaplan-Meier survival analysis with log-rank test in SPSS version 17.0 software was used to analyze the overall survival in osteosarcoma patients. Pearson's correlation coefficient in SPSS version 17.0 software was used to analyze the correlation between the expression of miR-148a and the protein levels of PTEN.
Results
Increased miR-148a expression in osteosarcoma
To investigate the miRNA expression profile in osteosarcoma, miRNA microarray analysis was applied to human osteosarcoma tissue and adjacent normal tissue. The microarray analysis of miRNA expression was performed by a service provider, Shanghai Biotechnology Corporation (Shanghai, China) (3). A set of miRNAs were observed to be deregulated in osteosarcoma tissue, including miR-31, miR-210 and miR-148a (the most upregulated miRNAs identified in the analysis), and miR-206, miR-1 and miR-133a (the most downegulated miRNAs identified in the analysis) (Fig. 1A). Of these miRNAs, the roles of miR-133a in osteosarcoma development were previously reported by the present authors (13). Therefore, the present study focused on the roles of miR-148a in human osteosarcoma progression. miR-148a expression in osteosarcoma was further evaluated in osteosarcoma cell lines, and it was observed to be increased in the human osteosarcoma cell lines MG63 and U2OS, compared with the normal osteoblastic cell line hFOB 1.19 (P<0.0001; Fig. 1B). Furthermore, in the 92 pairs of human clinical osteosarcoma and adjacent normal tissue specimens analyzed in the present study, miR-148a expression was determined to be significantly increased in osteosarcoma tissues, compared with normal tissues (Fig. 1C). Thus, the present data indicate that miR-148a expression is increased in osteosarcoma.
Figure 1.
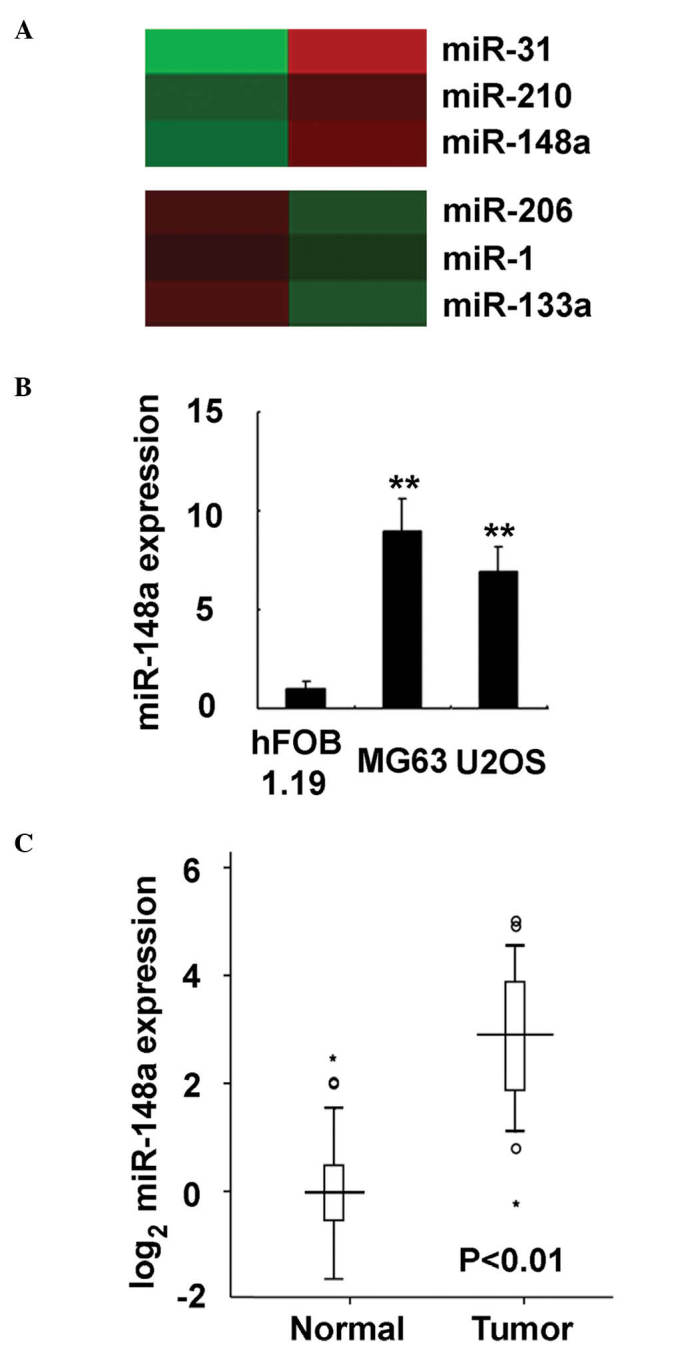
