Abstract
Understanding the events at a protein level that govern the progression from melanoma in situ to invasive melanoma are important areas of current research to be developed. Recent advances in the analysis of formalin-fixed, paraffin-embedded tissue by proteomics, particularly using the filter-aided sample preparation protocol, has opened up the possibility of studying vast archives of clinical material and associated medical records. In the present study, quantitative protein profiling was performed using tandem mass spectrometry, and the proteome differences between melanoma in situ and invasive melanoma were compared. Biological pathway analyses revealed several signalling pathways differing between melanoma in situ and invasive melanoma, including metabolic pathways and the phosphoinositide 3-kinase-Akt signalling pathway. Selected proteins of interest (14–3-3ε and fatty acid synthase) were subsequently investigated using immunohistochemical analysis of tissue microarrays. Identifying the key proteins that play significant roles in the establishment of a more invasive phenotype in melanoma may ultimately aid diagnosis and treatment decisions.
Keywords: melanoma, proteomics, FFPE, mass spectrometry, FASN, 14-3-3ε
Introduction
During the past 30 years, the annual incidence of melanoma has increased by >60% in the USA (1). Globally, according to estimates for 2008, there were almost 200,000 novel cases of invasive cutaneous malignant melanoma, and an estimated 46,000 mortalities from the disease (2). Although melanoma accounts for only 4% of all cutaneous malignancies, it is responsible for 80% of mortalities from skin cancer (3). Malignant melanoma is one of the most aggressive malignancies, due to its effective capacity to grow, invade and metastasise (4). Despite recent successes of targeted therapies, there are limited treatment options for metastatic melanoma (5). It is therefore of high priority to increase the understanding of disease progression, to predict response to specific therapeutic regimes (6) and to identify novel therapeutic targets (7). Significant advances have been made in the area of proteomics over the last two decades, undoubtedly improving our understanding of human biology at the cellular level, and identifying novel diagnostic molecules and potential therapeutic targets, all of which contribute to the current personalized medicine era (8–10). Various techniques, including mass spectrometry (MS), liquid chromatography (LC), fractionation and multiplex assays, have contributed to build protein expression knowledge and global protein analysis (11).
Possibly one of the most recent advances has been the application of proteomics technologies to the analysis of formalin-fixed, paraffin-embedded (FFPE) tissues (12). Due to the stability of these tissues following fixation, numerous hospitals have acquired vast archives of FFPE tissue with associated information regarding patient diagnosis and outcome (12). In particular, the filter-aided sample preparation (FASP) method has been demonstrated to be better, both qualitatively and quantitatively, than alternative methods for processing FFPE tissue for proteomics analysis (13,14).
The goal of the present study was to gain a more in-depth understanding of the differences in protein expression levels between in situ and invasive melanoma FFPE tissue biopsies. For that purpose, a FASP-FFPE tissue preparation protocol, LC-tandem MS (MS/MS), label-free quantification and pathway mapping were utilized to explore specific signalling network modulation. The present findings may facilitate a more comprehensive understanding of the adaptive processes activated during the in situ-to-invasive transition of melanoma, thus facilitating the identification of novel therapeutic targets and biomarkers of aggressive disease.
Materials and methods
FFPE tissue samples
The present research protocol was approved by the St. James's Hospital Ethics Committee (Dublin, Ireland) and informed consent was obtained from all participating patients. Tissue specimens were processed using standard procedures for formalin fixation, and were embedded in paraffin as normally performed for routine clinical analysis (Table I) (15). Upon completion of the histopathological diagnosis, additional 10-µm-thick sections were placed on glass slides, and were compared with haematoxylin and eosin (H&E)-stained slides from the same block to identify histologically distinct (tumour-rich) tissue regions. Tissue blocks from 14 patients diagnosed with melanoma in situ or invasive melanoma were selected from the biobank of clinical material at St James's Hospital; the samples were originally obtained at biopsy between January and December 2013. Table I describes the clinical information of the patients included.
Table I.
Clinical data.
| A, Melanoma in situ | ||||
|---|---|---|---|---|
| No. naevi | Age, years | Gender | ||
| 100 | 44 | 1 | ||
| 20 | 48 | 0 | ||
| 100 | 60 | 0 | ||
| 10 | 74 | 1 | ||
| 0 | 77 | 0 | ||
| B, Invasive melanoma | ||||
| No. naevi | Age, years | Gender | Stage | Breslow depth, mm |
| 100 | 58 | 1 | 1a | 0.59 |
| 8 | 44 | 1 | 1a | 0.36 |
| 0 | 69 | 0 | 1a | 0.16 |
| 50 | 52 | 0 | 1a | 0.93 |
| 50 | 65 | 0 | 3 | 1.90 |
| 50 | 61 | 1 | 1a | 0.40 |
| 100 | 46 | 0 | 1b | 0.76 |
| 50 | 18 | 0 | 3 | 0.77 |
| 0 | 51 | 1 | 1b | 0.78 |
Clinical information regarding formalin-fixed, paraffin-embedded tissues from patients (0, male; 1, female) with in situ or invasive melanoma.
FASP
Melanoma in situ or invasive melanoma tissue was identified by comparing the sections to those stained with H&E. These specific areas from the tissue sections were removed from the glass slides with a scalpel and placed in microtubes. Paraffin was removed from the tissue slices by successive incubations in xylene (2X) and absolute ethanol (2X), and the samples were then rehydrated in a series of 97, 80, 70 and 50% ethanol solutions in water. Following vacuum-drying, 10 mg of dried tissue was weighed out and resuspended in 200 µl of UPX Universal Protein Extraction Buffer, which was provided in the FFPE-FASP Protein Digestion kit (Expedeon, Swavesey, UK), and protein concentrations of the FFPE-FASP-treated samples were determined using the RC DC Protein Assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Next, detergent was depleted from the lysates (50 µl), and proteins were digested with trypsin. Briefly, to YM-30 filter units (Merck Millipore, Carrigtwohill, Ireland) containing protein concentrates, 200 µl of 8 M urea in 0.1 M Tris/HCl (pH 8.5) was added, and the samples were centrifuged at 14,000 × g at 20°C for 15 min. This step was repeated once. Then, 50 µl of 0.05 M iodoacetamide in 8 M urea was added to the filters, and the samples were incubated in the darkness for 20 min. The filters were washed twice with 100 µl of 8 M UA, followed by two washes with 100 µl of 50 mM NH4HCO3. Finally, trypsin (Promega Corporation, Madison, WI, USA) was added in 40 µl of 50 mM NH4HCO3 to each filter at an enzyme:protein ratio of 1:50. The samples were incubated overnight at 37°C, and peptides were collected by centrifugation (14,000 × g, 10 min at 4°C), followed by an additional wash with an elution solution [70% acetonitrile (ACN) and 1% formic acid]. Finally, the peptide mixture was brought to dryness and reconstituted in sample buffer (2% ACN and 0.1% formic acid). To ensure an even suspension of peptide populations, samples were briefly vortexed and sonicated (16).
