Abstract
Colorectal cancer (CRC) is one of the most prevalent malignancies worldwide and remains the third leading cause of cancer-associated mortality. The present study aimed to fully elucidate the pathogenesis of CRC and identify associated genes in tumor development. Microarray GSE44076, GSE41328 and GSE44861 datasets were downloaded from the Gene Expression Omnibus database and integrated with meta-analysis. Differentially-expressed genes (DEGs) were identified from CRC samples compared with adjacent non-cancerous controls using the Limma package in R, followed by functional analysis using the Database for Annotation, Visualization, and Integrated Discovery online tool. A protein-protein interaction (PPI) network of DEGs and linker genes was constructed using NetBox software and modules were also mined. Functional annotation was performed for modules with a maximum number of nodes. Subsequent to meta-analysis to pool the data, one dataset that included 327 samples involved in 11,081 genes was obtained. A total of 697 DEGs were identified between CRC samples and adjacent non-cancerous controls. In the PPI network, modules 1 and 5 contained the maximum number of nodes. Collagen, type I, α1 (COL1A1), COL1A2 and matrix metallopeptidase 9 (MMP9) in module 1 and UDP-glucose 6-dehydrogenase (UGDH), aldehyde dehydrogenase 1 family, member A1 (ALDH1A1), fatty acid binding protein 4 (FABP4) and monoglyceride lipase (MGLL) in module 5 exhibited a high degree of connectivity. Functional analysis indicated that the genes in module 1 were involved in extracellular matrix (ECM)-associated functions and that the genes in module 5 were involved in metabolism-related functions. Overall, significant DEGs and linker genes, namely COL1A1, COL1A2, MMP9, UGDH, ALDH1A1, FABP4 and MGLL, play a crucial role in the development of CRC via regulating the ECM and cell metabolism.
Keywords: colorectal cancer, linker gene, protein-protein interaction, module
Introduction
Colorectal cancer (CRC), also known as colon cancer, is one of the most prevalent malignancies worldwide and remains the third leading cause of cancer-associated mortality (1). In 2014, ~65,000 women and 71,830 men were estimated to be diagnosed with CRC (2).
Similar to the majority of other complex tumors, CRC has been the subject of multiple studies with regards to its pathogenesis, diagnosis and therapy. Much has been elucidated about the molecular mechanism of CRC in recent years. It is widely recognized that chromosomal instability is the most common genetic abnormality to occur in CRC and has been found in almost 85% of all CRC cases (3). Key genes involved in this pathway include Kirsten rat sarcoma viral oncogene homolog (KRAS), deleted in colorectal carcinoma (DCC), SMAD family member 2 (SMAD2) and SMAD4. KRAS is a proto-oncogene that plays a critical role in the transduction of intracellular signals. The activation of KRAS by binding to guanosine triphosphate could regulate downstream mediator mitogen-activated protein kinase, which is involved in cell division (4). Additionally, SMAD2 and SMAD4 play a vital role in the transforming growth factor-β signaling pathway, which in involved in the regulation of cell proliferation, differentiation and apoptosis (5). DCC has been shown to correlate with metastasis and a poor prognosis in CRC (6). Moreover, chronic inflammation has been shown to increase the incidence of bowel cancer (7). In the process of inflammation, cyclooxygenase-2 (COX2) is a key molecule highlighted by experiments (8). Previous studies into the downstream effects of COX have illustrated that basic fibroblast growth factor and vascular endothelial growth factor are activated by COX2 via prostaglandin E2, all of which are involved in the regulation of cell proliferation and angiogenesis contributing to tumor development (9–11). However, the pathogenesis of CRC is complex and multifactorial. The complete elucidation of its etiology remains to be defined.
The present study analyzed three microarray datasets, comparing between colon tumor samples and adjacent normal mucosa tissue samples. Differentially-expressed genes (DEGs) were identified and functional annotation was performed for significant genes, followed by protein-protein interaction (PPI) network construction. The study aimed to detect the molecular mechanisms and associated genes in the development of CRC.
