Summary
As sessile organisms, plants must cope with abiotic stress such as soil salinity, drought, and extreme temperatures. Core stress signaling pathways involve protein kinases related to the yeast SNF1 and mammalian AMPK, suggesting that stress signaling in plants evolved from energy sensing. Stress signaling regulates proteins critical for ion and water transport and for metabolic and gene-expression reprogramming to bring about ionic and water homeostasis and cellular stability under stress conditions. Understanding stress signaling and responses will increase our ability to improve stress resistance in crops to achieve agricultural sustainability and food security for a growing world population.
Introduction
Plants live in constantly changing environments that are often unfavorable or stressful for growth and development. These adverse environmental conditions include biotic stress, such as pathogen infection and herbivore attack, and abiotic stress, such as drought, heat, cold, nutrient deficiency, and excess of salt or toxic metals like aluminum, arsenate, and cadmium in the soil. Drought, salt, and temperature stresses are major environmental factors that affect the geographical distribution of plants in nature, limit plant productivity in agriculture, and threaten food security. The adverse effects of these abiotic stresses are exacerbated by climate change, which has been predicted to result in an increased frequency of extreme weather (Fedoroff et al., 2010). How plants sense stress signals and adapt to adverse environments are fundamental biological questions. Further, improving plant stress resistance is critical for agricultural productivity and also for environmental sustainability because crops with poor stress resistance consume too much water and fertilizers and thus greatly burden the environment.
For drought and salt stress, it is helpful to distinguish the primary stress signals from secondary signals caused by too little water or too much salt. The primary signal caused by drought is hyperosmotic stress, which is often referred to simply as osmotic stress because a hypo-osmotic condition typically is not a significant problem for plant cells. Salt stress has both osmotic and ionic or ion toxicity effects on cells. The secondary effects of drought and salt stresses are complex and include oxidative stress; damage to cellular components such as membrane lipids, proteins, and nucleic acids; and metabolic dysfunction. While some cellular responses ensue from primary stress signals, others arise primarily from secondary signals. Thus drought and salt have unique and overlapping signals. An important feature of drought and salt stress is that the hyperosmotic signal causes the accumulation of the phytohormone abscisic acid (ABA), which in turn elicits many adaptive responses in plants (Zhu, 2002).
Here, I review what is known about stress sensors and the roles of organelles in stress sensing and responses. I will also review recent developments in elucidating the signaling pathways for ionic stress, osmotic stress, ABA, and temperature stress. This review will focus on how SNF1/AMPK-related protein kinases and other kinases are activated and how these kinases may be connected to upstream sensing mechanisms and to downstream changes in gene expression, metabolism, physiology, growth, and development.
Stress sensing and putative sensors
Because plants show specific changes in gene expression, metabolism, and physiology in response to different environmental stress conditions, it is safe to presume that plant cells must be capable of sensing various environmental signals. However, despite much effort, only a few putative sensors have been identified. The difficulty seems to lie in the functional redundancy in genes encoding sensor proteins such that dysfunction in one gene does not cause significant phenotypes in stress responses. Alternatively, a sensor may be essential for the plant, such that loss-of-function mutants are lethal, precluding further studies. In addition, demonstrating that a protein or other macromolecule functions as a sensor for a physical signal (e.g., change in osmotic pressure, ion concentration, or temperature) is technically very challenging, and direct experimental evidence is absent even for widely accepted osmosensors or temperature sensors in bacterial, yeast, or mammalian systems.
A putative sensor for hyperosmotic stress is the Arabidopsis OSCA1 (reduced hyperosmolality-induced calcium increase 1) (Yuan et al., 2014). Osmotic stress agents, high salt, cold, and heat as well as oxidative stress, heavy metals, and ABA, can cause increases in the cytosolic free calcium concentration in plants, which can be detected by researchers using genetically encoded aequorin or other calcium reporters. Arabidopsis osca1 loss-of-function mutants display a reduced calcium spike compared to wild-type plants when treated with osmotic stress agents such as sorbitol and mannitol (Yuan et al., 2014). OSCA1 encodes a plasma membrane protein that forms hyperosmolality-gated calcium-permeable channels. Because these mutant plants do not display any drought or salt stress phenotypes, the physiological significance of OSCA1 under the stress conditions is unclear. We do not know how OSCA1 senses osmotic stress, which is expected to reduce turgor and thus may affect membrane stretching and membrane–cell wall interactions. A number of mechano-sensitive channels are known in non-plant systems, including TRP, MscS-like, Piezo, DEG/ENaC, and K2P (Arnadottir and Chalfie, 2010; Hedrich, 2012). Animal TRP channels are well-known calcium channels that can sense changes in the membrane caused by fluctuations in temperature or osmotic pressure (Arnadottir and Chalfie, 2010). Plants lack TRP and DEG/ENaC genes but contain a family of MscS-like proteins and one Piezo homolog (Hedrich, 2012). One of the MscS-like proteins in Arabidopsis, MSL8, is required for pollen to survive hypo-osmotic shock during hydration, suggesting MSL8 as a sensor of hypo-osmotic stress-induced membrane tension (Hamilton et al., 2015). Plants also have a large family of cyclic nucleotide-gated channels (CNGCs) as well as a family of glutamate receptor-like (GLR) channels that are potentially very important in generating cytosolic Ca2+ signals under stress (Swarbreck et al., 2013).
Another putative stress sensor is COLD1, which was recently reported to mediate cold stress sensing in rice. COLD1 is required for chilling (0–15°C) tolerance in the Nipponbare sub-species of rice (Ma et al., 2015). COLD1 is a transmembrane protein in the plasma membrane and ER and interacts with RGA1, the alpha subunit of the sole heterotrimeric G protein in plants. The authors speculated that COLD1 may regulate a calcium channel or is itself a cold-sensing calcium channel (Ma et al., 2015). It is unclear how COLD1-mediated calcium signaling leads to chilling tolerance.
Both cold and heat can change the fluidity of cellular phospholipid membranes (Sangwan et al., 2002). This change may be sensed by integral membrane proteins, including channels and various other transporters and membrane-anchored receptor-like kinases. Heat stress may also be sensed by molecular chaperones that bind to misfolded proteins caused by heat denaturation (Scharf et al., 2012). The binding of misfolded proteins would release heat stress transcription factors from the chaperones, allowing the heat stress transcription factors to activate heat-responsive genes. Recently, H2A.Z-containing nucleosomes were suggested to be a thermosensory apparatus in plants and yeast (Kumar and Wigge, 2010). The authors proposed that H2A.Z-containing nucleosomes wrap DNA more tightly than H2A-containing nucleosomes and that elevated temperatures increase the expression of heat shock proteins and other genes by dissociating H2A.Z nucleosomes to make DNA more accessible to Pol II transcription. This is an attractive hypothesis that remains to be further tested.
