Abstract
Evolution sculpts the olfactory nervous system in response to the unique sensory challenges facing each species. In vertebrates, dramatic and diverse adaptations to the chemical environment are possible because of the hierarchical structure of the olfactory receptor (OR) gene superfamily: rapid growth or pruning across the OR gene tree accompany major changes in habitat and lifestyle; independent selection on OR subfamilies can permit local adaptation or conserved chemical communication; and genetic variation in single OR genes among thousands can alter odor percepts and behaviors driven by precise chemical cues. However, this genetic flexibility contrasts with the relatively fixed neural architecture of the vertebrate olfactory system, whose slower rate of divergence mandates that new olfactory receptors integrate into segregated and functionally-distinct neural pathways. This organization allows evolution to couple critical chemical signals with selectively advantageous responses, but also constrains relationships between olfactory receptors and behavior. The coevolution of the OR repertoire and the structure of the olfactory system therefore reveals general principles of how the brain solves specific sensory problems and how it adapts to new ones.
Olfaction, the sense of smell, has evolved to detect signals from the chemical environment, which contains clues about where to move, what to eat, when to reproduce, and which stimuli to remember as rewarding or dangerous [1, 2]. How an animal responds to chemical cues is in part learned and in part innate, depending on how the olfactory nervous system has been shaped by both experience across a lifetime and evolution across generations.
Interacting with the chemical world presents a challenge unlike other sensory tasks. In contrast to light and sound waves, which vary continuously in wavelength and amplitude, chemical compounds differ discretely in an enormous number of dimensions. Compared to vision and audition, olfaction requires many more receptors – proteins expressed in sensory organs that convert a physical event into an electrochemical signal carried by neurons. A single photoreceptor pigment may interact with a range of wavelengths that is sufficiently informative for an animal, as with our dark-vision rhodopsin; however, biophysical constraints restrict chemoreceptor proteins to interact with only subsets of chemical space. The high dimensionality of chemical space and the diversity of the chemical stimuli an animal might encounter have therefore selected for genomes that encode enormous receptor repertoires, containing hundreds to thousands of olfactory receptor (OR) genes [3–6].
Here we examine how the olfactory receptor repertoire and the structure of the olfactory nervous system evolve in concert to sense and interpret chemical information. Adaptations in the genetic and neural architecture of olfaction reflect the unique chemosensory challenges faced along a species’ lineage — detecting novel odorants, discerning especially critical odors with high acuity, and responding behaviorally to molecules that acquire new meaning. We argue that the receptors underlying vertebrate olfaction possess two properties essential for the range of adaptations seen in vertebrate olfactory systems: a flexible and hierarchical pattern of evolution that allows receptor adaptation to both dramatic and subtle changes in the chemical environment; and access, through specific expression patterns, to a diverse array of neural pathways that govern both hardwired, instinctual behaviors as well as more flexible odor learning. In this way, OR families and subfamilies have evolved on larger scales to inform the animal about wide swaths of chemical space, while at the same time some individual receptors have become highly tuned to key odorants that elicit innate responses. We speculate that the unique ability to incorporate new and evolving OR genes has produced the substantial genetic and structural diversity of olfactory systems seen across the vertebrate lineage [7–9].
The Molecular, Cellular and Neural Architecture of Vertebrate Olfaction
Smell begins when odor molecules bind to OR proteins on the endings of sensory neurons. The set of odors an animal can detect therefore depends on the expression pattern and the protein structure encoded by each of its OR genes. In vertebrates, nearly all OR genes encode seven-pass transmembrane G-protein coupled receptors (GPCRs) that, upon ligand binding, signal through G-proteins and intracellular second messengers to ultimately open membrane ion channels; this depolarizes the sensory neuron to drive action potentials that are conducted along its axon into the olfactory bulb of the brain (Figure 1) [10].
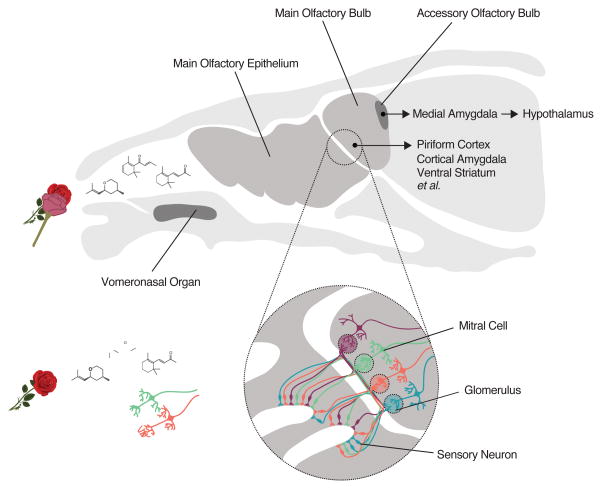
Figure 1.
Organization of the mouse olfactory system (partially adapted from Dulac & Torello, 2003). Odors are blends of molecular compounds inhaled into the nasal cavity, where they interact with olfactory receptor proteins expressed in the main olfactory epithelium (medium gray), the vomeronasal organ (dark gray), or one of several smaller sensory structures not pictured (upper). Each sensory neuron expresses a single olfactory receptor denoted by its color, and neurons expressing the same receptor project to insular structures in the olfactory bulb called glomeruli (lower). Glomeruli corresponding to a given receptor have stereotyped spatial positions across animals. Mitral cells in the olfactory bulb each send a dendrite into one glomerulus (main olfactory bulb) or multiple glomeruli corresponding to the same olfactory receptor (accessory olfactory bulb, not pictured), and project axons to a variety of central brain regions that mediate odor learning and innate odor-driven behaviors. The targets of main and accessory bulb mitral cell projections are largely distinct.
In vertebrates, most olfactory sensory neurons express a single olfactory receptor gene from one of five major GPCR families [11]. The evolutionary history of each family relates roughly to the region of chemical space it probes, as the odorant structures bound by family members are reminiscent of those bound by the specific ancestral non-olfactory GPCR from which each family derived (detailed below.) In the rodent olfactory system where they were first discovered, members of each OR family are expressed in a Pointilistic pattern within spatially restricted zones of the nasal epithelia. Most ciliated sensory neurons of the main olfactory epithelium (MOE) express receptor genes of the largest family, the “classical” mammalian olfactory receptors (mORs) [12, 13], while a minority of neurons instead express members of the much smaller trace amino-acid receptor (TAAR) family [14]. In the vomeronasal organ (VNO), microvillar sensory neurons in the apical epithelial layer express Vomeronasal type 1 Receptors (V1Rs) [15], but neurons in the basal layer each express one Vomeronasal type 2 Receptor (V2R) plus a chaperone V2R in characteristic combinations [16, 17] [18]. Finally, some neurons in both layers of the VNO instead express members of the small Formyl Peptide Receptor-related family (FPRs) [19, 20] (Figures 1, 2).
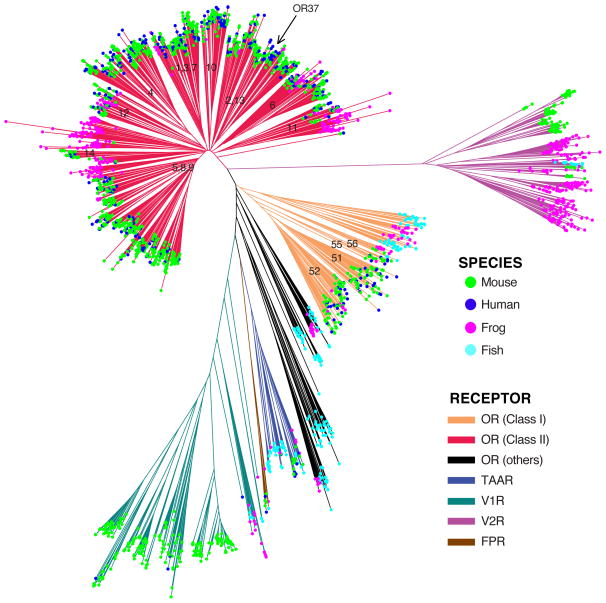
Figure 2.
Phylogenetic relationships of the vertebrate GPCR olfactory receptor repertoire. A representative sample of ~2700 GPCR functional OR genes from fish (Danio rerio), frog (Xenopus laevis), mouse (Mus musculus), and human (Homo sapiens) genomes was used to construct a phylogenetic tree (cf. Manzini & Korsching 2011). Notable features are visible, including large expansions of Class II ORs (red) in tetrapods, V2Rs (purple) in amphibians, TAARs (blue) in fishes, and V1Rs (teal) in mammals; significant losses of functional ORs in humans (blue dots) compared to mice (green dots); the monophyly of Class I (orange) and Class II (red) tetrapod ORs; and the subfamily structure of the “classical” ORs, with numeric labels corresponding to the subfamilies defined in Hayden et al. (2010). A Class II OR subfamily (2,13) itself contains a smaller subfamily, the “OR37” receptors, which has atypically expanded and been conserved at the protein sequence level in humans. Note that all mammalian and some frog Class I ORs form a monophyletic clade within the larger set of Class I ORs and other fish OR subfamilies that are neither Class I nor Class II.