Materials and methods
Microarray data
A total of 3 microarray datasets were downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/), including GSE44076 (12), GSE41328 (13) and GSE44861 (14). Expression data from GSE44076, which included colon tumor samples from 98 patients and adjacent paired normal mucosa tissues from 50 healthy donors, were obtained using platform GPL13667 (Affymetrix Human Genome U219 Arrays). Microarray data from GSE41328, which included 5 colorectal adenocarcinomas samples and 5 matched normal colon tissue, were generated with the [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array platform. Expression data from GSE44861, which included 56 tumor samples and 55 adjacent non-cancerous tissue samples, were obtained through the [HT_HG-U133A] Affymetrix HT Human Genome U133A Array platform.
Data preprocessing
Prior to analysis, probe identifications in each dataset were converted into standard gene symbols. For genes with more than one probe set in the array, the average value for the probes was obtained as the expression value of the gene. By contrast, the probe set was deleted when mapped to more than one gene. As the genes were different in the 3 datasets, meta-analysis was performed of these studies, pooling the microarray data across different platforms. In the combined process, batch effects are inevitable. To adjust the data for these batch effects, the surrogate variable analysis package (15) was applied, and normalization was performed using the preprocessCore package (16) in R.
Identification of DEGs in CRC
To identify significant DEGs in colon tumor samples compared with adjacent non-cancerous controls, preprocessed data were exported to Limma package in R language (17). An adjusted P-value was estimated using the Benjamini & Hochberg (BH) method (18). Significant DEGs were identified as those with |log 2 FC (fold-change)|>1 and an adjusted P-value of <0.05.
Functional annotation of DEGs in CRC
Testing for functional enrichment of DEGs in CRC was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) online tool (19). Categories analyzed included Gene Ontology (GO) terms (20) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (21). Data from the GO annotations was used to construct a functional enrichment network, which was visualized by the enrichment map plugin in Cytoscape (22). The BH correction for multiple testing was performed with a cutoff for an adjusted P-value of <0.05.
PPI network construction
NetBox software, which is written in the Java language, is used to store and establishment the Human Interaction Network based on public databases consisting of Reactome (23,24), the Human Protein Reference Database (25), Memorial Sloan-Kettering Cancer Center Cancer Cell Map (26) and the National Cancer Institute-Nature Pathway Interaction Database (27). Linker genes with statistical significance, which are not differentially-expressed in colon tumors, but interact with DEGs, were obtained through mapping DEGs onto the network. Cytoscape software (28) was used to visualize the molecular interaction. Besides the PPI network under the criteria, NetBox also divided the network into modules. The modules with the maximum number of nodes in the PPI network were subjected to GO terms and Swiss-Prot and Protein Information Resource Keywords enrichment analysis with the DAVID online tool.
Results
Preprocessed results and DEGs in CRC
Following meta-analysis of these 3 studies to pool microarray data across the different platforms, one dataset was obtained that included 327 samples and 11,081 genes. The dataset was preprocessed and normalized, followed by further analysis. The normalized results are shown in Fig. 1. A total of 697 genes were selected as DEGs, including 286 upregulated and 411 downregulated genes, between CRC samples and adjacent non-cancerous control.
Figure 1.
Results of normalization. The upper image represents data prior to normalization, the middle image represents the data adjusted for the batch effect and the image below represents data after normalization.