LC-MS/MS analysis
LC-MS/MS analysis of in situ vs. invasive FFPE tissue was performed using an UltiMate® 3000 Nano LC system (Dionex Corporation; Thermo Fisher Scientific, Inc., Waltham, MA, USA) coupled to a an LTQ Orbitrap XL™ mass spectrometre (Thermo Fisher Scientific, Inc.) in the Proteomics Facility of the National Institute for Cellular Biotechnology, Dublin City University (Dublin, Ireland). The optimized methodology has been previously described in detail (17). Peptide mixtures (5-µl volume) were loaded onto a C18 trap column [Acclaim™ PepMap™ 100 C18 LC column; 300 mm internal diameter (id) × 5 mm, 5 mm particle size, 100 Å pore size; Dionex Corporation; Thermo Fisher Scientific, Inc.). Desalting was achieved at a flow rate of 25 µl/min in 0.1% trifluoroacetic acid for 3 min. The trap column was switched online with an analytical Acclaim™ PepMap™ 100 C18 LC column (75 mm id × 500 mm, 3 mm particle size and 100 Å pore size; Dionex Corporation; Thermo Fisher Scientific, Inc.). The peptides generated from the digestion were eluted with the following binary gradients: Solvent A (2% ACN and 0.1% formic acid in LC-MS-grade water) and 0–25% solvent B (80% ACN and 0.08% formic acid in LC-MS-grade water) for 160 mins, followed by 25–50% solvent B for additional 20 min. The column flow rate was set to 350 nl/min. Data was acquired with Xcalibur™ software version 2.0.7 (Thermo Fisher Scientific, Inc.). The mass spectrometre was operated in data-dependent mode and externally calibrated. Survey MS scans were acquired in the LTQ Orbitrap XL™ in the 400–1,800 mass/charge (m/z) range, with the resolution set to a value of 30,000 at m/z=400 and the lock mass set to 445.120025 m/z. Collision-induced dissociation fragmentation was conducted in the linear ion trap with the three most intense ions per scan. Within 40 sec, a dynamic exclusion window was applied. A normalised collision energy of 35%, an isolation window of 2 m/z and one microscan were used to collect suitable MS/MS spectra.
Data analysis and quantitative profiling
Processing of the raw data generated from LC-MS/MS analysis was performed with Progenesis label-free LC-MS software version 3.1 (Nonlinear Dynamics, Newcastle upon Tyne, UK). Data alignment was based on the LC retention time of each sample (17). A reference sample was established, the retention times of all other replicates were aligned to this reference and peak intensities were then normalised. Prior to exporting the MS/MS output files to Mascot software (www.matrixscience.com) for protein identification, a number of criteria were employed to filter the data, including: i) Peptide features with analysis of variance (ANOVA) score of <0.05 between experimental groups, ii) mass peaks (features) with charge states +1, +2 and +3; and iii) data with >1 isotope/peptide. A Mascot generic file was generated from all the exported MS/MS spectra from the aforementioned Progenesis software, which was used for peptide identification with Mascot software version 2.2, and searched against the UniProtKB-SwissProt database (www.uniprot.org) with 16,638 proteins (taxonomy, Homo sapiens). The following search parameters were used for protein identification: i) MS/MS mass tolerance set at 0.5 Da; ii) peptide mass tolerance set to 20 ppm; iii) carbamidomethylation set as a fixed modification; iv) ≤2 missed cleavages were allowed; and v) methionine oxidation set as a variable modification. For further consideration and re-importation back into Progenesis label-free LC-MS software for further analysis, only peptides with ion scores ≥40 were selected. Importantly, the following criteria were applied to assign a plasma associated protein as properly identified: i) An ANOVA score between experimental groups of ≤0.05, ii) proteins with ≥2 peptides matched; and iii) a Mascot score of >40.
Immunohistochemistry (IHC)
Two proteins that were observed to be highly expressed in invasive melanoma vs. in situ melanoma were selected for validation by IHC analysis using commercially available melanoma cancer tissue microarrays (TMAs; ME208; US Biomax, Inc., Rockville, MD, USA), which comprised triplicate tissue cores from primary (n=30) and metastatic (n=30) melanoma. TMAs were stained for 14-3-3ε and fatty acid synthase (FASN) expression using specific anti-14-3-3ε (NBP1-32695; Novus Biologicals, Ltd., Cambridge, UK) and anti-FASN (C20G5; Cell Signalling Technology, Inc., Danvers, MA, USA) polyclonal antibodies. Deparaffinisation and antigen retrieval was performed using Dako Target Retrieval Solution, pH 6 (Dako, Glostrup, Denmark) and the PT Link system (Dako). Slides were heated to 97°C for 20 min and then cooled to 65°C. IHC analyses were performed using the Dako Autostainer Plus staining system according to the manufacturer's protocol. Dual Endogenous Enzyme Block (S2003; Dako) was applied to the TMAs to block endogenous peroxidase, pseudoperoxidase and alkaline phosphatase activities. The EnVision G|2 System/AP (K5355; Dako) was used for amplification and visualisation of bound anti-FASN and anti-14-3-3ε antibodies using an automated IHC staining apparatus (Autostainer Plus; Dako) according to the manufacturer's protocol. Briefly, primary antibodies were optimally diluted in REAL™ Antibody Diluent (14–3-3ε; 1:800; Dako) or SignalStain® Antibody Diluent (FASN; 1:50; Cell Signalling Technology, Inc.). Optimum primary antibody dilutions were predetermined using full-face sections of known positive-control tissues (invasive ductal breast carcinoma, FASN; and placenta, 14-3-3ε). Primary antibodies were applied for 30 min at room temperature, and then all the immunostained slides were counterstained with haematoxylin (Dako), rinsed in deionised water, washed in buffer and glass-mounted using Faramount mounting solution (Dako). Negative-control (where the primary antibody was omitted and replaced instead by antibody diluent) and appropriate positive-control slides were processed at the same time.
IHC scoring
Scores were assigned based on the staining intensity of 14-3-3ε and FASN. Stained cores were deemed negative (no staining), weakly positive, moderately positive or strongly positive. If a core was uninformative (lost during the procedure or very highly pigmented), the overall score was that of the remaining core(s). A consensus score was assigned where discordant scoring results (cores from same tumour with different scores) were obtained (for example, scores of positive, positive and weak assigned an overall score of positive).
Bioinformatics analysis of potential protein interactions
Standard bioinformatics software programmes (string-db.org/) were applied to catalog the clustering of molecular functions and to identify potential protein interactions of the MS-identified statistically significant proteins when comparing in situ vs. invasive melanoma FFPE tissues. Analyses were performed using STRING version 9.1 (http://string-db.org/), a database of known and predicted protein interactions that include direct physical and indirect functional protein associations (18).
Results
MS analysis
Label-free LC-MS/MS analysis revealed significant changes in 184 FFPE tissue-associated proteins from in situ vs. invasive melanoma. This number included identification based on only one matched peptide (data not shown). For the final establishment of the list of altered components, only proteins with ≥2 matched peptides and a Mascot score >40 were assigned as properly identified FFPE-tissue proteins. Excluding protein hits with only one matched peptide, 80 altered proteins were clearly identified when comparing FFPE tissue from in situ vs. invasive melanoma (Table II). As listed in Table II, 64 proteins exhibited an increased abundance, while 16 proteins exhibited a decreased abundance when comparing invasive vs. in situ melanoma. The most statistically significant protein observed to be increased in invasive melanoma was phosphatidylethanolamine-binding protein 1 (4.04-fold, P≤0.001), and the most significant decreased protein was hornerin (7.24-fold, P≤0.001).
