Abstract
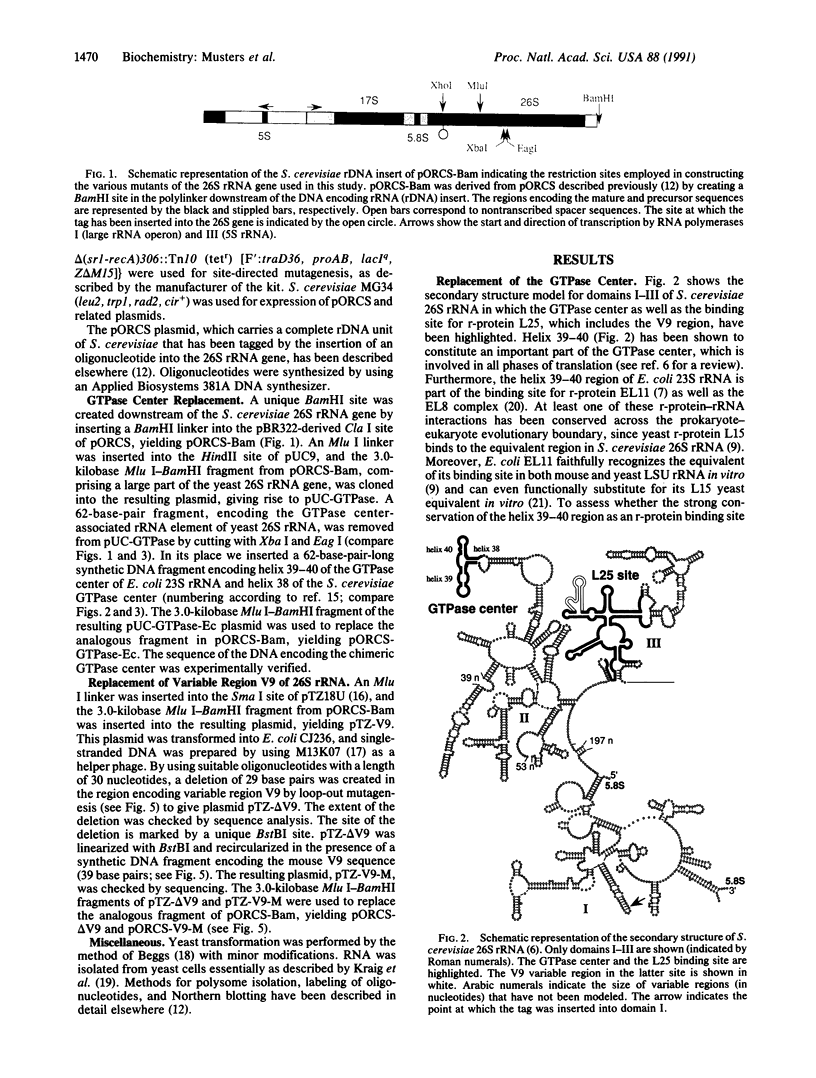
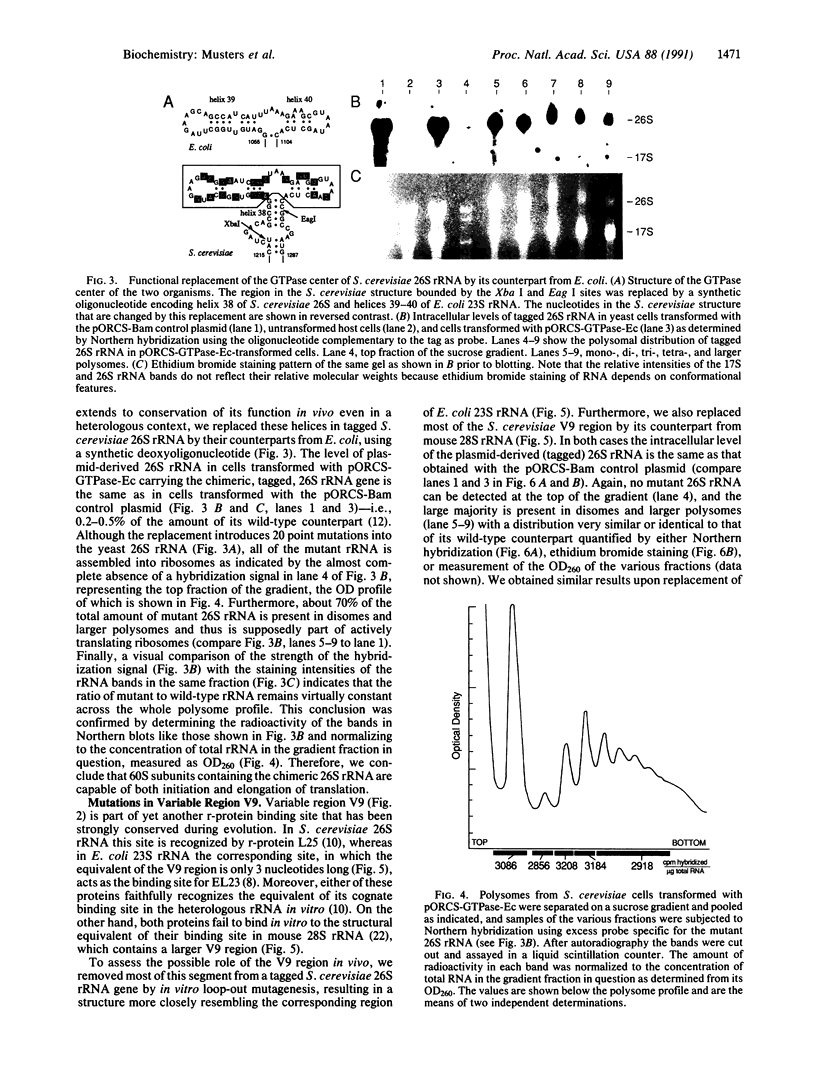
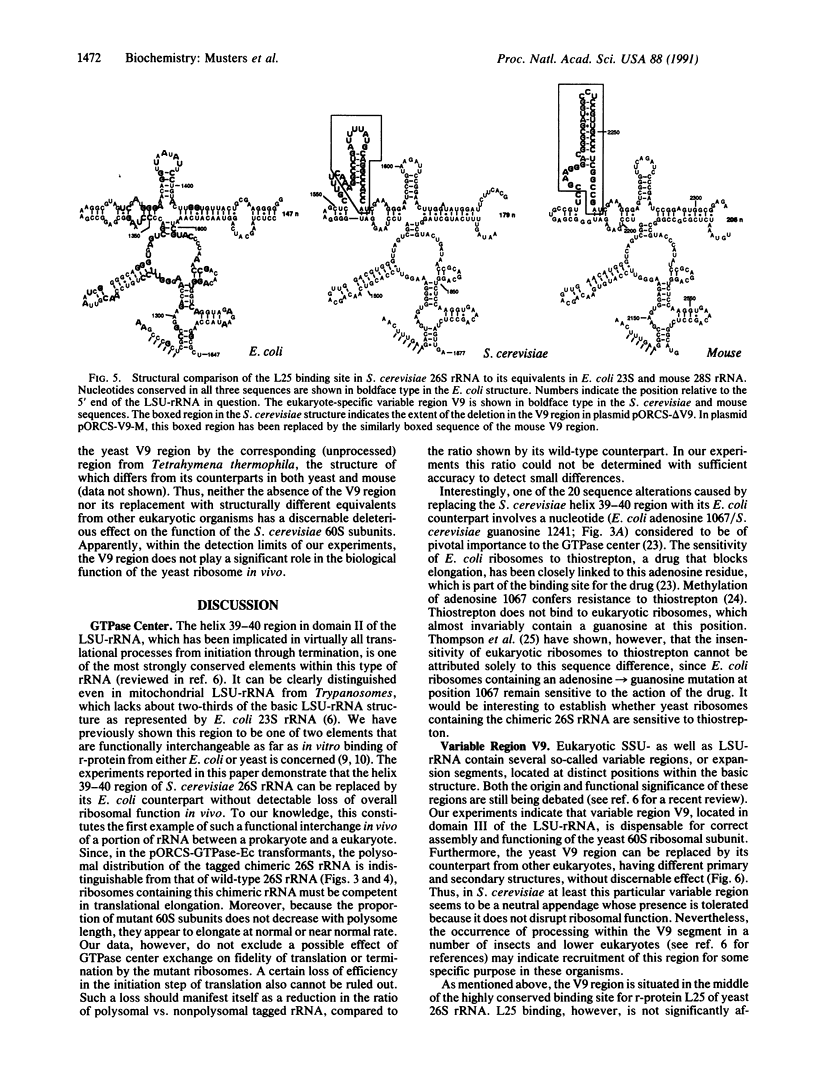
Using the "tagged" rRNA gene system, which allows in vivo mutational analysis of Saccharomyces cerevisiae rRNA, we studied the role of two distinct structural elements of 26S rRNA in ribosome biogenesis and function--namely, the evolutionarily highly conserved "GTPase center" located in domain II and the eukaroyote-specific variable region V9 in domain III. Replacement of the S. cerevisiae GTPase center with its counterpart from Escherichia coli did not affect the assembly of the mutant 26S rRNA into functional (as judged by their polysomal distribution) 60S subunits, indicating that the E. coli GTPase center functions efficiently in the context of the heterologous rRNA. Removal of most of the S. cerevisiae V9 region or replacement of this segment by the equivalent segment from mouse 28S rRNA also did not affect the formation of functional 60S subunits carrying the mutant 26S rRNA. Therefore, the V9 region does not seem to play a role in the biological functioning of the yeast 60S subunits, and these subunits appear to be able to accommodate V9 regions of various size and secondary structure without apparent loss of function.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beauclerk A. A., Cundliffe E., Dijk J. The binding site for ribosomal protein complex L8 within 23 s ribosomal RNA of Escherichia coli. J Biol Chem. 1984 May 25;259(10):6559–6563. [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Brimacombe R., Atmadja J., Stiege W., Schüler D. A detailed model of the three-dimensional structure of Escherichia coli 16 S ribosomal RNA in situ in the 30 S subunit. J Mol Biol. 1988 Jan 5;199(1):115–136. doi: 10.1016/0022-2836(88)90383-x. