Abstract
Understanding the role and mechanism of signaling pathways including Notch and Wnt in colorectal carcinogenesis is critical to the development of novel therapeutics. In the present study, we analyzed the cell proliferation, migration, G2/M percentage and the expression of molecules of signaling pathways in HCT-116 cells through the inhibition of Wnt and Notch pathways, and also investigated the effect of inhibitors of Wnt and Notch pathways on tumor growth in a transplantation tumor model. We observed that rDDK-1 (an inhibitor of the Wnt signaling pathway) and LY374973 (an inhibitor of the Notch signaling pathway) synergistically inhibited the proliferation, migration and G2/M percentage of HCT-116 cell lines, and could further synergistically inhibit the tumor volume and weight in the transplantation tumor model. In the cell line and the transplantation tumor model, rDDK-1 and LY374973 further synergistically inhibited the expression level of all detected Wnt and Notch pathway genes. Our results may pave the way for using inhibitors of Wnt and Notch signaling pathways together to treat colorectal cancer.
Keywords: Wnt signaling pathway, Notch signaling pathway, colorectal cancer
Introduction
Colorectal cancer (CRC) is a leading cause of cancer mortality worldwide, with over 1.2 million new cases and more than 600,000 mortalities every year (1). In China, the incidence has increased rapidly since the 1980s (2,3). Distant metastasis following surgery is the main cause of treatment failure. There are few effective strategies to treat CRC once first-line approaches have failed. Therefore, improved understanding of the role and mechanism of signaling pathways including Notch and Wnt in colorectal carcinogenesis is critical for the development of novel therapeutics.
A hallmark of tumors is the alteration of signaling pathways that control cellular differentiation during developmental processes, including the Wnt, Notch and Hedgehog pathways (4). Notably, there is evidence that crosstalk exists among these pathways at the molecular level, and the key nodes of intersection may provide opportunities for effective targeted therapies. Multiple intracellular signaling pathways including Wnt/β-catenin signaling, epidermal growth factor receptor/Ras signaling and Notch signaling play major roles and have demonstrated crosstalk in intestinal development and tumorigenesis (5). Notch and Wnt/β-catenin play key roles not only in maintaining the growth and proliferation of CRC but also in cellular drug resistance and cancer recurrence by regulating colon cancer stem cells (6–8). Therefore, understanding the effects of Wnt and Notch signaling pathway inhibitors on tumor progression is essential.
In the present study, we investigated the effects of the inhibitors of Wnt and Notch signaling pathway synergistically on the proliferation, migration and cell cycle of HCT-116 cells, and the effects on tumor growth in a transplantation tumor model. We further studied the expression of molecules in these two signaling pathways treated by inhibitors rDDK-1 and LY374973.
Materials and methods
Cell culture
Human HCT-116 cell lines were cultured in RPMI-1640 (Invitrogen Life Technologies, San Diego, CA, USA) supplemented with 10% fetal bovine serum at 37°C and 5% of CO2/air.
Proliferation assay, Transwell migration assay and cell cycle assay
3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay was used to estimate the cell viability. HCT-116 cells were seeded at an initial density of 5,000 cells per well in a flat-bottomed 96-well cell culture plate and allowed to grow for 48 h in a humidified 5% CO2, 95% air atmosphere in an incubator maintained at 37°C. Twenty microliters of MTT (5 mg/ml) solution (Sigma Chemical Co., St. Louis, MO, USA) were added to each well and then incubated for 4 h at 37°C. After the media were removed, 200 µl dimethyl sulfoxide was added to each well to dissolve the formazan formed. After 30 min incubation at room temperature, the plates were scanned with a microplate reader that was set at 490 nm for measuring the absorbance.
The migration of cells was assayed in Transwell cell culture chambers with 6.5-mm diameter polycarbonate membrane filters having an 8 µm pore size. Briefly, 4×104 cells in 100 µl serum-free medium were added to the upper chamber of the device, and the lower chamber was filled with 600 µl culture medium with 20% fetal bovine serum. After 10 h of incubation at 37°C, the cells of the lower chamber were analyzed.