Significant functions and pathways of DEGs
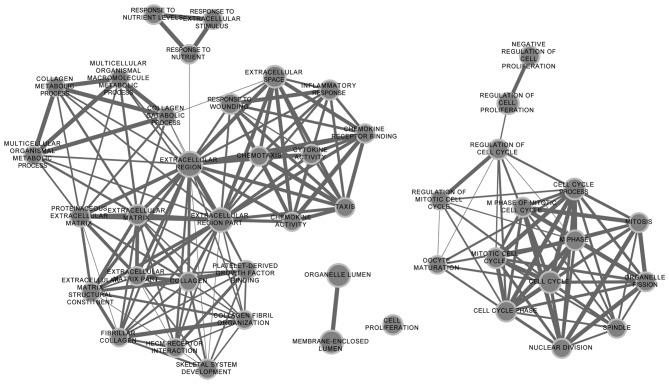
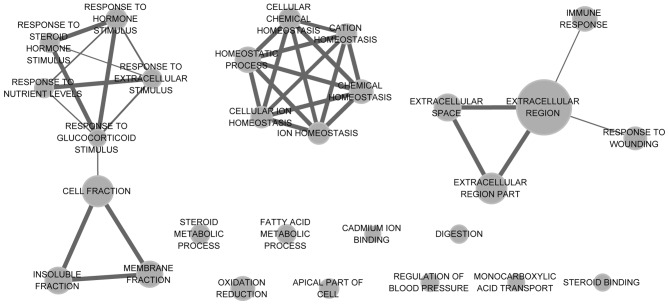
To annotate these DEGs in the tumor samples, DAVID was used for GO function and KEGG pathway analysis, with the threshold of the adjusted P-value at <0.05. Functional enrichment networks of upregulated and downregulated DEGs are shown in Figs. 2 and 3. The results showed that upregulated DEGs were significantly enriched in cell cycle-related functions, including the cell cycle process, the regulation of the mitotic cell cycle and the regulation of cell proliferation. Downregulated DEGs were mainly enriched in homeostasis-related functions, including chemical homeostasis and cellular ion homeostasis.
Figure 2.
Functional enrichment map of upregulated differentially-expressed genes (DEGs). The nodes represent the enriched functions of the upregulated genes. Edge thickness is proportional to the number of overlapped genes between the different functions. The size of the nodes is proportional to the number of DEGs.
Figure 3.
Functional enrichment map of downregulated differentially-expressed genes (DEGs). The nodes represent the enriched functions of the downregulated genes. Edge thickness is proportional to the number of overlapped genes between the different functions. The size of the nodes is proportional to the number of DEGs.
PPI network analysis
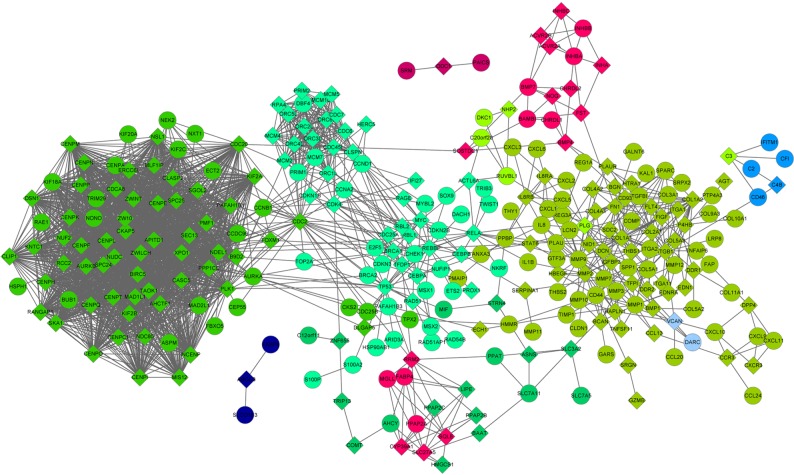
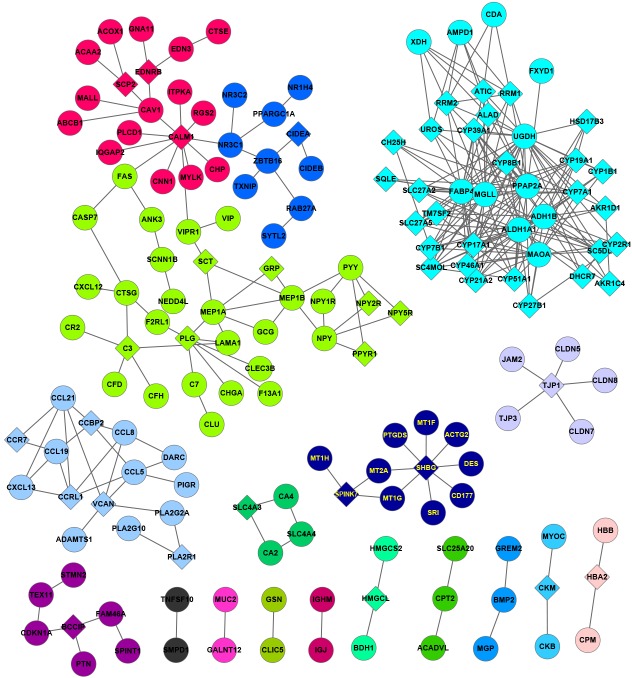
Significant DEGs and linker genes were used to construct the PPI network (Figs. 4 and 5). In the PPI network for upregulated DEGs (Fig. 4), there were 2,508 edges and 296 genes, including 140 DEGs and 156 linker genes. The network was divided into 9 modules by NetBox, in which module 1 contained the maximum number of nodes. Additionally, in the PPI network for downregulated DEGs, there were 301 edges and 165 genes, including 113 DEGs and 42 linker genes. The network was divided into 18 modules, in which module 5 contained the maximum number of nodes. In the PPI network, the hub genes were mined with the top-five degrees of connectivity in the different modules (Table I). The upregulated minichromosome maintenance complex component 7 gene in module 0, linker genes collagen, type I, α1 (COL1A1) and COL1A2, and differentially-expressed matrix