Table II.
List of differentially expressed proteins in in situ vs. invasive melanoma formalin-fixed, paraffin-embedded tissues.
| A, Upregulated proteins in invasive melanoma vs. in situ melanoma | |||||
|---|---|---|---|---|---|
| Gene name | Protein identification | Peptide count | Confidence scorea | ANOVA (P-value) | Fold-change |
| PEBP1 | Phosphatidylethanolamine-binding protein 1 | 2 | 118.45 | 0.0000 | 4.04 |
| HNRNPK | Heterogeneous nuclear ribonucleoprotein K | 4 | 273.85 | 0.0002 | 4.63 |
| EEF2 | Elongation factor 2 | 7 | 418.22 | 0.0002 | 5.50 |
| HSP90B1 | Endoplasmin | 3 | 221.46 | 0.0003 | 2.57 |
| PSAP | Proactivator polypeptide | 2 | 92.05 | 0.0005 | 5.83 |
| HSP90AB1 | Heat shock protein HSP 90-beta | 4 | 300.79 | 0.0006 | 2.58 |
| HSPA5 | 78 kDa glucose-regulated protein | 3 | 176.94 | 0.0007 | 2.45 |
| PPIA | Peptidyl-prolyl cis-trans isomerase A | 3 | 258.37 | 0.0008 | 2.91 |
| FLNA | Filamin-A | 5 | 260.43 | 0.0010 | 5.75 |
| PGK1 | Phosphoglycerate kinase 1 | 4 | 328.39 | 0.0012 | 2.80 |
| TPI1 | Triosephosphate isomerase | 2 | 185.99 | 0.0014 | 3.98 |
| GSTP1 | Glutathione S-transferase P | 4 | 301.10 | 0.0014 | 2.61 |
| VCP | Transitional endoplasmic reticulum ATPase | 3 | 238.53 | 0.0015 | 2.53 |
| KRT31 | Keratin, type I cuticular Ha1 | 3 | 150.13 | 0.0015 | 22.11 |
| YWHAB | 14-3-3 protein beta/alpha | 3 | 278.01 | 0.0016 | 1.65 |
| PKM | Pyruvate kinase isozymes M1/M2 | 6 | 405.02 | 0.0024 | 3.78 |
| FBN1 | Fibrillin-1 | 4 | 245.97 | 0.0030 | 9.02 |
| FABP5 | Fatty acid-binding protein, epidermal | 3 | 179.44 | 0.0047 | 4.03 |
| PLEC | Plectin | 3 | 175.04 | 0.0050 | 2.85 |
| MDH2 | Malate dehydrogenase, mitochondrial | 3 | 181.57 | 0.0053 | 2.52 |
| COL6A1 | Collagen alpha-1(VI) chain | 6 | 444.60 | 0.0053 | 3.83 |
| PDIA3 | Protein disulfide-isomerase A3 | 2 | 116.05 | 0.0054 | 3.27 |
| RPS12 | 40S ribosomal protein S12 | 2 | 99.78 | 0.0061 | 2.98 |
| ACTB | Actin, cytoplasmic 1 | 9 | 640.45 | 0.0066 | 3.14 |
| PCBP1 | Poly(rC)-binding protein 1 | 3 | 183.71 | 0.0067 | 2.54 |
| TUBB2B | Tubulin beta-2B chain | 7 | 487.19 | 0.0073 | 6.87 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 3 | 290.29 | 0.0075 | 2.64 |
| OLFML1 | Olfactomedin-like protein 1 | 3 | 180.42 | 0.0078 | 2.86 |
| PRDX6 | Peroxiredoxin-6 | 2 | 125.87 | 0.0086 | 3.32 |
| LGALS1 | Galectin-1 | 2 | 122.70 | 0.0088 | 5.05 |
| VIM | Vimentin | 4 | 257.04 | 0.0097 | 2.16 |
| TUBA1A | Tubulin alpha-1A chain | 2 | 152.25 | 0.0098 | 2.77 |
| TUBA4A | Tubulin alpha-4A chain | 3 | 232.50 | 0.0103 | 2.58 |
| ENO1 | Alpha-enolase | 4 | 352.14 | 0.0105 | 1.46 |
| COL6A2 | Collagen alpha-2(VI) chain | 4 | 330.08 | 0.0111 | 2.23 |
| DPYSL2 | Dihydropyrimidinase-related protein 2 | 3 | 284.36 | 0.0113 | 2.06 |
| FASN | Fatty acid synthase | 6 | 441.55 | 0.0134 | 3.19 |
| YWHAE | 14-3-3 protein epsilon | 2 | 135.71 | 0.0135 | 8.98 |
| PLIN3 | Perilipin-3 | 2 | 115.71 | 0.0150 | 1.66 |
| ACTC1 | Actin, alpha cardiac muscle 1 | 5 | 258.90 | 0.0150 | 1.91 |
| TUBB | Tubulin beta chain | 7 | 505.61 | 0.0151 | 1.81 |
| AMBP | Protein AMBP | 2 | 123.71 | 0.0155 | 2.56 |
| ALDOA | Fructose-bisphosphate aldolase A | 2 | 223.37 | 0.0161 | 1.85 |
| ALB | Serum albumin | 20 | 1632.42 | 0.0162 | 3.94 |
| PDIA6 | Protein disulfide-isomerase A6 | 2 | 125.31 | 0.0162 | 2.29 |
| HSPD1 | 60 kDa heat shock protein, mitochondrial | 2 | 197.72 | 0.0166 | 2.66 |
| YWHAZ | 14-3-3 protein zeta/delta | 3 | 166.90 | 0.0171 | 2.95 |
| HLA-A | Human leukocyte antigen class I | 2 | 150.32 | 0.0195 | 7.68 |
| ACTN1 | Alpha-actinin-1 | 3 | 167.03 | 0.0197 | 4.05 |
| UBA1 | Ubiquitin-like modifier-activating enzyme 1 | 2 | 106.53 | 0.0200 | 1.81 |
| ACTN4 | Alpha-actinin-4 | 4 | 284.64 | 0.0226 | 4.13 |
| SERPINF1 | Pigment epithelium-derived factor | 2 | 130.19 | 0.0247 | 2.24 |
| CALM1 | Calmodulin | 3 | 286.87 | 0.0255 | 1.55 |
| NPM1 | Nucleophosmin | 2 | 154.40 | 0.0278 | 2.32 |
| YWHAQ | 14-3-3 protein theta | 2 | 125.98 | 0.0287 | 7.12 |
| TUBB4A | Tubulin beta-4A chain | 7 | 497.08 | 0.0307 | 3.63 |