HCT-116 cells at a density of 2×106 cells/well were placed in 12-well plates and incubated with inhibitors of the Wnt and Notch signaling pathways for 48 h before the cells were harvested by centrifugation. The cells were then trypsinized, washed with phosphate-buffered saline (PBS) and treated with 50 µg/ml cold propidium iodide solution for 30 min in the dark. Flow cytometric analysis was performed on a FACSCalibur instrument (Becton-Dickinson, San Jose, CA, USA). The percentage of cells in the G0/G1, S and G2/M phases was determined by flow cytometry.
Immunofluorescence and microscopy
Cells were cultivated on cover glass slides in 24-well culture plates. After washing with PBS four times, cells were fixed in 4% paraformaldehyde for 30 min. All of the following washing and incubation steps were carried out with PBS/Tween-20 (PBST) on a rocker: cells were washed for 5 min four times and permeabilized with 0.05% saponin for 5 min. After another three washing steps, cells were fixed with pre-chilled ice-cold 100% acetone for 2 min on ice and immediately washed three times. Blocking with 4% bovine serum albumin in PBST for 30 min was followed by overnight incubation at 4°C with the primary antibody. The next day cells were washed three times and incubated with the secondary antibody for 1 h. For double-staining, the procedure was repeated with the next antibodies. Cells were imaged with a fluorescence microscope (Axiovert 40C; Carl Zeiss Microscopy, Inc., Thornwood, NY, USA).
Xenografts in BALB/c mice
A total of 24, 6–8-week-old, female BALB/c mice (Center of Experimental Animal, Kunming Medical University, Kunming, China) were subcutaneously injected with 1×106 HCT-116 cells/animal. All animals were housed under a controlled room humidity (50±10%), and maintained under a 12-h light/dark cycle with free access to water and food. Upon tumor formation, the animals were divided into four groups: i) control group (n=6); ii) rDDK-1 group (5 mg/kg, administered once weekly, all three times, n=6); iii) LY374973 group (4 mg/kg, administered once weekly, all three times, n=6); and iv) rDDK-1+LY374973 group (n=6). The study was approved by the ethics committee of Guangdong General Hospital (Guangzhou, China; approval number 2015268A).
Quantitative polymerase chain reaction (qPCR)
qPCR was used to detect the mRNA expression of molecules in the Wnt and Notch signaling pathways. The PCR reactions were performed in a total volume of 20 µl, including 10 µl 2X Power SYBR®-Green PCR master mix (Applied Biosystems, Warrington, UK), 2 µl cDNA (5 ng/µl) and 1 µl primer mix (10 µM each). The PCR amplification and detection were carried out using the LightCycler 480 II (Roche Applied Science, Basel, Switzerland) as follows: an initial denaturation at 95°C for 10 min; 40 cycles of 95°C for 15 sec and 60°C for 1 min. The relative gene expression was calculated using the comparative CT method (9). The gene expression of the target gene normalized to an endogenous reference (GAPDH) and relative to the calibrator was given by the formula 2−ΔΔCq (10).ΔCq was calculated by subtracting the average GAPDH Cq from the average Cq of the gene of interest. The ratio defines the level of relative expression of the target gene to that of GAPDH.
Western blot analysis
Cells were washed with ice-cold PBS and lysed in RIPA buffer [50 mmol/l Tris (pH 7.5), 150 mmol/l NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS)] containing phenylmethylsulfonyl fluoride (PMSF; 1 mmol/l) and protease inhibitors (2 g/ml; Protease inhibitor cocktail set III, Calbiochem, Billerica, MA, USA) on ice for 30 min. The lysates were clarified by centrifugation at 13,000 × g for 30 min at 4°C. The total protein concentration was estimated using a Protein Assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Proteins were separated by sodium dodecyl sulphate-polyacrylamide gel, transferred to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA), blocked and probed with antibodies against β-catenin (1:1,000; sc-65480; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), c-myc (1:1,000; sc-40; Santa Cruz Biotechnology, Inc.), Jagged (1:1,000; sc-390177; Santa Cruz Biotechnology, Inc.), Notch1 (1:1,000; sc-373891; Santa Cruz Biotechnology, Inc.), Notch2 (1:1,000; sc-5545; Santa Cruz Biotechnology, Inc.), DLL4 (1:1,000; ab7280; Abcam, Cambridge, MA, USA) and Pra-1 (1:1,000; ab76413; Abcam). Upon washing, blots were incubated with horseradish peroxidase-conjugated secondary antibodies and visualized by super enhanced chemiluminescence detection reagent (Applygen, Beijing, China).