Sensing of organellar stress
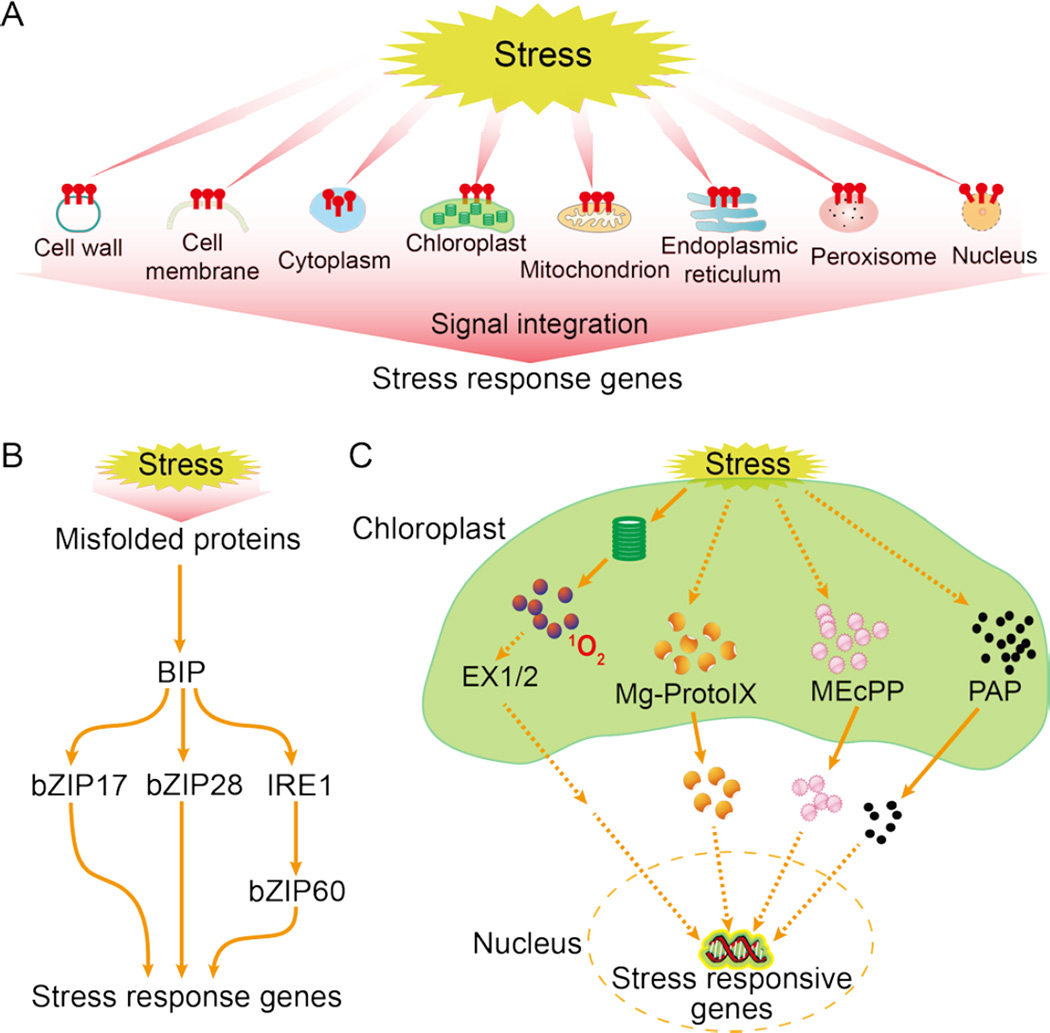
Stress sensing is often compared to ligand perception, so it is frequently thought to occur on the cell surface or at the cell membrane. Then, the signal would be relayed to various subcellular locations such as the nucleus. Theoretically, physical stress signals, particularly temperature signals, may be sensed anywhere in the cell, as long as the stress signal causes a change in the status of the cellular component (protein, DNA, RNA, carbohydrate, or lipid) or compartment (e.g., metabolic reaction in a compartment), and as long as that change then affects other cellular components or activities. Stress-induced changes in protein folding in the ER are now widely recognized as an important cellular response to stress conditions, referred to as ER stress. Similarly, there is stress associated with other organelles such as the chloroplast, mitochondrion, peroxisome, nucleus, and cell wall, and the stress-generated signals from all organelles are integrated to regulate stress responsive gene expression and other cellular activities to restore cellular homeostasis (Figure 1A).
Figure 1.
Stress sensing and signaling in different cell organelles. A. Model of dispersed stress sensing by organelles. Stress causes perturbations in various organelles, generating signals that are integrated to regulate nuclear gene expression and other cellular activities, which helps to restore cellular homeostasis. B. ER stress sensing and signaling. C. Chloroplast stress sensing and signaling. Dashed lines indicate postulated regulation.
ER stress
Both biotic and abiotic stress can cause protein misfolding or the accumulation of unfolded proteins, which is sensed as ER stress by specific sensor proteins in the ER membrane. This sensing leads to the expression of genes encoding chaperones and other proteins important for enhancing protein folding capacity, ER-associated degradation (ERAD), or protein translation suppression to reduce the amount of synthesized proteins loaded to ER via the PKR-like ER eIF2a kinase (Walter and Ron, 2011). These changes help restore ER homeostasis, i.e., the equilibrium between protein-folding demands and folding capacity, and are known as the unfolded protein response (UPR), a conserved stress response in eukaryotes (Walter and Ron, 2011). Two main types of sensors of ER stress have been identified in plants: ER membrane-associated transcription factors and an RNA splicing factor (Liu and Howell, 2016) (Figure 1B). The basic leucine zipper bZIP28 may sense heat and other ER stress agents through its interaction with the chaperone protein BIP (binding immunoglobulin protein) in the ER. Unfolded or misfolded proteins accumulate under stress, and these proteins can interact with BIP, which releases bZIP28 for transport to the Golgi, where it is cleaved. Its cytosolic portion then relocates to the nucleus to activate the expression of stress-response genes to restore ER homeostasis. bZIP28 may also sense other changes that promote its release from BIP, including alterations in energy charge levels and redox status or interactions between BIP and the DNAJ domain-containing protein chaperone (Liu and Howell, 2016). bZIP17 can be activated by salt stress in a similar manner. In addition, several ER- or plasma membrane-associated NAC transcription factors can be activated by ER stress and contribute to UPR (Liu and Howell, 2016). The second type of ER stress sensor in plants is IRE1, a splicing factor conserved from yeast to metazoans. Presumably, plant IRE1 proteins bind to unfolded proteins and sense ER stress in a manner similar to their yeast homolog. Activated IRE1 in Arabidopsis recognizes and splices bZIP60 mRNA and perhaps other target mRNAs. The splicing of bZIP60 mRNA by IRE1 results in a bZIP60 variant that can enter the nucleus to activate UPR genes (Liu and Howell, 2016).
Chloroplast stress
The chloroplast is an organelle where photosynthetic electron transport and many metabolic reactions take place; the metabolic balance in chloroplasts is easily perturbed by environmental stresses. A disturbance in chloroplast homeostasis is communicated to the nucleus through retrograde signals so that all cellular activities can be adjusted and coordinated according to the ability of the stressed chloroplast to supply sugar and other compounds (Figure 1C). The chloroplast is a major site for the production of ROS including superoxide anion, hydrogen peroxide, hydroxyl radical, and singlet oxygen (Mignolet-Spruyt et al., 2016). Various environmental stresses, particularly high light stress, exacerbate ROS production, which overwhelms ROS-managing systems and generates various secondary messengers.
Singlet oxygen triggers a signaling pathway that requires EXECUTER (EX1) and EX2, two nuclear-encoded proteins in the thylakoid membrane of chloroplasts (Wagner et al., 2004). The Arabidopsis fluorescent (flu) mutant produces a burst of singlet oxygen at dark-to-light transitions, because it accumulates the chlorophyll precursor protochlorophyllide. The accumulation of singlet oxygen triggers dramatic changes in nuclear gene expression and causes chlorosis and cell death in wild type but not in ex mutants (Wagner et al., 2004). Whether EX1 and EX2 directly sense singlet oxygen and how these proteins relay the singlet oxygen signal to the nucleus need resolution. Singlet oxygen triggered signal also occurs independently of EX1 and EX2, possibly through nonenzymatic oxidative breakdown products of β-carotene (Ramel et al., 2012).
High light and other stresses can also cause increases in the level of the plastid metabolite methylerythritol cyclodiphosphate (MEcPP), a precursor of isoprenoids. MEcPP functions as a retrograde signal to activate the expression of stress-responsive nuclear genes that encode plastid proteins (Xiao et al., 2012). Another plastid metabolite important for stress responses is the phosphonucleotide 3'-phosphoadenosine 5'-phosphate (PAP). PAP levels increase following drought and high light stress (Estavillo et al., 2011). SAL1/FRY1 is a bifunctional phosphatase that can dephosphorylate both inositol phosphates and PAP (to AMP), and its dysfunction causes PAP accumulation. PAP inhibits 5’ to 3’ exoribonucleases, contributing to the enhanced expression of a subset of drought- and high light-responsive genes and to altered drought resistance (Estavillo et al., 2011). In addition to PAP and MEcPP, other chloroplast metabolites, such as the tetrapyrroles (Norén et al., 2016) and oxidative breakdown products of β-carotene (Ramel et al., 2012), have been proposed as retrograde signals. How stress modulates the levels of MEcPP, PAP, tetrapyrroles, and other retrograde signaling metabolites is not well understood.
Mitochondrion and peroxisome stress
Like chloroplasts, mitochondria and perhaps even peroxisomes can generate retrograde signals that are important for stress responses. Mitochondria and peroxisomes produce ROS and many metabolites, some of which may serve as retrograde signals (Ng et al., 2014). Dysfunction in a mitochondrial DEXH box RNA helicase or mitochondrial pentatricopeptide repeat protein causes ROS accumulation and altered responses to the plant stress hormone ABA (He et al., 2012). A defect in the mitochondrial complex I also enhances ROS accumulation and causes the mutant plants to have reduced expression of cold-responsive genes and to exhibit chilling and freezing sensitivity (Lee et al., 2002). Similarly, mutations in CHY1, which encodes a peroxisomal beta-hydroxyisobutyryl (HIBYL)-CoA hydrolase needed for valine catabolism and fatty acid beta-oxidation, also cause ROS accumulation and impair cold-responsive gene expression and freezing tolerance (Dong et al., 2009). ROS signals from mitochondria and peroxisomes may modulate cold stress responses by affecting calcium signaling.