| CLEC3B | Tetranectin | 2 | 114.64 | 0.0319 | 4.37 |
| ACSL1 | Long-chain-fatty-acid-CoA ligase 1 | 2 | 120.91 | 0.0327 | 3.10 |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 | 2 | 204.90 | 0.0350 | 2.17 |
| COL14A1 | Collagen alpha-1(XIV) chain | 2 | 147.41 | 0.0375 | 2.20 |
| IGHG3 | Ig gamma-3 chain C region | 2 | 128.39 | 0.0402 | 1.54 |
| TF | Serotransferrin | 7 | 626.01 | 0.0405 | 3.70 |
| BGN | Biglycan | 2 | 119.89 | 0.0433 | 2.49 |
| EZR | Ezrin | 2 | 111.07 | 0.0458 | 12.78 |
| B, Downregulated proteins in invasive melanoma vs. in situ melanoma | |||||
| Gene name | Protein identification | Peptide count | Confidence scorea | ANOVA (P-value) | Fold-change |
| HRNR | Hornerin | 2 | 144.28 | 0.0003 | 7.24 |
| ITIH1 | Inter-alpha-trypsin inhibitor heavy chain H1 | 3 | 219.20 | 0.0021 | 1.78 |
| DSP | Desmoplakin | 16 | 956.60 | 0.0030 | 6.70 |
| KRT9 | Keratin, type I cytoskeletal 9 | 31 | 3005.85 | 0.0030 | 4.25 |
| KRT1 | Keratin, type II cytoskeletal 1 | 34 | 2801.68 | 0.0042 | 3.04 |
| DCD | Dermcidin | 2 | 136.27 | 0.0047 | 4.12 |
| LGALS7 | Galectin-7 | 3 | 395.32 | 0.0058 | 1.78 |
| CAT | Catalase | 3 | 223.78 | 0.0089 | 1.62 |
| ATP5A1 | ATP synthase subunit alpha, mitochondrial | 3 | 258.83 | 0.0101 | 2.11 |
| IGHA1 | Ig alpha-1 chain C region | 2 | 188.63 | 0.0108 | 2.24 |
| CALML5 | Calmodulin-like protein 5 | 2 | 175.25 | 0.0113 | 3.72 |
| KRT2 | Keratin, type II cytoskeletal 2 epidermal | 24 | 2034.24 | 0.0113 | 3.23 |
| KRT10 | Keratin, type I cytoskeletal 10 | 19 | 1719.86 | 0.0147 | 2.54 |
| FLG2 | Filaggrin-2 | 2 | 129.72 | 0.0229 | 2.75 |
| HSPB1 | Heat shock protein beta-1 | 2 | 125.22 | 0.0242 | 1.61 |
| MSN | Moesin | 2 | 109.96 | 0.0479 | 2.09 |
Confidence score obtained from Mascot. ANOVA, analysis of variance.
Protein-protein interactions
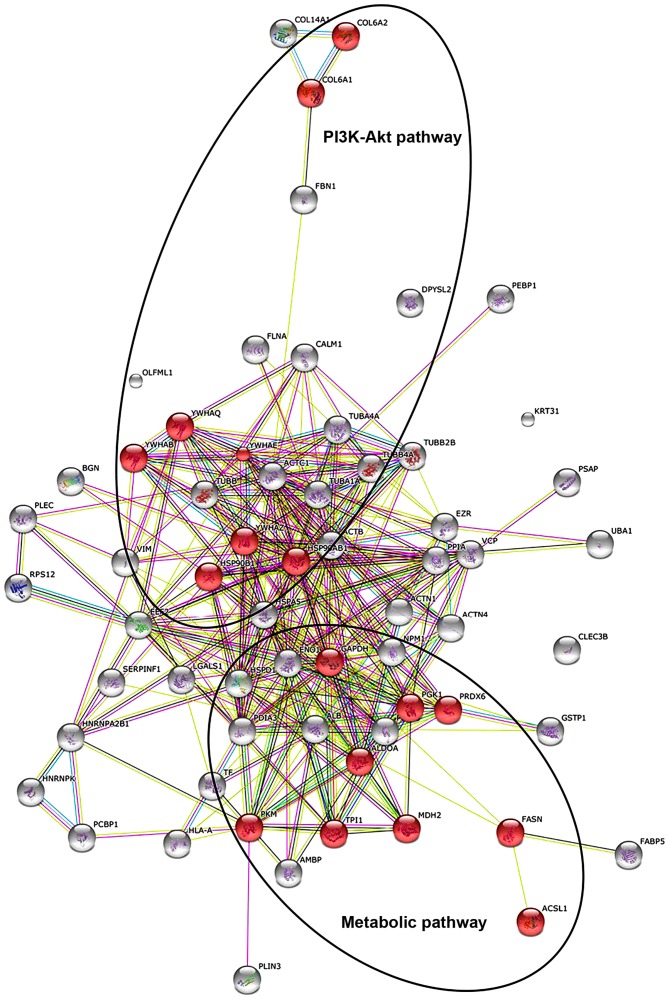
The existence of potential interactions between proteins with a changed abundance in in situ vs. invasive FFPE melanoma tissue, as identified by the proteomics survey presented in the current study, was evaluated using the STRING database of direct physical and indirect functional protein interactions. The bioinformatic analysis resulted in a complex interaction map (Fig. 1). Main interaction nodes included proteins involved in metabolism (triosephosphate isomerase, peroxiredoxin-6, fructose-bisphosphate aldolase A, long-chain-fatty-acid-CoA ligase 1, malate dehydrogenase, mitochondrial, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase 1, pyruvate kinase isozymes M1/M2 and FASN), which were discovered to be elevated in invasive melanoma compared with in situ melanoma. In addition, proteins associated with the phosphoinositide 3-kinase (PI3K)-Akt signalling pathway were also increased in invasive melanoma FFPE tissues compared with in situ melanoma FFPE tissues, including 14-3-3ε, 14-3-3ζ, endoplasmin, 14-3-3α/β, 14-3-3ζ/δ, heat shock protein HSP 90-β, collagen α-2(VI) chain and collagen α-1(VI) chain).
Figure 1.
STRING analysis of metabolic and PI3K-Akt signalling pathways. STRING analysis, generating functional protein association networks, displays 64 proteins discovered to be upregulated in invasive melanoma compared with in situ melanoma, as listed in Table II. Highlighted in red are nine proteins that participate in metabolism (TPI1, PRDX6, ALDOA, ACSL1, MDH2, GAPDH, PGK1, PKM and FASN) and eight proteins that participate in the PI3K-Akt signalling pathway (YWHAE, YWHAQ, HSP90B1, YWHAB, YWHAZ, HSP90AB1, COL6A2 and COL6A1). PI3K, phosphoinositide 3-kinase; TPI1, triosephosphate isomerase; PRDX6, peroxiredoxin-6; ALDOA, fructose-bisphosphate aldolase A; ACSL1, acyl-CoA synthetase long-chain family member 1; MDH2, malate dehydrogenase, mitochondrial; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PGK1, phosphoglycerate kinase 1; PKM, pyruvate kinase isozymes M1/M2; FASN, fatty acid synthase; YWHAE, 14-3-3ε; YWHAQ, 14-3-3ζ; HSP90B1, endoplasmin; YWHAB, 14-3-3α/β; YWHAZ, 14-3-3ζ/δ; HSP90AB1, heat shock protein HSP 90-β; COL6A2, collagen α-2(VI) chain; COL6A1, collagen α-1(VI) chain.