Statistical analysis
Statistical analyses were conducted using Student's t-tests with the statistical software SPSS 15.0 (SPSS, Inc., Chicago, IL, USA). A corresponding two-sided P-value <0.05 was considered to indicate a statistically significant difference.
Results
Effect of inhibition of Wnt and Notch signaling pathway on proliferation, migration and cell cycle of HCT-116 cells
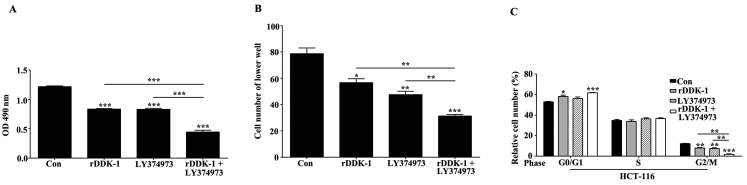
To investigate the effects of inhibition of the Wnt and Notch signaling pathway on the proliferation, migration and cell cycle of HCT-116 cells, we used the specific inhibitor rDDK-1 for the Wnt signaling pathway and LY374973 for the Notch signaling pathway. We observed that rDDK-1 and LY374973 reduced the proliferation of HCT-116 cells when used separately, and synergistically inhibited the proliferation (Fig. 1A). We also revealed that rDDK-1 and LY374973 reduced the migration ability and percentage of G2/M phase cells. rDDK-1 and LY374973 could further synergistically inhibit migration and the G2/M percentage (Fig. 1B and C).
Figure 1.
Effect of inhibition of Wnt (inhibitor, rDDK-1) and Notch (inhibitor, LY374973) signaling pathway on proliferation, migration and cell cycle of HCT-116 cells. (A) Effects of rDDK-1 or LY374973 separately and of the two