miR-148a is upregulated in osteosarcoma tissues and cell lines. (A) miRNA microarray analysis of paired osteosarcoma tissues and adjacent normal tissues. The three most upregulated and downregulated miRNAs identified in the analysis are indicated. (B) miR-148a expression was examined in osteosarcoma cell lines using RT-qPCR (**P<0.01). (C) In 92 paired clinical human osteosarcoma tissues and adjacent normal tissues, miR-148a expression was examined by RT-qPCR. Data are presented as the mean ± standard deviation (n=4) in cell lines, or as horizontal lines (median) with boxes (interquartile range) and whiskers (corresponding to the 2.5th and 97.5th percentiles) in human samples (*data out of scope). miR/miRNA, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Increased miR-148a expression is significantly correlated with progression and prognosis of osteosarcoma
As miR-148a expression was observed to be significantly increased in osteosarcoma, the correlation between miR-148a upregulation and osteosarcoma progression was further investigated. miR-148a expression was significantly positive-correlated with tumor stages of osteosarcoma in the human samples, according to the Spearman's rank correlation coefficient (P<0.0001; Fig. 2A). Furthermore, miR-148a upregulation in osteosarcoma tissues was significantly correlated with poor overall survival of osteosarcoma patients by Kaplan-Meier survival analysis (Fig. 2B). Taken together, these data suggest that miR-148a upregulation may be important in osteosarcoma progression and prognosis, and determination of miR-148a expression may be used in the pathological identification and prediction of prognosis in osteosarcoma patients.
Figure 2.
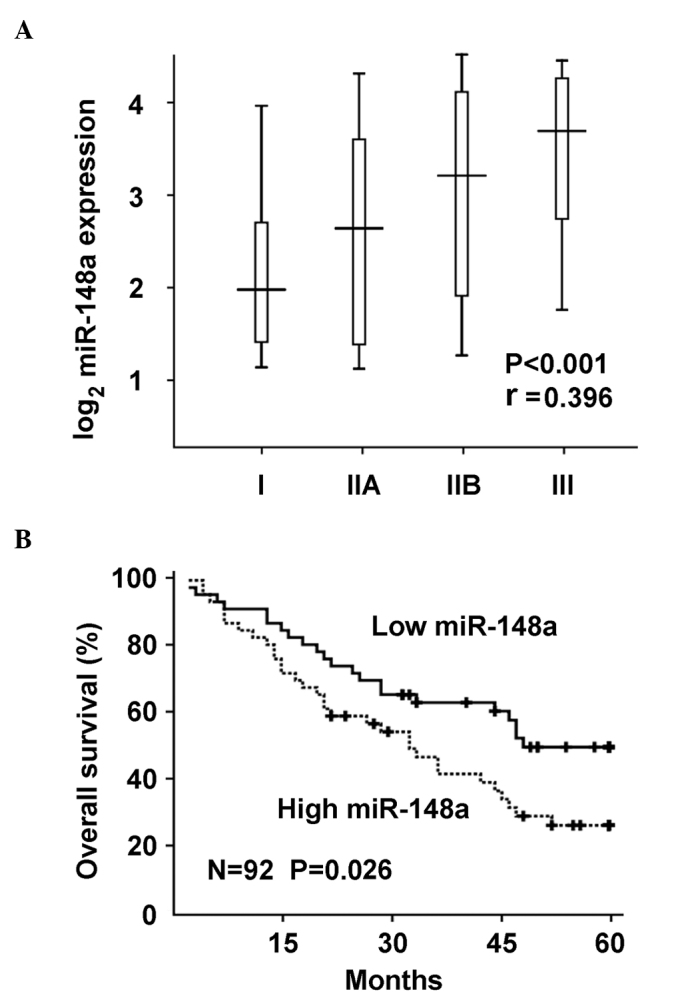
miR-148a upregulation is correlated with osteosarcoma progression and patient prognosis. (A) Correlation between miR-148a expression and tumor stages I–III was analyzed by Spearman's rank correlation coefficient. The r and P-values obtained for the 92 human osteosarcoma tissues included in the present study are indicated. (B) Kaplan-Meier survival curve of overall survival in the 92 osteosarcoma patients analyzed, according to their miR-148a expression levels in tumor tissues. The P-value obtained is indicated. The median value of miR-148a expression in the tumor tissues was selected as the cut-off value. miR, microRNA.
