Abstract
The FLOWERING LOCUS T (FT) protein is a central component of a mobile flowering signal (florigen) that is transported from leaves to the shoot apical meristem (SAM). Two FT monomers and two DNA-binding bZIP transcription factors interact with a dimeric 14-3-3 protein bridge to form a hexameric protein complex. This complex, designated as “florigen activation complex” (FAC), plays a critical role in flowering. The wheat homolog of FT, designated FT1 (= VRN3), activates expression of VRN1 in the leaves and the SAM, promoting flowering under inductive long days. In this study, we show that FT1, other FT-like proteins, and different FD-like proteins, can interact with multiple wheat and barley 14-3-3 proteins. We also identify the critical amino acid residues in FT1 and FD-like proteins required for their interactions, and demonstrate that 14-3-3 proteins are necessary bridges to mediate the FT1-TaFDL2 interaction. Using in vivo bimolecular fluorescent complementation (BiFC) assays, we demonstrate that the interaction between FT1 and 14-3-3 occurs in the cytoplasm, and that this complex is then translocated to the nucleus, where it interacts with TaFDL2 to form a FAC. We also demonstrate that a FAC including FT1, TaFDL2 and Ta14-3-3C can bind to the VRN1 promoter in vitro. Finally, we show that relative transcript levels of FD-like and 14-3-3 genes vary among tissues and developmental stages. Since FD-like proteins determine the DNA specificity of the FACs, variation in FD-like gene expression can result in spatial and temporal modulation of the effects of mobile FT-like signals.
Keywords: Triticum aestivum, Hordeum vulgare, flowering, florigen activation complex, FT, FDL, 14-3-3, VRN1
INTRODUCTION
Florigen is a systemic signal that is produced in leaves in response to the stimulus of inductive day length and is transported to the shoot apex to induce flowering (Zeevaart, 2006). Recent studies in model plant species Arabidopsis and rice have revealed that FLOWERING LOCUS T (FT) (= HEADING DATE 3 (HD3A) in rice) is the main component of florigen (Corbesier et al., 2007; Tamaki et al., 2007). In both species, FT transcription is activated in the leaf vascular tissue and the encoded protein is transported to the shoot apical meristem (SAM) through the phloem (Corbesier et al., 2007; Tamaki et al., 2007).
FT encodes a RAF-kinase-inhibitor-like protein with a unique phosphatidylethanolamine-binding protein (PEBP) domain. Crystal structure studies in mammals suggest that PEBP domain proteins contain an anion-binding pocket composed of highly conserved residues, which can bind to anions, phosphate groups and phospholipids (Banfield et al., 1998; Serre et al., 1998; reviewed in Granovsky et al., 2008). Both FT and HD3A proteins have a conserved anion-binding pocket. Although it is unclear whether the anion-binding pocket binds to anions or phospholipids in plants, a tyrosine (Y85 in FT and Y87 in HD3A) in the pocket region has been shown to be critical for flowering regulation (Hanzawa et al., 2005; Ahn et al., 2006; Taoka et al., 2013).
Wheat FT1 (=VRN3) is the closest homolog to Arabidopsis FT (Yan et al., 2006). FT1 is transcribed in the leaves under long days, and its expression is closely correlated with the transcription of its target gene, the meristem identity gene VRN1 in the apex, suggesting that FT1 also encodes a mobile signal in wheat (Yan et al., 2006; Li and Dubcovsky, 2008; Pearce et al., 2013).
In Arabidopsis, FD and FD PARALOG (FDP) are basic leucine zipper (bZIP) transcription factors that interact with FT and activate the expression of the floral meristem identity gene APETALA1 (AP1) (Abe et al., 2005; Wigge et al., 2005). The C-terminus of FD has a short motif that is the potential target of calcium-dependent protein kinases (CDPKs), and a single amino acid substitution (a serine/threonine substituted by an alanine) in this motif disrupts FD protein function (Abe et al., 2005). Other members of the bZIP protein family, including OsFD1 from rice (Taoka et al., 2011), TaFDL2 and TaFDL6 from wheat (Li and Dubcovsky, 2008) and DLF1 from maize (Muszynski et al., 2006) also have been shown to interact with FT orthologs (HD3A in rice, FT1 in wheat and ZCN8 in maize).
Many of the FT-interacting bZIP proteins were identified by yeast two-hybrid assays. However, a recent study in rice suggests that these yeast two-hybrid interactions are likely mediated by endogenous yeast 14-3-3 proteins, since no direct interaction between OsFD1 and HD3A could be detected by NMR titration assays, isothermal titration calorimetry or pull-down assays (Taoka et al., 2011). Indeed, 14-3-3 proteins have been identified as FT-interacting proteins in a previous study in tomato (Pnueli et al., 2001). Taoka et al. (2011) demonstrated that HD3A first interacts with 14-3-3 proteins in the cytoplasm, and that the resulting protein complex then translocates to the nucleus where it binds to OsFD1. The resultant protein complex, henceforth the florigen activation complex (FAC), then interacts with the promoter of the meristem identity gene OsMADS15 and induces its transcription (Taoka et al., 2011).
The crystal structure of the rice FAC is predicted to be a deep W-shaped structure with two 14-3-3 molecules forming a W-shaped dimer via their N-terminal regions. One HD3A monomer binds to the C-terminal region of each 14-3-3 monomer, extending the W-shape structure. Two positively charged pockets formed at the corners at the inner base of the ‘W’ then interact with the phosphorylated C terminus of OsFD1proteins (Taoka et al., 2011). The resulting hexameric complex is essential for HD3A-mediated flowering promotion in rice and provides a mechanistic basis for florigen function in flowering (Taoka et al., 2011).
14-3-3 proteins are present in all eukaryotes and regulate diverse cellular functions through protein–protein interactions. Most plant species have multiple 14-3-3 paralogs that provide sequence and functional diversity (Paul et al., 2012). Plant 14-3-3 proteins can interact with a wide range of proteins including kinases, transcription factors, structural proteins, ion channels and pathogen defense-related proteins. Through these interactions, plant 14-3-3 proteins can modulate enzyme activity, protein structure, stability, and subcellular localization, affecting multiple physiological and developmental processes (reviewed in Denison et al., 2011).
To understand better how different FT, FD and 14-3-3 proteins regulate flowering in the temperate cereals, we characterized their interactions by both yeast two-hybrid and in vivo bimolecular fluorescent complementation (BiFC) assays. We identified the critical amino acids in FT1 and FD-like proteins required for their interactions with 14-3-3 proteins, and demonstrated that 14-3-3 proteins are necessary to mediate FT1-TaFDL2 interaction. BiFC assays showed that FT1-14-3-3 interactions occur in the cytoplasm, and that the tetrameric complex is then translocated to the nucleus where it interacts with TaFDL2 to form the hexameric FAC. Finally, we established that the FAC including FT1, TaFDL2 and Ta14-3-3C can bind to the VRN1 promoter.
RESULTS
Wheat FT1 protein interacts with multiple 14-3-3 proteins
Sequences of five previously identified barley 14-3-3 paralogs A to E (Schoonheim et al., 2007) were used to search and clone their wheat orthologs, which were assigned the same letters as the corresponding barely genes. In addition, an unclassified 14-3-3 protein (GenBank accession number BAK01024) with high sequence similarity to rice Os14-3-3H (also known as OsGF14h or UniProt Q2R1D5.1) was identified and cloned from barley, and was designated Hv14-3-3H. We did not identify a sequence for this gene in T. aestivum, but found a copy in a T. urartu transcriptome database (Krasileva et al., 2013). Tu14-3-3H, Hv14-3-3H and Os14-3-3H are placed in a separate cluster from the other five 14-3-3 proteins in a phylogenetic analysis (Fig. S1), indicating that they diverged from the other 14-3-3 early in the evolution of this gene family.