The one-receptor-per-neuron organization shapes how odor representations propagate through the olfactory system. The axons of primary sensory neurons expressing the same receptor gene coalesce onto insular structures called glomeruli in the outer layer of the olfactory bulb, with the positions of the glomeruli corresponding to a given OR approximately fixed from one animal to the next [21] (Figure 1). A given odorant binds to a stereotyped subset of olfactory receptors and thereby activates a characteristic subset of glomeruli – like keys pressed together as a chord on a piano – such that odors elicit unique spatiotemporal activity patterns across the olfactory bulb [22].
Odor responses in the olfactory bulb drive innate and learned behaviors via neural projections to more central brain regions. Mitral and tufted (MT) cells of the main olfactory bulb each send a single dendrite into one glomerulus, and extend parallel axon branches into multiple higher olfactory targets, including the piriform cortex – which promotes associations between odors and rewards or punishments – and the cortical amygdala – which mediates instinctual responses to some predator odors (Figure 1) [23–25]. In contrast, mitral cells in the accessory olfactory bulb each extend a dendrite into multiple glomeruli corresponding to the same VR and project to amygdalar, hypothalamic, and basal forebrain nuclei involved in reproductive and stress responses (Figure 1) [26]. The targets of main and accessory bulb mitral cells are mostly distinct, and thus higher-order olfactory brain regions have likely adapted to receive information about odors sensed by different olfactory receptors that release distinct behaviors.
Olfactory brain regions, especially the olfactory bulb, vary tremendously in relative size across vertebrate species; furthermore, in many species, the accessory olfactory bulb and the vomeronasal organ are absent [7, 8]. We therefore focus primarily on the conserved genetic architecture of vertebrate olfaction, which likely arose in the common ancestor of all ray-fined fishes and tetrapods (lived ~460-430 MYA). The genome of the zebrafish contains multiple members of four of the five major GPCR OR families (ORs, TAARs, V2Rs, and several V1Rs) (Figure 2) [27–29] and its sensory neurons generally express these receptors in the same “one receptor, one sensory neuron” pattern as in mice [30–32]. Zebrafish sensory neurons project to insular glomeruli within the olfactory bulb, where innervation from different sensory neuron types (expressing distinct OR families) is at least partially segregated [33, 34]. Although the pattern of convergence from sensory neurons to glomeruli has not been assessed for most species, this conservation across zebrafish and rodents suggests that the neural organization of the olfactory system forms a stable background on which to compare the changes in OR gene repertoire. The molecular mechanism for choosing a single OR to be expressed in each sensory neuron – which engages both cell type-specific transcription factors as well as stochastic DNA-DNA interactions [11] – seems to have directly linked the genetic evolution of OR families with the functional evolution of odor perception. This process has allowed the olfactory system to flexibly incorporate or discard chemosensors, as each OR gene stakes out its own population of sensory neurons in the olfactory epithelium and its own glomerular territory in the olfactory bulb without disrupting the expression or function of other receptors.
The Birth-and-Death Mechanism of Gene Family Evolution Allows Specific Adaptation to Chemical Space
Singular OR expression potentially affords each newly arising OR gene a neural pathway to inform the brain about the odorants it binds. Evolution of the OR gene repertoire should therefore reflect the salient features of an animal’s chemical environment. To test this hypothesis, genomic observations of OR evolution must be combined with knowledge of what molecules individual receptors bind and how different animals respond to these odors. Unfortunately, because of the enormous number of OR proteins and the difficulty of measuring their molecular tuning in vitro, we know only a small fraction of OR-odorant interactions [35].
Nevertheless, thanks to the mechanism of OR repertoire evolution, it is possible to compare finer portions of the OR gene tree as they grow or die off on multiple evolutionary timescales. The membership of olfactory receptor families evolves through a characteristic birth-and-death process. Changes in the number of OR genes result from duplications of contiguous family members during unequal meiotic crossing over, resulting in a redundancy and functional independence of the new genes that allows them to mutate and acquire new ligand-binding properties [36]. A side-effect of the relaxed selection pressure on recently duplicated genes is the accumulation of nonsense or frameshift mutations that terminate receptor function, producing pseudogenes embedded in rapidly duplicating OR gene clusters [37]. This birth-and-death process further explains the low proportion of orthologous receptors found among species, as many new OR genes can arise in the time since the divergence of even closely related lineages [38]. Other modes of gene family evolution have occasionally homogenized the OR repertoire, such as gene conversion between tightly-linked subfamily members and across-species conservation of single ORs that function outside the olfactory system [38–40]; however it is clear that birth-and-death and relaxed selection are the dominant processes by which different species evolve markedly different sets of olfactory receptors [41, 42].
The birth-and-death mechanism allows natural selection to act independently at multiple levels of the OR gene tree, including each of the five olfactory GPCR families, more recently diverged subfamilies within each OR family, and individual OR genes (Figure 2). We argue below that progressively more subtle adaptations in the habitat, lifestyle, and behavior of vertebrate lineages correspond to increasingly fine-scale growth and death on branches of the OR tree. We then discuss how the olfactory nervous system might take advantage of these changes in the receptor repertoire to generate new innate and learned olfactory behaviors.
Olfactory Overhaul: Widespread Reshaping of the OR Repertoire in New Sensory Environments
As some vertebrates have adapted to new habitats or shifted to lifestyles that rely less on smell, entire OR families and olfactory sensory organs have become dispensable. For example, as its visual system has expanded dramatically, the old world primate lineage has seen a massive loss of functional olfactory receptor genes, as well as a near total loss of the vomeronasal receptor repertoire, accompanied by pseudogenization of an important VNO chemotransduction channel gene TrpC2 (Figure 2) [43, 44]. Selection on primate OR genes may have relaxed following the duplication of the opsin gene encoding the long-wavelength retinal photopigment, as the genomes of trichromat primates – all old world monkeys and one species of new world monkey, the howler monkey – have significantly higher ratios of pseudogenized to intact OR genes than dichromat new world monkeys [45]. Trichromacy may have allowed color vision to take over some olfactory tasks, like discriminating the ripeness of leaves by their appearance along the red-green axis [46, 47]. This view is controversial, however, because the proportion of OR pseudogenes in a mammalian genome does not correlate with the number of intact ORs and thus poorly estimates olfactory function [38]; moreover, loss of intact ORs in primates may have begun before the divergence of new world monkeys and old world monkeys [48]. Nevertheless, the growth of the visual nervous system in old world primates likely reduced reliance on olfaction for many social and foraging behaviors, which may represent a general pattern: in multiple cases the enhancement of other senses like vision, echolocation, and electroreception coincides with a smaller OR repertoire [49] [50].
Changes in habitat that broadly alter chemical ecology appear to have also resized and reshaped the OR families dramatically, though directly demonstrating causal relationships remains a challenge. The divergence of the tetrapod lineage from its aquatic relatives ~420 MYA was followed by an order-of-magnitude expansion in the major vertebrate OR gene family (represented by the mORs in mammals), as hundreds to thousands of these “classical” OR genes are found in individual amphibian and terrestrial vertebrate genomes, whereas fish genomes have, at most, 150 (Figure 2) [51]. The idea that having more OR genes is adaptive on land is strengthened by converse evidence from multiple independent mammalian lineages that returned to aquatic life. In these animals (e.g., whales, seals, and manatees), the number of intact OR genes has dropped since they diverged from their terrestrial relatives (e.g. cattle, dogs, and elephants) [52].
The classical OR gene family has two major branches, the so-called Class I and Class II ORs; however, only the Class II branch shows a massive increase in membership as a possible result of adapting to land. This receptor subfamily is expansive in the genomes of tetrapods (amphibians and terrestrial vertebrates) but is essentially absent in fishes, including one more closely related to tetrapods than teleosts – the coelacanth (Figure 2) [53, 54]. Why might a particular OR subfamily have expanded in a given vertebrate lineage? The simplest hypothesis is that the ancestral family members were useful for sensing type of chemical structure common or critical in that animal’s chemical environment. Having more OR genes with somewhat diversified ligand-binding regions could have provided a selective advantage – for instance by increasing perceptual range, sensitivity, or discriminative power in this region of chemical space. The evolutionary history and functional tests of the distinction between Class I and Class II ORs support this model. First identified in the amphibian Xenopus laevis genome, Class I genes cluster phylogenetically with fish ORs, while Class II genes cluster with the majority of mammalian ORs (Figure 2) [55]. Furthermore, Class I genes are expressed exclusively in the lateral diverticulum of the frog nasal cavity, which senses waterborne but not airborne odorants from the outside world, while Class II genes are expressed only in the medial diverticulum, which opens only when the frog is in air [55]. Functional characterization supports the hypothesis that Xenopus Class I genes are “fish-like”, as when expressed in heterologous cells they detect highly water-soluble ligands like charged amino acids – important water-borne chemosignals for fish. Conversely, Class II Xenopus ORs detect some airborne volatile compounds like the aroma of coffee [56]. The ligands of mammalian Class I ORs are also on average more polar (and thus more water-soluble) than Class II ligands [35]; but the relative size of the Class I repertoire has remained stable across mammals, suggesting that even in purely terrestrial environments these receptors bind odorants of ecological significance [38]..