metallopeptidase 9 (MMP9) in module 1, and linker genes polo-like kinase 1 and exportin 1 in module 2 exhibited a connectivity degree of >20. Downregulated genes UDP-glucose 6-dehydrogenase (UGDH), aldehyde dehydrogenase 1 family, member A1 (ALDH1A1), fatty acid binding protein 4, adipocyte (FABP4) and monoglyceride lipase (MGLL) in module 5 exhibited a connectivity degree of >20.
Figure 4.
Protein-protein interaction network of up-regulated genes and linker genes. The coloration of the nodes is representative of the genes in different modules. Circular nodes represent differentially-expressed genes and rhombic nodes represent linker genes.
Figure 5.
Protein-protein interaction network of downregulated genes and linker genes. The coloration of the nodes is representative of the genes in different modules. Circular nodes represent differentially-expressed genes and rhombic nodes represent linker genes.
Table I.
Connectivity degree of hub genes in the top-five modules.
| Module no. | Hub gene | Degree |
|---|---|---|
| Upregulated gene modules | ||
| 0 | MCM7 | 22 |
| 0 | TP53 | 21 |
| 0 | ORC1L | 21 |
| 0 | ORC4L | 20 |
| 0 | CDC45L | 20 |
| 1 | COL1A1 | 25 |
| 1 | COL1A2 | 20 |
| 1 | MMP9 | 20 |
| 1 | FN1 | 19 |
| 1 | ITGB1 | 18 |
| 2 | PLK1 | 71 |
| 2 | XPO1 | 68 |
| 2 | CDC20 | 66 |
| 2 | BIRC5 | 64 |
| 2 | PAFAH1B1 | 63 |
| 4 | RRM2 | 11 |
| 4 | SLC27A5 | 9 |
| 4 | CYP39A1 | 8 |
| 4 | SQLE | 7 |
| 4 | LIPE | 6 |
| 5 | BMP7 | 7 |
| 5 | INHBA | 7 |
| 5 | BMP4 | 6 |
| 5 | INHBB | 5 |
| 5 | BAMBI | 5 |
| Downregulated gene modules | ||
| 5 | UGDH | 26 |
| 5 | ALDH1A1 | 22 |
| 5 | FABP4 | 21 |
| 5 | MGLL | 21 |
| 5 | PPAP2A | 21 |
| 6 | PLG | 7 |
| 6 | MEP1A | 6 |
| 6 | MEP1B | 6 |
| 6 | C3 | 5 |
| 6 | NPY | 5 |
| 8 | SHBG | 8 |
| 8 | SPINK7 | 3 |
| 8 | MT1G | 2 |
| 8 | MT2A | 2 |
| 8 | MT1F | 1 |
| 9 | CCRL1 | 5 |
| 9 | VCAN | 5 |
| 9 | CCL5 | 5 |
| 9 | CCL21 | 5 |
| 9 | CCL19 | 4 |
| 12 | CALM1 | 11 |
| 12 | CAV1 | 5 |
| 12 | SCP2 | 3 |
| 12 | EDNRB | 3 |
| 12 | EDN3 | 2 |
The functional annotation results showed that the DEGs in module 1 were mainly enriched in extracellular region-related functions and extracellular matrix (ECM)-associated functions (Table II). Downregulated DEGs in module 5 were significantly enriched in metabolic process and biosynthetic process-related functions (Table III).