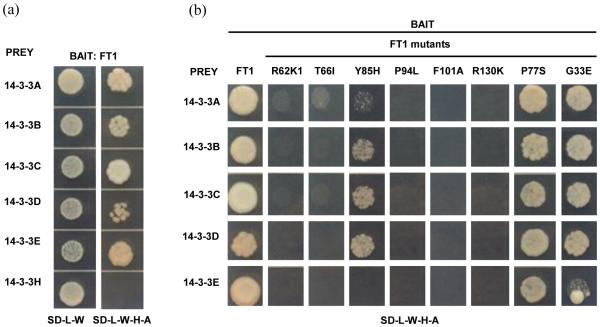
Full-length coding regions of these six 14-3-3 proteins were cloned into yeast vector pGADT7 as preys, and FT1 was cloned into pGBKT7 as bait. Yeast two-hybrid assays revealed that FT1 interacts with 14-3-3 proteins A to E, but not with 14-3-3H (Fig. 1A), suggesting some functional differentiation between the two 14-3-3 clades (Fig. S1).
Figure 1. Interactions between FT1 and 14-3-3 proteins.
(a). Yeast two-hybrid assays between FT1 and six 14-3-3 proteins. (b) Mutations of critical amino acid residues in FT1 protein abolish protein interactions with 14-3-3 proteins in yeast two-hybrid assays. FT1 and mutants were used as baits and 14-3-3 proteins were used as preys. SD medium lacking Leucine and Tryptophan (−L−W) was used to select for yeast transformants containing both bait and prey vectors. Interaction strength was tested on SD media lacking Leucine, Tryptophan, Histidine and Adenine (−L−W−H−A).
Identification of the critical amino acid residues in FT1 protein responsible for interactions with 14-3-3 proteins
Two amino acid residues Y85 and R132 of the Arabidopsis FT protein have been previously shown to be critical for flowering (Hanzawa et al., 2005; Ahn et al., 2006) and for interaction with 14-3-3 proteins (Taoka et al., 2011). We created point mutations in wheat FT1 by site-directed mutagenesis at these two positions (Y85 and R130), at four positions predicted as surface-exposed residues (R62, T66, P94 and F101, Taoka et al., 2011), and at two additional sites (P77S and G33E) that have been recently shown to delay flowering when mutated in wheat (Lv, et al., 2014). Yeast two-hybrid assays showed that FT1 mutations R62K, T66I, P94L, F101A, and R130K abolished the interactions with 14-3-3 proteins. Y85H abolished interaction with Ta14-3-3E and reduced interactions with Ta14-3-3A-D proteins (Fig. 1B). The G33E and P77S mutations did not affect interactions with the 14-3-3 proteins, suggesting that their effect on wheat flowering is not mediated by the disruption of these protein-protein interactions (Fig. 1B).
Interactions between FD-like and 14-3-3 proteins in wheat and barley
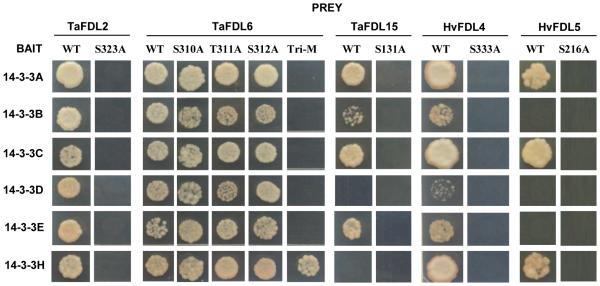
Wheat TaFDL2, TaFDL6, TaFDL13 and TaFDL15 were identified from EST database TGI (The Gene Indices, http://compbio.dfci.harvard.edu/tgi/) and have been characterized in a previous study (Li and Dubcovsky, 2008). We show in this study that the previously cloned TaFDL2 and TaFDL6 proteins can interact as prey with all six 14-3-3 proteins in yeast two-hybrid assays (Fig. 2). Previously cloned TaFDL15 interacted strongly with Ta14-3-3A, B and C, weakly with Ta14-3-3E, and not at all with Ta14-3-3D and Hv14-3-3H. Previously cloned TaFDL13 did not interact with any 14-3-3 proteins (Fig.2).
Figure 2. Interactions between FD-like proteins and 14-3-3 proteins.
14-3-3 proteins were used as baits, wild type and mutant TaFDL2, TaFDL6, TaFDL15, HvFDL4, and HvFDL5 were used as preys. SD-L-W-H-A medium was used to test interactions. Tri-M= triple mutant S310A-T311A-S312A.
Re-analyses of these four FDL genes using the wheat genomic sequences released by the International Wheat Genome Sequencing Consortium (IWGSC 2014; Appendix S1) showed that the previously cloned TaFDL2 cDNA was a partial transcript missing the 5’ terminal end and that TaFDL13 is an alternative splicing form of TaFDL15. We cloned the full-length coding region of TaFDL2 and confirmed by yeast two-hybrid assays that the full length TaFDL2 protein also interacts with all six 14-3-3 proteins (Fig. S2). Comparison of the TaFDL13 transcript and genomic sequences revealed that the TaFDL13 transcript is an alternative splicing form generated by the retention of the single intron present in TaFDL15. The alternative splice variant includes a premature stop codon that results in the truncation of the last 30 amino acids (Appendix S1), which may explain the inability of the encoded TaFDL13 protein to interact with any of the 14-3-3 proteins.
In addition, we identified two barley FD-like proteins from the GenBank sequence database, which are not orthologous to any of the previously characterized wheat FD-like proteins, and designated them as HvFDL4 (GenBank BAK05116) and HvFDL5 (GenBank BAJ92629). Yeast two-hybrid assays showed that HvFDL4 interacts with all six 14-3-3 proteins (Fig. 2), whereas HvFDL5 interacts only with Ta14-3-3A, Ta14-3-3C and Hv14-3-3H (Fig. 2). The wheat homologs of HvFDL4 and HvFDL5 (TaFDL4 and TaFDL5) were also obtained from the IWGSC and their sequences are described in Appendix S1. The relationships among the different FDL proteins described in this study are summarized in a phylogenetic analysis generated from the conserved bZIP and CDPK protein regions (Fig. S3B).
The C-terminal regions of the proteins encoded by the canonical FD-like transcripts all contain a short motif targeted by CDPKs (Fig. S3A), and an alanine substitution of a serine/threonine residue within this motif disrupts FD function in Arabidopsis (Abe et al., 2005). We generated substitution mutations for the five FD-like proteins and confirmed by yeast two-hybrid assays that this phosphorylation site was required for interactions between FD-like proteins and 14-3-3 proteins in wheat and barley (Fig. 2). An alanine substitution of the serine residue was enough to disrupt all interactions between FD-like and 14-3-3 proteins (Fig. 2) except TaFDL6, for which a S310A/T311A/S312A triple mutation was required to abolish TaFDL6 interactions with Ta14-3-3A to E (Fig. 2). The interaction between TaFDL6 and Hv14-3-3H was not affected by any of the mutations, presented alone or in combination (Fig. 2).