Cell wall stress
In plants, the primary cell wall consists of cellulose fibrils interconnected by hemicellulose tethers, such as xyloglucan and arabinoxylan, and embedded in a pectin gel (Tenhaken, 2014). The wall also contains phenolics; peroxidases, pectin esterases, and other enzymes; extensins, expansins, and other proteins; and Ca2+. Salt, drought, and other osmotic stress treatments can cause ROS accumulation and other changes in the cell wall (Tenhaken, 2014). Accumulation of ROS can cause crosslinking of phenolics and cell wall glycoproteins such as extensins, resulting in cell wall stiffening. On the other hand, stress upregulates the expression of expansins and xyloglucan-modifying enzymes that can remodel the wall (Tenhaken, 2014). In addition, salt stress can also cause the wall to lose Ca2+. Recently, cell wall stress has been proposed to trigger dedicated signaling pathways analogous to the fungal cell wall integrity (CWI) pathway (Voxeur and Hofte, 2016; Jendretzki et al., 2011). In yeast, cell wall stress is sensed by the wall-associated plasma membrane proteins Wsc1-3, Mid2, and Mtl1 (Jendretzki et al., 2011). How these sensors detect cell wall deformations is not known. Since these proteins all contain a large O-mannosylated extracellular domain rich in serine and threonine residues, these proteins may function as mechanosensors. Downstream of these sensors are GDP/GTP exchange factors, small GTPase Rho1, protein kinase C, and a MAPK cascade (Jendretzki et al., 2011). The identities of plant cell wall stress sensors are unclear. However, plants have hundreds of receptor-like kinases and other kinases with an extracellular domain, a transmembrane domain, and cytoplasmic kinase domain, so many candidates exist to sense cell wall perturbations.
Cell wall perturbations greatly affect plant stress resistance. A subtle cell wall defect caused by dysfunction in the putative pectin biosynthesis enzyme AtCSLD5 in the Arabidopsis sos6 (salt overly sensitive 6) mutant results in oxidative stress and greatly increased sensitivity to osmotic treatments and salt and drought stress (Zhu et al., 2010). Recently, two proteins were identified in the cellular synthase complex, which help connect the complex to microtubules and are important for plant growth under salt stress (Endler et al., 2015). Much remains to be elucidated about stressinduced physical and chemical alterations in the cell wall, how these alterations are sensed and signals are transduced, and the output of the CWI pathway in plants.
Ionic stress signaling
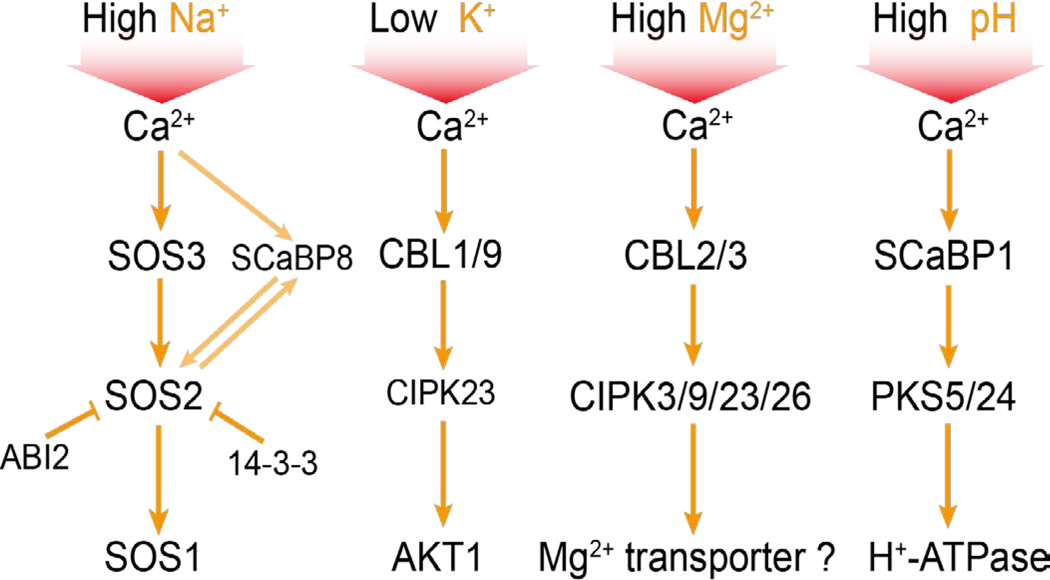
Soil salinity affects a substantial percentage of cultivated land and is a significant factor limiting agricultural productivity worldwide. High salt levels cause ion toxicity (mainly Na+), hyperosmotic stress, and secondary stresses such as oxidative damage (Zhu, 2002). It is not known how Na+ is sensed in any cellular system. In the yeast S. cerevisiae, the calcineurin pathway plays a major role in Na+ stress signaling and tolerance (Thewes, 2014). Na+ stress-triggered cytosolic calcium binds to the EF-hand calcium-binding proteins calmodulin and the B subunit of calcineurin (CnB). Ca2+-CnB and Ca2+-calmodulin activate the phosphatase catalytic subunit of calcineurin, CnA. The activated phosphatase dephosphorylates the zinc finger transcription factor CRZ1, which then moves to the nucleus to activate the expression of ENA1 and other target genes. ENA1 encodes a Na+-ATPase that pumps the toxic Na+ out of the cell, thus restoring ion homeostasis. Plant genomes do not encode any calcineurin proteins, even though the name calcineurin B-like (CBL) has been widely used to refer to a family of plant EF-hand calcium-binding proteins (Yu et al., 2014). Plants instead use a calcium-dependent protein kinase pathway known as the Salt-Overly-Sensitive (SOS) pathway for salt stress signaling and Na+ tolerance (Zhu, 2002) (Figure 2). In this pathway, the EF-hand calcium-binding protein SOS3 senses the cytosolic calcium signal elicited by salt stress. SOS3 interacts with and activates SOS2, a serine/threonine protein kinase. SOS3 is preferentially expressed in the root, and an SOS3 paralog, SCaBP8/CBL10 mainly expressed in the shoot, performs an equivalent role as SOS3 (Quan et al., 2007). The activated SOS2 phosphorylates and activates SOS1, a Na+/H+ antiporter at the plasma membrane (Zhu, 2002).
Figure 2.
The Ca2+-CBL-CIPK module mediates signaling of various ionic stresses. High Na+, low K+, excess Mg2+ and high pH (low H+) conditions cause cytosolic Ca2+ signals, which activate SOS3 (CBL4)/SCaBP8 (CBL10)-SOS2 (CIPK24), CBL1/9-CIPK23, CBL2/3-CIPK3/9/23/26 and SCaBP1 (CBL2)-PKS5/24 (CIPK11/14) to phosphorylate and regulate the activity of SOS1 (Na+/H+ antiporter), AKT1 (K+ channel), a putative Mg2+ transporter and H+-ATPase, respectively. Also shown is ABI2 and 14-3-3 inhibition of SOS2 and SOS2 regulation of SCaBP8 by phosphorylation. Arrows indicate activation, and bars indicate inhibition.
SOS1 is expressed in root epidermal cells and xylem parenchyma cells, so that activated SOS1 can extrude Na+ into the soil solution and load Na+ into the xylem for long distance transport to leaves by the transpirational stream (Shi et al., 2002; Zhu et al., 2016). The long-distance role of SOS1 is only partly understood, because SOS1 is also expressed in the xylem parenchyma in leaves where its function is unclear. Perhaps there is a Casparin strip-like structure in the xylem parenchyma cells in leaves that prevents the xylem stream from directly entering the apoplastic space of mesophyll cells. As a consequence, SOS1 may function to extrude Na+ from the xylem parenchyma cells into the apoplastic space of mesophyll cells. A prediction of this model is that SOS1 may preferentially localize on the side of the cell facing the mesophyll apoplastic space. HKT1 is another important transporter with a critical role in long-distance Na+ transport (Mäser et al., 2002). HKT1 in Arabidopsis is a Na+ importer also expressed in xylem parenchyma cells and other cells in the vasculature throughout the plant (Mäser et al., 2002). In roots, HKT1 may unload Na+ from the xylem to restrict the amount of Na+ in the transpirational stream. In leaves, HKT1 is proposed to load Na+ into the phloem for recirculation back to the root. Dysfunction in any of the SOS genes including SOS1 greatly increases the sensitivity of the mutant plants to salt stress (Zhu, 2000). In fact, the SOS genes were discovered because of the salt overly sensitive phenotype of these mutants (Zhu, 2000). Mutations in HKT1 cause obvious salt stress sensitivity in transpiring plants but can suppress the salt sensitivity of sos mutants grown in culture media where there is minimal transpiration (Rus et al., 2004). How the apparently antagonistic functions of SOS1 and HKT1 are coordinated remains an important question in salt stress research. It is unclear whether SOS1 and HKT1 are expressed in exactly the same cells in the vascular system. Overall, SOS1 seems to promote Na+ transport from roots to leaves, whereas HKT1 seems to restrict this movement. The importance of these two transporters in long-distance Na+ transport may depend on the severity of salt stress. Under mild salt stress, it may be advantageous for the plant to activate SOS1 in xylem parenchyma cells to deliver more Na+ to leaves where Na+ can be stored in the large vacuoles of mesophyll cells for cellular osmotic adjustment to facilitate growth. Under severe salt stress, however, high levels of Na+ delivered to leaves may exceed storage capacities, and thus Na+ must be unloaded from the root xylem and also recirculated back from leaves to roots using HKT1.