Skipping the Classics: Particular Habitats May Have Favored Expansion of Distinct OR Families
The presence of smaller “classical” OR families in aquatic animals does not necessarily mean that olfaction is less important in water than in air. Similar to the above cases of Class I and Class II ORs, the aquatic environment may have instead favored the use of other OR families because their ancestral ligand-binding properties gave them a head-start in detecting more common aqueous odorants. For instance, the TAAR family has expanded rapidly in teleost fishes (Figure 2). The zebrafish genome encodes over 100 functional TAARs, almost an order of magnitude more than any vertebrate outside the teleost clade. This expansion includes the derivation of an entirely new TAAR subfamily under strong positive selection in the teleost lineage, which was first thought not to encode amine receptors (like the other TAARs) because a canonical amine-binding motif is lost in many subfamily members [27]. It has now been shown, however, that these teleost TAARs initially evolved a second amine-binding motif, changing their specificity to diamines over monoamines; later, a subset of these secondarily lost the original motif, resulting in TAARs that use a distinct mechanism for amine binding [57]. Together this pattern of TAAR duplication and diversification suggests that the outsized importance of amine chemosensation drove this particular OR family to prominence in the teleost lineage. This notion is consistent with the important role known to be played by amino acids and their diamine decarboxylation products in the chemical ecology of zebra fish [58].
The ancestral TAAR was likely the orthologue of the mammalian TAAR1 and was therefore probably a receptor for internally produced trace monoamines like TAAR1 ligands tyramine, octopamine, and tryptamine [59]. Thus, the expansion of the TAAR family in teleosts may have been biased from the start, since only a few mutations in new TAAR genes would allow better sensing of amines, a region of chemical space apparently important for the behavior of these fishes. For example, cadaverine, a diamine signature of putrefaction, is detected by the zebrafish-specific receptor TAAR13c [58] and elicits innate aversion. This link between a novel receptor and amine-driven behavior may be a lineage-specific adaptation, as another teleost, the goldfish, is attracted to cadaverine (perhaps via the action of distinct TAAR receptors) (Figure 3) [57, 58, 60].
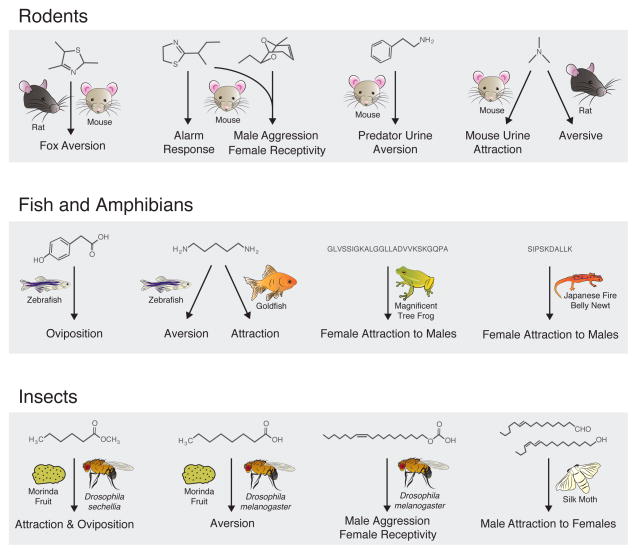
Figure 3.
A sample of monomolecular odorants that drive innate, species-specific behaviors across vertebrates and insects. Instinctual reproductive, protective, and feeding behaviors are released by a variety of chemical structures, including volatile airborne small molecules in rodents (top row); waterborne acids, bases, and peptides in fishes and amphibians (middle row); and volatile hydrocarbons in insects (bottom row). Chemical structures (receptor responsible for behavior, if known), left to right: top row 2,4,5-trimethythiazoline, 2-sec-butyl-4,5- dihydrothiazole [SBT], 2,3-dehydro-exo-brevicomin [DHB] [76], phenethylamine (Mus musculus TAAR4) [84], trimethylamine (Mus musculus TAAR5) [85]; middle row 4-hydroxyphenylacetic acid (Danio rerio ORA1 [a V1R]) [95], cadaverine (Danio rerio TAAR13c) [58], splendiferin (peptide pheromone for Litoria splendida), sodefrin (peptide pheromone for Cynops pyrrhogaster) [63]; bottom row methylhexanoate (D. sechellia Or22a), octanoic acid [78], cis-11-vaccenyl acetate (D. melanogaster Or67d) [79], (10E,12Z)-hexadeca-10,12-dien-1-al [upper] and (10E,12Z)-hexadeca-10,12-dien-1-ol [lower] (aliases bombykal, receptor Bombyx mori BmOr3, and bombykol, receptor B. mori BmOr1, respectively) [80]. Behavioral responses are sometimes enhanced by combinations of odorants, as with SBT and DHB in mice and bombykal and bombykol in the silk moth. Moreover, responses to the same compound(s) may differ drastically between species or between sexes of the same species.
The evolutionary trajectory of the TAARs raises the possibility that that along different lineages, specific ancestral receptors have “gotten lucky” by binding chemical structures similar to those with wider ecological significance. This line of reasoning could explain why the V2R family expanded dramatically and reached its largest membership in the amphibian lineage (Figure 2) [61]. The V2Rs are derived from an ancestral GPCR that senses the amino acid glutamate and have diversified to allow broader detection of amino acids and longer polypeptides [17]. The amphibian V2R family is expressed in the amphibian vomeronasal organ, which plays an essential role in sensing waterborne peptide sex signals and coordinating complex mating behavior [62, 63].
Given these examples suggesting that the ecological importance of a given class of chemicals is reflected by the size of the OR family that detects that chemical class, it is intriguing to consider its converse: whether the apparent expansion of a specific OR family can reveal the types of chemical cues that are most relevant to a given species. Consider the case of the semiaquatic platypus, one of only two extant members of the monotreme lineage, which was initially thought not to rely on strong olfactory abilities given the high proportion of pseudogenes in its classical OR family and its use of a unique electroreceptive “bill sense” for locating prey in brackish water; however, it was later found that the platypus genome contains more functional V1R family members than any other vertebrate, reflecting an unparalleled expansion of this particular gene family in the monotreme lineage [64]. Indeed, the platypus possesses one of the most complex vomeronasal organs described, suggesting that odor detection through the “accessory” olfactory system has become its main mode of chemosensation. Because V1Rs detect many steroid hormones that regulate reproductive and mating behavior [65], combing this region of chemical space may reveal cues critical to the behavior of the platypus and ecological reasons that this particular receptor family expanded.
The Finer Things in Life: Olfactory Specialization Through OR Subfamily Selection
OR birth and death operates locally within the genome, producing closely-related receptor subfamilies (Figure 2) [5, 37, 42] that may evolve independently of one another. There is evidence that natural selection can operate on the OR repertoire with this finer-scale flexibility to probe niche-specific chemical cues with more precision. For example, Hayden et al. (2010) found that the relative sizes of each mammalian OR subfamily were more representative of a species’ ecotype – aquatic, semi-aquatic, terrestrial, or flying – than of its ancestry within the phylogenetic tree of mammals [52]. These results suggest that particular receptor subfamilies are better suited to probe different environments.
Testing this hypothesis requires identifying the odors sensed by these receptors and determining whether larger subfamilies actually confer a selective advantage in discerning these cues. Observations of subfamily selection on shorter timescales, correlated with even finer niche adaptation, may help hone in on the sensory demands driving receptor evolution. For example, in an analysis of OR evolution in bats, the relative sizes of two subfamilies correlate positively and negatively with independent adaptations to a purely frugivorous diet [66]. A similar investigation of bird and reptile OR repertoires identified a subfamily associated with carnivory and several Class I and Class II subfamilies enlarged in aquatic and terrestrial feeders, respectively [67]. Members of these OR subfamilies may have become tuned to volatile compounds found in the foods sought by individual species, and their expression in the olfactory system may reveal how selective neural pathways adapt to influence foraging and food selection.
Similarly, distinct subfamilies of the mouse V2R tree appear tuned to labile, innate fear-producing chemosignals (“kairomones”) derived from different types of mouse predators (e.g. snakes, birds, rats, and cats) [65]. The observation that the family of Major Urinary Proteins (MUP) acts through V2Rs to control both interspecies defensive behaviors and intraspecies aggressive behaviors suggests that V2R-MUP pairing or binding affinities may be particular targets of lineage-specific subfamily expansion and selection [68–70]. Furthermore, if different V2R subfamilies are in fact tuned to the chemical signatures of particular predators, then their sensory signals could remain segregated as they propagate through the nervous system – which might allow unique defensive responses to evolve for each.
Finally, the modular nature of receptor gene evolution allows small subfamilies to retain critical chemosensory roles, even against a backdrop of widespread receptor loss. One peculiar mammalian subfamily, called “OR37”, is surprisingly conserved in both mice and humans, even though humans have lost more than half of their functional OR genes overall (Figure 2) [50, 71]. Indeed, this subfamily has expanded independently in the human lineage, suggesting it has not lost its utility like other human subfamilies (Figure 2) [72]. Together these data suggest that humans may still detect a set of informative odors through the OR37 family, such as the long-chain fatty aldehyde ligands sensed by mouse OR37 members [73]. Intriguingly, mouse mitral cell projections from OR37 glomeruli target several hypothalamic nuclei rather than the cortical regions typical of most main olfactory bulb projections (Figure 5) [74, 75]. Thus the OR37 family may have evolved to link the sensation of a certain chemical class to regulation of innate behaviors or internal state in a way that has remained crucial across species. The overall pattern of birth-and-death OR evolution therefore demonstrates lineage-specific adaptation to the chemical environment.