Dimerization of 14-3-3 proteins
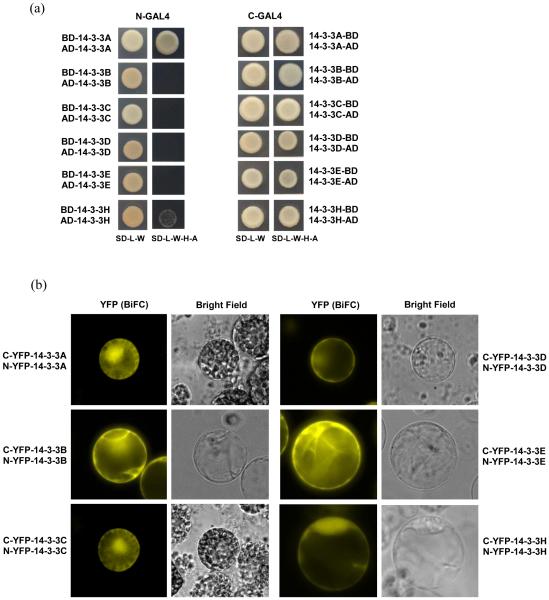
We then investigated the dimer formation ability of wheat and barley 14-3-3 proteins by yeast two-hybrid assays and tested the position effect of the GAL4 DNA binding domain (BD) and activation domain (AD) on 14-3-3 dimerization. When the GAL4-BD and GAL4-AD were placed at the N-terminus of the fusion proteins (pGBKT7 and pGADT7 vectors), only Ta14-3-3A and Hv14-3-3H were able to form homodimers, whereas the other four 14-3-3 proteins (B to E) did not (Fig. 3A). By contrast, when the GAL4-BD and GAL4-AD were placed at the C-terminus of the fusion proteins (pGBKCg and pGADCg vectors), all 14-3-3 proteins were able to form homodimers (Fig. 3A). These results suggest that the addition of the GAL4-BD or AD to the N-terminal regions of the 14-3-3 proteins interferes with the formation of homodimers.
Figure 3. Dimerization of 14-3-3 proteins.
(a) Homodimer formation tests by yeast two-hybrid assays. Bait is listed first in each pair combination. (b) Homodimer formation of 14-3-3 proteins by BiFC in rice protoplasts. The split YFP system was used in BiFC assays.
Among all the heterodimer combinations, only six were able to form heterodimers when the GAL4-BD and AD were placed at the N terminus (Fig. S4). Ta14-3-3A and Ta14-3-3C, which did not form heterodimers when the GAL4 domains were placed at the N-terminus, were able to heterodimerize when the GAL4 domains were placed at the C-terminus (Fig. S4). Taken together these results suggest that the N-terminus GAL4 interfered with the formation of 14-3-3 homodimers and heterodimers in yeast. We did not test the position effect of the C-terminus GAL4 on all possible 14-3-3 heterodimer pairs in yeast because those interactions were confirmed by BiFC assays in rice protoplasts, as described below.
BiFC assays were conducted using a split yellow fluorescent protein (YFP) system (Bracha-Drori, et al., 2004) in which 14-3-3 proteins were fused to the N-terminal half of YFP (N-YFP) and the C-terminal half of YFP (C-YFP), respectively. Construct pairs containing one N-YFP fusion and one C-YFP fusion were co-transformed into rice protoplasts. All six 14-3-3 proteins were able to form homodimers (Fig. 3B) and heterodimers with each other (Fig. S4) in rice protoplasts, suggesting limited specificity in 14-3-3 dimerization. Although both N-YFP and C-YFP were placed at the N-terminus of all fusion proteins, we did not observe any interference on 14-3-3 dimerization in protoplasts.
Subcellular localization and in vivo interaction of FT1, 14-3-3 and TaFDL2
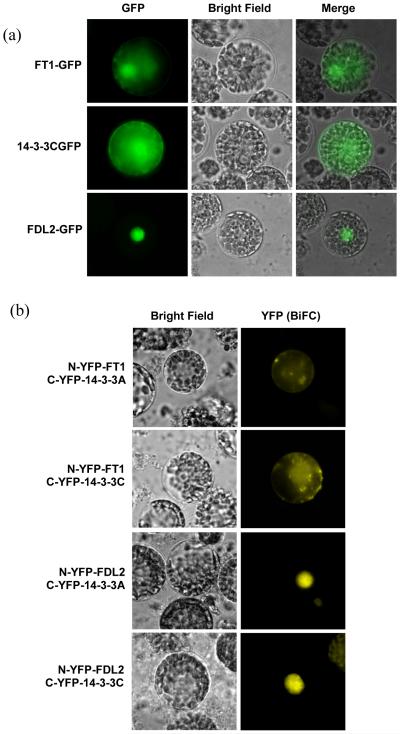
We fused FT1, TaFDL2 and Ta14-3-3C proteins with the green fluorescent protein (GFP) and determined the subcellular localization of these proteins in wheat protoplasts. Microscopy analysis detected fluorescence from FT1-GFP in both the cytoplasm and nucleus; and from Ta14-3-3C-GFP predominantly in the cytoplasm and weakly in the nucleus (Fig. 4A), suggesting that the interactions between FT1 and 14-3-3 proteins are most likely to occur in the cytoplasm. Fluorescence from TaFDL2-GFP was only detected in the nucleus (Fig. 4A).
Figure 4. Subcellular localization and in vivo interaction of FT1, 14-3-3 and TaFDL2.
(a) Subcellular localization images of FT1-GFP, TaFDL2-GFP and Ta14-3-3C-GFP in wheat protoplasts. (b) BiFC assays confirming interactions of FT1-Ta14-3-3A, FT1-14-3-3C, TaFDL2-Ta14-3-3A and TaFDL2-Ta14-3-3C in wheat protoplasts. The split YFP system was used in BiFC assays.
To validate the protein interactions among FT1, TaFDL2 and 14-3-3 proteins, we conducted BiFC assays using wheat protoplasts. Among the six 14-3-3 proteins, Ta14-3-3A and Ta14-3-3C showed stronger interactions in yeast with both FT1 and FD-like proteins, and therefore were selected for the BiFC assays. FT1 and TaFDL2 were fused to N-YFP and Ta14-3-3A and Ta14-3-3C were fused to C-YFP. The reconstituted YFP signal by FT1-14-3-3 interaction was detected in the cytoplasm, where 14-3-3 proteins were mainly localized (Fig. 4B). The TaFDL2-14-3-3 BiFC signal was mainly detected in the nucleus (Fig. 4B), indicating that the FT1-14-3-3 complex needs to be translocated from the cytoplasm to the nucleus to interact with TaFDL2.
14-3-3 proteins are required for the FT-FDL2 interaction
We showed previously that the TaFDL2 protein can interact with FT1 in a yeast two-hybrid assay and can bind to the VRN1 promoter in vitro (Li and Dubcovsky, 2008). Transgenic wheat plants over-expressing FT1 showed parallel increases in VRN1 transcripts both in leaves and shoot apical meristem, suggesting that FT1 provides transcriptional activation of VRN1 through its protein interaction with TaFDL2 (Li and Dubcovsky, 2008; Pearce et al., 2013). However, we were not able to detect a direct interaction between an FT1-TaFDL2 protein complex and the VRN1 promoter (Li and Dubcovsky, 2008). Since both FT1 and TaFDL2 were able to interact with multiple 14-3-3 proteins (Fig. 1 and 2), we decided to test if the FT1-TaFDL2 interaction detected before in yeast two-hybrid assays was mediated by endogenous yeast 14-3-3 proteins. We cloned one of the yeast 14-3-3 proteins (Sc14-3-3, GenBank EDN60443) and tested its interaction with FT1 and TaFDL2. Indeed, both FT1 and TaFDL2 were able to interact with the cloned yeast 14-3-3 protein (Fig. S5).