Table II.
Functional annotation of genes in module 1.
| Category | Term | Count | Bonferroni |
|---|---|---|---|
| Annotation cluster 1 | Enrichment score: 42.348195504843254 | ||
| GOTERM_CC_FAT | GO:0044421~extracellular region part | 63 | 2.07×10−45 |
| SP_PIR_KEYWORDS | Secreted | 65 | 8.47×10−42 |
| GOTERM_CC_FAT | GO:0005576~extracellular region | 71 | 2.53×10−35 |
| Annotation Cluster 2 | Enrichment score: 41.212870504723476 | ||
| SP_PIR_KEYWORDS | Signal | 80 | 6.46×10−42 |
| UP_SEQ_FEATURE | Signal peptide | 80 | 2.31×10−41 |
| GOTERM_CC_FAT | GO:0005576~extracellular region | 71 | 2.53×10−35 |
| Annotation cluster 3 | Enrichment score: 26.15393329757373 | ||
| SP_PIR_KEYWORDS | Extracellular matrix | 32 | 3.45×10−33 |
| GOTERM_CC_FAT | GO:0031012~extracellular matrix | 35 | 8.66×10−28 |
| GOTERM_CC_FAT | GO:0005578~proteinaceous extracellular matrix | 33 | 3.76×10−26 |
| GOTERM_CC_FAT | GO:0044420~extracellular matrix part | 15 | 1.57×10−11 |
GO, Gene Ontology; SP_PIR_KEYWORDS, Swiss-Prot and Protein Information Resource Keywords; CC, cellular component.
Table III.
Functional annotation of genes in module 5.
| Category | Term | Count | Benjamini |
|---|---|---|---|
| Annotation cluster 1 | Enrichment score: 28.81018243601853 | ||
| SP_PIR_KEYWORDS | Oxidoreductase | 28 | 1.58×10−31 |
| GOTERM_BP_FAT | GO:0055114~oxidation reduction | 28 | 2.57×10−25 |
| GOTERM_BP_FAT | GO:0008202~steroid metabolic process | 21 | 5.77×10−25 |
| Annotation cluster 2 | Enrichment score: 17.61044037865203 | ||
| GOTERM_BP_FAT | GO:0008202~steroid metabolic process | 21 | 5.77×10−25 |
| SP_PIR_KEYWORDS | Nadp | 14 | 3.94×10−17 |
| GOTERM_BP_FAT | GO:0016125~sterol metabolic process | 12 | 2.20×10−13 |
| GOTERM_BP_FAT | GO:0008203~cholesterol metabolic process | 10 | 1.82×10−10 |
| Annotation cluster 3 | Enrichment score: 10.153502326316204 | ||
| GOTERM_BP_FAT | GO:0006694~steroid biosynthetic process | 15 | 5.19×10−20 |
| GOTERM_BP_FAT | GO:0016125~sterol metabolic process | 12 | 2.20×10−13 |
| GOTERM_BP_FAT | GO:0008610~lipid biosynthetic process | 15 | 6.44×10−12 |
| SP_PIR_KEYWORDS | Steroid biosynthesis | 8 | 1.29×10−11 |
| GOTERM_BP_FAT | GO:0016126~sterol biosynthetic process | 7 | 2.68×10−8 |
| KEGG_PATHWAY | hsa00100:steroid biosynthesis | 7 | 1.05×10−8 |
| SP_PIR_KEYWORDS | Sterol biosynthesis | 6 | 1.17×10−8 |
| SP_PIR_KEYWORDS | Lipid synthesis | 7 | 2.69×10−7 |
| SP_PIR_KEYWORDS | Cholesterol biosynthesis | 3 | 2.98×10−3 |
| GOTERM_BP_FAT | GO:0006695~cholesterol biosynthetic process | 3 | 4.89×10−2 |
GO, Gene Ontology; SP_PIR_KEYWORDS, Swiss-Prot and Protein Information Resource Keywords; BP, biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
Using a meta-analysis approach to group 3 microarray datasets, including GSE44076, GSE41328 and GSE44861, DEGs were identified in CRC mucosa compared with adjacent normal mucosa samples. The results suggested that there were 697 DEGs, including 286 upregulated genes. Functional annotation results showed that the upregulated DEGs were involved in cell cycle-related functions, in comparison with the downregulated DEGs, which were enriched in homeostasis-associated functions. In the PPI network, the linker genes COL1A1 and COL1A2, and the DEGs MMP9, UGDH, ALDH1A1, FABP4 and MGLL, which exhibited a connectivity degree of >20, participated in the development of CRC.