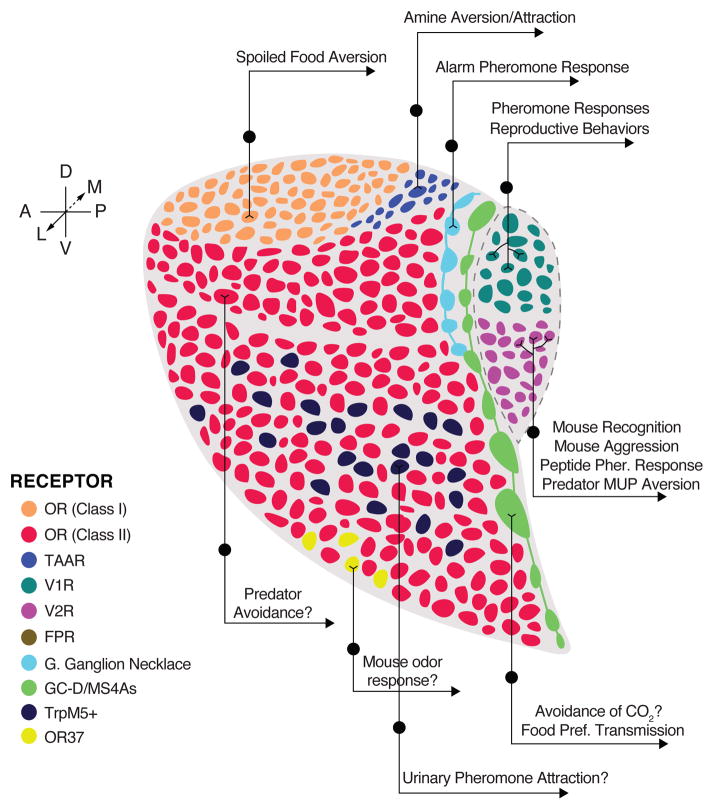
Figure 5.
Spatial and molecular organization of projection targets and behavioral responses downstream of distinct mouse olfactory bulb glomeruli. Circumscribed zones of glomeruli are innervated by sensory neurons expressing each of the four largest GPCR OR families or the MS4As. In addition, Class I and Class II OR glomeruli occupy the dorsal-most and more ventral zones, respectively; and projections from Grueneberg Ganglion sensory neurons form their own “necklace” (light blue) anterior to the GC-D/Ms4a “necklace” (green). Dorsal Class II OR glomeruli form a molecularly defined zone, which includes glomeruli necessary for innate aversion of some predator odorants. Within the ventral zone of Class II OR glomeruli, some regions are enriched for the glomeruli of TrpM5- or OR37 subfamily-expressing sensory neurons, which have second-order projections to the “vomeronasal” amygdala and the hypothalamus, respectively.
The Evolution of Single OR Genes: Direct Links Between Sensation and Action
Adaptation through change in the number of OR genes is only possible if the olfactory nervous system has ways to employ the new receptor genes that emerge as particular subfamilies grow and diversify. Because natural odor blends activate only a subset of receptors [22], expressing newly duplicated and diversified subfamily members may provide a more nuanced neural representation of the regions of chemical space they sense. This could in turn allow finer resolution in discriminating the makeup or concentrations of the odors sensed by the ancestral receptors.
What this model does not explain is how an animal’s interpretation of a given odor may evolve. Many vertebrates exhibit innate patterns of behavior that can be released by precise chemical stimuli, including even some monomolecular odorants outside the context of their natural odor blends (Figure 3) [58, 63, 76, 77]. It is not obvious how evolution could couple these blend constituents to appropriate innate behaviors via changes in olfactory receptor number. Animals may instead interpret these chemosignals through single, precisely- and sensitively-tuned olfactory receptors that tap into hardwired, behavior-evoking neural circuits. This pattern of receptor tuning and neural organization is prominent in insects, perhaps because their olfactory receptor repertoires are much smaller (and likely less redundant) than in vertebrates. The compounds that elicit innate responses in both insects and vertebrates are essential for survival and reproduction – sex pheromones, attractive or aversive food odors, and predator signatures – explaining why olfactory neurons have evolved to detect them with high sensitivity and to couple directly to neural circuits that drive innate behaviors (Figure 3) [78–81]. For instance, the exocrine-secreted peptides ESP1 and ESP22 act exclusively through V2Rp5 and an unknown V2R to promote female sexual receptivity (the “lordosis” posture) and to block male sexual activity toward juveniles below reproductive age, respectively [82, 83]. Similarly phenethylamine found in the urine of carnivores, acts by binding to the receptor TAAR4 to promote avoidance behavior [84, 85]. These individual receptors therefore play non-redundant roles in shaping odor perception and interpretation, suggesting that even in olfactory systems that use hundreds or thousands of receptors, single members of these gene families may evolve outsized ecological significance and privileged neural access to specific central circuits [84].
This model also predicts that, in animal lineages in which the role of olfaction in driving stereotyped reproductive and defensive behaviors has waned, there will be few cases where individual receptor genes are under strong selection pressure. This seems to be borne out in humans, for whom there is almost no evidence for positive selection in the coding sequences of single OR genes since the human-chimpanzee divergence ~7 MYA. It is therefore unlikely that particular OR genes have conferred a selective advantage during human evolution [86].
That said, genetic variation in single human ORs – under selective pressure or not – can produce large differences in odor perception. Detection thresholds and (un)pleasantness ratings of many odors have been linked to common polymorphisms in and around human OR genes, including a receptor coding mutation that alters sensitivity to and valence of the male hormones androstenone and androstenedione [87]; a mutation that accounts for nearly all observed variation in the perception of β-ionone and dramatically alters preference for foods emiting this odor [88]; and a polymorphism closely linked to a cluster of OR genes that tracks with the aversive “soapiness” of cilantro [89]. Furthermore, as in other mammals, gland secretions can in a laboratory setting can bias sexual attraction and nipple-orienting behavior in adult and infant humans, respectively, raising the possibility that we employ unidentified pheromones [90] [91]. If these chemosignals follow the pattern of other social and suckling pheromones, they are likely to include monomolecular odorants acting through high-affinity, specialist receptors [77, 92]. Even if they are not under strong selection, then, it remains possible that single human ORs contribute meaningfully to behavior through unknown mechanisms.
Segregated Olfactory Pathways As Substrates For Innate Olfactory Behaviors
Defined odorants can drive innate responses or preferences through a few receptors, even against a background of hundreds or thousands – many of which may be active simultaneously during typical sensation of complex odor blends [93]. How, then, are particular chemical cues coupled precisely to appropriate odor-driven behaviors? It appears that the olfactory system begins to sort odor information in the peripheral sense organs. Several hardwired, parallel pathways – from sensory neurons to subregions of the olfactory bulb to distinct central brain targets – arose early in the evolution of vertebrates, using distinct receptor families and subfamilies to convey different categories of chemical signal. Although zebrafish, unlike mice, have only a single olfactory epithelium, the primary sensory neurons that express ORs and TAARs are molecularly, morphologically, and anatomically distinct from those that express amino acid-sensing V2Rs; these two cell types (ciliated and microvillar sensory neurons, respectively) are found in different layers of the olfactory epithelium and project to separate regions of the olfactory bulb, presaging the separation of mammalian ciliated neurons (in the MOE) and microvillar neurons (in the VNO) and their projections to distinct main and accessory olfactory bulbs [33, 34]. The zebrafish epithelium also contains a third cell type (crypt neurons) that expresses one of the few zebrafish V1R genes in most cells and projects to a unique glomerulus [29, 32, 94]. In addition, a closely related receptor has recently been found to detect a pheromone that releases egg-laying behavior, reminiscent of the reproductive behaviors regulated by mammalian V1Rs (Figure 3) [95]. This “ancestral” organization of the olfactory periphery therefore suggests that discrete subsystems and the receptors they express are hardwired to yield different behavioral responses to odor (Figure 4 top).