We then examined whether FT1 mutants FT1- Interactions between FT1 and FT1-P94L were still able to interact with TaFDL2 by yeast two-hybrid assays. These two mutant proteins lost their ability to interact with 14-3-3 proteins (Fig. 1), and were also unable to interact with TaFDL2 (Fig. 5). Similarly, when TaFDL2-14-3-3 interaction was disrupted by the TaFDL2-S323A mutation, TaFDL2 lost its ability to interact with FT1 (Fig. 5). These data suggest that a 14-3-3 bridge is essential to mediate FT1-TaFDL2 interactions and, therefore, for the formation of the FACs.
Figure 5. Yeast two-hybrid interactions between FT1 and TaFDL2 and the effect of mutations on their interaction.
SD-L-W was used to select for yeast transformants and SD-L-W-H-A was used to test interactions. Dilution factors from left to right are 1, 1:10, and 1:100.
The FAC including FT1, Ta14-3-3C and TaFDL2 can bind to the VRN1 promoter
Using Electrophoretic Mobility Shift Assays (EMSAs), we have shown before that the TaFDL2 protein binds to the VRN1 promoter in vitro (Li and Dubcovsky, 2008). However, no super-shift was observed in the presence of both FT1 and TaFDL2 proteins (Li and Dubcovsky, 2008). Here, we performed additional EMSAs with Ta14-3-3C protein included in the binding reactions. TaFDL2 protein alone was sufficient to bind to VRN1 promoter, whereas FT1 or Ta14-3-3C alone did not bind to DNA (Fig. 6). When both TaFDL2 and Ta14-3-3C were added to the binding reaction, a super-shift was detected, and the presence of all three proteins (TaFDL2, Ta14-3-3C and FT1) was able to further delay the mobility of the protein-DNA complex (Fig. 6). These data indicate that the FAC including FT1, Ta14-3-3C, and TaFDL2 has the ability to bind the VRN1 promoter in vitro.
Figure 6. Interactions between FAC component proteins and VRN1 promoter by Electrophoretic Mobility Shift Assays.
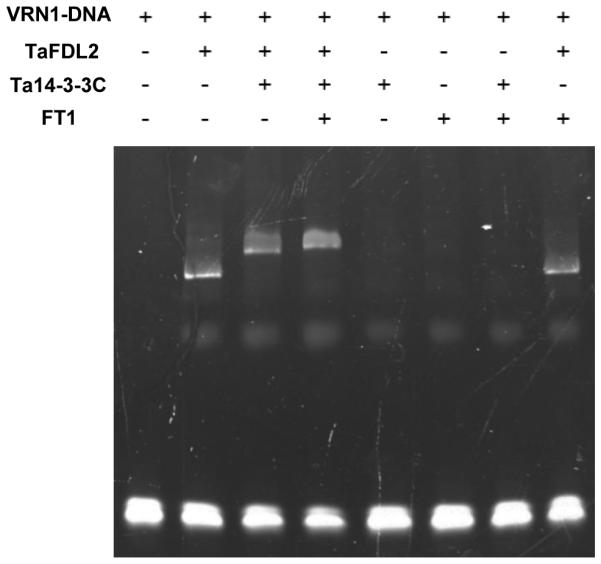
The DNA probe is a fragment of the VRN1 promoter containing bZIP binding sites CACGTA and CACGTC (Li and Dubcovsky, 2008). The proteins used in the DNA-protein binding reactions include purified FT1-GST, TaFDL2-GST and Ta14-3-3C-GST. A delay in the electrophoretic mobility of the DNA fragment indicates binding to the protein.
In the absence of Ta14-3-3C, the binding reaction including both FT1 and TaFDL2 resulted in a DNA shift similar to the one observed when only the TaFDL2 protein was incubated with the DNA probe (Fig. 6). This result provides additional evidence that FT1 and TaFDL2 cannot interact in the absence of the 14-3-3 protein bridge.
Different FT-like proteins interact with different subsets of 14-3-3 proteins
In addition to FT1, five additional FT-like genes were described before in barley and wheat, and their phylogenetic relationship were also characterized (Lv, et al., 2014). To test the interactions between the different FT-like and 14-3-3 proteins, we cloned TaFT2 from wheat, and HvFT3 (DQ411319), HvFT4 (DQ411320), HvFT5 (EF012202) from barley. We also cloned TaTFL1 from wheat and HvCEN1 from barley, which are orthologs of the related Arabidopsis TERMINAL FLOWER 1 (TFL1) and Antirrhinum CENTRORADIALIS (CEN), respectively. The results from these yeast two-hybrid assays are summarized in Table 1. The different FT-like proteins showed differences in their interactions with the wheat 14-3-3 proteins. HvFT3 and HvFT5 interacted only with Ta14-3-3C, whereas HvFT4 interacted with Ta14-3-3A, B, C and D proteins. Surprisingly, wheat TaFT2, which is the closest homolog of FT1, did not interact with any of the tested 14-3-3 proteins (Table 1). TaTFL1 showed a very weak interaction with Ta14-3-3C and no interactions with the other 14-3-3 proteins. HvCEN1 did not interact with any of the wheat 14-3-3 proteins (Table 1).
Table 1.
Yeast two-hybrid interactions of different 14-3-3 with FT1 and FT-like proteins
| PREY | FT1 | TaFT2 | HvFT3 | HvFT4 | HvFT5 | TaTFL1 | HvCEN1 |
|---|---|---|---|---|---|---|---|
| BAIT | |||||||
| 14-3-3A | YES* | NO | NO | YES | NO | NO | NO |
| 14-3-3B | YES | NO | NO | YES | NO | NO | NO |
| 14-3-3C | YES* | NO | YES | YES | YES | Weak | NO |
| 14-3-3D | YES | NO | NO | YES | NO | NO | NO |
| 14-3-3E | YES | NO | NO | NO | NO | NO | NO |
| 14-3-3H | NO | NO | NO | NO | NO | NO | NO |
These interactions were validated by BiFC (Fig. 4).
FDL and 14-3-3 transcript levels in different tissues and developmental stages
Previous studies have shown that FT-like genes (FT1 to FT5) in barley and wheat are expressed predominantly in the leaves, and have limited expression in the SAM (Faure et al., 2007; Lv, et al., 2014). We show by qRT-PCR that FD-like and 14-3-3 genes are expressed at relatively high levels in both leaves and SAM under both long day (LD, 16 h light) and short day (SD, 8 h light) photoperiod (Figs. S6 A-B), and also at different stages of SAM development (Fig. S6 C-D). Analyses of previously published RNAseq datasets (Choulet et al., 2014) show expression of FDL and 14-3-3genes in roots, leaves, stems, spikes, and grains (Fig. S6 E-F).