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E. The functional role of ribosomal RNA in protein synthesis. Cell. 1989 May 19;57(4):525–529. doi: 10.1016/0092-8674(89)90122-0. [DOI] [PubMed] [Google Scholar]
- Egebjerg J., Douthwaite S., Garrett R. A. Antibiotic interactions at the GTPase-associated centre within Escherichia coli 23S rRNA. EMBO J. 1989 Feb;8(2):607–611. doi: 10.1002/j.1460-2075.1989.tb03415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui A., Jhurani P., de Boer H. A. Directing ribosomes to a single mRNA species: a method to study ribosomal RNA mutations and their effects on translation of a single messenger in Escherichia coli. Methods Enzymol. 1987;153:432–452. doi: 10.1016/0076-6879(87)53070-1. [DOI] [PubMed] [Google Scholar]
- Juan-Vidales F., Sánchez Madrid F., Saenz-Robles M. T., Ballesta J. P. Purification and characterization of two ribosomal proteins of Saccharomyces cerevisiae. Homologies with proteins from eukaryotic species and with bacterial protein EC L11. Eur J Biochem. 1983 Nov 2;136(2):275–281. doi: 10.1111/j.1432-1033.1983.tb07738.x. [DOI] [PubMed] [Google Scholar]
- Kraig E., Haber J. E., Rosbash M. Sporulation and rna2 lower ribosomal protein mRNA levels by different mechanisms in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Oct;2(10):1199–1204. doi: 10.1128/mcb.2.10.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk T., Planta R. J. Analysis of the protein composition of yeast ribosomal subunits by two-dimensional polyacrylamide gel electrophoresis. Mol Biol Rep. 1974 Sep;1(7):409–415. doi: 10.1007/BF00385674. [DOI] [PubMed] [Google Scholar]