After the upregulated and downregulated networks were divided into multiple modules, modules 1 and 5 with the maximum number of nodes were subjected to functional annotation. In module 1, the linker genes COL1A1 and COL1A2, and the DEG MMP9 exhibited the highest degree of connectivity. COL1A1 and COL1A2, two type I collagen members, are major components of the ECM. Growing evidence has shown that the ECM plays a critical role in promoting epithelial-to-mesenchymal transition (EMT), which is associated with tumor invasion and metastasis (29). Additionally, EMT is indicated to confer tumor cell resistance to apoptosis and to promote the escape of tumor cells from the senescence process (30,31). Moreover, a pioneer study uncovered the fact that EMT has the capacity of endowing tumor cells with cancer stem cell-like characteristics, which could promote tumor development and chemoresistance (32). MMP9 (also known as gelatinase B), a member of the MMP family, has been proven to degrade various components of the ECM, including type I collagen (33). Notably, an elevated level of MMP9 has been found in CRC (34), which is consistent with the present analysis. Numerous studies have shown that MMP9 plays crucial roles in invasion, metastasis, cell proliferation and angiogenesis (35,36). Angiogenesis and cell proliferation are critically important for tumor development and metastatic spreading (37). From the results of the functional annotation in the present study, the three important genes in module 1 were mainly enriched in ECM-related functions and the ECM-receptor interaction pathway. Accordingly, COL1A1, COL1A2 and MMP9 are involved in CRC tumorigenesis and metastasis via regulation of ECM-associated functions.
In module 5, UGDH, ALDH1A1, FABP4 and MGLL were downregulated in colorectal tumor samples and were significantly involved in metabolism-related functions. UGDH is the four-electron transfer enzyme and is associated with the biosynthesis of hyaluronan (HA), which participants in tissue organization, development and cell proliferation (38). A previous study showed that elevated levels of HA are directly involved in the progression of various cancers, and UGDH has been proposed as a biomarker for prostate cancer (39). In parallel, ALDH1A1, which belongs to a superfamily of enzymes, has been identified as a crystalline in the lens and cornea (40). Notably, ALDH1A1 also plays a critical role in regulating lipid metabolism and gluconeogenesis (41). In addition, FABP4, known as a new adipokine, is involved in fatty acid trafficking from the cytoplasm to the nucleus and in lipid metabolism (42). Furthermore, FABP4 is considered as a candidate biomarker of lipodystrophy and metabolic syndrome (43). It is well known that MGLL is a member of the serine hydrolase superfamily, which hydrolyze intracellular triglyceride and cholesteryl ester into free fatty acid as an important fuel in mammals (44). More recently, MGLL was found to be abnormally expressed in aggressive human cancer, and to promote cell proliferation and tumor growth (45). It is now clear that the conversion of cells from a normal to cancerous state requires metabolic alterations, including changes in lipid metabolism and gluconeogenesis, in order to support tumor growth and survival. As a result, UGDH, ALDH1A1, FABP4 and MGLL play a key role in metabolism-related functions and regulate the tumorigenesis of CRC.