Several SOS3-like calcium-binding proteins are phosphorylated by their interacting SOS2-like protein kinases, and the phosphorylation appears important for the activation of the kinases by the calcium-binding proteins (Du et al., 2011). In the resting state or under non-stress conditions, SOS2 interacts with 14-3-3 proteins to ensure that SOS2 is inactive (Zhou et al., 2014). In addition, SOS2 interacts with the type 2C protein phosphatase ABI2, which may keep SOS2 inactive (Ohta et al., 2003).
The SOS pathway was the first abiotic stress signaling pathway established in plants (Zhu, 2000). The central signaling component, SOS2, represents a large family of protein kinases whose catalytic domain is similar to the yeast sucrose nonfermenting 1 (SNF1) and mammalian AMP-activated kinase (AMPK). In Arabidopsis, these proteins are generally referred to as SNF1-related kinases (SnRKs). SnRKs have 3 members in subfamily 1 (SnRK1s), 10 members in subfamily 2 (SnRK2s), and 25 members in subfamily 3 (SnRK3s) (Hrabak et al., 2003). Each of the 25 SnRK3s (aka PKSs or CIPKs), which include the founding member SOS2, interacts with one or more members of the family of 10 SOS3-like calcium-binding proteins (SCaBPs, aka CBLs) (Guo et al., 2001). The interaction is mediated through a common motif known as FISL in the N-terminal regulatory region of the kinases (Guo et al., 2001). Deletion of the FISL motif or the entire regulatory region leads to constitutive activation of the kinases (Guo et al., 2001). The large number of possible SCaBP/CBL-PKS/CIPK combinations suggests that the Ca2+-SOS3-SOS2 signaling principle is widely used in plants. Indeed, the CBL-CIPK module is important for various abiotic stress signaling pathways where calcium serves as a second messenger and particularly where regulation of ion transporter activities is involved (Figure 2). For example, low potassium stress, which presumably triggers a cytosolic calcium signal, activates CIPK23 via CBL1 and CBL9, to phosphorylate and activate the potassium channel AKT1 (Xu et al., 2006). SCaBP1 interacts with and activates PKS5/CIPK11 and PKS24/CIPK14, which phosphorylate and inhibit H+-ATPase on the plasma membrane. This inhibition is important for cellular pH regulation (Fuglsang et al., 2007). CBL2 and CBL3 function redundantly with their interacting proteins CIPK3/9/23/26 to regulate Mg2+ sequestration in the vacuole that is important for tolerance to high Mg2+ stress (Tang et al., 2015). Other CBL-CIPK combinations are known to regulate various transporters important for plant responses to nitrate, ABA, or other abiotic stresses (Yu et al., 2014).
Osmotic stress signaling
Plants have large families of MAP kinase pathway components, which can combine to form thousands of MAP kinase modules. For example, Arabidopsis contains over 60 MAP kinase kinase kinases (MAP3K), 10 MAP kinase kinases (MAP2K), and 20 MAP kinases (MAPK) (de Zelicourt et al., 2016). The rapid activation of multiple MAPKs, including MAPK3, 4, and 6, have long been observed in plants in response to biotic as well as abiotic stimuli such as salt, drought, cold, heat, and wounding, and in response to growth and developmental signals (de Zelicourt et a., 2016). The challenge in defining MAPK signaling pathways for abiotic stress remains in the identification of upstream sensor proteins, in the identification of MAP3Ks and MAK2Ks that are responsible for the MAPK activation, and in connecting the kinase activation to downstream effector proteins and physiological outputs. It is still unclear whether a MAPK pathway similar to the yeast high osmolarity glycerol (HOG) MAPK pathway (Hohmann, 2002) may function to mediate osmoregulation in plants. Perhaps the salt- and drought-activated MAPKs are activated mainly by stress-triggered secondary signals rather than by the primary osmotic stress signal.
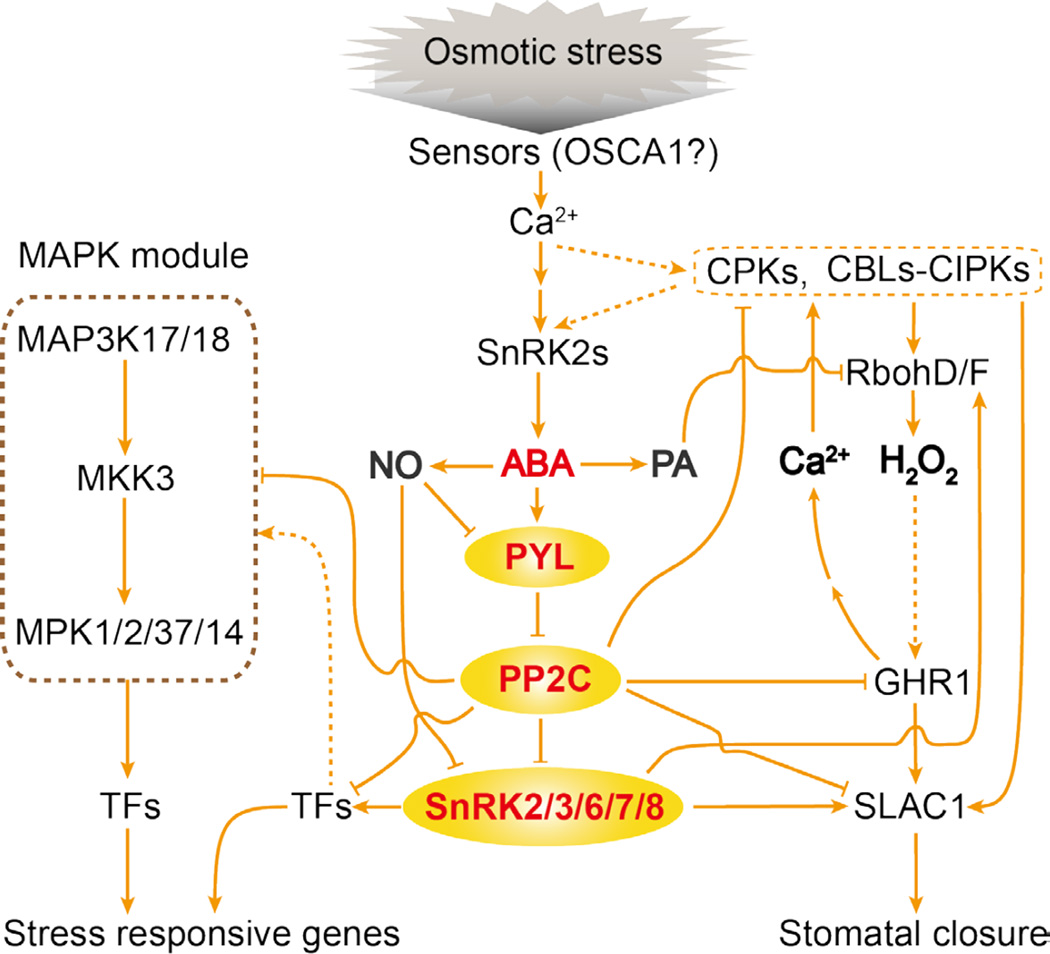
Salt, drought, and osmotic stress treatments also rapidly activate the SnRK2 family of protein kinases. In Arabidopsis, all 10 SnRK2s except SnRK2.9 are activated by osmotic stress, and SnRK2.2/3/6/7/8 are also activated by ABA (Boudsocq et al., 2004). Although the mechanism by which ABA activates the SnRK2s has been elucidated (see section below), how osmotic stress activates the kinases is not known. Genetic evidence shows that tolerance to osmostic stress requires SnRK2s, since decuple Arabidopsis mutant plants with all 10 SnRK2s disrupted are very sensitive to osmotic stress-inhibition of growth (Fujii et al., 2011). The decuple snrk2 mutant plants are impaired in osmotic stress-regulation of a large number of genes and in the accumulation of ABA, the compatible osmolyte proline, and the second messenger IP3, but they are unaffected in the osmotic stress-induced accumulation of ROS. These results suggest that SnRK2 activation is upstream of ABA accumulation and that this activation also controls osmotic adjustment and other adaptive responses to osmotic stress (Figure 3). Future efforts should be directed at finding upstream components important for SnRK2 activation under osmotic stress conditions and at identifying substrate proteins of osmotic stress-activated SnRK2s. Because osmotic stress triggers a cytosolic calcium signal and because the calcium channel OSCA1 is a putative osmosensor (Yuan et al., 2014), candidate factors upstream of the SnRK2s include calcium-responsive kinases such as CPKs and SCaBP/CBL-PKS/CIPKs (Figure 3). In the moss Physcomitrella patens, recent work showed that a Raf-like kinase is critical for SnRK2 activation by both osmotic stress and ABA (Saruhashi et al., 2015). It will be of interest to determine whether and how similar kinases may integrate osmotic stress and ABA signaling in higher plants.
Figure 3.