- Krzyzosiak W., Denman R., Nurse K., Hellmann W., Boublik M., Gehrke C. W., Agris P. F., Ofengand J. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into a functional 30S ribosome. Biochemistry. 1987 Apr 21;26(8):2353–2364. doi: 10.1021/bi00382a042. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Musters W., Venema J., van der Linden G., van Heerikhuizen H., Klootwijk J., Planta R. J. A system for the analysis of yeast ribosomal DNA mutations. Mol Cell Biol. 1989 Feb;9(2):551–559. doi: 10.1128/mcb.9.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raué H. A., Klootwijk J., Musters W. Evolutionary conservation of structure and function of high molecular weight ribosomal RNA. Prog Biophys Mol Biol. 1988;51(2):77–129. doi: 10.1016/0079-6107(88)90011-9. [DOI] [PubMed] [Google Scholar]
- Rutgers C. A., Schaap P. J., van 't Riet J., Woldringh C. L., Raué H. A. In vivo and in vitro analysis of structure-function relationships in ribosomal protein L25 from Saccharomyces cerevisiae. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):74–79. doi: 10.1016/0167-4781(90)90144-q. [DOI] [PubMed] [Google Scholar]
- Schmidt F. J., Thompson J., Lee K., Dijk J., Cundliffe E. The binding site for ribosomal protein L11 within 23 S ribosomal RNA of Escherichia coli. J Biol Chem. 1981 Dec 10;256(23):12301–12305. [PubMed] [Google Scholar]