miR-148a promotes osteosarcoma growth in vitro and in vivo
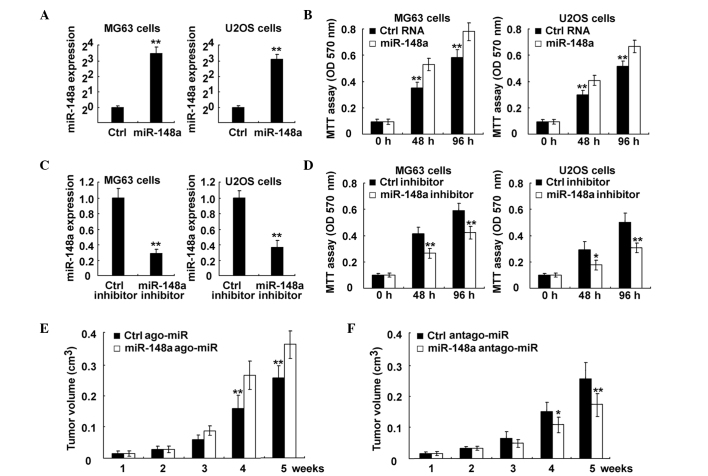
Since the expression of miR-148a was increased in osteosarcoma and correlated with poor prognosis, it was next investigated whether miR-148a functioned as an oncogene in osteosarcoma. When miR-148a was overexpressed in osteosarcoma MG63 and U2OS cells, it promoted cell proliferation in both cell lines (Fig. 3A and B). In addition, transfection of miR-148a inhibitor significantly suppressed miR-148a expression, and miR-148a inhibition suppressed proliferation in osteosarcoma cells (Fig. 3C and D). Furthermore, osteosarcoma cell growth in vivo was promoted by administration of miR-148a ago-miR, while it was inhibited by its antago-miR (Fig. 3E and F, respectively). Therefore, these data suggest that miR-148a may be an oncogene that promotes osteosarcoma growth both in vitro and in vivo, and inhibition of miR-148a bears considerable therapeutic potential in osteosarcoma.
Figure 3.
Inhibition of miR-148a suppressed osteosarcoma growth both in vitro and in vivo. (A and B) MG63 and U2OS cells were transfected with control RNA or miR-148a mimics, as indicated. (A) miR-148a expression was examined using RT-qPCR. The data are the relative values of miR-148 expression normalized to the internal control of U6 expression. (B) Cell proliferation was measured using MTT assay at the indicated time points following transfection. (C and D) MG63 and U2OS cells were transfected with control RNA or miR-148a inhibitor, as indicated. (C) miR-148a expression was examined using RT-qPCR. The data are the relative values of miR-148 expression normalized to the internal control of U6 expression. (D) Cell proliferation was measured using MTT assay at the indicated time points following transfection. (E and F) MG63 cells were injected subcutaneously into the posterior flank of nude mice. Two weeks later, cholesterol-conjugated small RNA (E) ago-miR or (F) antago-miR of miR-148a were locally injected into the tumor mass, and tumor growth was measured. The growth curves obtained are indicated in the image. Data are presented as the mean ± standard deviation (n=4), or correspond to one representative experiment, whereby similar results were obtained in three independent experiments. *P<0.05; **P<0.01. miR, microRNA; Ctrl, control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; OD, optical density; ago, agonist; antago, antagonist.