inhibitors together on proliferation of HCT-116 cells using MTT assay. (B) Effects on migration using Transwell assay. (C) Effects on cell cycle using flow cytometry assay. *P<0.05, **P<0.01, ***P<0.001 vs. control.
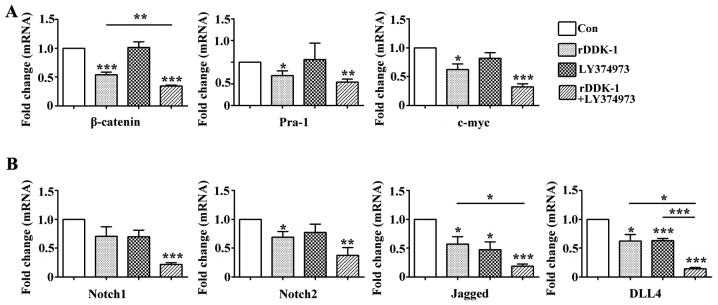
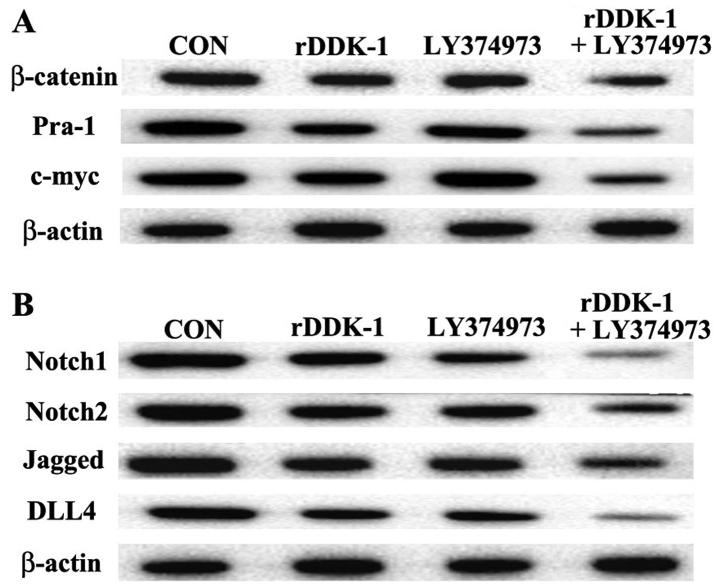
Effect of inhibitors rDDK-1 and LY374973 on Wnt and Notch pathway molecules in HCT-116 cells
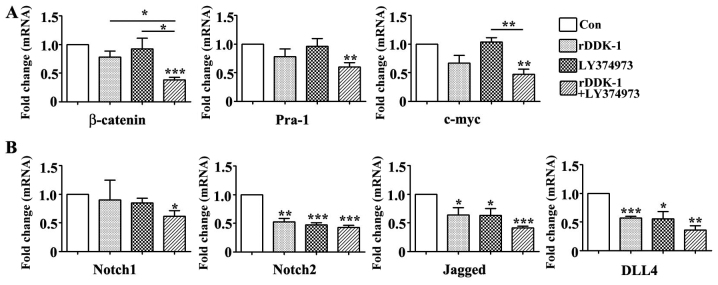
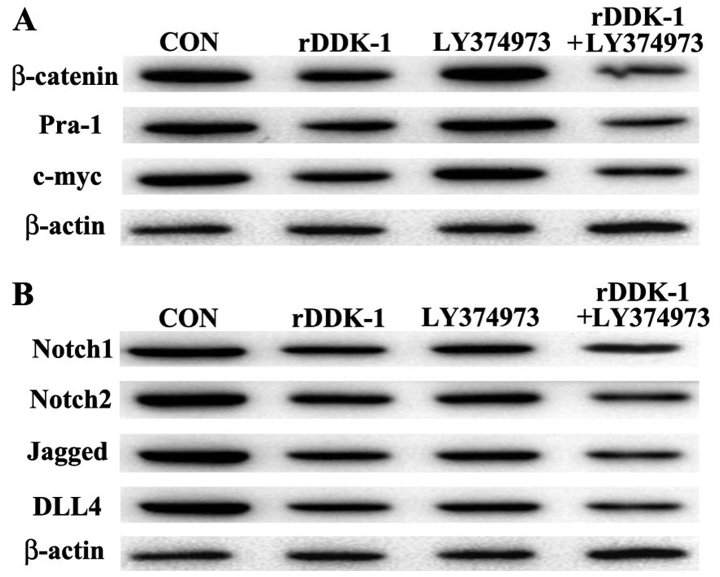
Our results revealed that rDDK-1 reduced the mRNA and protein expression of Wnt signaling pathway molecules β-catenin, Pra-1 and c-myc, and also inhibited the expression of Notch signaling pathway genes Notch2, Jagged and DLL4 (Figs. 2 and 3). LY374973 reduced the mRNA and protein expression level of Jagged and DLL4, but had no effect on Wnt signaling pathway molecules Pra-1 and c-myc (Figs. 2 and 3). With regard to the mRNA expression level, LY374973 further increased the rDDK-1 inhibition of the mRNA expression of β-catenin, Jagged and DLL4 but not Pra-1, c-myc, Notch1 and Notch2 compared with rDDK-1 alone (Fig. 2). Inconsistently, with regard to the protein expression level, LY374973 together with rDDK-1 reduced the protein expression level of all detected Wnt and Notch pathway genes (Fig. 3).
Figure 2.