Osmotic stress and ABA sensing and signaling. The Ca2+ channel OSCA1 may participate in osmosensing. The resulting Ca2+ signal may activate CPKs and CBLs-CIPKs. Eventually, SnRK2s are activated, which leads to ABA accumulation. ABA binds to PYLs, which then interact with and inhibit class A PP2Cs, resulting in the activation of SnRK2.2/3/6/7/8. The activated SnRK2s phosphorylate effector proteins including TFs (transcription factors), SLAC1 and RbohD/F. RbohD/F generates H2O2, which elicits a Ca2+ signal through GHR1. This Ca2+ signal activates CPKs and CBLs-CIPKs that also phosphorylate effector proteins such as SLAC1. In addition to Ca2+, ABA also induces the second messengers NO (nitric oxide), and PA (phosphatidic acid) and other phospholipids. NO inhibits SnRK2s and PYLs, and PA regulates proteins like Rbohs. Also depicted is ABA activation of a MAP kinase module. Components of the core ABA signaling pathway are colored in red. Arrows indicate activation, bars indicate inhibition, and dashed lines indicate postulated regulation.
Osmotic as well as temperature stress causes the generation of various lipid signals including phosphatidic acid, phosphoinositides, sphingolipids, lysophospholipids, oxylipins, N-acylethanolamines, and many others (Hou et al., 2016). How stress regulates biosynthesis enzymes to generate the lipid signals is not well understood. In general, the lipid molecules bind to signaling proteins and affect their activity and membrane associations.
ABA signaling
The ABA signaling pathway is central to drought and salt stress responses in plants (Zhu, 2002). One of the most important advances in stress signaling in the past decade has been the identification of ABA receptors and the elucidation of the core ABA signaling pathway. Chemical genetics and protein interaction studies led to the identification of the PYR/PYL/RCAR (hereafter referred to as PYL) family of soluble, START domain proteins as receptors for ABA (Park et al., 2009; Ma et al., 2009). PYLs bind ABA with Kds in the micromolar range, though the binding affinity is increased by nearly 100-fold in the presence of clade A PP2Cs, such as ABI1, ABI2, HAB1, and PP2CA. Therefore, these PP2Cs can be considered as co-receptors (Ma et al., 2009). In the absence of ABA, the PP2Cs are associated with SnRK2 kinases including SnRK2.2, 2.3, and SnRK2.6 (aka OST1), which keeps the kinases inactive by blocking their catalytic cleft and by dephosphorylating the activation loop (Soon et al., 2012). ABA enters the central hydrophobic pocket of PYLs and induces the gate and latch loops to close and lock the pocket, creating a binding surface for the PP2Cs (Melcher et al., 2009). Inside the PYL-ABA-PP2C complex, a tryptophan residue in PP2C is inserted into the ABA-binding pocket and locks ABA tightly in place. The protein phosphatase activity of PP2C in the complex is inhibited by the ABA-PYL (Park et al., 2009). This binding and inhibition of the PP2Cs by ABA-PYLs releases the SnRK2s from association with and inhibition by the PP2Cs. The released SnRK2s are activated through autophosphorylation, and can then phosphorylate many downstream effectors (Fujii et al., 2009) (Figure 3). The small GTPase ROP11 interacts with and protects ABI1 from inhibition by PYL9 (Li et al., 2012). In turn, ABI1 and the other PP2Cs protect the GTP exchange factor RopGEF1 from ABA-induced degradation, thus forming a RopGEF-ROP-PP2C control loop that may counteract leaky ABA signaling by monomeric PYLs in the absence of stress (Li et al., 2016).
Much redundancy exists in the functions of Arabidopsis PYLs, although each PYL may have unique biochemical properties and expression patterns. Knockout of PYL8 leads to ABA insensitivity in lateral root growth recovery from stress inhibition (Zhao et al., 2014). The role of PYL8 in regulating lateral root growth is independent of the core ABA signaling pathway, because PYL8 directly interacts with MYB77, and this ABA-induced interaction enhances MYB77-dependent transcription of auxin-responsive genes (Zhao et al., 2014). Similarly, PYL6 interacts with MYC2, a key transcription factor in jasmonate responses, and thus connects ABA and jasmonate pathways (Aleman et al., 2016). Single-gene mutations in most other PYLs do not result in drastic ABA phenotypes. In contrast, pyr1prl1ply2pyl4 quadruple mutant plants are insensitive to ABA in germination and seedling growth (Park et al., 2009), and pyr1pyl1pyl2prl4ply5pyl8 mutant plants are even more resistant to ABA in not only germination and seedling growth but also in stomatal closure (Gonzalez-Guzman et al., 2012).
ABA strongly activates SnRK2.2, SnRK2.3, and SnRK2.6/OST1, and weakly activates SnRK2.7 and SnRK2.8 (Boudsocq et al., 2004). The snrk2.2/3/6 triple mutant in Arabidopsis is extremely insensitive to ABA in seed germination, seedling growth, stomatal closure, and gene regulation (Fujii and Zhu, 2009). Many effector proteins of ABA responses are direct substrates of SnRK2 kinases. bZIP transcription factors such as ABI5 and ABFs (ABA responsive element binding factors) are phosphorylated by the SnRK2s (Furihata et al., 2006). Much of the ABA signaling occurs at the plasma membrane. The association of PYLs with the plasma membrane is mediated by their interaction with C2 domain proteins (Rodriguez et al., 2014). Plasma membrane proteins such as the anion channel SLAC1 are SnRK2 substrates that mediate ABA-induced stomatal closure and reduce transpirational water loss under drought stress (Geiger et al., 2009). Recent phosphoproteomic studies identified dozens of additional SnRK2 substrate proteins including several proteins important for chloroplast function, flowering time control, miRNA and chromatin regulation, and RNA splicing (Wang et al., 2013). The PYL-PP2C-SnRK2 core ABA signaling module activates a MAPK cascade comprised of the MAP3Ks MAP3K17/18, the MAP2K MKK3, and the MAPKs MPK1/2/7/14, which may regulate many ABA effector proteins through phosphorylation (de Zelicourt et a., 2016).
ABA-activated SnRK2s also phosphorylate the plasma membrane NADPH oxidase RbohF, which when phosphorylated generates O2− in the apoplastic space. The O2− subsequently forms H2O2, a signaling molecule that mediates various ABA responses including stomatal closure (Sirichandra et al., 2009). ABA induction of ROS in guard cells is impaired in Arabidopsis pip2;1 mutant plants, indicating that apoplastic H2O2 can enter cells via the aquaporin PIP2;1 (Grondin et al., 2015). The MAP kinases MPK9 and MPK12 function redundantly in anion channel regulation in guard cells downstream of ROS, and affect ABA regulation of stomatal closure (Jammes et al., 2009). Another important component that connects ABA signaling and ROS is the plasma membrane receptor-like kinase GHR1 (Hua et al., 2012) (Figure 3). GHR1 interacts with and activates SLAC1. GHR1 is critical for ABA and ROS regulation of stomatal closure. Interestingly, GHR1 function is antagonized by ABI2 but not by ABI1 (Hua et al., 2012). H2O2 could also modulate the calcium signal to affect ABA responses, and GHR1 is required for H2O2-activation of plasma membrane Ca2+ channel (Hua et al., 2012). Calcium signaling is critical for ABA regulation of stomatal closure, and mutant plants defective in four redundant calcium-dependent protein kinases, CPK5, CPK6, CPK11, and CPK23, are incapable of closing stomata in response to ABA (Brandt et al., 2015). Like the SnRK2s, CPKs can phosphorylate effectors (including SLAC1) of ABA responses in guard cells (Geiger et al., 2010; Brandt et al., 2015). Furthermore, ABA-triggered calcium signal can activate the CBL1/9-CIPK26 module to cause phosphorylation of effector proteins such as RbohF (Drerup et al., 2013). In addition to inducing H2O2 and calcium signals, ABA triggers the generation of nitric oxide (NO) and phospholipids such as phosphatidic acid (Hou et al., 2016) (Figure 3). NO causes the S-nitrosylation of a cysteine residue adjacent to the catalytic site of SnRK2s, resulting in inactivation of the kinases (Wang et al., 2015). NO also causes tyrosine nitration and S-nitrosylation of cysteine residues in PYLs (Castillo et al., 2015). Tyrosine nitration inhibits PYLs activity and also accompanies polyubiquitylation and proteasome-mediated degradation of PYLs. Phosphatidic acid contributes to ABA signaling by binding to and activating RbohD and RbohF (Zhang et al., 2009). Therefore, the in vivo regulation of SLAC1, Rbohs and other effectors of ABA responses in plants requires a network of signaling pathways that include not only the PYL-PP2C-SnRK2 core pathway but also other pathways that involve calcium, ROS, NO, phospholipids, and the other kinases described above (Figure 3).