Interestingly, relative abundances among FDL genes varied across tissues and developmental stages (Fig. S6A, C and E). For example, transcript levels of TaFDL6 were higher than those of TaFDL2 in leaves (LD and SD), stems, roots and grains, but not in the SAM and spikes, where TaFDL2 transcript levels were higher than those of TaFDL6 (Fig. S6A, C and E). TaFDL15 also showed marked differences in transcript levels, varying from being the most highly expressed FDL gene in developing spikes, stems, and leaves (RNAseq, Zadok sage 23), to having relative low transcript levels in young leaves and SAM (Fig. S6A), roots, and grain (Fig. S6E).
The relative transcript levels of the wheat 14-3-3 genes also showed some variability across tissues and developmental stages but the differences were less evident than for the FDL genes (Fig. S6B, D and F). Ta14-3-3A was consistently the most highly expressed 14-3-3 gene in all the qRT-PCR and RNA-seq studies, and Ta14-3-3B was the second highest, except for roots and leaves (Zadok 23) in the RNAseq study (Fig. S6B, D, and F). The other three wheat 14-3-3 genes were expressed at relatively lower levels and showed limited variability across tissues and developmental stages (Figs. 6B and D).
DISCUSSION
Differential functions of FT-like proteins
In addition to FT, the FT-like family in Arabidopsis includes five other members: TWIN SISTER OF FT (TSF), TERMINAL FLOWER 1 (TFL1), BROTHER OF FT AND TFL1 (BFT), MOTHER OF FT AND TFL1 (MFT), and Arabidopsis thaliana CENTRORADIALIS homolog (ATC). TSF functions as a florigenic activator (Kardailsky et al., 1999; Kobayashi et al., 1999; Yamaguchi et al., 2005; Andres and Coupland, 2012), whereas TFL1, ATC and BFT function as floral repressors (Bradley et al., 1997; Mimida et al., 2001; Yoo et al., 2010). MFT is involved in seed germination (Xi et al., 2010). Some of these FT-like genes have been shown to function through interactions with FD-like proteins. For examples, TFL1 has been shown to modulate the transcriptional repression of its target genes in an FD-dependent manner (Hanano and Goto, 2011), and Arabidopsis BFT and Chrysanthemum Anti-florigenic protein (CsAFT) have been shown to compete with FT for FD binding and to function as floral repressors (Ryu et al., 2013; Higuchi et al., 2013).
In rice, both HD3A and RICE FLOWERING LOCUS T1 (RFT1) encode mobile flowering signals that function as flowering promoters (Komiya et al., 2009; Tsuji et al., 2011). Overexpression of barley HvFT1, HvFT2 and HvFT3 in rice led to early flowering, indicating that these three FT-like genes can act as flowering promoters (Kikuchi et al., 2009). Similarly, overexpression of FT1 and FT2 Brachypodium orthologs (Bradi2g07070 and Bradi1g48830) resulted in extremely early flowering in transgenic Brachypodium plants (Wu et al., 2013). Transgenic wheat plants overexpressing FT1 show an upregulation of other FT-like genes suggesting that FT1 acts as a positive regulator of other FT-like genes (Lv, et al., 2014). In spite of its importance, FT1 does not seem to be essential for flowering in wheat. Combined mutations in FT-A1 and FT-B1 in tetraploid wheat or downregulation of all FT1 homoeologs by RNA interference in hexaploid wheat, delay flowering only 2-4 weeks under LD (Lv, et al., 2014). Therefore, it is likely that other FT-like genes have overlapping roles in the promotion of wheat flowering.
Potential competitive interactions among FT-like, FD-like and 14-3-3 proteins
The interactions of some FT-like proteins with the same 14-4-3 proteins (Fig. S7) can result in competitive interactions. For example, FT1, HvFT3, HvFT4, HvFT5 and TaTFL1 all interact with Ta14-3-3C (Table 1 and Fig. S7) and may compete for this interaction. Interestingly, Ta14-3-3C is also able to interact with all the FDL proteins tested so far creating the opportunity for additional competitive interactions (Fig. 2 and Fig. S7). Proteins Ta14-3-3A, Ta14-3-3B and Ta14-3-3D also interact with more than one FT-like proteins (FT1 and HvFT4) and may also compete for these interactions (Table 1 and Fig. S7). FT1 and HvFT4 may be less affected by these competitive interactions because they can interact with a large number of different 14-3-3 proteins (Table 1 and Fig. S7). By contrast, the related HvFT3 and HvFT5 proteins (80 to 84% identical in sequence) interact only with 14-3-3C suggesting that they can be very sensitive to competition for interactions with this particular protein.
FT1 and TaFT2 share high protein identity (~78%), but they do not seem to compete for common interactors. These two proteins have similar functions as strong flowering activators (Lv, et al., 2014; Wu et al., 2013) but show no overlaps in their 14-3-3 protein interaction profiles. FT1 interacted with five out of the six 14-3-3 proteins tested so far, whereas TaFT2 showed no interactions with any of these 14-3-3 proteins. This result suggests that some functional differentiation may exist between these two proteins.
The functions of the other FT-like proteins in wheat and barley are still not well characterized, so it is currently not possible to predict the effect of the different 14-3-3 interactions. A detailed functional characterization of FT-like proteins and their protein partners including 14-3-3 and FD-like proteins will help determine whether they act redundantly or antagonistically to FT1 in wheat flowering regulation.
Dimerization among 14-3-3 proteins
The selectivity of the interactions between FT-like and 14-3-3 proteins contrasts with the less selective interactions between 14-3-3 monomers. Our BiFC results in rice protoplasts showed no selectivity in the interactions among wheat 14-3-3 proteins. Therefore, dimerization between different 14-3-3 proteins is likely to depend on their relative affinities and abundances. In mammalian and yeast systems it has been demonstrated that the dimerization of 14-3-3 proteins is not completely random, and that there are preferences for certain dimer combinations (Paul et al., 2012). For example, the mammalian 14-3-3σ shows a preference to form homodimers, whereas 14-3-3β is present primarily as heterodimers (Wilker et al., 2005). The two yeast 14-3-3 proteins also show an inclination to form heterodimers rather than homodimers (Chaudheri et al., 2003).
In plants, 14-3-3 dimerization is complex and still not well understood. Among the eight Arabidopsis 14-3-3 proteins tested so far, all can form homodimers as well as heterodimers with each other in yeast two-hybrid assays and native gel western blot analysis (Wu et al., 1997). Proteomic profiling results indicate that 14-3-3ω can dimerize with at least 10 of the 12 14-3-3 proteins expressed in Arabidopsis (Chang et al., 2009). In cotton, none of the six cotton 14-3-3 proteins were able to form homodimers in yeast two-hybrid, and only some combinations of heterodimers were detected (Zhang et al., 2010). However, the yeast two-hybrid vectors used in the cotton study (pGBKT7 and pGADT7) have both GAL4-BD and AD at the N-terminus of the fusion proteins, which could have interfered with the interactions between 14-3-3 proteins, as demonstrated in this study (Fig. 3A). Additional yeast two-hybrid experiments with the GAL4-BD and AD at the C-terminus of the fusion proteins will be required to interpret the cotton results correctly.
Even though the interactions among 14-3-3 proteins were not very selective, their interactions with different FT and FD-like proteins showed some specificity (Table 1 and Fig. S7) and can play a role in the modulation of the effects of different FT-like mobile proteins. To understand these effects better we characterized the spatial and temporal expression of the different components of the FACs.