IHC
IHC analysis of FASN in a cohort of primary melanoma tumour samples (n=28) and a cohort of metastatic melanoma tumours (n=29) revealed that a higher percentage (72%) of metastatic melanomas exhibited FASN immunoreactivity (weak/moderate) compared with primary tissue (50%), (Fig. 2 and Table III). Furthermore, a greater number of primary tumours (50%) exhibited negative FASN immunoreactivity, compared with metastatic tumours (28%). IHC results also indicate that 14-3-3ε, a member of the 14-3-3 family of proteins, may be associated with melanoma disease progression, since a greater number of metastatic tumours exhibited strong 14-3-3ε immunoreactivity (57%) compared with primary melanomas (32%) (Fig. 2 and Table III).
Figure 2.
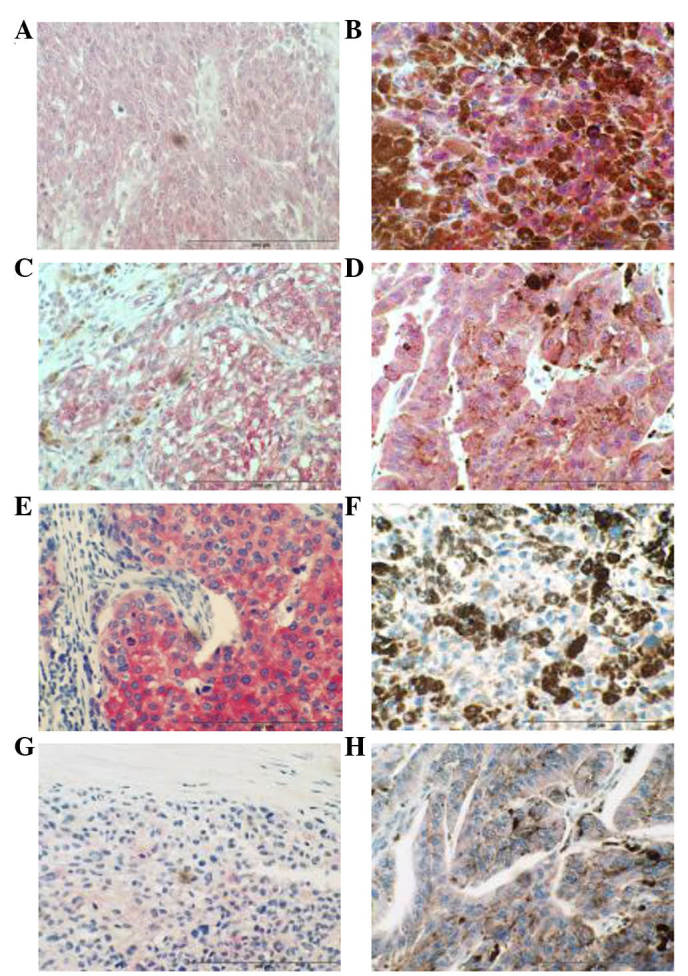
IHC analysis of 14-3-3ε and FASN in primary and metastatic melanoma. (A) Representative photomicrograph showing decreased 14-3-3ε immunoreactivity in primary melanoma tumour tissue compared with metastatic melanoma tissue. (B) Representative photomicrograph showing increased 14-3-3ε immunoreactivity in metastatic melanoma tumour tissue compared with primary melanoma tissue. (C) Representative photomicrograph showing decreased 14-3-3ε immunoreactivity in primary melanoma tumour tissue compared with metastatic melanoma tissue. (D) Representative photomicrograph showing increased 14-3-3ε immunoreactivity in metastatic melanoma tumour tissue compared with primary melanoma tissue. (E) Positive control. Strong FASN immunoreactivity is observed in invasive ductal breast carcinoma. (F) Representative photomicrograph showing increased FASN immunoreactivity in metastatic melanoma tumour tissue compared with primary melanoma tissue. (G) Representative photomicrograph showing decreased FASN immunoreactivity in primary melanoma tumour tissue. (H) Representative photomicrograph showing increased FASN immunoreactivity in metastatic melanoma tumour tissues compared with primary melanoma tissue. Magnification, ×400. Scale bar=200 µm. FASN, fatty acid synthase.
Table III.
Immunohistochemistry analysis of FASN and 14-3-3ε in normal, primary and metastatic melanoma tissues.
| A, FASN | Intensity scores | ||
|---|---|---|---|
| Normal (n=3) | Negative | 3/3 | |
| Primary melanomas (n=28) | Negative | 14/28 | |
| Weak | 9/28 | ||
| Moderate | 5/28 | ||
| Strong | 0/28 | ||
| Metastatic melanomas (n=29) | Negative | 8/29 | |
| Weak | 11/29 | ||
| Moderate | 10/29 | ||
| Strong | 0/29 | ||
| B, 14-3-3ε | Intensity scores | ||
| Normal (n=3) | Moderate | 3/3 | |
| Primary melanomas (n=28) | Weak | 6/28 | |
| Moderate | 13/28 | ||
| Strong | 9/28 | ||
| Metastatic melanomas (n=30) | Negative | 1/30 | |
| Weak | 2/30 | ||
| Moderate | 10/30 | ||
| Strong | 17/30 | ||
FASN, fatty acid synthase.
Discussion
Molecular pathways regulating melanoma initiation and progression may represent potential targets of therapeutic development for this aggressive cancer. To gain additional understanding into the mechanism by which melanoma in situ, which is confined to the epidermis, progresses into invasive melanoma at the protein level, label-free MS analysis of FFPE tissue using the FASP methodology was applied in the present study to identify proteins differentially expressed between these two stages of melanoma. The FASP method presents numerous advantages over alternative approaches for analysing FFPE tissues, particularly the ability to buffer-exchange the sample into a more digestion-compatible solution, thus facilitating the classification of large numbers of disease-associated proteins (19).
Using this approach, a number of proteins were identified that were significantly increased in invasive melanoma compared with melanoma in situ. Proteins associated with alterations in the majority of cancer cells use glucose at high levels and convert it into lactate instead of relying on mitochondrial oxidative phosphorylation to generate energy, even with adequate oxygen levels, a phenomenon termed the ‘Warburg effect’ (20). A decisive consequence of this altered metabolism is the amplification of lactate production in tumour cells, resulting in a low pH created by extracellular acidification (21). This process provides a favorable microenvironment for the activation of proteases, which induce extracellular matrix degradation and assist tumour cells to metastasize (22). The enzymes involved in glycolysis also play a vital part in tumour migration and invasion (23). Pyruvate kinase M2 (PKM2), an embryonic isoform predominantly expressed in malignant cells, catalyzes the conversion of phosphoenolpyruvate to pyruvate (24). In the present study, PKM2 was observed to be significantly elevated in invasive melanoma tissue compared with in situ melanoma tissue. Clinical evidence of PKM2 contribution to tumourigenesis includes reported increased serum levels of this protein in colon and breast cancer patients (25,26). The elevation of PKM2 is associated with a poor prognosis in small cell lung cancer and in head and neck cancer (24,27,28). High expression of PKM2 and two other glycolytic proteins has been associated with a poor outcome in stage-2 melanoma (29). This finding of increased levels of PKM2 further implicates a potential role for this glycolytic protein in melanoma disease progression.