Cold and heat stress signaling
Cold stress greatly affects plant metabolism and transcriptomes. The effect on plant metabolism arises from both direct inhibition of metabolic enzymes by cold temperatures and reprogramming of gene expression (Chinnusamy et al., 2007). In temperate plants, an exposure to low, nonfreezing temperatures enhances the tolerance to subsequent freezing temperatures, a process known as cold acclimation. Cold stress rapidly induces the expression of many transcription factors, including the AP2-domain proteins CBFs, which then activate the expression of numerous downstream cold responsive (COR) genes (Chinnusamy et al., 2007). The CBF genes are controlled by upstream transcription factors, including the bHLH transcription factor ICE1. ICE1 is subjected to sumoylation, and polyubiquitylation and subsequent proteasomal degradation, mediated by the SUMO E3 ligase SIZ1 and ubiquitin E3 ligase HOS1, respectively (Chinnusamy et al., 2007). Cold stress induction of the CBF and COR genes is gated by the circadian clock, whose activity is modulated by diurnal oscillation of the plastid retrograde signal tetrapyrroles (Norén et al., 2016).
Ding et al. (2015) recently reported that cold stress activates SnRK2.6/OST1 and that SnRK2.6 interacts with and phosphorylates ICE1 to activate the CBF-COR gene expression cascade and freezing tolerance. Furthermore, cold-activated SnRK2.6 disrupts the interaction between ICE1 and HOS1 to prevent ICE1 degradation. The reported cold stress activation of SnRK2.6 is independent of ABA, although the activation is negatively regulated by ABI1 and other PP2Cs. Therefore, the activation presumably does not involve PYL ABA receptors. There is discrepancy in the reported cold activation of SnRK2.6, since Boudsocq et al. (2004) reported that none of the 10 Arabidopsis SnRK2s is activated by cold stress.
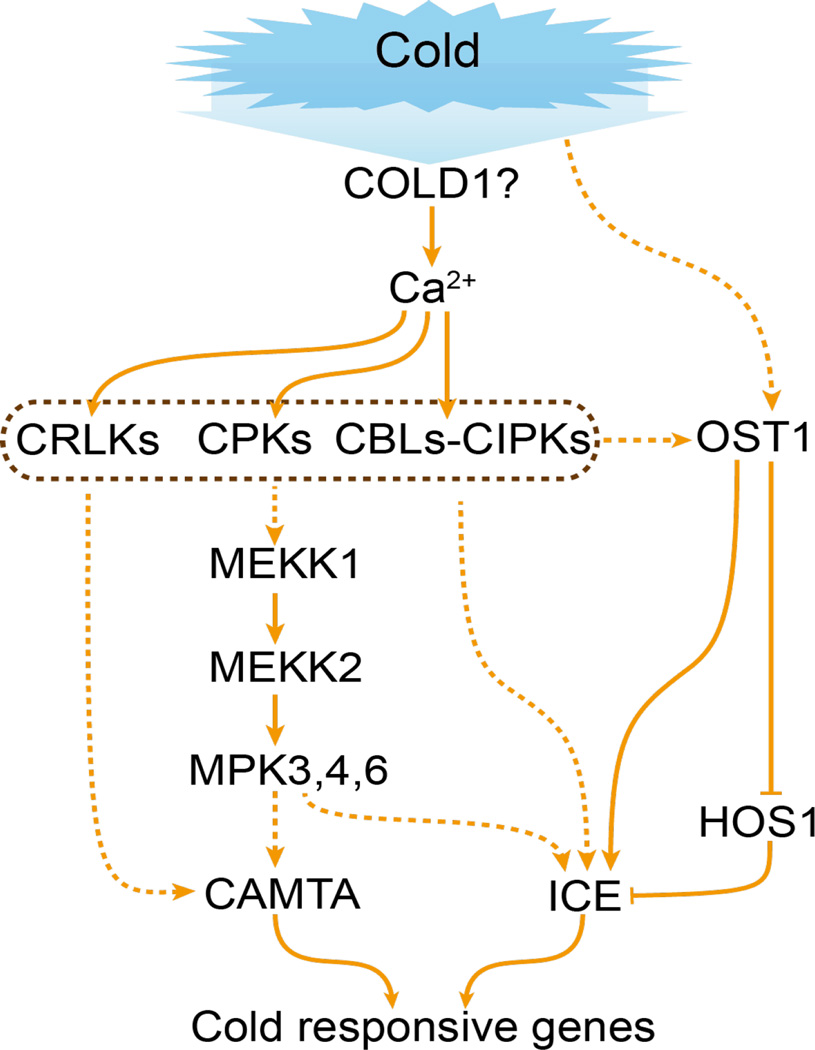
Teige et al. (2004) showed that the Arabidopsis MAP2K MKK2 is activated by cold and salt stress and controls COR gene expression and plant tolerance to freezing and salt stress. MKK2 is downstream of the MAP3K MEKK1 and upstream of MPK4 and MPK6, which are activated by various stresses including cold temperatures. Pharmacological studies indicate that membrane fluidity and cytoskeleton and calcium influx are involved in cold stress regulation of COR genes and MAPKs (Sangwan et al., 2002). The receptor-like kinase CRLK1 may connect cold stress-induced calcium signaling with the MAPK cascade, because CRLK1 binds to calcium and calmodulin, interacts with MEKK1, and is requisite for cold stress activation of MAPK activities (Yang et al., 2010). It appears that cold stress affects membrane fluidity, which may be sensed by plasma membrane proteins such as calcium channels or associated proteins, resulting in calcium influx and activation of calcium-responsive protein kinases (CPKs, CIPKs, and CRLK1) and the MAPK cascade, which regulate COR gene expression (Figure 4). It will be necessary to clarify the roles of these kinases, and the relationships among them and between them and SnRK2.6 in cold stress signaling through genetic, molecular, and biochemical analysis in the future.
Figure 4.
Cold stress sensing and signaling. Cold stress is sensed by membrane proteins such as COLD1, leading to a cytosolic Ca2+ signal. CPKs and CBLs-CIPKs may mediate the Ca2+ signal to activate a MAP kinase cascade. The activated MPKs are postulated to phosphorylate TFs such as CAMTAs and ICE1/2, which then activate cold responsive genes. Through an unknown mechanism, cold stress also activates OST1 (SnRK2.6), which inhibits HOS1, and phosphorylates and activates ICE1. Arrows indicate activation, bars indicate inhibition, and dashed lines indicate postulated regulation.
Heat stress induces the expression of heat shock proteins (HSPs), many of which function as molecular chaperones to prevent protein denaturation and maintain protein homeostasis (Scharf et al., 2012). Like mammalian heat stress transcription factors (HSFs), plant HSFs are released from association with and inhibition by the HSP70 and HSP90 chaperones as a result of the binding of the chaperones to misfolded proteins caused by heat stress, and thus the HSFs are made available to activate heat stress responses (Scharf et al., 2012). Heat stress also activates MAPKs, which modulate HSP gene expression (Sangwan et al., 2002). The MAPK activation may be linked to heat-induced changes in membrane fluidity and calcium signaling that are important for HSP gene expression and thermotolerance (Sangwan et al., 2002). Common features between cold and heat stress signaling are not limited to membrane fluidity changes, calcium signaling, and MAPK activation, but also include the involvement of ROS, NO, phospholipid signals, protein sumoylation, and proteosomal degradation (Chinnusamy et al., 2007; Scharf et al., 2012).