miR-148a promotes phosphoinositide 3-kinase (PI3K) signaling pathway activation by targeting PTEN expression
The mechanism responsible for the growth-promoting effect of miR-148a on osteosarcoma was next investigated. The intracellular cell signaling pathways in MG63 and U2OS cells overexpressing miR-148a was examined, including PI3K, mitogen-activated protein kinase, nuclear factor-κB and signal transducer and activator of transcription signaling pathways, and it was observed that only the activation of the PI3K signaling pathway was significantly promoted by miR-148a overexpression (Fig. 4A and data not shown). In addition, the inhibition of miR-148a suppressed PI3K activation in osteosarcoma cells (Fig. 4B). As PI3K activation and downstream AKT phosphorylation are critical in osteosarcoma growth and progression (18,19), it was concluded that miR-148a may promote osteosarcoma growth by enhancing the activation of the PI3K signaling pathway.
Figure 4.
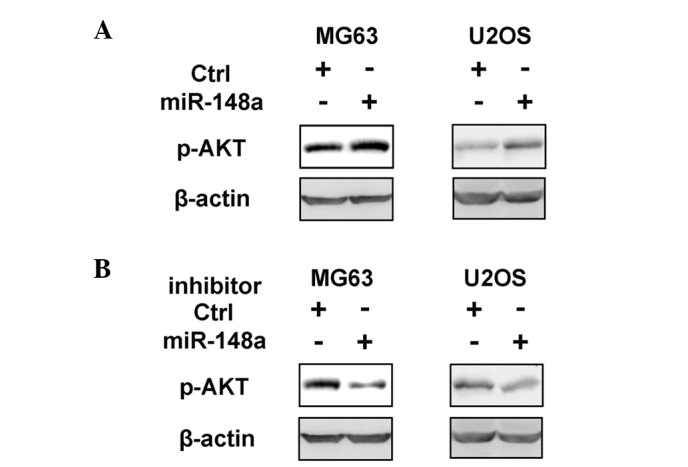
miR-148a promotes the activation of the phosphoinositide 3-kinase signaling pathway in osteosarcoma. MG63 and U2OS cells were transfected with (A) miR-148a mimics or (B) miR-148a inhibitor. The phosphorylation of AKT at Ser 473 was examined by western blotting. Data correspond to one representative experiment. Similar results were obtained in three independent experiments (P<0.01). miR, microRNA; Ctrl, control; p-, phosphorylated.
Since miRNAs mainly function through post-transcriptional inhibition of their target mRNAs, the target genes of miR-148a, which may be responsible for the activated PI3K signaling pathway in osteosarcoma cells, were next investigated. The predicted target genes of miR-148a were retrieved from TargetScan (http://www.targetscan.org), and subjected to the PI3K signaling pathway analysis of the Kyoto Encyclopedia of Genes and Genomes pathway database (http://www.genome.jp/kegg/). Notably, PTEN, an important PI3K inhibitor, was observed to contain two putative target sites in the 3′-UTR sequence of its messenger (m)RNA (Fig. 5A). To verify the potential interaction between miR-148a and PTEN, a dual-luciferase reporter system was performed by co-transfection of miR-148a and luciferase reporter plasmids containing the 3′-UTR sequence of human PTEN, or bearing deletions of the putative miR-148a target sites. As represented in Fig. 5B, miR-148a co-transfection with the above plasmids inhibited the luciferase activity of wild-type PTEN 3′-UTR reporter, but failed to inhibit that of the reporter containing deletion of both target sites. These results indicate that miR-148a is able to target directly PTEN mRNA 3′-UTR sequences. Furthermore, PTEN expression was suppressed by miR-148a overexpression, while it was enhanced by miR-148a inhibition in MG63 and U2OS cells (Fig. 5C). Thus, these data further demonstrate that PTEN expression is directly targeted by miR-148a, which may in turn promote the activation of the PI3K signaling pathway in osteosarcoma.
Figure 5.