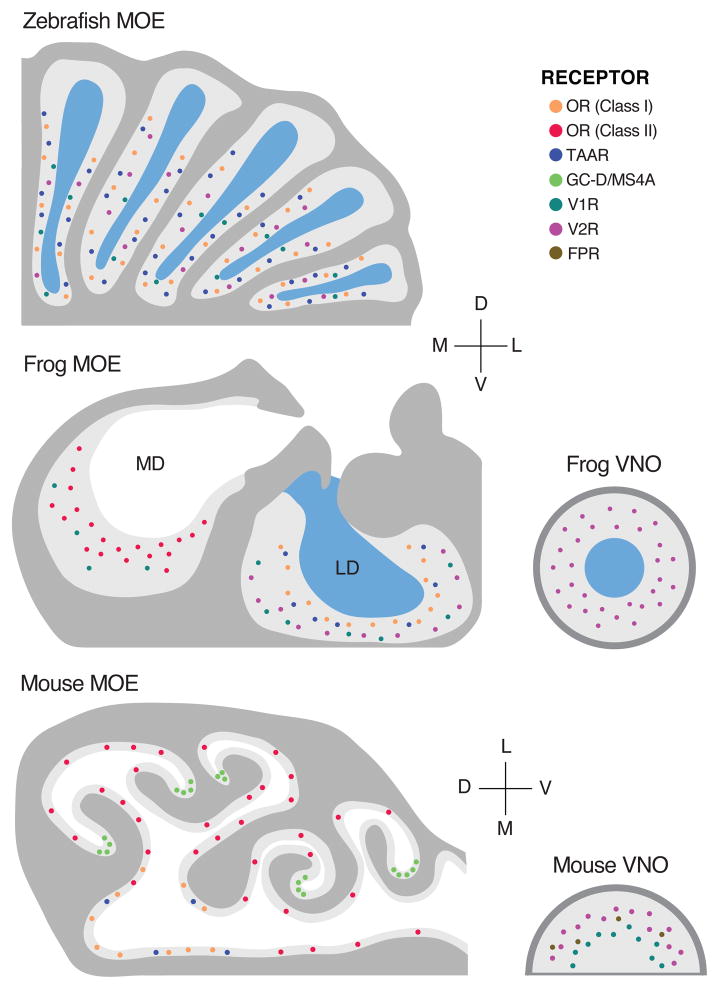
Figure 4.
Peripheral organization of OR family expression across vertebrates. Top: A horizontal section of a quarter of one olfactory rosette from the zebrafish (Danio rerio). Sensory neurons are embedded in a main olfactory epithelium (MOE) with a water-filled lumen. Members of four of the five major GPCR OR families are expressed in different sensory neuron types: TAARs and Class I “classical” ORs [here denoting also fish ORs that belong to neither Class I nor Class II] are expressed in ciliated cells occupying the basal layers of the epithelium; V2Rs in microvillar cells in the middle layer; and at least one V1R in “crypt cells” of the upper layer. Axons of the three cell types project to different, spatially segregated, and stereotyped glomeruli of the zebrafish olfactory bulb. Middle: A coronal section of one half of the main olfactory epithelium and the vomeronasal organ (VNO) of the western clawed frog (Xenopus laevis). An air-filled medial diverticulum (MD) houses neurons expressing Class II “classical” ORs and several V1Rs; a water-filled lateral diverticulum (LD), Class I “classical” ORs, several TAARs, and ancestral V2Rs; the water-filled (VNO), newer members of the expanded V2R family. Bottom: A coronal section of the MOE and VNO of the house mouse (Mus musculus). Receptor family expression is segregated (several exceptions not pictured), with Class I ORs and TAARs expressed in the dorsal MOE, Class II ORs in the ventral MOE (and some in the dorsal MOE, not pictured), V1Rs in the apical VNO, V2Rs in the basal VNO, and FPRs in both VNO layers. Some sensory neurons in the “cul-de-sacs” of the MOE express the receptor guanylate cyclase GC-D and multiple members of a non-GPCR family of four-pass transmembrane chemoreceptor family, Ms4a.
Consistent with this possibility, a major olfactory adaptation along the amphibian and terrestrial branches of the vertebrate tree includes the establishment of segregated neuroanatomical structures that express more recently-derived ORs. Unlike fish, amphibians have evolved a separate vomeronasal organ that detects attractive peptide sex pheromones (specific to amphibians) through unusually large and recently expanded subfamilies of V2Rs, as well as an air-filled MOE compartment the expresses the massively enlarged Class II OR repertoire described above; evolutionarily more ancient V2Rs tuned to single amino acids remain expressed in the water-filled MOE compartment along with V1Rs, Class I ORs, and TAARs [62, 96, 97] (Figure 4 middle). The observation that newer V2R genes have acquired both a novel expression pattern and a novel role in innate reproductive behaviors suggests that odor interpretation depends not only on what ligands are detected, but also on the developmental identities and specialized sensory pathways of the cells that detect them [98].
The segregation of the olfactory periphery has proceeded even further in rodents. Mice, for example, have separate epithelial layers for V1Rs and V2Rs within the VNO (Figure 4 bottom); distinct dorsoventral zones and molecular machinery for the expression of TAARs, Class I ORs, and Class II ORs in the MOE [99]; a group of neurons at the tip of the nose, the Grueneberg ganglion, which is thought to detect alarm pheromones and structurally related predator odorants [100, 101]; a patch of olfactory neurons on the nasal septum (the “septal organ”) that largely express a single broadly-tuned OR and may convey information about breathing [102, 103]; and within cul-de-sacs of the MOE, a set of neurons which are thought to mediate the social acquisition of food preference and which express both the receptor guanylate cyclase GC-D and a recently-described family of non-GPCR olfactory receptors (Figure 4 bottom) [104–106].
The olfactory bulb appears to organize this variety of incoming sensory information into domains larger than a single glomerulus. For instance, the sensory neurons that express class I OR-, class II OR-, TAAR-, or GC-D are developmentally constrained to innervate circumscribed and nonintersecting regions of the olfactory bulb (Figure 5) [99] [107]. Furthermore, there is substantial evidence that activity in spatially and molecularly segregated glomeruli drives different patterns of stereotyped behavior via hardwired projections to central brain regions. First, an “innate aversion” domain for predator odor sensation can be found at the ventral subdivision of the dorsal olfactory bulb, while the dorsal-most Class I-expressing domain is instead required for aversive responses to spoiled food odorants [108]. Next, the ventral olfactory bulb houses the glomeruli of molecularly distinct neurons (expressing components of an alternative, TrpM5-dependent chemotransduction pathway) that preferentially respond to urine-derived pheromones and project to the “vomeronasal amygdala”, as well as the cluster of OR37 glomeruli and their unusual mitral cell projections to the hypothalamus [75, 77, 109, 110]. Finally, the TAAR domain of the olfactory bulb is required for innate behavioral responses to several ecologically important amines, even though these same odorants can be detected by receptors in the main olfactory system (Figure 5) [84]. Recent data also suggest a global organization to the main olfactory bulb, in which glomeruli that reside more rostrally evoke investigatory behaviors, while those that are more caudal evoke aversion [111]. Characterizing the mitral cell projections downstream of specific glomeruli may reveal different capacities to influence a variety of central processing or behavioral centers. This “switchboard” organization could allow each OR gene to adapt its chemosensory tuning according to its domain’s function without altering the circuits downstream of the receptor.
However, the interpretation of an odor is not determined simply by whether it recruits neural activity within a specific glomerular domain. For instance within the TAAR domain, the TAAR4 glomerulus is required for aversion to phenethylamine, but the nearby TAAR5 glomerulus is highly tuned to the attractive mouse pheromone trimethylamine [84, 112]. Thus even closely related receptors and their closely apposed glomeruli — within a similar subdomain of the olfactory bulb – may drive opposite behaviors. Combinatorial effects of multiple (TAAR and non-TAAR) glomeruli may produce the different behavioral responses to high-affinity TAAR4 and TAAR5 ligands. Consistent with this possibility, the effect of TAAR5 activation appears to be inverted in rats, for which trimethylamine is aversive (Figure 3) [112]. However it is also possible that, within the TAAR domain of the bulb, TAAR4 and TAAR5 glomeruli are connected to distinct downstream circuits that drive fundamentally different behaviors. Determining whether the differential effects of TAAR4 and TAAR5 ligands in mice relies upon glomerulus-specific projections to higher brain regions, the coordinated activity of groups of glomeruli, or on circuit activity within the TAAR domain will therefore reveal key principles of how odor representations in the olfactory bulb govern perception and behavior.
The appeal of the “domain” hypothesis for coupling odors to behavior lies in its relative developmental simplicity. In this model the olfactory system could take advantage of developmentally-specific gradients of morphogenetic and axon guidance molecules to build sub-circuits within the olfactory bulb that connect to distinct higher brain regions. How might the olfactory system create a system in which individual glomeruli within a given domain are differentially connected to higher brain regions? One highly speculative possibility is that the olfactory bulb’s output layer of mitral cells is, like the retinal ganglion cell layer of the retina, tiled with molecularly distinct cell types that convey different types of information via their projection or spiking patterns. Hardwired synaptic connections to these mitral cell types might then be biased by the complement of cell surface proteins on OSN axons, which are themselves determined by both their developmental identity and the OR gene stochastically chosen for expression [113]. This sort of coupling between ORs and central circuits could explain why many odors appear to have a stereotyped attractive or aversive valence in mice and humans, even when the ORs responsible for their detection are evolving neutrally [114, 115]. In other words, behavioral biases could be substrates for adaptation (e.g. through changes in the expression of cell surface proteins given a particular OR choice) but would also necessarily exist by chance. In this model, the multiglomerular domains of the olfactory bulb may determine which downstream targets are available (e.g. medial amygdala from the accessory bulb and cortical amygdala from the main bulb) but leave enough flexibility for receptor-specific behaviors to evolve. The loss of several subsystems in humans may explain why single OR genes are not under strong selective pressure in our lineage, as a highly-tuned receptor would have no specialized circuitry to tap into to generate an adaptive behavioral response.