Effect of inhibitors rDDK-1 and LY374973 on Wnt and Notch pathway molecules in HCT-116 cells detected by quantitative polymerase chain reaction assay. (A) Effects of rDDK-1 and LY374973 on Wnt pathway molecules β-catenin, Pra-1 and c-myc. (B) Effects of rDDK-1 and LY374973 on Notch pathway molecules Notch 1, Notch2, Jagged and DLL4. *P<0.05, **P<0.01, ***P<0.001 vs. control.
Figure 3.
Effects of inhibitors rDDK-1 and LY374973 on Wnt and Notch pathway molecules in HCT-116 cells detected by western blot assay. (A) Effects of rDDK-1 and LY374973 on Wnt pathway molecules β-catenin, Pra-1 and c-myc. (B) Effects of rDDK-1 and LY374973 on Notch pathway molecules Notch 1, Notch2, Jagged and DLL4.
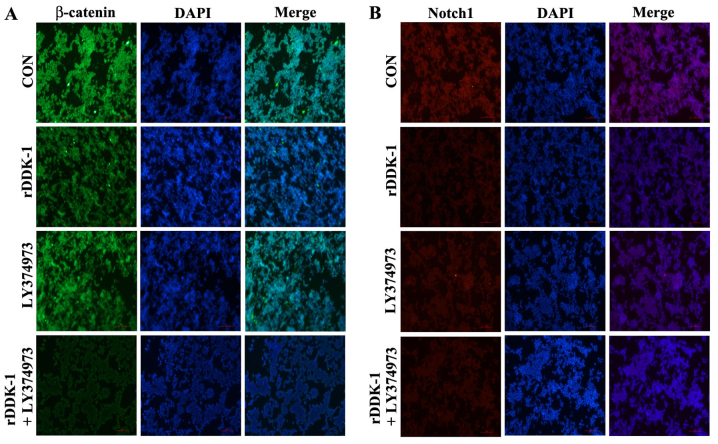
In the immunofluorescence assay, rDDK-1 was able to inhibit the expression of β-catenin and Notch1, and LY374973 increased this effect on β-catenin. However, LY374973 could only slightly inhibit the expression of Notch1 but not β-catenin (Fig. 4).
Figure 4.
Effect of inhibitors rDDK-1 and LY374973 on Wnt and Notch pathway molecules in HCT-116 cells detected by immunofluorescence assay. (A) Effects of rDDK-1 and LY374973 on Wnt pathway molecule β-catenin. (B) Effects of rDDK-1 and LY374973 on Notch pathway molecule Notch1.
Effect of inhibition of Wnt and Notch signaling pathway on tumor growth in transplantation tumor model
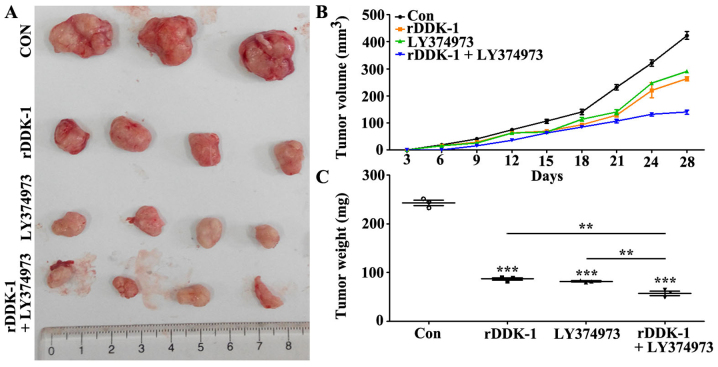
We further investigated the effects of inhibition of Wnt and Notch signaling pathway in a transplantation tumor model. The results revealed that rDDK-1 and LY374973 decreased the tumor volume and weight when used separately, and could further synergistically inhibit the tumor volume and weight (Fig. 5B and C).
Figure 5.
Effect of inhibition of Wnt (inhibitor, rDDK-1) and Notch (inhibitor, LY374973) signaling pathway on tumor growth in transplantation tumor model. (A) Effects on tumor size. (B) Effects on tumor volume. (C) Effects on tumor weight. **P<0.01, ***P<0.001 vs. control.