- Sweeney R., Yao M. C. Identifying functional regions of rRNA by insertion mutagenesis and complete gene replacement in Tetrahymena thermophila. EMBO J. 1989 Mar;8(3):933–938. doi: 10.1002/j.1460-2075.1989.tb03454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Cundliffe E., Dahlberg A. E. Site-directed mutagenesis of Escherichia coli 23 S ribosomal RNA at position 1067 within the GTP hydrolysis centre. J Mol Biol. 1988 Sep 20;203(2):457–465. doi: 10.1016/0022-2836(88)90012-5. [DOI] [PubMed] [Google Scholar]
- Thompson J., Schmidt F., Cundliffe E. Site of action of a ribosomal RNA methylase conferring resistance to thiostrepton. J Biol Chem. 1982 Jul 25;257(14):7915–7917. [PubMed] [Google Scholar]
- Vester B., Garrett R. A. Structure of a protein L23-RNA complex located at the A-site domain of the ribosomal peptidyl transferase centre. J Mol Biol. 1984 Nov 5;179(3):431–452. doi: 10.1016/0022-2836(84)90074-3. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- el-Baradi T. T., Raué H. A., de Regt V. C., Verbree E. C., Planta R. J. Yeast ribosomal protein L25 binds to an evolutionary conserved site on yeast 26S and E. coli 23S rRNA. EMBO J. 1985 Aug;4(8):2101–2107. doi: 10.1002/j.1460-2075.1985.tb03898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Baradi T. T., de Regt V. C., Einerhand S. W., Teixido J., Planta R. J., Ballesta J. P., Raué H. A. Ribosomal proteins EL11 from Escherichia coli and L15 from Saccharomyces cerevisiae bind to the same site in both yeast 26 S and mouse 28 S rRNA. J Mol Biol. 1987 Jun 20;195(4):909–917. doi: 10.1016/0022-2836(87)90494-3. [DOI] [PubMed] [Google Scholar]
- el-Baradi T. T., de Regt V. C., Planta R. J., Nierhaus K. H., Raué H. A. Interaction of ribosomal proteins L25 from yeast and EL23 from E. coli with yeast 26S and mouse 28S rRNA. Biochimie. 1987 Sep;69(9):939–948. doi: 10.1016/0300-9084(87)90227-6. [DOI] [PubMed] [Google Scholar]