Concluding Remarks
How evolution sculpts the olfactory system is a response to both the general problem of probing a wide swath of chemical space, as well as the unique sensory challenges facing each animal species. In vertebrates, both dramatic and subtle adaptations to the chemical environment are possible because of the vast, hierarchical structure of the olfactory receptor (OR) gene families. First, rapid growth or pruning of large OR families accommodate major changes in habitat and lifestyle, as with ocean-to-terrestrial shifts and derived primacy of the visual sense. Second, selection on individual OR families or subfamilies permits niche adaptation or conserved chemical communication between conspecifics. Third, genetic variation in single OR genes, among thousands, can alter behaviors driven by precise chemical cues. We speculate that the molecular logic and neuroanatomical architecture of the vertebrate olfactory nervous system, in which newly arising olfactory receptor genes can tap into a number of segregated pathways that influence both innate and learned behaviors, may have favored this dramatic mode of olfactory receptor evolution. In this model OR genes form the marble from which the vertebrate olfactory system is sculpted, specifying the ability both to detect odors and to implement appropriate survival- and reproduction-promoting responses. Peering into this dynamic process reveals a direct link between the evolution of an animal’s genome and the specific tasks solved by its nervous system.
Acknowledgments
This review was intended to address core themes in the evolution of vertebrate odor receptors rather than to be comprehensive; we apologize to the many researchers whose work we could not cite due to limitations of space. We thank Hannah Somhegyi for help designing figures and Taralyn Tan for contributing figure illustrations. DMB is supported by a National Science Foundation predoctoral fellowship and a Sackler Scholarship in Psychobiology. JML is supported by the European Molecular Biology Organization (ALTF 379-2011), the Human Frontiers Science Program (LT001086/2012), and the Belgian American Educational Foundation. HEH is an investigator of the Howard Hughes Medical Institute. SRD is supported by fellowships from the Burroughs Wellcome Fund, the Vallee Foundation, and by grant RO11DC011558 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wyatt TD. Introduction to Chemical Signaling in Vertebrates and Invertebrates. In: Mucignat-Caretta C, editor. Neurobiology of Chemical Communication. Boca Raton (FL): 2014. [PubMed] [Google Scholar]
- 2.Meister M. On the dimensionality of odor space. Elife. 2015;4:e07865. doi: 10.7554/eLife.07865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargmann CI. Comparative chemosensation from receptors to ecology. Nature. 2006;444:295–301. doi: 10.1038/nature05402. [DOI] [PubMed] [Google Scholar]
- 4.Hansson BS, Stensmyr MC. Evolution of insect olfaction. Neuron. 2011;72:698–711. doi: 10.1016/j.neuron.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 6.Touhara K, Vosshall LB. Sensing odorants and pheromones with chemosensory receptors. Annu Rev Physiol. 2009;71:307–332. doi: 10.1146/annurev.physiol.010908.163209. [DOI] [PubMed] [Google Scholar]
- 7.Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- 8.Meisami E, Bhatnagar KP. Structure and diversity in mammalian accessory olfactory bulb. Microsc Res Tech. 1998;43:476–499. doi: 10.1002/(SICI)1097-0029(19981215)43:6<476::AID-JEMT2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 9.Yopak KE, Lisney TJ, Collin SP. Not all sharks are “swimming noses”: variation in olfactory bulb size in cartilaginous fishes. Brain Struct Funct. 2015;220:1127–1143. doi: 10.1007/s00429-014-0705-0. [DOI] [PubMed] [Google Scholar]
- 10.Manzini I, Korsching S. The peripheral olfactory system of vertebrates: molecular, structural and functional basics of the sense of smell. e-Neuroforum. 2011;2:68–77. [Google Scholar]
- 11.Dalton RP, Lomvardas S. Chemosensory receptor specificity and regulation. Annu Rev Neurosci. 2015;38:331–349. doi: 10.1146/annurev-neuro-071714-034145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 13.Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 14.Johnson MA, Tsai L, Roy DS, Valenzuela DH, Mosley C, Magklara A, Lomvardas S, Liberles SD, Barnea G. Neurons expressing trace amine-associated receptors project to discrete glomeruli and constitute an olfactory subsystem. Proc Natl Acad Sci U S A. 2012;109:13410–13415. doi: 10.1073/pnas.1206724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 16.Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- 17.Ryba NJ, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- 18.Silvotti L, Moiani A, Gatti R, Tirindelli R. Combinatorial co-expression of pheromone receptors, V2Rs. J Neurochem. 2007;103:1753–1763. doi: 10.1111/j.1471-4159.2007.04877.x. [DOI] [PubMed] [Google Scholar]
- 19.Riviere S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature. 2009;459:574–577. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- 20.Liberles SD, Horowitz LF, Kuang D, Contos JJ, Wilson KL, Siltberg-Liberles J, Liberles DA, Buck LB. Formyl peptide receptors are candidate chemosensory receptors in the vomeronasal organ. Proc Natl Acad Sci U S A. 2009;106:9842–9847. doi: 10.1073/pnas.0904464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- 22.Lin da Y, Shea SD, Katz LC. Representation of natural stimuli in the rodent main olfactory bulb. Neuron. 2006;50:937–949. doi: 10.1016/j.neuron.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 23.Sosulski DL, Bloom ML, Cutforth T, Axel R, Datta SR. Distinct representations of olfactory information in different cortical centres. Nature. 2011;472:213–216. doi: 10.1038/nature09868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi GB, Stettler DD, Kallman BR, Bhaskar ST, Fleischmann A, Axel R. Driving opposing behaviors with ensembles of piriform neurons. Cell. 2011;146:1004–1015. doi: 10.1016/j.cell.2011.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Root CM, Denny CA, Hen R, Axel R. The participation of cortical amygdala in innate, odour-driven behaviour. Nature. 2014;515:269–273. doi: 10.1038/nature13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohedano-Moriano A, Pro-Sistiaga P, Ubeda-Banon I, Crespo C, Insausti R, Martinez-Marcos A. Segregated pathways to the vomeronasal amygdala: differential projections from the anterior and posterior divisions of the accessory olfactory bulb. Eur J Neurosci. 2007;25:2065–2080. doi: 10.1111/j.1460-9568.2007.05472.x. [DOI] [PubMed] [Google Scholar]
- 27.Hussain A, Saraiva LR, Korsching SI. Positive Darwinian selection and the birth of an olfactory receptor clade in teleosts. Proc Natl Acad Sci U S A. 2009;106:4313–4318. doi: 10.1073/pnas.0803229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saraiva LR, Ahuja G, Ivandic I, Syed AS, Marioni JC, Korsching SI, Logan DW. Molecular and neuronal homology between the olfactory systems of zebrafish and mouse. Sci Rep. 2015;5:11487. doi: 10.1038/srep11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saraiva LR, Korsching SI. A novel olfactory receptor gene family in teleost fish. Genome Res. 2007;17:1448–1457. doi: 10.1101/gr.6553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weth F, Nadler W, Korsching S. Nested expression domains for odorant receptors in zebrafish olfactory epithelium. Proc Natl Acad Sci U S A. 1996;93:13321–13326. doi: 10.1073/pnas.93.23.13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato Y, Miyasaka N, Yoshihara Y. Hierarchical regulation of odorant receptor gene choice and subsequent axonal projection of olfactory sensory neurons in zebrafish. J Neurosci. 2007;27:1606–1615. doi: 10.1523/JNEUROSCI.4218-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oka Y, Saraiva LR, Korsching SI. Crypt neurons express a single V1R-related ora gene. Chem Senses. 2012;37:219–227. doi: 10.1093/chemse/bjr095. [DOI] [PubMed] [Google Scholar]
- 33.Sato Y, Miyasaka N, Yoshihara Y. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J Neurosci. 2005;25:4889–4897. doi: 10.1523/JNEUROSCI.0679-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braubach OR, Fine A, Croll RP. Distribution and functional organization of glomeruli in the olfactory bulbs of zebrafish (Danio rerio) J Comp Neurol. 2012;520:2317–2339. doi: 10.1002/cne.23075. Spc2311. [DOI] [PubMed] [Google Scholar]
- 35.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Firestein S. Genomics of olfactory receptors. Results Probl Cell Differ. 2009;47:25–36. doi: 10.1007/400_2008_28. [DOI] [PubMed] [Google Scholar]