A second metabolic-associated protein discovered to be significantly elevated in invasive melanoma is FASN, a key protein involved in the metabolism of cancer cells (30). In order to validate this finding and other selected proteins, the expression levels of FASN and 14-3-3ε were further investigated in melanoma samples. 14-3-3ε, one of the members of the 14-3-3 family of proteins that is considered to participate in the PI3K-Akt signalling pathway (31), was upregulated in invasive melanoma. A number of proteins involved in the PI3K-Akt signalling pathway were also observed to be increased in invasive melanoma. An ideal follow-up validation study would have involved IHC analysis of protein expression in archival material from the same patients used for the current FASP proteomics study; however, this was not feasible due to the limited amount of tissue available. In addition, at the time of the study, it was not possible to obtain FFPE slides of in situ melanoma tumours. Thus, IHC analysis was performed using commercially sourced TMAs of primary invasive melanoma tumours and metastatic melanoma tumours as a surrogate for the initial discovery-phase analysis. As well as validating the expression of candidate proteins in invasive melanoma tumour tissues, this approach would enable to confirm if these proteins were associated with a metastatic melanoma phenotype.
FASN is the key enzyme responsible for the synthesis of fatty acids, catalyzing the conversion of acetyl-CoA and malonyl-CoA into long-chain fatty acids (32). Normal cells express low levels of FASN because they use fatty acids from dietary lipids, with the exception of those that are lipogenic (33). As expected, no FASN immunoreactivity was observed in normal skin archival samples. FASN expression was reported to be upregulated in several human cancers, including prostate, breast, ovarian and lung cancer (34–38). FASN was significantly elevated in invasive melanoma in the current study. In cutaneous melanomas, FASN protein expression was previously associated with Breslow's thickness, and thus, with a poorer prognosis (39,40). Orlistat (also known as tetrahydrolipstatin) is a FASN inhibitor that reduces metastases and angiogenesis in B-16-F10 melanoma cells (41). In the present study, IHC analysis of FASN in a cohort of primary melanoma tumour samples (n=28) and a cohort of metastatic melanoma tumours (n=29) revealed that a higher proportion of metastatic melanomas exhibited FASN immunoreactivity. Furthermore, a greater number of primary tumours (14/28) exhibited negative FASN immunoreactivity, compared with metastatic tumours (8/29). This result provides further evidence that FASN is potentially involved in the progression of the malignant melanoma phenotype. The metabolic products of the FASN complex are rapidly consumed by actively dividing cells, and previous studies demonstrated that FASN expression is important for tumour growth and survival, indicating that FASN is a metabolic oncogene (42). FASN overexpression and the Warburg effect are widespread metabolic abnormalities in cancer, and are likely to contribute to the in situ-to-invasive transition evaluated in the current study. FASN expression in adipocyte/3T3 cells as well as in breast, prostate and ovarian cancer cells has previously been suggested to be regulated by the PI3K/Akt signalling cascade (43–46). Proteins involved in the PI3K/Akt pathway were also observed to be significantly increased in invasive melanoma tissue, suggesting the involvement of this signalling pathway in the observed increased FASN protein expression. However, while the present validation studies observed FASN protein overexpression, they did not provide any information regarding FASN activity.
In melanoma, both the Ras/Raf/MEK/ERK and PI3K-Akt signalling pathways are constitutively activated through multiple mechanisms, and thus exert a number of key functions in melanoma development and progression (47–49). The ubiquitously expressed 14-3-3 proteins regulate multiple pathways involved in transformation, including the PI3K-Akt pathway (50). Phosphorylation-dependent association with binding partners forms the mechanistic basis for the essential role for 14-3-3 proteins in modulating kinase signalling pathways (51). The family of 14-3-3 proteins comprises seven small molecules termed α/β, ε, γ, σ, ζ/δ, τ/θ and η, which are expressed in a diversity of tissues types (52). Four family members were observed to be significantly elevated in invasive melanoma FFPE tissue compared with in situ melanoma tissue, including YWHAE (14-3-3ε), YWHAQ (14-3-3τ/θ), YWHAB (14-3-3α/β) and YWHAZ (14-3-3ζ/δ). Neal et al reported that 14-3-3ζ overexpression was significantly associated with increased Akt phosphorylation in human breast tumours, and that 14-3-3ζ overexpression combined with strong Akt phosphorylation was significantly associated with increased recurrence in cancer patients (53). The authors also observed that knockdown of 14-3-3ζ expression by small interfering RNA in cancer cell lines and in tumour xenografts reduced Akt phosphorylation (53). Schultz et al demonstrated that epigenetic silencing of 14-3-3σ may contribute to tumour progression in malignant melanoma via loss of cell cycle control, impaired cellular senescence program and support of migratory capacity (54), a process regulated by high levels of gene methylation (55). The IHC studies presented in the current report indicate that 14-3-3ε may be associated with melanoma disease progression, since a significant number of metastatic tumours (17/30) exhibited strong 14-3-3ε immunoreactivity compared with primary melanomas (9/28). 14-3-3ε has been reported to upregulate focal adhesion kinase (FAK) via nuclear factor-κB activation in hepatocellular carcinoma (56). The PI3K-Akt pathway is one of the downstream pathways implicated in FAK viability signalling (57).
In summary, the use of FFPE tissue to study protein changes during tumourogenesis is now a very powerful and reproducible approach to identify the key molecules involved in disease progression. In particular, MS and IHC analysis of FASN and 14-3-3ε in normal, primary and metastatic melanoma tissues demonstrated significant changes in the abundance levels of these proteins, which play significant roles in metabolic pathways and in PI3K-Akt signalling, respectively, and are key events in the establishment of a more invasive/aggressive phenotype in melanoma.