Effect of inhibitors rDDK-1 and LY374973 on Wnt and Notch pathway molecules in transplantation tumor model
Our results indicated that LY374973 could inhibit the mRNA and protein expression of Notch2, Jagged and DLL4, but had no effect on Wnt signaling pathway genes in tumor tissues (Figs. 6 and 7). With regard to the protein expression level, rDDK-1 together with LY374973 could further synergistically inhibit the expression level of all detected Wnt and Notch pathway genes compared with rDDK-1 or LY374973 alone (Fig. 7).
Figure 6.
Effect of inhibitors rDDK-1 and LY374973 on Wnt and Notch pathway molecules in transplantation tumor model detected by quantitative polymerase chain reaction assay. (A) Effects of rDDK-1 and LY374973 on Wnt pathway molecules β-catenin, Pra-1 and c-myc. (B) Effects of rDDK-1 and LY374973 on Notch pathway molecules Notch1, Notch2, Jagged and DLL4. *P<0.05, **P<0.01, ***P<0.001 vs. control.
Figure 7.
Effect of inhibitors rDDK-1 and LY374973 on Wnt and Notch pathway molecules in transplantation tumor model detected by western blot assay. (A) Effects of rDDK-1 and LY374973 on Wnt pathway molecules β-catenin, Pra-1 and c-myc. (B) Effects of rDDK-1 and LY374973 on Notch pathway molecules Notch1, Notch2, Jagged and DLL4.
Discussion
Understanding the role and mechanism of signaling pathways including Notch and Wnt in colorectal carcinogenesis is likely to be critical for the development of novel therapeutics. Previous studies revealed that Wnt/β-catenin and Notch signaling play major roles and had crosstalk in intestinal development and tumorigenesis (5). In addition, increased Notch/Wnt signaling also promoted the early onset of adenoma formation in APC-Cidn1 mice (11).
Previous studies have revealed that Wnt and Notch signaling pathways worked together with transcription factors including Slug, Snail and Twist to suppress E-cadherin, and finally to increase tumor progression and migration (12). Gopalakrishnan et al reported that β-catenin interacted significantly with Notch intracellular domain in adenoma and adenocarcinoma compared with normal tissues. This interaction activated CyclinD1 and Hes1 and finally promoted cell proliferation (13). Membrane-bound Notch is physically associated with unphosphorylated β-catenin in stem and colon cancer cells and negatively regulates post-translational accumulation of β-catenin protein by altering the endocytic adaptor protein Numb and lysosomal activity (14). Bordonaro et al revealed that the Notch ligand Delta-like 1 augmented the activity of the Wnt signaling pathway and transcriptionally upregulated the connective tissue growth factor gene and promoted cell growth in colon cancer (15). Zhu et al divided CRC into three transcriptional subtypes, and identified driver networks or pathways for each group. Genomic alterations in the Wnt signaling pathway were common among all three subtypes, however. Unique combinations of pathway alterations including Wnt, VEGF and Notch drove distinct molecular and clinical phenotypes in different CRC subtypes (16). Prasetyanti et al observed that Notch signals coordinated self-renewal and lineage determination not only in normal cells, but also at the adenoma and carcinoma stage in humans and mice (17). Notably, the Wnt pathway exhibited a heterogeneous activity pattern that determined stemness in all stages of disease, whereas it was previously predicted to be constitutively active in adenomas and carcinomas.
Notch and Wnt/β-catenin signaling also intersect in stem and progenitor cells and regulate each other transcriptionally. Notch and Wnt signaling function together to regulate colonic progenitor cell division and differentiation. Reedijk et al reported that Notch signaling was required for adenoma formation in response to elevated Wnt pathway signaling that occurred in an APCMin mouse model of human adenomatous polyposis coli (18). DLL4 and JAG1 were the Notch ligand genes. Katoh et al observed that JAG1 was widely expressed in a number of cancers including colon cancer, head and neck cancer and gastric cancer. JAG1 was a Wnt-dependent Notch signaling activator and was the key molecule maintaining the homeostasis of stem and progenitor cells (19). Wnt and Notch pathways regulate the self-renewal of normal stem cells (20).