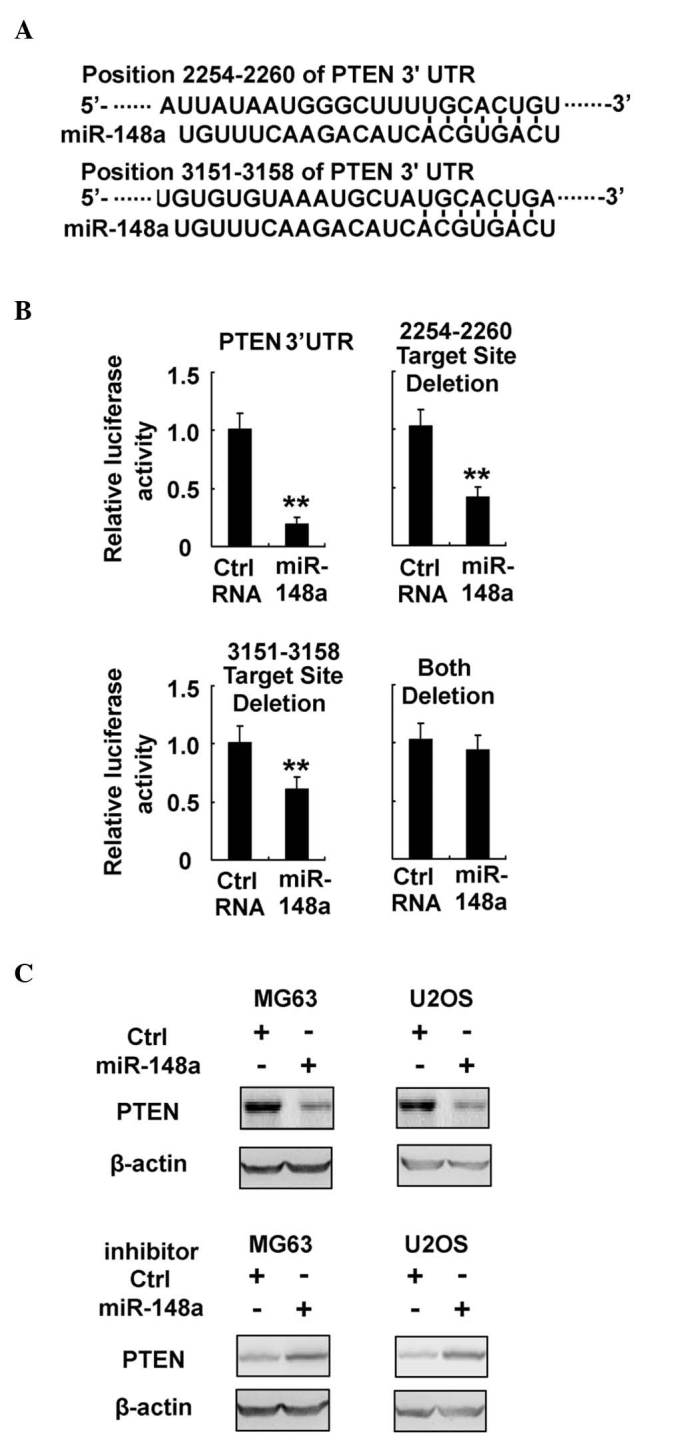
miR-148a directly targets PTEN expression in osteosarcoma. (A) Sequence alignment of miR-148a and its predictive target sites in the 3′-UTR region of PTEN. (B) MG63 cells were co-transfected with PTEN 3′-UTR firefly luciferase reporter plasmid, or with a reporter construct whereby the miR-148a target sites had been deleted, together with Renilla luciferase plasmids, control RNA or miR-148a mimics, as indicated. Following 48 h, luciferase activity was measured and normalized. (C) MG63 and U2OS cells were transfected with miR-148a mimics or miR-148a inhibitor, as indicated. Protein expression of PTEN and β-actin (which served as an internal control) were detected by western blotting. Data are presented as the mean ± standard deviation (n=4) of one representative experiment. Similar results were obtained in three independent experiments. **P<0.01. miR, microRNA; Ctrl, control; UTR, untranslated region; PTEN, phosphatase and tensin homolog.