- 38.Niimura Y, Matsui A, Touhara K. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 2014;24:1485–1496. doi: 10.1101/gr.169532.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharon D, Glusman G, Pilpel Y, Khen M, Gruetzner F, Haaf T, Lancet D. Primate evolution of an olfactory receptor cluster: diversification by gene conversion and recent emergence of pseudogenes. Genomics. 1999;61:24–36. doi: 10.1006/geno.1999.5900. [DOI] [PubMed] [Google Scholar]
- 40.Neuhaus EM, Zhang W, Gelis L, Deng Y, Noldus J, Hatt H. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J Biol Chem. 2009;284:16218–16225. doi: 10.1074/jbc.M109.012096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niimura Y, Nei M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Hum Genet. 2006;51:505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adipietro KA, Mainland JD, Matsunami H. Functional evolution of mammalian odorant receptors. PLoS Genet. 2012;8:e1002821. doi: 10.1371/journal.pgen.1002821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liman ER. Changing senses: chemosensory signaling and primate evolution. Adv Exp Med Biol. 2012;739:206–217. doi: 10.1007/978-1-4614-1704-0_13. [DOI] [PubMed] [Google Scholar]
- 44.Liman ER, Innan H. Relaxed selective pressure on an essential component of pheromone transduction in primate evolution. Proc Natl Acad Sci U S A. 2003;100:3328–3332. doi: 10.1073/pnas.0636123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilad Y, Przeworski M, Lancet D. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2:E5. doi: 10.1371/journal.pbio.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucas PW, Dominy NJ, Riba-Hernandez P, Stoner KE, Yamashita N, Loria-Calderon E, Petersen-Pereira W, Rojas-Duran Y, Salas-Pena R, Solis-Madrigal S, et al. Evolution and function of routine trichromatic vision in primates. Evolution. 2003;57:2636–2643. doi: 10.1111/j.0014-3820.2003.tb01506.x. [DOI] [PubMed] [Google Scholar]
- 47.Dominy NJ, Lucas PW. Ecological importance of trichromatic vision to primates. Nature. 2001;410:363–366. doi: 10.1038/35066567. [DOI] [PubMed] [Google Scholar]
- 48.Matsui A, Go Y, Niimura Y. Degeneration of olfactory receptor gene repertories in primates: no direct link to full trichromatic vision. Mol Biol Evol. 2010;27:1192–1200. doi: 10.1093/molbev/msq003. [DOI] [PubMed] [Google Scholar]
- 49.Zhao H, Xu D, Zhang S, Zhang J. Widespread losses of vomeronasal signal transduction in bats. Mol Biol Evol. 2011;28:7–12. doi: 10.1093/molbev/msq207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci U S A. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayden S, Bekaert M, Crider TA, Mariani S, Murphy WJ, Teeling EC. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 2010;20:1–9. doi: 10.1101/gr.099416.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niimura Y. Evolutionary dynamics of olfactory receptor genes in chordates: interaction between environments and genomic contents. Hum Genomics. 2009;4:107–118. doi: 10.1186/1479-7364-4-2-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freitag J, Ludwig G, Andreini I, Rossler P, Breer H. Olfactory receptors in aquatic and terrestrial vertebrates. J Comp Physiol A. 1998;183:635–650. doi: 10.1007/s003590050287. [DOI] [PubMed] [Google Scholar]
- 55.Freitag J, Krieger J, Strotmann J, Breer H. Two classes of olfactory receptors in Xenopus laevis. Neuron. 1995;15:1383–1392. doi: 10.1016/0896-6273(95)90016-0. [DOI] [PubMed] [Google Scholar]
- 56.Mezler M, Fleischer J, Breer H. Characteristic features and ligand specificity of the two olfactory receptor classes from Xenopus laevis. J Exp Biol. 2001;204:2987–2997. doi: 10.1242/jeb.204.17.2987. [DOI] [PubMed] [Google Scholar]
- 57.Li Q, Tachie-Baffour Y, Liu Z, Baldwin MW, Kruse AC, Liberles SD. Non-classical amine recognition evolved in a large clade of olfactory receptors. Elife. 2015;4:e10441. doi: 10.7554/eLife.10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hussain A, Saraiva LR, Ferrero DM, Ahuja G, Krishna VS, Liberles SD, Korsching SI. High-affinity olfactory receptor for the death-associated odor cadaverine. Proc Natl Acad Sci U S A. 2013;110:19579–19584. doi: 10.1073/pnas.1318596110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liberles SD. Trace amine-associated receptors: ligands, neural circuits, and behaviors. Curr Opin Neurobiol. 2015;34:1–7. doi: 10.1016/j.conb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rolen SH, Sorensen PW, Mattson D, Caprio J. Polyamines as olfactory stimuli in the goldfish Carassius auratus. J Exp Biol. 2003;206:1683–1696. doi: 10.1242/jeb.00338. [DOI] [PubMed] [Google Scholar]
- 61.Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 62.Syed AS, Sansone A, Nadler W, Manzini I, Korsching SI. Ancestral amphibian v2rs are expressed in the main olfactory epithelium. Proc Natl Acad Sci U S A. 2013;110:7714–7719. doi: 10.1073/pnas.1302088110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kikuyama S, Yamamoto K, Iwata T, Toyoda F. Peptide and protein pheromones in amphibians. Comp Biochem Physiol B Biochem Mol Biol. 2002;132:69–74. doi: 10.1016/s1096-4959(01)00534-6. [DOI] [PubMed] [Google Scholar]
- 64.Grus WE, Shi P, Zhang J. Largest vertebrate vomeronasal type 1 receptor gene repertoire in the semiaquatic platypus. Mol Biol Evol. 2007;24:2153–2157. doi: 10.1093/molbev/msm157. [DOI] [PubMed] [Google Scholar]
- 65.Isogai Y, Si S, Pont-Lezica L, Tan T, Kapoor V, Murthy VN, Dulac C. Molecular organization of vomeronasal chemoreception. Nature. 2011;478:241–245. doi: 10.1038/nature10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hayden S, Bekaert M, Goodbla A, Murphy WJ, Davalos LM, Teeling EC. A cluster of olfactory receptor genes linked to frugivory in bats. Mol Biol Evol. 2014;31:917–927. doi: 10.1093/molbev/msu043. [DOI] [PubMed] [Google Scholar]
- 67.Khan I, Yang Z, Maldonado E, Li C, Zhang G, Gilbert MT, Jarvis ED, O’Brien SJ, Johnson WE, Antunes A. Olfactory Receptor Subgenomes Linked with Broad Ecological Adaptations in Sauropsida. Mol Biol Evol. 2015;32:2832–2843. doi: 10.1093/molbev/msv155. [DOI] [PubMed] [Google Scholar]
- 68.Yang H, Shi P, Zhang YP, Zhang J. Composition and evolution of the V2r vomeronasal receptor gene repertoire in mice and rats. Genomics. 2005;86:306–315. doi: 10.1016/j.ygeno.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 69.Papes F, Logan DW, Stowers L. The vomeronasal organ mediates interspecies defensive behaviors through detection of protein pheromone homologs. Cell. 2010;141:692–703. doi: 10.1016/j.cell.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chamero P, Marton TF, Logan DW, Flanagan K, Cruz JR, Saghatelian A, Cravatt BF, Stowers L. Identification of protein pheromones that promote aggressive behaviour. Nature. 2007;450:899–902. doi: 10.1038/nature05997. [DOI] [PubMed] [Google Scholar]
- 71.Hoppe R, Lambert TD, Samollow PB, Breer H, Strotmann J. Evolution of the “OR37” subfamily of olfactory receptors: a cross-species comparison. J Mol Evol. 2006;62:460–472. doi: 10.1007/s00239-005-0093-4. [DOI] [PubMed] [Google Scholar]
- 72.Hoppe R, Breer H, Strotmann J. Organization and evolutionary relatedness of OR37 olfactory receptor genes in mouse and human. Genomics. 2003;82:355–364. doi: 10.1016/s0888-7543(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 73.Bautze V, Bar R, Fissler B, Trapp M, Schmidt D, Beifuss U, Bufe B, Zufall F, Breer H, Strotmann J. Mammalian-specific OR37 receptors are differentially activated by distinct odorous fatty aldehydes. Chem Senses. 2012;37:479–493. doi: 10.1093/chemse/bjr130. [DOI] [PubMed] [Google Scholar]
- 74.Strotmann J, Conzelmann S, Beck A, Feinstein P, Breer H, Mombaerts P. Local permutations in the glomerular array of the mouse olfactory bulb. J Neurosci. 2000;20:6927–6938. doi: 10.1523/JNEUROSCI.20-18-06927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bader A, Klein B, Breer H, Strotmann J. Connectivity from OR37 expressing olfactory sensory neurons to distinct cell types in the hypothalamus. Front Neural Circuits. 2012;6:84. doi: 10.3389/fncir.2012.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dulac C, Torello AT. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat Rev Neurosci. 2003;4:551–562. doi: 10.1038/nrn1140. [DOI] [PubMed] [Google Scholar]
- 77.Lin DY, Zhang SZ, Block E, Katz LC. Encoding social signals in the mouse main olfactory bulb. Nature. 2005;434:470–477. doi: 10.1038/nature03414. [DOI] [PubMed] [Google Scholar]
- 78.Dekker T, Ibba I, Siju KP, Stensmyr MC, Hansson BS. Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D. sechellia. Curr Biol. 2006;16:101–109. doi: 10.1016/j.cub.2005.11.075. [DOI] [PubMed] [Google Scholar]