Taken together, the present results suggest that COL1A1, COL1A2 and MMP9 in module 1, and UGDH, ALDH1A1, FABP4 and MGLL in module 5 serve as key hub genes in CRC development, where the genes regulate ECM and cell metabolism-associated functions that are important for tumor growth. However, additional experiments will be required to confirm the bioinformatic results.
References
- 1.Kemp Z, Thirlwell C, Sieber O, Silver A, Tomlinson I. An update on the genetics of colorectal cancer. Hum Mol Genet. 2004;13:R177–R185. doi: 10.1093/hmg/ddh247. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, DeSantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 3.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 5.Bellam N, Pasche B. Tgf-beta signaling alterations and colon cancer. Cancer Treat Res. 2010;155:85–103. doi: 10.1007/978-1-4419-6033-7_5. [DOI] [PubMed] [Google Scholar]
- 6.Chang SC, Lin JK, Yang SH, Wang HS, Li AF, Chi CW. Relationship between genetic alterations and prognosis in sporadic colorectal cancer. Int J Cancer. 2006;118:1721–1727. doi: 10.1002/ijc.21563. [DOI] [PubMed] [Google Scholar]
- 7.Harpaz N, Polydorides AD. Colorectal dysplasia in chronic inflammatory bowel disease: Pathology, clinical implications and pathogenesis. Arch Pathol Lab Med. 2010;134:876–895. doi: 10.5858/134.6.876. [DOI] [PubMed] [Google Scholar]
- 8.Zisman TL, Rubin DT. Colorectal cancer and dysplasia in inflammatory bowel disease. World J Gastroenterol. 2008;14:2662–2669. doi: 10.3748/wjg.14.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Dubois RN. Prostaglandins and cancer. Gut. 2006;55:115–122. doi: 10.1136/gut.2004.047100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisinger AL, Prescott SM, Jones DA, Stafforini DM. The role of cyclooxygenase-2 and prostaglandins in colon cancer. Prostaglandins Other Lipid Mediat. 2007;82:147–154. doi: 10.1016/j.prostaglandins.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 11.Doherty GA, Byrne SM, Molloy ES, Malhotra V, Austin SC, Kay EW, Murray FE, Fitzgerald DJ. Proneoplastic effects of PGE2 mediated by EP4 receptor in colorectal cancer. BMC Cancer. 2009;9:207. doi: 10.1186/1471-2407-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanz-Pamplona R, Berenguer A, Cordero D, Molleví DG, Crous-Bou M, Sole X, Paré-Brunet L, Guino E, Salazar R, Santos C, et al. Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer. Mol Cancer. 2014;13:46. doi: 10.1186/1476-4598-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin G, He X, Ji H, Shi L, Davis RW, Zhong S. Reproducibility probability score-incorporating measurement variability across laboratories for gene selection. Nat Biotechnol. 2006;24:1476–1477. doi: 10.1038/nbt1206-1476. [DOI] [PubMed] [Google Scholar]
- 14.Ryan BM, Zanetti KA, Robles AI, Schetter AJ, Goodman J, Hayes RB, Huang WY, Gunter MJ, Yeager M, Burdette L, et al. Germline variation in NCF4, an innate immunity gene, is associated with an increased risk of colorectal cancer. Int J Cancer. 2014;134:1399–1407. doi: 10.1002/ijc.28457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leek JT, Johnson WE, Parker HS, Fertig EJ, Jaffe AE, Storey JD. Package ‘SVA’: Surrogate Variable Analysis. https://www.bioconductor.org/packages/devel/bioc/manuals/sva/man/sva.pdf R package version 3. 2013
- 16.Bolstad BM. Package ‘preprocessCore’: A collection of pre-processing functions. https://www.bioconductor.org/packages/devel/bioc/manuals/preprocessCore/man/preprocessCore.pdf R package version 1. 2013
- 17.Smyth GK. Bioinformatics and computational biology solutions using R and Bioconductor. Springer; NY: 2005. Limma: Linear models for microarray data; pp. 397–420. [DOI] [Google Scholar]
- 18.Ferreira JA. The Benjamini-Hochberg method in the case of discrete test statistics. Int J Biostat. 2007;3:11. doi: 10.2202/1557-4679.1065. [DOI] [PubMed] [Google Scholar]