PTEN expression was further compared in osteosarcoma MG63 and U2OS cells vs. normal osteoblastic hFOB 1.19 cells, as increased miR-148a expression was identified in osteosarcoma. As represented in Fig. 6A, PTEN expression was inhibited in osteosarcoma cells. Additionally, the correlation between miR-148a and PTEN expression in human osteosarcoma tissues was examined by RT-qPCR and western blot analysis. PTEN expression inversely correlated with miR-148a expression in osteosarcoma tissues by Pearson's correlation coefficient (Fig. 6B). Thus, these results further suggest that increased miR-148a expression may inhibit PTEN expression, which in turn promotes PI3K signaling pathway activation in osteosarcoma.
Figure 6.
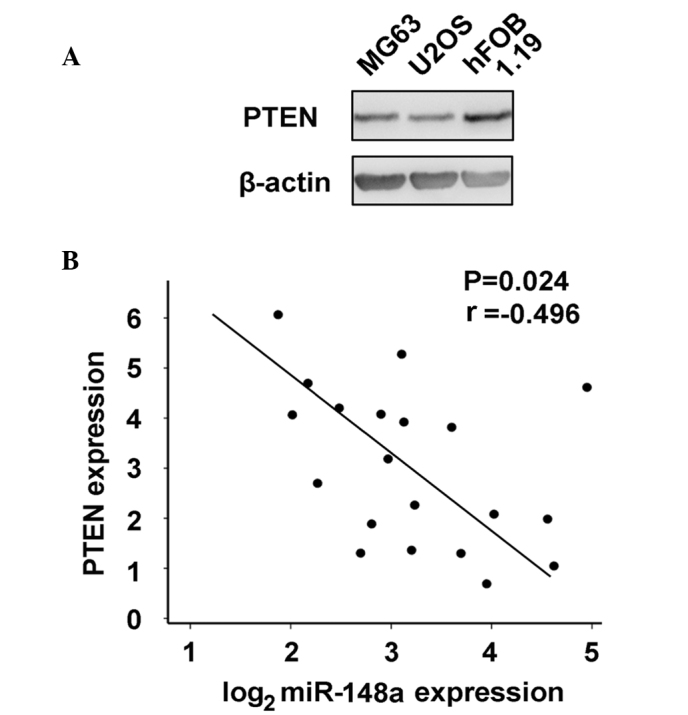
Correlation between the expression of miR-148a and the protein levels of PTEN in osteosarcoma. (A) PTEN expression in osteoblastic hFOB 1.19 cells vs. osteosarcoma MG63 and U2OS cells was detected by western blotting. (B) Correlation between PTEN and miR-148a expression levels in osteosarcoma tissues was statistically analyzed by Pearson's correlation coefficient. PTEN and miR-148a expression levels are presented as standardized values. The r and P-values obtained are indicated in the image. miR, microRNA; PTEN, phosphatase and tensin homolog.
Discussion
Osteosarcoma is characterized by an aggressive clinical course, and is the most common primary malignant bone tumor (5–7). Recently, molecular mechanisms responsible for osteosarcoma carcinogenesis and progression have attracted much attention in osteosarcoma development research, and deregulated miRNAs have been identified to be important in osteosarcoma pathogenesis (8–13). In the present study, miR-148a expression was observed to be significantly upregulated in osteosarcoma, and miR-148a could promote osteosarcoma growth by inhibiting PTEN expression, thus promoting PI3K signaling pathway activation. Furthermore, miR-148a upregulation in osteosarcoma appeared to be correlated with osteosarcoma progression and prognosis, and inhibiting miR-148a using antago-miR in vivo could suppress osteosarcoma growth. Thus, the present results suggest that miR-148a may be a novel prognosis predictor and therapeutic target in osteosarcoma.
Previously, miR-148a expression was suggested to be deregulated in other types of cancer, including downregulated in gastric cancer, hepatocellular carcinoma and pancreatic cancer, and upregulated in glioblastoma and chordomas (20–24). The present study has determined that miR-148a is an onco-miR, and its expression is upregulated in osteosarcoma. In combination with previous reports of miR-148a in cancer biology, it is possible to assume that miR-148a may play different roles in different types of cancer, and the target genes and corresponding mechanisms are also likely to be different depending on the type of cancer. However, the roles and underlying mechanism of miR-148a deregulation in carcinogenesis and tumor progression require to be further investigated in different types of cancer.