Systemic signaling
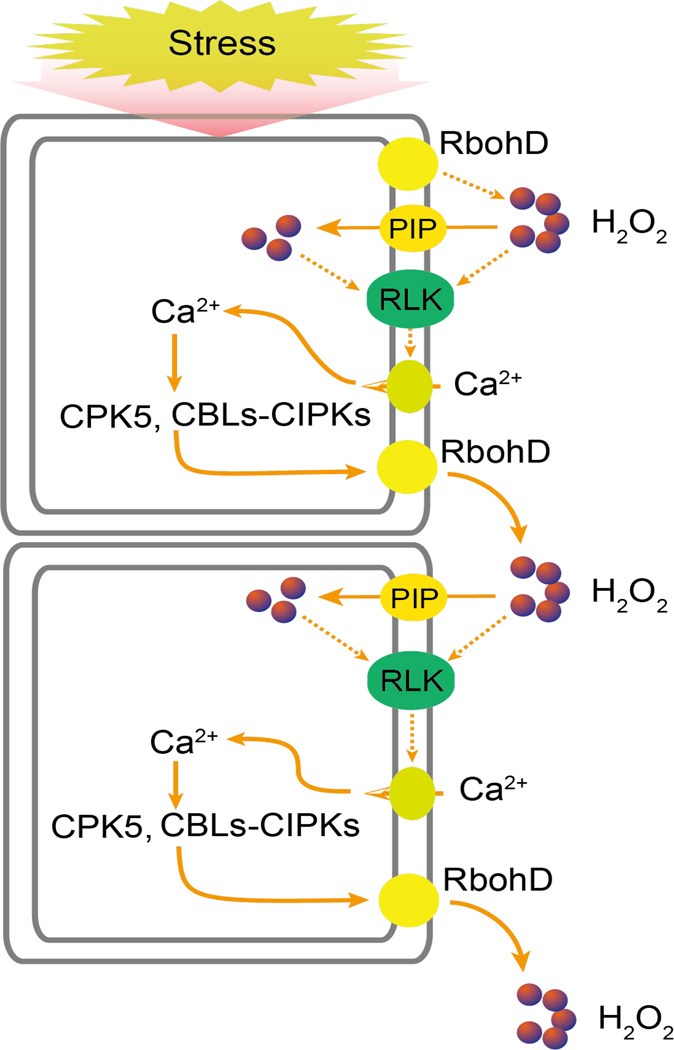
Pathogen infection and wounding trigger systemic responses in plants. Similarly, abiotic stresses such as drought, salt, cold, heat, and high light also elicit systemic responses such that locally applied stress causes responses not only locally but also in distal tissues, resulting in systemic acquired acclimation (SAA). SAA involves long-distance electrical and hydraulic signals as well as calcium and ROS waves (Choi et al., 2014; Miller et al., 2009). Stress-triggered calcium and ROS waves can move at speeds that exceed 1000 µm per second as visualized in transgenic plants expressing calcium-sensitive fluorescent protein (Choi et al., 2014) and luciferase reporter driven by a ROS-responsive promoter (Miller et al., 2009), respectively. Calcium and ROS waves have been shown to cause transcriptional responses in distal tissues (Choi et al., 2014; Miller et al., 2009). ROS waves require the plasma membrane NADPH oxidase RbohD (Miller et al., 2009), while calcium waves depend on the vacuolar ion channel TPC1, which may be involved in calcium-induced calcium release (Choi et al., 2014). RbohD is phosphorylated by the calcium-dependent protein kinase CPK5, which can be activated by ROS and is required for systemic defense responses (Dubiella et al., 2013). A model that emerges is that ROS generated by NADPH oxidases triggers a cytosolic calcium signal to activate more NADPH oxidases via calcium-responsive kinases, thus generating a self-propagating mutual activation circuit between ROS and calcium signals (Figure 5). H2O2 generated by RbohD may enter cells through a plasma membrane intrinsic protein/water channel (PIP) (Grondin et al., 2015), and its activation of the plasma membrane calcium channel likely requires GHR1 (Hua et al., 2012) or a similar receptor like kinase (RLK). The plasma membrane calcium channel that is activated by ROS has not been identified. In addition, it is unclear how the calcium and ROS waves may be linked to long-distance electrical signals.
Figure 5.
Model of systemic stress signaling. Local exposure to stress generates H2O2 and Ca2+ signals. The Ca2+ signal can activate CPKs and CBLs-CIPKs, which phosphorylate and activate RbohD. Activated RbohD generates H2O2 that diffuses through the cell wall to neighoring cells, where it induces a Ca2+ signal, through RLKs like GHR1. H2O2 may activate Ca2+ signaling at the cell surface and may also enter the cell through PIP water channels and then activate Ca2+ signaling intracellularly. The mutual activation between the Ca2+ and H2O2 signals generates a self propagating calcium and ROS waves that can travel to distant tissues to cause systemic acquired acclimation responses. Dashed lines indicate postulated regulation.
Conclusions and perspectives
Cell signaling in response to salt, drought, and the stress hormone ABA largely depends on the SnRK family of protein kinases in plants. SnRKs are related to the yeast SNF1 and mammalian AMPK, which are key sensors of cellular energy status (Hardie et al., 2016). In plants, abiotic stresses diminish the energy supply by inhibiting photosynthesis and energy-releasing catabolic reactions. So, SNF1/AMPK-related kinases proliferated and diversified through evolution to mediate the signaling of various abiotic stresses. SnRK1s are SNF1/AMPK orthologs that function in regulating metabolism in plants. All SnRK2s participate in osmotic stress and ABA signaling, while SnRK3s are key regulators of ion homeostasis required to cope with salt and nutrient stress in soil. Many of these stress signaling pathways also involve the calcium-dependent protein kinase CPKs, which share homology to SnRKs in their kinase domains (Hrabak et al., 2003). In addition, virtually all of the stress pathways also involve MAPKs, which is a conserved feature of stress signaling in organisms from fungi to plants and metazoans. Other conserved features include the widespread use of calcium, ROS, NO, and lipid molecules as second messengers, although the generation and signal transduction of the second messengers are different in plants.
Identifying stress sensors remains an important but challenging goal for abiotic stress research in plants. Efficient gene editing technologies and chemical genetic approaches will help overcome the gene redundancy problems that prevent the genetic identification of stress sensors. The growing appreciation of the importance of various cell organelles in stress sensing and responses and the dispersed stress sensing model (Figure 1) will also help us understand stress sensing and stress resistance, although the integration of signals from perturbed organelles is still poorly understood. Because plant stress responses must be coordinated with growth and development, it is important to understand the crosstalk between stress signaling pathways and hormonal as well as growth and developmental signaling pathways. Similarly, more attention should be directed toward plant responses to simultaneous, multiple abiotic stresses, and to the crosstalk between abiotic and biotic stress signaling because much of the abiotic stress research thus far has been carried out on sterile plants grown in culture media in the laboratory; in nature, however, plants co-exist with insects and microorganisms. The root and shoot microbiomes presumably include many beneficial bacteria and fungi that help plants resist stress. Understanding how bacteria and fungi enhance plant stress resistance should increase our ability to use these beneficial organisms and should also increase our understanding of stress resistance in plants.
Acknowledgments
I thank Drs. Mike Hasegawa, Zhizhong Gong and Yan Guo for helpful suggestions and Dr. Changsong Zhou for assistance with preparing figures. Work in my lab was supported by the Chinese Academy of Sciences and by US National Institutes of Health Grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aleman F, Yazaki J, Lee M, Takahashi Y, Kim AY, Li Z, Kinoshita T, Ecker JR, Schroeder JI. An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: A putative link of ABA and JA signaling. Sci. Rep. 2016;6:28941. doi: 10.1038/srep28941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnadottir J, Chalfie M. Eukaryotic mechanosensitive channels. Annu. Rev. Biophys. 2010;39:111–137. doi: 10.1146/annurev.biophys.37.032807.125836. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- Brandt B, Munemasa S, Wang C, Nguyen D, Yong T, Yang PG, Poretsky E, Belknap TF, Waadt R, Aleman F, et al. Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. Elife. 2015;4:e03599. doi: 10.7554/eLife.03599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo MC, Lozano-Juste J, Gonzalez-Guzman M, Rodriguez L, Rodriguez PL, Leon J. Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Sci. Signal. 2015;8:ra89. doi: 10.1126/scisignal.aaa7981. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Choi WG, Toyota M, Kim SH, Hilleary R, Gilroy S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA. 2014;111:6497–6502. doi: 10.1073/pnas.1319955111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zelicourt A, Colcombet J, Hirt H. The Role of MAPK Modules and ABA during Abiotic Stress Signaling. Trends Plant Sci. 2016;21:677–685. doi: 10.1016/j.tplants.2016.04.004. [DOI] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell. 2015;32:278–289. doi: 10.1016/j.devcel.2014.12.023. [DOI] [PubMed] [Google Scholar]
- Dong CH, Zolman BK, Bartel B, Lee BH, Stevenson B, Agarwal M, Zhu JK. Disruption of Arabidopsis CHY1 reveals an important role of metabolic status in plant cold stress signaling. Mol. Plant. 2009;2:59–72. doi: 10.1093/mp/ssn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup MM, Schlücking K, Hashimoto K, Manishankar P, Steinhorst L, Kuchitsu K, Kudla J. The Calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant. 2013;6:559–569. doi: 10.1093/mp/sst009. [DOI] [PubMed] [Google Scholar]
- Du W, Lin H, Chen S, Wu Y, Zhang J, Fuglsang AT, Palmgren MG, Wu W, Guo Y. Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in Arabidopsis. Plant Physiol. 2011;156:2235–2243. doi: 10.1104/pp.111.173377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA. 2013;110:8744–8749. doi: 10.1073/pnas.1221294110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Kesten C, Schneider R, Zhang Y, Ivakov A, Froehlich A, Funke N, Persson S. A Mechanism for Sustained Cellulose Synthesis during Salt Stress. Cell. 2015;162:1353–1364. doi: 10.1016/j.cell.2015.08.028. [DOI] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23:3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff NV, Battisti DS, Beachy RN, Cooper PJ, Fischhoff DA, Hodges CN, Knauf VC, Lobell D, Mazur BJ, Molden D, et al. Radically rethinking agriculture for the 21st century. Science. 2010;327:833–834. doi: 10.1126/science.1186834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+ -ATPase by preventing interaction with 14-3-3 protein. Plant Cell. 2007;19:1617–1634. doi: 10.1105/tpc.105.035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. USA. 2011;108:1717–1722. doi: 10.1073/pnas.1018367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernandez MA, Holdsworth MJ, Perez-Amador MA, Kollist H, et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012;24:2483–2496. doi: 10.1105/tpc.112.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin A, Rodrigues O, Verdoucq L, Merlot S, Leonhardt N, Maurel C. Aquaporins Contribute to ABA-Triggered Stomatal Closure through OST1-Mediated Phosphorylation. Plant Cell. 2015;27:1945–1954. doi: 10.1105/tpc.15.