- 79.Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446:542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- 80.Sakurai T, Mitsuno H, Mikami A, Uchino K, Tabuchi M, Zhang F, Sezutsu H, Kanzaki R. Targeted disruption of a single sex pheromone receptor gene completely abolishes in vivo pheromone response in the silkmoth. Sci Rep. 2015;5:11001. doi: 10.1038/srep11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stensmyr MC, Dweck HK, Farhan A, Ibba I, Strutz A, Mukunda L, Linz J, Grabe V, Steck K, Lavista-Llanos S, et al. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell. 2012;151:1345–1357. doi: 10.1016/j.cell.2012.09.046. [DOI] [PubMed] [Google Scholar]
- 82.Haga S, Hattori T, Sato T, Sato K, Matsuda S, Kobayakawa R, Sakano H, Yoshihara Y, Kikusui T, Touhara K. The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor. Nature. 2010;466:118–122. doi: 10.1038/nature09142. [DOI] [PubMed] [Google Scholar]
- 83.Ferrero DM, Moeller LM, Osakada T, Horio N, Li Q, Roy DS, Cichy A, Spehr M, Touhara K, Liberles SD. A juvenile mouse pheromone inhibits sexual behaviour through the vomeronasal system. Nature. 2013;502:368–371. doi: 10.1038/nature12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dewan A, Pacifico R, Zhan R, Rinberg D, Bozza T. Non-redundant coding of aversive odours in the main olfactory pathway. Nature. 2013;497:486–489. doi: 10.1038/nature12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferrero DM, Lemon JK, Fluegge D, Pashkovski SL, Korzan WJ, Datta SR, Spehr M, Fendt M, Liberles SD. Detection and avoidance of a carnivore odor by prey. Proc Natl Acad Sci U S A. 2011;108:11235–11240. doi: 10.1073/pnas.1103317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gimelbrant AA, Skaletsky H, Chess A. Selective pressures on the olfactory receptor repertoire since the human-chimpanzee divergence. Proc Natl Acad Sci U S A. 2004;101:9019–9022. doi: 10.1073/pnas.0401566101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007;449:468–472. doi: 10.1038/nature06162. [DOI] [PubMed] [Google Scholar]
- 88.Jaeger SR, McRae JF, Bava CM, Beresford MK, Hunter D, Jia Y, Chheang SL, Jin D, Peng M, Gamble JC, et al. A Mendelian trait for olfactory sensitivity affects odor experience and food selection. Curr Biol. 2013;23:1601–1605. doi: 10.1016/j.cub.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 89.Eriksson N, Wu S, Do CB, Kiefer AK, Tung JY, Mountain JL, Hinds DA, Francke U. A genetic variant near olfactory receptor genes influences cilantro preference. Flavour. 2012;1:1. [Google Scholar]
- 90.Gelstein S, Yeshurun Y, Rozenkrantz L, Shushan S, Frumin I, Roth Y, Sobel N. Human tears contain a chemosignal. Science. 2011;331:226–230. doi: 10.1126/science.1198331. [DOI] [PubMed] [Google Scholar]
- 91.Varendi H, Porter RH. Breast odour as the only maternal stimulus elicits crawling towards the odour source. Acta Paediatr. 2001;90:372–375. [PubMed] [Google Scholar]
- 92.Schaal B, Coureaud G, Langlois D, Ginies C, Semon E, Perrier G. Chemical and behavioural characterization of the rabbit mammary pheromone. Nature. 2003;424:68–72. doi: 10.1038/nature01739. [DOI] [PubMed] [Google Scholar]
- 93.Rokni D, Hemmelder V, Kapoor V, Murthy VN. An olfactory cocktail party: figure-ground segregation of odorants in rodents. Nat Neurosci. 2014;17:1225–1232. doi: 10.1038/nn.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ahuja G, Ivandic I, Salturk M, Oka Y, Nadler W, Korsching SI. Zebrafish crypt neurons project to a single, identified mediodorsal glomerulus. Sci Rep. 2013;3:2063. doi: 10.1038/srep02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Behrens M, Frank O, Rawel H, Ahuja G, Potting C, Hofmann T, Meyerhof W, Korsching S. ORA1, a zebrafish olfactory receptor ancestral to all mammalian V1R genes, recognizes 4-hydroxyphenylacetic acid, a putative reproductive pheromone. J Biol Chem. 2014;289:19778–19788. doi: 10.1074/jbc.M114.573162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Syed AS, Sansone A, Roner S, Bozorg Nia S, Manzini I, Korsching SI. Different expression domains for two closely related amphibian TAARs generate a bimodal distribution similar to neuronal responses to amine odors. Sci Rep. 2015;5:13935. doi: 10.1038/srep13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Date-Ito A, Ohara H, Ichikawa M, Mori Y, Hagino-Yamagishi K. Xenopus V1R vomeronasal receptor family is expressed in the main olfactory system. Chem Senses. 2008;33:339–346. doi: 10.1093/chemse/bjm090. [DOI] [PubMed] [Google Scholar]
- 98.Gliem S, Syed AS, Sansone A, Kludt E, Tantalaki E, Hassenklover T, Korsching SI, Manzini I. Bimodal processing of olfactory information in an amphibian nose: odor responses segregate into a medial and a lateral stream. Cell Mol Life Sci. 2013;70:1965–1984. doi: 10.1007/s00018-012-1226-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bozza T, Vassalli A, Fuss S, Zhang JJ, Weiland B, Pacifico R, Feinstein P, Mombaerts P. Mapping of class I and class II odorant receptors to glomerular domains by two distinct types of olfactory sensory neurons in the mouse. Neuron. 2009;61:220–233. doi: 10.1016/j.neuron.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brechbuhl J, Moine F, Klaey M, Nenniger-Tosato M, Hurni N, Sporkert F, Giroud C, Broillet MC. Mouse alarm pheromone shares structural similarity with predator scents. Proc Natl Acad Sci U S A. 2013;110:4762–4767. doi: 10.1073/pnas.1214249110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brechbuhl J, Klaey M, Broillet MC. Grueneberg ganglion cells mediate alarm pheromone detection in mice. Science. 2008;321:1092–1095. doi: 10.1126/science.1160770. [DOI] [PubMed] [Google Scholar]
- 102.Grosmaitre X, Fuss SH, Lee AC, Adipietro KA, Matsunami H, Mombaerts P, Ma M. SR1, a mouse odorant receptor with an unusually broad response profile. J Neurosci. 2009;29:14545–14552. doi: 10.1523/JNEUROSCI.2752-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grosmaitre X, Santarelli LC, Tan J, Luo M, Ma M. Dual functions of mammalian olfactory sensory neurons as odor detectors and mechanical sensors. Nat Neurosci. 2007;10:348–354. doi: 10.1038/nn1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Greer PL, Bear DM, Lassance JM, Bloom ML, Tsukahara T, Pashkovski SL, Masuda FK, Nowlan AC, Kirchner R, Hoekstra HE, et al. A Family of non-GPCR Chemosensors Defines an Alternative Logic for Mammalian Olfaction. Cell. 2016;165:1734–1748. doi: 10.1016/j.cell.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Munger SD, Leinders-Zufall T, McDougall LM, Cockerham RE, Schmid A, Wandernoth P, Wennemuth G, Biel M, Zufall F, Kelliher KR. An olfactory subsystem that detects carbon disulfide and mediates food-related social learning. Curr Biol. 2010;20:1438–1444. doi: 10.1016/j.cub.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hu J, Zhong C, Ding C, Chi Q, Walz A, Mombaerts P, Matsunami H, Luo M. Detection of near-atmospheric concentrations of CO2 by an olfactory subsystem in the mouse. Science. 2007;317:953–957. doi: 10.1126/science.1144233. [DOI] [PubMed] [Google Scholar]
- 107.Pacifico R, Dewan A, Cawley D, Guo C, Bozza T. An olfactory subsystem that mediates high-sensitivity detection of volatile amines. Cell Rep. 2012;2:76–88. doi: 10.1016/j.celrep.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450:503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- 109.Lin W, Margolskee R, Donnert G, Hell SW, Restrepo D. Olfactory neurons expressing transient receptor potential channel M5 (TRPM5) are involved in sensing semiochemicals. Proc Natl Acad Sci U S A. 2007;104:2471–2476. doi: 10.1073/pnas.0610201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Thompson JA, Salcedo E, Restrepo D, Finger TE. Second-order input to the medial amygdala from olfactory sensory neurons expressing the transduction channel TRPM5. J Comp Neurol. 2012;520:1819–1830. doi: 10.1002/cne.23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kermen F, Midroit M, Kuczewski N, Forest J, Thevenet M, Sacquet J, Benetollo C, Richard M, Didier A, Mandairon N. Topographical representation of odor hedonics in the olfactory bulb. Nat Neurosci. 2016;19:876–878. doi: 10.1038/nn.4317. [DOI] [PubMed] [Google Scholar]
- 112.Li Q, Korzan WJ, Ferrero DM, Chang RB, Roy DS, Buchi M, Lemon JK, Kaur AW, Stowers L, Fendt M, et al. Synchronous evolution of an odor biosynthesis pathway and behavioral response. Curr Biol. 2013;23:11–20. doi: 10.1016/j.cub.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Serizawa S, Miyamichi K, Takeuchi H, Yamagishi Y, Suzuki M, Sakano H. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 2006;127:1057–1069. doi: 10.1016/j.cell.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 114.Saraiva LR, Kondoh K, Ye X, Yoon KH, Hernandez M, Buck LB. Combinatorial effects of odorants on mouse behavior. Proc Natl Acad Sci U S A. 2016;113:E3300–3306. doi: 10.1073/pnas.1605973113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haddad R, Medhanie A, Roth Y, Harel D, Sobel N. Predicting odor pleasantness with an electronic nose. PLoS Comput Biol. 2010;6:e1000740. doi: 10.1371/journal.pcbi.1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]