References
- 1.Masters GA, Krilov L, Bailey HH, Brose MS, Burstein H, Diller LR, Dizon DS, Fine HA, Kalemkerian GP, Moasser M, et al. Clinical cancer advances 2015: Annual report on progress against cancer from the American society of clinical oncology. J Clin Oncol. 2015;33:786–809. doi: 10.1200/JCO.2014.59.9746. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 4.Boyle GM. Therapy for metastatic melanoma: An overview and update. Expert Rev Anticancer Ther. 2011;11:725–737. doi: 10.1586/era.11.25. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan R, LoRusso P, Boerner S, Dummer R. Achievements and challenges of molecular targeted therapy in melanoma. Am Soc Clin Oncol Educ Book. 2015;2015:177–186. doi: 10.14694/EdBook_AM.2015.35.177. [DOI] [PubMed] [Google Scholar]
- 6.Kalia M. Biomarkers for personalized oncology: Recent advances and future challenges. Metabolism. 2015;64(3):S16–S21. doi: 10.1016/j.metabol.2014.10.027. Suppl 1. [DOI] [PubMed] [Google Scholar]
- 7.Moschos SJ, Pinnamaneni R. Targeted therapies in melanoma. Surg Oncol Clin N Am. 2015;24:347–358. doi: 10.1016/j.soc.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Huang C, Nice E. Recent advances in proteomics: Towards the human proteome. Biomed Chromatogr. 2014;28:848–857. doi: 10.1002/bmc.3157. [DOI] [PubMed] [Google Scholar]
- 9.Bougnoux AC, Solassol J. The contribution of proteomics to the identification of biomarkers for cutaneous malignant melanoma. Clin Biochem. 2013;46:518–523. doi: 10.1016/j.clinbiochem.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Byrum SD, Larson SK, Avaritt NL, Moreland LE, Mackintosh SG, Cheung WL, Tackett AJ. Quantitative proteomics identifies activation of hallmark pathways of cancer in patient melanoma. J Proteomics Bioinform. 2013;6:43–50. doi: 10.4172/jpb.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manzini MC, Perez KR, Riske KA, Bozelli JC, Jr, Santos TL, da Silva MA, Saraiva GK, Politi MJ, Valente AP, Almeida FC, et al. Peptide: lipid ratio and membrane surface charge determine the mechanism of action of the antimicrobial peptide BP100. Conformational and functional studies. Biochim Biophys Acta. 2014;1838:1985–1999. doi: 10.1016/j.bbamem.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Gustafsson OJ, Arentz G, Hoffmann P. Proteomic developments in the analysis of formalin-fixed tissue. Biochim Biophys Acta. 2015;1854:559–580. doi: 10.1016/j.bbapap.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Quesada-Calvo F, Bertrand V, Longuespée R, Delga A, Mazzucchelli G, Smargiasso N, Baiwir D, Delvenne P, Malaise M, De Pauw-Gillet MC, et al. Comparison of two FFPE preparation methods using label-free shotgun proteomics: Application to tissues of diverticulitis patients. J Proteomics. 2015;112:250–261. doi: 10.1016/j.jprot.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Tanca A, Abbondio M, Pisanu S, Pagnozzi D, Uzzau S, Addis MF. Critical comparison of sample preparation strategies for shotgun proteomic analysis of formalin-fixed, paraffin-embedded samples: Insights from liver tissue. Clin Proteomics. 2014;11:28. doi: 10.1186/1559-0275-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linge A, Kennedy S, O'Flynn D, Beatty S, Moriarty P, Henry M, Clynes M, Larkin A, Meleady P. Differential expression of fourteen proteins between uveal melanoma from patients who subsequently developed distant metastases versus those who did not. Invest Opthalmol Vis Sci. 2012;53:4634–4643. doi: 10.1167/iovs.11-9019. [DOI] [PubMed] [Google Scholar]
- 16.Ostasiewicz P, Zielinska DF, Mann M, Wiśniewski JR. Proteome, phosphoproteome, and N-glycoproteome are quantitatively preserved in formalin-fixed paraffin-embedded tissue and analyzable by high-resolution mass spectrometry. J Proteome Res. 2010;9:3688–3700. doi: 10.1021/pr100234w. [DOI] [PubMed] [Google Scholar]
- 17.Dowling P, Hughes DJ, Larkin AM, Larkin AM, Meiller J, Henry M, Meleady P, Lynch V, Pardini B, Naccarati A, et al. Elevated levels of 14-3-3 proteins, serotonin, gamma enolase and pyruvate kinase identified in clinical samples from patients diagnosed with colorectal cancer. Clin Chim Acta. 2015;441:133–141. doi: 10.1016/j.cca.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen K, Sun J, Cao X, Zhou D, Li J. Comparison of different buffers for protein extraction from formalin-fixed and paraffin-embedded tissue specimens. PLoS One. 2015;10:e0142650. doi: 10.1371/journal.pone.0142650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warburg O, Wind F, Negelein E. THE metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Böhme I, Bosserhoff AK. Acidic tumor microenvironment in human melanoma. Pigment Cell Melanoma Res. 2016 May 27; doi: 10.1111/pcmr.12495. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 22.Han T, Kang D, Ji D, Wang X, Zhan W, Fu M, Xin HB, Wang JB. How does cancer cell metabolism affect tumor migration and invasion? Cell Adh Migr. 2013;7:395–403. doi: 10.4161/cam.26345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu QF, Liu Y, Fan Y, Hua SN, Qu HY, Dong SW, Li RL, Zhao MY, Zhen Y, Yu XL, et al. Alpha-enolase promotes cell glycolysis, growth, migration, and invasion in non-small cell lung cancer through FAK-mediated PI3K/AKT pathway. J Hematol Oncol. 2015;8:22. doi: 10.1186/s13045-015-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356:184–191. doi: 10.1016/j.canlet.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Eigenbrodt E, Basenau D, Holthusen S, Mazurek S, Fischer G. Quantification of tumor type M2 pyruvate kinase (Tu M2-PK) in human carcinomas. Anticancer Res. 1997;17:3153–3156. [PubMed] [Google Scholar]
- 26.Lüftner D, Mesterharm J, Akrivakis C, Geppert R, Petrides PE, Wernecke KD, Possinger K. Tumor type M2 pyruvate kinase expression in advanced breast cancer. Anticancer Res. 2000;20:5077–5082. [PubMed] [Google Scholar]
- 27.Karachaliou N, Papadaki C, Lagoudaki E, Trypaki M, Sfakianaki M, Koutsopoulos A, Mavroudis D, Stathopoulos E, Georgoulias V, Souglakos J. Predictive value of BRCA1, ERCC1, ATP7B, PKM2, TOPOI, TOPΟ-IIA, TOPOIIB and C-MYC genes in patients with small cell lung cancer (SCLC) who received first line therapy with cisplatin and etoposide. PLoS One. 2013;8:e74611. doi: 10.1371/journal.pone.0074611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desai S, Ding M, Wang B, Lu Z, Zhao Q, Shaw K, Yung WK, Weinstein JN, Tan M, Yao J. Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers. Oncotarget. 2014;5:8202–8210. doi: 10.18632/oncotarget.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falkenius J, Lundeberg J, Johansson H, Tuominen R, Frostvik-Stolt M, Hansson J, Brage S Egyhazi. High expression of glycolytic and pigment proteins is associated with worse clinical outcome in stage III melanoma. Melanoma Res. 2013;23:452–460. doi: 10.1097/CMR.0000000000000027. [DOI] [PubMed] [Google Scholar]