Recently, the identification of molecular biomarkers correlating with progression and/or predicting prognosis of cancer patients has attracted considerable attention (3). Previously, the present authors reported that downregulation of miR-133a expression in osteosarcoma is correlated with cancer stages and prognosis of osteosarcoma patients (13). In the present study, increased miR-148a expression in osteosarcoma was also demonstrated to be correlated with cancer stages and able to predict prognosis. Therefore, the combined detection of deregulated miRNAs, including miR-148a, miR-133a or other miRNAs and coding genes, may be valuable to identify pathological stages and predict prognosis of osteosarcoma more accurately than the detection of single miRNA.
In conclusion, the present study has validated both in vitro and in vivo that miR-148a promotes osteosarcoma growth, while inhibition of miR-148a could suppress tumor progression. In particular, cholesterol-conjugated RNA antago-miR of miR-148a could significantly inhibit osteosarcoma growth in vivo. These data strongly suggest that inhibition of miR-148a bears considerable therapeutic potential for the treatment of osteosarcoma, as the enforced expression or inhibition of miRNAs in vivo has been demonstrated to be promising in cancer therapy (25). Therefore, future studies on in vivo administration of miRNA modulators in cancer therapy may lead to important advances on cancer treatment, particularly for those patients who respond poorly to radiotherapy or chemotherapy.
Acknowledgements
The authors would like to thank Professor Zhiwei Wang and Dr Yue Wang for their helpful discussion, and Ms. Jianfang Chen and Liqing Fu for their excellent technical assistance. The present study was supported by grants from the National Natural Science Foundation of China (Beijing, China; grant nos. 30973019, 81272942 and 81202122), the Key Biomedicine Research Program of the Science and Technology Commission of Shanghai (Shanghai, China; grant no. 10411956000) and the Shanghai Natural Science Foundation (Shanghai, China; grant no. 064119605).
References
- 1.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 3.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi E, Hornicek FJ, Duan Z. MicroRNA involvement in osteosarcoma. Sarcoma. 2012;2012:359739. doi: 10.1155/2012/359739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longhi A, Errani C, De Paolis M, Mercuri M, Bacci G. Primary bone osteosarcoma in the pediatric age: State of the art. Cancer Treat Rev. 2006;32:423–436. doi: 10.1016/j.ctrv.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Provisor AJ, Ettinger LJ, Nachman JB, Krailo MD, Makley JT, Yunis EJ, Huvos AG, Betcher DL, Baum ES, Kisker CT, Miser JS. Treatment of nonmetastatic osteosarcoma of the extremity with preoperative and postoperative chemotherapy: A report from the Children's Cancer Group. J Clin Oncol. 1997;15:76–84. doi: 10.1200/JCO.1997.15.1.76. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson WS, Goorin AM. Current treatment of osteosarcoma. Cancer Invest. 2001;19:292–315. doi: 10.1081/CNV-100102557. [DOI] [PubMed] [Google Scholar]
- 8.Luo XJ, Tang DG, Gao TL, Zhang YL, Wang M, Quan ZX, Chen J. MicroRNA-212 inhibits osteosarcoma cells proliferation and invasion by down-regulation of Sox4. Cell Physiol Biochem. 2014;34:2180–2188. doi: 10.1159/000369661. [DOI] [PubMed] [Google Scholar]