Differential expression profiles of FT-like, FD-like and 14-3-3 genes
Previous studies have shown differences in the relative levels of FT-like genes expression at different photoperiods (Lv, et al., 2014). When plants of the winter wheat variety Jagger were grown under LD for five weeks without vernalization, FT3 transcript levels were significantly lower than those of the other FT-like genes. By contrasts, when plants from the same variety were grown under SD, transcript levels of FT3 (and also FT4 and FT5) were significantly higher than those of FT1 and FT2 (Lv, et al., 2014). These results suggest a specific regulatory role of FT3 (and possibly FT4 and FT5) under short days.
Mobile FT-like proteins can reach multiple plant tissues via the phloem. Therefore, tissue specific responses likely depend on the other two members of the FAC. The FDL members are particularly important since they determine the potential DNA targets of the FAC. For example, the ectopic expression of FD in the leaves of transgenic Arabidopsis plants, results in high expression levels of its AP1 target in the leaves, where these two genes are not normally expressed (Wigge et al., 2005). The wheat FDL2 gene is normally expressed in both apices and leaves (Fig. S6), and its VRN1 target is also expressed in both tissues (when FT1 is present).
In this study we show that the relative abundances of FDL transcripts vary significantly across tissues and developmental stages (Figs. S6A, C and E). These differences are likely to be reflected in the relative abundance of FAC complexes and can contribute to the spatial and temporal modulation of the effects of the mobile FT-like signals. The relative transcript levels of the different wheat 14-3-3 genes also show some temporal and spatial variability (Fig. S6B, D and E), which may also contribute to the modulation of the effects of the FT-like mobile proteins across tissues and developmental stages.
FD-like transcription factors determine the DNA target of florigen activation complexes
We show in this study that among the three components of the particular FAC that interacts with the VRN1 promoter, only TaFDL2 is essential for DNA binding (Fig. 6). This agrees with previous results in rice, which indicated that the FDL recruited to the FAC is critical in the determination of the DNA specificity of the complex (Taoka et al., 2011). This has been demonstrated for rice proteins OsFD1 and OsFD2, both of which can interact with HD3A and 14-3-3 proteins to form FACs (Tsuji et al., 2013). If OsFD1 is recruited, the resulting FAC acts to promote flowering; whereas if OsFD2 is recruited, the resulting FAC functions in rice leaf development (Tsuji et al., 2013). This result demonstrates that the presence of different FD-like genes has the potential to determine which genes are activated or repressed by their interacting FT-like proteins. It also shows that some of the FAC can be involved in the regulation of processes other than flowering. The presence of different FT-like proteins in the complex may determine the promoting or repressing role of the FAC or affect the intensity of their responses. In a previous study, we showed that both TaFDL2 and TaFDL6 proteins interact with FT1, but only TaFDL2 was able to bind to the VRN1 promoter (Li and Dubcovsky, 2008). These results suggested that TaFDL6 may target other genes, a possibility reinforced here by the observation that TaFDL6 interacts with all six 14-3-3 proteins (Fig. 2) and that therefore, can be part of different FACs.
Additional FD-like proteins TaFDL15, HvFDL4 and HvFDL5 also interact with multiple 14-3-3 proteins (Fig. 2). Therefore, the combinations of different FT-like, FD-like and 14-3-3 proteins can potentially generate a large number of different FACs that plants can use to decode the information carried by FT-like proteins. We are currently screening for mutations of individual FD-like and FT-like proteins to understand better the specific and overlapping functions of the different FACs.
Potential downstream targets of florigen activation complexes
The wheat VRN1 gene encodes a MADS-box transcription factor that is important but not essential for wheat flowering (Chen and Dubcovsky, 2012). This gene is closely related to Arabidopsis paralogous genes APETALA 1 (AP1), CAULIFLOWER (CAL) and FRUITFULL (FUL) (Yan et al., 2003; Trevaskis et al., 2003), which have overlapping functions in the induction of flowering (Ferrándiz et al., 2000). In the temperate cereals, there are two additional MADS-box genes similar to VRN1, designated as FUL2 (= HvMADS8 = OsMADS15) and FUL3 (= HvMADS3 = OsMADS18) (Preston et al., 2006), but their role in the regulation of flowering is currently unknown.
In this study we show that a FAC including FT1, Ta14-3-3C and TaFDL2 can bind to the VRN1 promoter in vitro (Fig. 6). In rice, the FAC including HD3A, OsFD1 and GF14c (=14-3-3C) proteins binds to the promoter of OsMADS15, which is the rice ortholog of FUL2 (Taoka et al., 2011). This result suggests that wheat FUL2 (and may be FUL3) promoters could also be potential downstream targets of some wheat FACs.
A recent study has demonstrated that the induction of FT1 is associated with the up-regulation of GA biosynthetic genes and the downregulation of GA catabolic genes in the shoot apical meristems of wheat (Pearce et al., 2013). Therefore, it would be interesting to investigate if the promoters of any of the biosynthetic GA20ox or catabolic GA2ox genes are targets of FACs including FT1.
In summary, our study shows that the multiple FT-like, FDL and 14-3-3 wheat proteins have the potential to generate a very large number of different FACs. These multiple factorial protein combinations provide the complexity and flexibility required to modulate differential responses to mobile FT-like signals in different tissue and developmental stages.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
Rice japonica variety Kitaake and wheat variety Chinese Spring were used for protoplast extraction. Rice plants were grown in the dark at 28°C for 2 weeks before leaves and stems were harvested for protoplast extraction following procedures described before (Bart, et al., 2006). We adapted the rice protoplast extraction protocol to wheat protoplast extraction, but modified the growth conditions as following: wheat plants were grown under LD (16h light/8h dark) at temperatures of 22 °C during the day and 18 °C during the night for 2 weeks before protoplast extraction.
For cloning, wheat variety Chinese Spring and barley Golden Promise plants were grown under LD at temperatures of 22 °C during the day and 18 °C during the night for 4 weeks before leaves were collected for total RNA extraction and cDNA synthesis.
Phylogenetic analysis
Phylogenetic analysis of the 14-3-3 proteins was performed using full-length proteins. A neighbor-joining tree was constructed using the program MEGA 5.0 (Tamura et al. 2011). Bootstrap confidence values were calculated based on 1000 iterations and only values larger than 50 are shown in the respective nodes.
GenBank accession numbers used in phylogenetic analysis include AtGF14chi= GRF1 (At4g09000), AtGF14omega= GRF2 (At1g78300), AtGF14psi= GRF3 (At5g38480), AtGF14phi= GRF4 (At1g35160), AtGF14upsilon= GRF5 (At5g16050), AtGF14 lambda = GRF6 (At5g10450), AtGF14nu= GRF7 (At3g02520), AtGF14kappa= GRF8 (At5g65430), AtGF14 mu= GRF9 (At2g42590), AtGF14epsilon = GRF10 (At1g22300), OsGF14a (Os08g0480800), OsGF14b (Os04g0462500), OsGF14c (Os08g0430500), OsGF14d (Os11g0546900), OsGF14e (Os02g0580300), OsGF14f (Os03g0710800), OsGF14g (Os01g0209200), OsGF14h (Q2R1D5), Ta14-3-3A (AAR89812), Ta14-3-3B (AGA84102), Ta14-3-3C (AEQ53933), Ta14-3-3D (BAB11740), Ta14-3-3E (BAB11739), Hv14-3-3A (CAA44259), Hv14-3-3B (CAA63658), Hv14-3-3C (CAA74592), HvGF14D (DQ295785), HvGF14E (DQ295786) and Hv14-3-3H (BAK01024). The two letters in front of gene or protein names indicate the organism from which the sequence or clone was obtained (At= Arabidopsis thaliana, Ta= Triticum aestivum, Hv= Hordeum vulgare, Os= Oryza sativa).