- 19.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merico D, Isserlin R, Bader GD. Visualizing gene-set enrichment results using the Cytoscape plug-in enrichment map. Methods Mol Biol. 2011;781:257–277. doi: 10.1007/978-1-61779-276-2_12. [DOI] [PubMed] [Google Scholar]
- 23.Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, et al. Reactome: A knowledgebase of biological pathways. Nucleic Acid Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acid Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasad TK, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database-2009 update. Nucleic Acid Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somwar R, Erdjument-Bromage H, Larsson E, Shum D, Lockwood WW, Yang G, Sander C, Ouerfelli O, Tempst PJ, Djaballah H, Varmus HE. Superoxide dismutase 1 (SOD1) is a target for a small molecule identified in a screen for inhibitors of the growth of lung adenocarcinoma cell lines. Proc Natl Acad Sci USA. 2011;108:16375–16380. doi: 10.1073/pnas.1113554108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: The pathway interaction database. Nucleic Acid Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 29.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 30.Valdes F, Alvarez AM, Locascio A, Vega S, Herrera B, Fernández M, Benito M, Nieto MA, Fabregat I. The epithelial mesenchymal transition confers resistance to the apoptotic effects of transforming growth factor Beta in fetal rat hepatocytes. Mol Cancer Res. 2002;1:68–78. [PubMed] [Google Scholar]
- 31.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J Clin Oncol. 2009;27:5287–5297. doi: 10.1200/JCO.2009.23.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turpeenniemi-Hujanen T. Gelatinases (MMP-2 and −9) and their natural inhibitors as prognostic indicators in solid cancers. Biochimie. 2005;87:287–297. doi: 10.1016/j.biochi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Deryugina EI, Quigley JP. Pleiotropic roles of matrix metalloproteinases in tumor angiogenesis: Contrasting, overlapping and compensatory functions. Biochim Biophys Acta. 2010;1803:103–120. doi: 10.1016/j.bbamcr.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278:16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 37.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viola M, Vigetti D, Genasetti A, Rizzi M, Karousou E, Moretto P, Clerici M, Bartolini B, Pallotti F, De Luca G, Passi A. Molecular control of the hyaluronan biosynthesis. Connect Tissue Res. 2008;49:111–114. doi: 10.1080/03008200802148405. [DOI] [PubMed] [Google Scholar]
- 39.Huang D, Casale GP, Tian J, Lele SM, Pisarev VM, Simpson MA, Hemstreet GP., III Udp-glucose dehydrogenase as a novel field-specific candidate biomarker of prostate cancer. Int J Cancer. 2010;126:315–327. doi: 10.1002/ijc.24820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Koppaka V, Thompson DC, Vasiliou V. Focus on molecules: ALDH1A1: From lens and corneal crystallin to stem cell marker. Exp Eye Res. 2012;102:105–106. doi: 10.1016/j.exer.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiefer FW, Orasanu G, Nallamshetty S, Brown JD, Wang H, Luger P, Qi NR, Burant CF, Duester G, Plutzky J. Retinaldehyde dehydrogenase 1 coordinates hepatic gluconeogenesis and lipid metabolism. Endocrinology. 2012;153:3089–3099. doi: 10.1210/en.2011-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wootan MG, Bernlohr DA, Storch J. Mechanism of fluorescent fatty acid transfer from adipocyte fatty acid binding protein to membranes. Biochemistry. 1993;32:8622–8627. doi: 10.1021/bi00084a033. [DOI] [PubMed] [Google Scholar]
- 43.Karakas SE, Almario RU, Kim K. Serum fatty acid binding protein 4, free fatty acids, and metabolic risk markers. Metabolism. 2009;58:1002–1007. doi: 10.1016/j.metabol.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]