- 9.Xu N, Li Z, Yu Z, Yan F, Liu Y, Lu X, Yang W. MicroRNA-33b suppresses migration and invasion by targeting c-Myc in osteosarcoma cells. PLoS One. 2014;9:e115300. doi: 10.1371/journal.pone.0115300. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Chong Y, Zhang J, Guo X, Li G, Zhang S, Li C, Jiao Z, Shao M. MicroRNA-503 acts as a tumor suppressor in osteosarcoma by targeting L1CAM. PLoS One. 2014;9:e114585. doi: 10.1371/journal.pone.0114585. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Yuan J, Lang J, Liu C, Zhou K, Chen L, Liu Y. The expression and function of miRNA-451 in osteosarcoma. Med Oncol. 2015;32:324. doi: 10.1007/s12032-014-0324-x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Cai X, Wang Y, Tang H, Tong D, Ji F. microRNA-143, down-regulated in osteosarcoma, promotes apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol Rep. 2010;24:1363–1369. doi: 10.3892/or_00000994. [DOI] [PubMed] [Google Scholar]
- 13.Ji F, Zhang H, Wang Y, Li M, Xu W, Kang Y, Wang Z, Wang Z, Cheng P, Tong D, et al. MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 2013;56:220–226. doi: 10.1016/j.bone.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Creighton CJ, Fountain MD, Yu Z, Nagaraja AK, Zhu H, Khan M, Olokpa E, Zariff A, Gunaratne PH, Matzuk MM, Anderson ML. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906–1915. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai H, Lin L, Cai H, Tang M, Wang Z. Prognostic evaluation of microRNA-210 expression in pediatric osteosarcoma. Med Oncol. 2013;30:499. doi: 10.1007/s12032-013-0499-6. [DOI] [PubMed] [Google Scholar]
- 16.Bao YP, Yi Y, Peng LL, Fang J, Liu KB, Li WZ, Luo HS. Roles of microRNA-206 in osteosarcoma pathogenesis and progression. Asian Pac J Cancer Prev. 2013;14:3751–3755. doi: 10.7314/APJCP.2013.14.6.3751. [DOI] [PubMed] [Google Scholar]
- 17.Novello C, Pazzaglia L, Cingolani C, Conti A, Quattrini I, Manara MC, Tognon M, Picci P, Benassi MS. miRNA expression profile in human osteosarcoma: Role of miR-1 and miR-133b in proliferation and cell cycle control. Int J Oncol. 2013;42:667–675. doi: 10.3892/ijo.2012.1717. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Zhu LB, Peng AF, Wang TF, Long XH, Gao S, Zhou RP, Liu ZL. LY294002 inhibits the malignant phenotype of osteosarcoma cells by modulating the phosphatidylinositol 3 kinase/Akt/fatty acid synthase signaling pathway in vitro. Mol Med Rep. 2015;11:1352–1357. doi: 10.3892/mmr.2014.2787. [DOI] [PubMed] [Google Scholar]
- 19.Jin S, Pang RP, Shen JN, Huang G, Wang J, Zhou JG. Grifolin induces apoptosis via inhibition of PI3K/AKT signalling pathway in human osteosarcoma cells. Apoptosis. 2007;12:1317–1326. doi: 10.1007/s10495-007-0062-z. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing C, Liu Z. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg. 2010;14:1170–1179. doi: 10.1007/s11605-010-1202-2. [DOI] [PubMed] [Google Scholar]
- 21.Liffers ST, Munding JB, Vogt M, Kuhlmann JD, Verdoodt B, Nambiar S, Maghnouj A, Mirmohammadsadegh A, Hahn SA, Tannapfel A. MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B. Lab Invest. 2011;91:1472–1479. doi: 10.1038/labinvest.2011.99. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng X, Zhu Z, Jiao H, Lin J, Jiang K, et al. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest. 2013;123:630–645. doi: 10.1172/JCI64265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Zhang Y, Skalski M, Hayes J, Kefas B, Schiff D, Purow B, Parsons S, Lawler S, Abounader R. microRNA-148a is a prognostic oncomiR that targets MIG6 and BIM to regulate EGFR and apoptosis in glioblastoma. Cancer Res. 2014;74:1541–1553. doi: 10.1158/0008-5472.CAN-13-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayrak OF, Gulluoglu S, Aydemir E, Ture U, Acar H, Atalay B, Demir Z, Sevli S, Creighton CJ, Ittmann M, et al. MicroRNA expression profiling reveals the potential function of microRNA-31 in chordomas. J Neurooncol. 2013;115:143–151. doi: 10.1007/s11060-013-1211-6. [DOI] [PubMed] [Google Scholar]
- 25.Mishra PJ, Merlino G. MicroRNA reexpression as differentiation therapy in cancer. J Clin Invest. 2009;119:2119–2123. doi: 10.1172/JCI40107. [DOI] [PMC free article] [PubMed] [Google Scholar]