Deregulation of Notch and Wnt signaling pathways plays a significant role in normal and cancer stem cells (CSCs) (21). These signaling pathways that are involved in proliferation and maintenance of CSCs lead to the development of CRC (22). A number of promising targets including Wnt/β-catenin and Notch signaling have been identified as useful targets to prevent or therapeutically inhibit CRC development (1,23). Singh et al revealed that green tea catechin epigallocatechin-3-gallate blocks carcinogenesis by affecting a wide array of signal transduction pathways, including Notch and Wnt (24).
Okuhashi et al reported that the Notch inhibitors γ-secretase inhibitors together with the Wnt inhibitor quercetin (Qu) could suppress the growth of DND-41 T-cell acute lymphoblastic leukemia cells synergistically, and that Qu treatment reduced the levels of Notch1 protein and its active fragment in DND-41 cells (24). However, there were no reports about the synergistic anti-tumor effects of Notch and Wnt inhibitors together in other types of tumors, including CRC. Our study demonstrated that rDDK-1 (an inhibitor of the Wnt signaling pathway) and LY374973 (an inhibitor of the Notch signaling pathway) synergistically inhibited the proliferation, migration and G2/M percentage of HCT-116 cell lines, and could further synergistically inhibit the tumor volume and weight in a transplantation tumor model. Our results may pave to way for using inhibitors of the Wnt and Notch signaling pathways together to treat CRC.
Further study should be conducted to explore the mechanism of Wnt and Notch signaling pathway crosstalk in CRC and to validate the effect of inhibition of the Wnt and Notch signaling pathways in other animals.
References
- 1.Curtin JC. Novel drug discovery opportunities for colorectal cancer. Expert Opin Drug Discov. 2013;8:1153–1164. doi: 10.1517/17460441.2013.807249. [DOI] [PubMed] [Google Scholar]
- 2.Lei T, Chen WQ, Zhang SW, Lei TH, Ying Q, He ZY, Wang XH. Prevalence trend of colorectal cancer in 10 cities and counties in China from 1988 to 2002. Zhonghua Zhong Liu Za Zhi. 2009;31:428–433. (In Chinese) [PubMed] [Google Scholar]
- 3.Li HL, Gao YT, Zheng Y, Zhang W, Gao LF, Xu B, Xiang YB. Incidence trends of colorectal cancer in urban Shanghai, 1973-2005. Zhonghua Yu Fang Yi Xue Za Zhi. 2009;43:875–879. (In Chinese) [PubMed] [Google Scholar]
- 4.Bertrand FE, Angus CW, Partis WJ, Sigounas G. Developmental pathways in colon cancer: crosstalk between WNT, BMP, Hedgehog and Notch. Cell Cycle. 2012;11:4344–4351. doi: 10.4161/cc.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu WK, Wang XJ, Cheng AS, Luo MX, Ng SS, To KF, Chan FK, Cho CH, Sung JJ, Yu J. Dysregulation and crosstalk of cellular signaling pathways in colon carcinogenesis. Crit Rev Oncol Hematol. 2013;86:251–277. doi: 10.1016/j.critrevonc.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Saif MW, Chu E. Biology of colorectal cancer. Cancer J. 2010;16:196–201. doi: 10.1097/PPO.0b013e3181e076af. [DOI] [PubMed] [Google Scholar]
- 7.Mishra L, Banker T, Murray J, Byers S, Thenappan A, He AR, Shetty K, Johnson L, Reddy EP. Liver stem cells and hepatocellular carcinoma. Hepatology. 2009;49:318–329. doi: 10.1002/hep.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ischenko I, Seeliger H, Schaffer M, Jauch KW, Bruns CJ. Cancer stem cells: how can we target them? Curr Med Chem. 2008;15:3171–3184. doi: 10.2174/092986708786848541. [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Brennan C, Rook M, Wolfe JL, Leo C, Chin L, Pan H, Liu WH, Price B, Makrigiorgos GM. Balanced-PCR amplification allows unbiased identification of genomic copy changes in minute cell and tissue samples. Nucleic Acids Res. 2004;32:e76. doi: 10.1093/nar/gnh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Pope JL, Ahmad R, Bhat AA, Washington MK, Singh AB, Dhawan P. Claudin-1 overexpression in intestinal epithelial cells enhances susceptibility to adenamatous polyposis coli-mediated colon tumorigenesis. Mol Cancer. 