- 30.Rossato FA, Zecchin KG, La Guardia PG, Ortega RM, Alberici LC, Costa RA, Catharino RR, Graner E, Castilho RF, Vercesi AE. Fatty acid synthase inhibitors induce apoptosis in non-tumorigenic melan-a cells associated with inhibition of mitochondrial respiration. PLoS One. 2014;9:e101060. doi: 10.1371/journal.pone.0101060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta. 2011;1813:1938–1945. doi: 10.1016/j.bbamcr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Hopperton KE, Duncan RE, Bazinet RP, Archer MC. Fatty acid synthase plays a role in cancer metabolism beyond providing fatty acids for phospholipid synthesis or sustaining elevations in glycolytic activity. Exp Cell Res. 2014;320:302–310. doi: 10.1016/j.yexcr.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 33.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 34.Swinnen JV, Roskams T, Joniau S, Van Poppel H, Oyen R, Baert L, Heyns W, Verhoeven G. Overexpression of fatty acid synthase is an early and common event in the development of prostate cancer. Int J Cancer. 2002;98:19–22. doi: 10.1002/ijc.10127. [DOI] [PubMed] [Google Scholar]
- 35.Rossi S, Graner E, Febbo P, Weinstein L, Bhattacharya N, Onody T, Bubley G, Balk S, Loda M. Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol Cancer Res. 2003;1:707–715. [PubMed] [Google Scholar]
- 36.Wang Y, Kuhajda FP, Li JN, Pizer ES, Han WF, Sokoll LJ, Chan DW. Fatty acid synthase (FAS) expression in human breast cancer cell culture supernatants and in breast cancer patients. Cancer Lett. 2001;167:99–104. doi: 10.1016/S0304-3835(01)00464-5. [DOI] [PubMed] [Google Scholar]
- 37.Bauerschlag DO, Maass N, Leonhardt P, Verburg FA, Pecks U, Zeppernick F, Morgenroth A, Mottaghy FM, Tolba R, Meinhold-Heerlein I, Bräutigam K. Fatty acid synthase overexpression: Target for therapy and reversal of chemoresistance in ovarian cancer. J Transl Med. 2015;13:146. doi: 10.1186/s12967-015-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerne D, Zitnik IP, Sok M. Increased fatty acid synthase activity in non-small cell lung cancer tissue is a weaker predictor of shorter patient survival than increased lipoprotein lipase activity. Arch Med Res. 2010;41:405–409. doi: 10.1016/j.arcmed.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Innocenzi D, Alò PL, Balzani A, Sebastiani V, Silipo V, La Torre G, Ricciardi G, Bosman C, Calvieri S. Fatty acid synthase expression in melanoma. J Cutan Pathol. 2003;30:23–28. doi: 10.1034/j.1600-0560.2003.300104.x. [DOI] [PubMed] [Google Scholar]
- 40.Kapur P, Rakheja D, Roy LC, Hoang MP. Fatty acid synthase expression in cutaneous melanocytic neoplasms. Mod Pathol. 2005;18:1107–1112. doi: 10.1038/modpathol.3800395. [DOI] [PubMed] [Google Scholar]
- 41.Seguin F, Carvalho MA, Bastos DC, Agostini M, Zecchin KG, Alvarez-Flores MP, Chudzinski-Tavassi AM, Coletta RD, Graner E. The fatty acid synthase inhibitor orlistat reduces experimental metastases and angiogenesis in B16-F10 melanomas. Br J Cancer. 2012;107:977–987. doi: 10.1038/bjc.2012.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flavin R, Peluso S, Nguyen PL, Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6:551–562. doi: 10.2217/fon.10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang YA, Han WF, Morin PJ, Chrest FJ, Pizer ES. Activation of fatty acid synthesis during neoplastic transformation: Role of mitogen-activated protein kinase and phosphatidylinositol 3-kinase. Exp Cell Res. 2002;279:80–90. doi: 10.1006/excr.2002.5600. [DOI] [PubMed] [Google Scholar]
- 44.Van de Sande T, De Schrijver E, Heyns W, Verhoeven G, Swinnen JV. Role of the phosphatidylinositol 3′-kinase/PTEN/Akt kinase pathway in the overexpression of fatty acid synthase in LNCaP prostate cancer cells. Cancer Res. 2002;62:642–646. [PubMed] [Google Scholar]
- 45.Wang D, Sul HS. Insulin stimulation of the fatty acid synthase promoter is mediated by the phosphatidylinositol 3-kinase pathway. Involvement of protein kinase B/Akt. J Biol Chem. 1998;273:25420–25426. doi: 10.1074/jbc.273.39.25420. [DOI] [PubMed] [Google Scholar]
- 46.Wang HQ, Altomare DA, Skele KL, Poulikakos PI, Kuhajda FP, Di Cristofano A, Testa JR. Positive feedback regulation between AKT activation and fatty acid synthase expression in ovarian carcinoma cells. Oncogene. 2005;24:3574–3582. doi: 10.1038/sj.onc.1208463. [DOI] [PubMed] [Google Scholar]
- 47.Chen B, Tardell C, Higgins B, Packman K, Boylan JF, Niu H. BRAFV600E negatively regulates the AKT pathway in melanoma cell lines. PLoS One. 2012;7:e42598. doi: 10.1371/journal.pone.0042598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, Kester M, Sandirasegarane L, Robertson GP. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 49.Meier F, Schittek B, Busch S, Garbe C, Smalley K, Satyamoorthy K, Li G, Herlyn M. The RAS/RAF/MEK/ERK and PI3K/AKT signaling pathways present molecular targets for the effective treatment of advanced melanoma. Front Biosci. 2005;10:2986–3001. doi: 10.2741/1755. [DOI] [PubMed] [Google Scholar]
- 50.Daitoku H, Sakamaki J, Fukamizu A. Regulation of FoxO transcription factors by acetylation and protein-protein interactions. Biochim Biophys Acta. 2011;1813:1954–1960. doi: 10.1016/j.bbamcr.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Freeman AK, Morrison DK. 14-3-3 proteins: Diverse functions in cell proliferation and cancer progression. Semin Cell Dev Biol. 2011;22:681–687. doi: 10.1016/j.semcdb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tzivion G, Gupta VS, Kaplun L, Balan V. 14-3-3 proteins as potential oncogenes. Semin Cancer Biol. 2006;16:203–213. doi: 10.1016/j.semcancer.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 53.Neal CL, Xu J, Li P, Mori S, Yang J, Neal NN, Zhou X, Wyszomierski SL, Yu D. Overexpression of 14-3-3ζ in cancer cells activates PI3K via binding the p85 regulatory subunit. Oncogene. 2012;31:897–906. doi: 10.1038/onc.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultz J, Ibrahim SM, Vera J, Kunz M. 14-3-3sigma gene silencing during melanoma progression and its role in cell cycle control and cellular senescence. Mol Cancer. 2009;8:53. doi: 10.1186/1476-4598-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vera J, Schultz J, Ibrahim S, Raatz Y, Wolkenhauer O, Kunz M. Dynamical effects of epigenetic silencing of 14-3-3sigma expression. Mol Biosyst. 2010;6:264–273. doi: 10.1039/B907863K. [DOI] [PubMed] [Google Scholar]
- 56.Ko BS, Jan YJ, Chang TC, Liang SM, Chen SC, Liu TA, Wu YM, Wang J, Liou JY. Upregulation of focal adhesion kinase by 14-3-3ε via NFκB activation in hepatocellular carcinoma. Anticancer Agents Med Chem. 2013;13:555–562. doi: 10.2174/1871520611313040004. [DOI] [PubMed] [Google Scholar]
- 57.Xia H, Nho RS, Kahm J, Kleidon J, Henke CA. Focal adhesion kinase is upstream of phosphatidylinositol 3-kinase/Akt in regulating fibroblast survival in response to contraction of type I collagen matrices via a beta 1 integrin viability signaling pathway. J Biol Chem. 2004;279:33024–33034. doi: 10.1074/jbc.M313265200. [DOI] [PubMed] [Google Scholar]