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu JK. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant cell. 2001;13:1383–1400. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton ES, Jensen GS, Maksaev G, Katims A, Sherp AM, Haswell ES. Mechanosensitive channel MSL8 regulates osmotic forces during pollen hydration and germination. Science. 2015;350:438–441. doi: 10.1126/science.aac6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Schaffer BE, Brunet A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends Cell Biol. 2016;26:190–201. doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Duan Y, Hua D, Fan G, Wang L, Liu Y, Chen Z, Han L, Qu LJ, Gong Z. DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell. 2012;24:1815–1833. doi: 10.1105/tpc.112.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R. Ion channels in plants. Physiol. Rev. 2012;92:1777–1811. doi: 10.1152/physrev.00038.2011. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Q, Ufer G, Bartels D. Lipid signalling in plant responses to abiotic stress. Plant Cell. Environ. 2016;39:1029–1048. doi: 10.1111/pce.12666. [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua D, Wang C, He J, Liao H, Duan Y, Zhu Z, Guo Y, Chen Z, Gong Z. A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell. 2012;24:2546–2561. doi: 10.1105/tpc.112.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, Cho D, Lee S, Giordo R, Sritubtim S, et al. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. USA. 2009;106:20520–20525. doi: 10.1073/pnas.0907205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendretzki A, Wittland J, Wilk S, Straede A, Heinisch JJ. How do I begin? Sensing extracellular stress to maintain yeast cell wall integrity. Eur. J. Cell Biol. 2011;90:740–744. doi: 10.1016/j.ejcb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Lee BH, Lee H, Xiong L, Zhu JK. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell. 2002;14:1235–1251. doi: 10.1105/tpc.010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Waadt R, Schroeder JI. Release of GTP Exchange Factor Mediated Down-Regulation of Abscisic Acid Signal Transduction through ABA-Induced Rapid Degradation of RopGEFs. PLoS Biol. 2016;14:e1002461. doi: 10.1371/journal.pbio.1002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Li Z, Gao X, Chinnusamy V, Bressan R, Wang ZX, Zhu JK, Wu JW, Liu D. ROP11 GTPase negatively regulates ABA signaling by protecting ABI1 phosphatase activity from inhibition by the ABA receptor RCAR1/PYL9 in Arabidopsis. J. Integr. Plant Biol. 2012;54:180–188. doi: 10.1111/j.1744-7909.2012.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016;211:418–428. doi: 10.1111/nph.13915. [DOI] [PubMed] [Google Scholar]
- Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, et al. COLD1 confers chilling tolerance in rice. Cell. 2015;160:1209–1221. doi: 10.1016/j.cell.2015.01.046. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002;531:157–161. doi: 10.1016/s0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignolet-Spruyt L, Xu E, Idanheimo N, Hoeberichts FA, Muhlenbock P, Brosche M, Van Breusegem F, Kangasjarvi J. Spreading the news: subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016;67:3831–3844. doi: 10.1093/jxb/erw080. [DOI] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2009;2:ra45. doi: 10.1126/scisignal.2000448. [DOI] [PubMed] [Google Scholar]
- Ng S, De Clercq I, Van Aken O, Law SR, Ivanova A, Willems P, Giraud E, Van Breusegem F, Whelan J. Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol. Plant. 2014;7:1075–1093. doi: 10.1093/mp/ssu037. [DOI] [PubMed] [Google Scholar]
- Norén L, Kindgren P, Stachula P, Rühl M, Eriksson ME, Hurry V, Strand Å. Circadian and Plastid Signaling Pathways Are Integrated to Ensure Correct Expression of the CBF and COR Genes during Photoperiodic Growth. Plant Physiol. 2016;171:1392–1406. doi: 10.1104/pp.16.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Guo Y, Halfter U, Zhu JK. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA. 2003;100:11771–11776. doi: 10.1073/pnas.2034853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell. 2007;19:1415–1431. doi: 10.1105/tpc.106.042291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA. 2012;109:5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez L, Gonzalez-Guzman M, Diaz M, Rodrigues A, Izquierdo-Garcia AC, Peirats-Llobet M, Fernandez MA, Antoni R, Fernandez D, Marquez JA, et al. C2-domain abscisic acid-related proteins mediate the interaction of PYR/PYL/RCAR abscisic acid receptors with the plasma membrane and regulate abscisic acid sensitivity in Arabidopsis. Plant Cell. 2014;26:4802–4820. doi: 10.1105/tpc.114.129973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Lee BH, Munoz-Mayor A, Sharkhuu A, Miura K, Zhu JK, Bressan RA, Hasegawa PM. AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 2004;136:2500–2511. doi: 10.1104/pp.104.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan V, Orvar BL, Beyerly J, Hirt H, Dhindsa RS. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 2002;31:629–638. doi: 10.1046/j.1365-313x.2002.01384.x. [DOI] [PubMed] [Google Scholar]
- Saruhashi M, Kumar Ghosh T, Arai K, Ishizaki Y, Hagiwara K, Komatsu K, Shiwa Y, Izumikawa K, Yoshikawa H, Umezawa T, et al. Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc. Natl. Acad. Sci. USA. 2015;112:E6388–E6396. doi: 10.1073/pnas.1511238112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta. 2012;1819:104–119. doi: 10.1016/j.bbagrm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009;583:2982–2986. doi: 10.1016/j.febslet.2009.08.033. [DOI] [PubMed] [Google Scholar]
- Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science. 2012;335:85–88. doi: 10.1126/science.1215106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck SM, Colaço R, Davies JM. Plant calcium-permeable channels. Plant Physiol. 2013;163:514–522. doi: 10.1104/pp.113.220855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang RJ, Zhao FG, Garcia VJ, Kleist TJ, Yang L, Zhang HX, Luan S. Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc Natl Acad Sci USA. 2015;112:3134–3139. doi: 10.1073/pnas.1420944112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell. 2004;15:141–152. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Tenhaken R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2014;5:771. doi: 10.3389/fpls.2014.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thewes S. Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryot. Cell. 2014;13:694–705. doi: 10.1128/EC.00038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur A, Hofte H. Cell wall integrity signaling in plants: "To grow or not to grow that's the question". Glycobiology. 2016 doi: 10.1093/glycob/cww029. In press. [DOI] [PubMed] [Google Scholar]
- Wagner D, Przybyla D, Op den Camp R, Kim C, Landgraf F, Lee KP, Wursch M, Laloi C, Nater M, Hideg E, et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang P, Du Y, Hou YJ, Zhao Y, Hsu CC, Yuan F, Zhu X, Tao WA, Song CP, Zhu JK. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. USA. 2015;112:613–618. doi: 10.1073/pnas.1423481112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, Zhu JK. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA. 2013;110:11205–11210. doi: 10.1073/pnas.1308974110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EE, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell. 2012;149:1525–1535. doi: 10.1016/j.cell.2012.04.038. [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006;125:1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Yang T, Shad Ali G, Yang L, Du L, Reddy AS, Poovaiah BW. Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal. Behav. 2010;5:991–994. doi: 10.4161/psb.5.8.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, An L, Li W. The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 2014;33:203–214. doi: 10.1007/s00299-013-1507-1. [DOI] [PubMed] [Google Scholar]
- Yuan F, Yang H, Xue Y, Kong D, Ye R, Li C, Zhang J, Theprungsirikul L, Shrift T, Krichilsky B, et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature. 2014;514:367–371. doi: 10.1038/nature13593. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhu H, Zhang Q, Li M, Yan M, Wang R, Wang L, Welti R, Zhang W, Wang X. Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell. 2009;21:2357–2377. doi: 10.1105/tpc.108.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xing L, Wang X, Hou YJ, Gao J, Wang P, Duan CG, Zhu X, Zhu JK. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Science Signal. 2014;7:ra53. doi: 10.1126/scisignal.2005051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lin H, Chen S, Becker K, Yang Y, Zhao J, Kudla J, Schumaker KS, Guo Y. Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell. 2014;26:1166–1182. doi: 10.1105/tpc.113.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Lee BH, Dellinger M, Cui X, Zhang C, Wu S, Nothnagel EA, Zhu JK. A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 2010;63:128–140. doi: 10.1111/j.1365-313X.2010.04227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 2000;124:941–948. doi: 10.1104/pp.124.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Ann. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Shabala L, Cuin TA, Huang X, Zhou M, Munns R, Shabala S. Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. J. Exp. Bot. 2016;67:835–844. doi: 10.1093/jxb/erv493. [DOI] [PMC free article] [PubMed] [Google Scholar]