2014;13:167. doi: 10.1186/1476-4598-13-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umar S. Enteric pathogens and cellular transformation: bridging the gaps. Oncotarget. 2014;5:6573–6575. doi: 10.18632/oncotarget.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopalakrishnan N, Saravanakumar M, Madankumar P, Thiyagu M, Devaraj H. Colocalization of β-catenin with Notch intracellular domain in colon cancer: a possible role of Notch1 signaling in activation of CyclinD1-mediated cell proliferation. Mol Cell Biochem. 2014;396:281–293. doi: 10.1007/s11010-014-2163-7. [DOI] [PubMed] [Google Scholar]
- 14.Kwon C, Cheng P, King IN, Andersen P, Shenje L, Nigam V, Srivastava D. Notch post-translationally regulates β-catenin protein in stem and progenitor cells. Nat Cell Biol. 2011;13:1244–1251. doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bordonaro M, Tewari S, Atamna W, Lazarova DL. The Notch ligand Delta-like 1 integrates inputs from TGFbeta/Activin and Wnt pathways. Exp Cell Res. 2011;317:1368–1381. doi: 10.1016/j.yexcr.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, Wang J, Shi Z, Franklin JL, Deane NG, Coffey RJ, Beauchamp RD, Zhang B. Deciphering genomic alterations in colorectal cancer through transcriptional subtype-based network analysis. PLoS One. 2013;8:e79282. doi: 10.1371/journal.pone.0079282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasetyanti PR, Zimberlin CD, Bots M, Vermeulen L, Fde S Melo, Medema JP. Regulation of stem cell self-renewal and differentiation by Wnt and Notch are conserved throughout the adenoma-carcinoma sequence in the colon. Mol Cancer. 2013;12:126. doi: 10.1186/1476-4598-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reedijk M, Odorcic S, Zhang H, Chetty R, Tennert C, Dickson BC, Lockwood G, Gallinger S, Egan SE. Activation of Notch signaling in human colon adenocarcinoma. Int J Oncol. 2008;33:1223–1229. doi: 10.3892/ijo_00000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh M, Katoh M. Notch ligand, JAG1, is evolutionarily conserved target of canonical WNT signaling pathway in progenitor cells. Int J Mol Med. 2006;17:681–685. [PubMed] [Google Scholar]
- 20.Korkaya H, Wicha MS. Selective targeting of cancer stem cells: a new concept in cancer therapeutics. BioDrugs. 2007;21:299–310. doi: 10.2165/00063030-200721050-00002. [DOI] [PubMed] [Google Scholar]
- 21.Karamboulas C, Ailles L. Developmental signaling pathways in cancer stem cells of solid tumors. Biochim Biophys Acta. 2013;1830:2481–2495. doi: 10.1016/j.bbagen.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Roy S, Majumdar AP. Signaling in colon cancer stem cells. J Mol Signal. 2012;7:11. doi: 10.1186/1750-2187-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandurangan AK, Esa NM. Dietary non-nutritive factors in targeting of regulatory molecules in colorectal cancer: an update. Asian Pac J Cancer Prev. 2013;14:5543–5552. doi: 10.7314/APJCP.2013.14.10.5543. [DOI] [PubMed] [Google Scholar]
- 24.Singh BN, Shankar S, Srivastava RK. Green tea catechin, epigallocatechin-3-gallate (EGCG): mechanisms, perspectives and clinical applications. Biochem Pharmacol. 2011;82:1807–1821. doi: 10.1016/j.bcp.2011.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]