The phylogenetic analysis of the FDL proteins was performed using only the conserved region of the proteins including the bZIP domain and the CDPK target region (Fig. S3A). The resulting neighbor-joining tree is presented in Fig. S3B and the complete FDL sequences (DNA and protein) are presented in Appendix 1. The rice FD-like proteins (and their corresponding genes) included in the phylogeny analysis are: OsFD2 (Os06g0720900), OsFD3 (Os02g0833600), OsFD4 (Os08g0549600), Os05g048970, Os01g0813100 and OsJ_10665 (CM000140).
Yeast two-hybrid assays
Yeast vectors pGBKT7 (GAL4 DNA-binding domain, BD), pGADT7 (GAL4 activation domain, AD) and yeast strain Y2HGold (Clontech, Mountain View, CA, USA) were used in the yeast two-hybrid assays. Two additional pairs of modified Gateway (Invitrogen) bait/prey vectors were also used in this study: (1) N-terminal GAL4 fusion vectors pLAW10 and pLAW11, which are the Gateway versions of pGBKT7 and pGADT7, respectively (Cantu, et al., 2013); (2) C-terminal GAL4 fusion vectors pGBKCg and pGADCg (Stellberger et al., 2010). For all Gateway compatible clonings, pDONR/Zeo (Life Technologies, Grand Island, NY, USA,) was used to generate the entry vectors. Primer sequences and cloning strategies are listed in Table S1. All constructs were verified by digests and sequencing. Yeast two-hybrid assays were performed according to the manufacturer’s instructions (Clontech). Transformants were selected on SD medium lacking leucine (−L) and tryptophan (−W) plates and re-plated on SD medium lacking −L, −W, histidine (−H) and adenine (−A) to test the interactions.
Site-directed mutagenesis
QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) was used to generate single amino acid substitution mutants for FT and FD-like proteins. All the mutant constructs were verified by sequencing. Primers used to generate the mutated genes are listed in Table S2.
Protein subcellular localization
To reduce plasmid size and increase the protoplast transfection efficiency, we engineered a smaller Gateway destination vector containing the GFP reporter. The 2X 35S promoter-GFP6-Gateway cassette from pMDC43 (Arabidopsis Biological Resource Center, stock # CD3-741) was inserted in pUC57-KAN between restriction sites EcoRI and HindIII to generate Gateway destination vector pUC57-GFP. Entry vectors generated for yeast two-hybrid assays using pDONR/Zeo system (Table S1) were used to transfer FT1, TaFDL2 or Ta14-3-3C into pUC57-GFP by site-specific recombination to generate GFP-FT1, GFP-TaFDL2 and GFP-Ta14-3-3C fusions. The resultant constructs were used in the transfection assays to determine the subcellular localization of these proteins in wheat protoplasts. Wheat protoplast isolation and transfection were conducted as described before (Bart, et al., 2006).
Bimolecular fluorescence complementation assays
Bimolecular fluorescence complementation assays using split YFP system were conducted using protocols described before (Bracha-Drori, et al., 2004). The pDONR entry vectors generated for yeast two-hybrid (Table S1) were used to generate BiFC constructs: each protein pair was recombined into the Gateway destination vectors pY736 and pY735 to generate N-terminal YFP and C-terminal YFP fusion proteins. All constructs were verified by sequencing. Rice and wheat protoplast isolation and transfection were conducted as described (Bart, et al., 2006).
Electrophoretic mobility shift assays (EMSA)
The generation of FT1-GST and TaFDL2–GST fusion constructs was described previously (Li and Dubcovsky, 2008). Entry vector pDONR/Zeo-Ta14-3-3C generated for yeast two-hybrid was used to transfer Ta14-3-3C into pDEST15 (Gateway compatible, Invitrogen) to generate the Ta14-3-3C-GST fusion construct. Protocols for GST-tagged protein expression and purification and for PCR procedures to amplify the VRN1 promoter fragment containing two hybrid bZIP binding sites were described before (Li and Dubcovsky, 2008). DNA-protein binding reactions were carried out as described by Taoka et al. (2011) and separated on 5% polyacrylamide gels in 0.5X TBE buffer and stained with ethidium bromide.
Gene expression analysis by qRT-PCR
For the qRT-PCR studies, plants from diploid wheat (T. monococcum L., genome Am) accessions DV92 and PI 349049 were grown for eight weeks under short day (SD, 8h of light) or for six weeks under SD followed by 2 weeks under long day (LD, 16 h of light). Both accessions have a spring growth habit and carry the same vrn2 allele, but differ in their VRN1 alleles. DV92 carries the vrn1 allele that is not expressed under SD, and PI34049 carries the natural mutant allele Vrn1g that has a 34-bp deletion in the promoter region and is expressed under SD resulting in faster SAM development (Dubcovsky et al., 2006 and Pearce et al., 2013).
RNA samples extracted from leaves and apices were used to compare the relative transcript levels of five FDL and five 14-3-3 genes. Comparisons between leaves and apices were performed only in DV92, whereas comparisons across SAM developmental stages included samples from both DV92 and PI 349049. Plant growth conditions, apex dissection, RNA extraction, cDNA synthesis and qRT-PCR were conducted as described before (Pearce et al., 2013). Briefly, qRT-PCR was performed using SYBR Green® and a 7500 Fast Real-Time PCR system (Applied Biosystems), and transcript levels were expressed as linearized fold-ACTIN levels calculated by the formula 2(ACTIN CT – TARGET CT). Primer sequences used in the qRT-PCR studies are listed in Table S3.
Published RNA-seq datasets (Choulet et al., 2014) including different tissues (spike, leaf, grain, stem, and roots) and developmental stages (Zadoks et al. 1974) were used to generate expression profiles of FDL and 14-3-3 genes among different tissues and developmental stages. Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values and their respective standard errors were obtained from one of the developmental stages for each of the five tissues and were used to generate the graphs presented in Figs. S6E and F.
Supplementary Material
SIGNIFICANCE STATEMENT.
This study shows that wheat mobile FT-like florigenic proteins can interact with different members of the FDL and 14-3-3 protein families to form a large diversity of florigen activation complexes (FACs). It also shows that different FDL genes vary in their spatial and temporal expression providing a possible mechanism for the FACs to modulate the effects of the mobile FT-like protein signals in different tissues and developmental stages.
ACKNOWLEDGMENTS
This project was supported by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation and by NRI grant numbers 2011-67013-30077 and 2011-68002-30029 from the USDA National Institute of Food and Agriculture. The authors thank Dr. Stephen Pearce and Mr. Hans Vasquez-Gross (University of California, Davis) for their help with expression analyses of the FDL and 14-3-3 genes.
Footnotes
CONFLICT OF INTEREST: The authors have no conflict of interest to declare.
SHORT LEGENDS FOR SUPPORTING INFORMATION
Figure S1. Phylogenetic analysis of 14-3-3 proteins.
Figure S2. Interactions between full-length TaFDL2 and 14-3-3 proteins.
Figure S3. Phylogenetic analysis of FDL proteins.
Figure S4. Dimerization analysis of 14-3-3 proteins by yeast two-hybrid and BiFC in rice protoplasts.
Figure S5. Yeast two-hybrid interactions among FT1, TaFDL2 and yeast 14-3-3 (Sc14-3-3).
Figure S6. Expression analysis of FDL and 14-3-3 genes in different tissues and developmental stages by qRT-PCR and RNAseq.
Figure S7. Interaction map summarizing the interactions among FT-like, FD-like and 14-3-3 proteins identified in yeast two-hybrid assays.
Table S1. Primers used for yeast two-hybrid constructs.
Table S2. Primers used in mutant generation by site-directed mutagenesis.
Table S3. Primers used in the Q-RT-PCR analysis
Appendix S1. Sequence annotations of the wheat (A and B genomes) and barley FD-like genes used in phylogenetic analysis.
REFERENCES
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006;25:605–614. doi: 10.1038/sj.emboj.7600950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Genet. 2012;13:628–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Banfield MJ, Barker JJ, Perry AC, Brady RL. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure. 1998;6:1245–1254. doi: 10.1016/s0969-2126(98)00125-7. [DOI] [PubMed] [Google Scholar]
- Bart R, Chern M, Park C, Bartley L, Ronald PC. A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods. 2006;2:13. doi: 10.1186/1746-4811-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N. Detection of protein-protein interactions in plants using biomolecular fluorescence complementation. Plant J. 2004;40:419–427. doi: 10.1111/j.1365-313X.2004.02206.x. [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997;275:80–83. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- Cantu D, Yang B, Ruan R, Li K, Menzo V, Fu D, Chern M, Ronald PC, Dubcovsky J. Comparative analysis of protein-protein interactions in the defense response of rice and wheat. BMC Genomics. 2013;14:166. doi: 10.1186/1471-2164-14-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang IF, Curran A, Woolsey R, Quilici D, Cushman JC, Mittler R, Harmon A, Harper JF. Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics. 2009;9:2967–2985. doi: 10.1002/pmic.200800445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhri M, Scarabel M, Aitken A. Mammalian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo. Biochem. Biophys. Res. Commun. 2003;300:679–685. doi: 10.1016/s0006-291x(02)02902-9. [DOI] [PubMed] [Google Scholar]
- Chen A, Dubcovsky J. heat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 2012;8:e1003134. doi: 10.1371/journal.pgen.1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulet F, Alberti A, Theil S, et al. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014;345:1249721. doi: 10.1126/science.1249721. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Serle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Denison FC, Paul A, Zupanska AK, Ferl RJ. 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 2011;22:720–727. doi: 10.1016/j.semcdb.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L. Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Molecular Biology. 2006;60:469–480. doi: 10.1007/s11103-005-4814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA. The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare) Genetics. 2007;176:599–609. doi: 10.1534/genetics.106.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- Granovsky AE, Rosner MR. Raf kinase inhibitory protein: a signal transduction modulator and metastasis suppressor. Cell Res. 2008;18:452–457. doi: 10.1038/cr.2008.43. [DOI] [PubMed] [Google Scholar]
- Hanano S, Goto K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell. 2011;23:3172–3184. doi: 10.1105/tpc.111.088641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y, Money T, Bradley D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA. 2005;102:7748–7753. doi: 10.1073/pnas.0500932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y, Nayumi T, Oda A, Nakano Y, Sumitomo K, Fukai S, Hisamatsu T. The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proc. Natl. Acad. Sci. USA. 2013;110:17137–17142. doi: 10.1073/pnas.1307617110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Kawahigashi H, Ando T, Tonooka T, Handa H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 2009;149:1341–1353. doi: 10.1104/pp.108.132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimanoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;135:767–774. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Buffalo V, Bailey P, Pearce S, Ayling S, Tabbita F, Soria M, Wang S, IWGS consortium. Akhunov E, Uauy C, Dubcovsky J. Separating homeologs by phasing in the tetraploid wheat transcriptome. Genome Biology. 2013;14:R66. doi: 10.1186/gb-2013-14-6-r66. doi:10.1186/gb-2013-14-6-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Dubcovsky J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 2008;55:543–554. doi: 10.1111/j.1365-313X.2008.03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B, Nitcher R, Han X, Wang S, Ni F, Li K, Pearce S, Wu J, Dubcovsky J, Fu D. Characterization of FLOWERING LOCUS T1 (FT1) gene in Brachypodium and wheat. PLoS One. 2014;9:e94171. doi: 10.1371/journal.pone.0094171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimida N, Goto K, Kobayashi Y, Araki T, Ahn JH, Weigel D, Murata M, Motoyoshi F, Sakamoto W. Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells. 2001;6:327–336. doi: 10.1046/j.1365-2443.2001.00425.x. [DOI] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON. delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 2006;142:1523–1536. doi: 10.1104/pp.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A, Denison FC, Schultz E, Zupanska AK, Ferl RJ. 14-3-3 phosphoprotein interaction networks – does isoform diversity present functional interaction specification? Front. Plant Sci. 2012;3:190. doi: 10.3389/fpls.2012.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce S, Vanzetti LS, Dubcovsky J. Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION 1. Plant Physiol. 2013;163:1433–1445. doi: 10.1104/pp.113.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell. 2001;13:2687–2702. doi: 10.1105/tpc.010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA. Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae) Genetics. 2006;174:421–437. doi: 10.1534/genetics.106.057125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JY, Lee HJ, Seo PJ, Jung JH, Ahn JH, Park CM. The Arabidopsis floral repressor BFT delays flowering by competing with FT for FD binding under high salinity. Mol. Plant. 2013;7:377–387. doi: 10.1093/mp/sst114. [DOI] [PubMed] [Google Scholar]
- Schoonheim PJ, Veiga H, Pereira Dda C, Friso G, van Wijk KJ, de Boer AH. A comprehensive analysis of the 14-3-3 interactome in barley leaves using a complementary proteomics and two-hybrid approach. Plant Physiol. 2007;143:670–683. doi: 10.1104/pp.106.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre L, Vallée B, Bureaud N, Schoentgen F, Zelwer C. Crystal structure of the phosphatidylethanolamine-binding protein from bovine brain: a novel structural class of phospholipid-binding proteins. Structure. 1998;6:1255–1265. doi: 10.1016/s0969-2126(98)00126-9. [DOI] [PubMed] [Google Scholar]
- Stellberger T, Häuser R, Baiker A, Pothineni VR, Haas J, Uetz P. Improving the yeast two-hybrid system with permutated fusions proteins: the Varicella Zoster Virus interactome. Proteome Sci. 2010;8:8. doi: 10.1186/1477-5956-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri Y, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Kojima C, Shimamoto K. Structure and function of florigen and the receptor complex. Trends in Plant Sci. 2013;18:287–294. doi: 10.1016/j.tplants.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. USA. 2003;100:13099–13104. doi: 10.1073/pnas.1635053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Taoka K, Shimamoto K. Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 2011;14:45–52. doi: 10.1016/j.pbi.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Tsuji H, Nakamura H, Taoka K, Shimamoto K. Functional diversification of FD transcription factors in rice, components of florigen activation complexes. Plant Cell Physiol. 2013;54:385–397. doi: 10.1093/pcp/pct005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Wilker EW, Grant RA, Artim SC, Yaffe MB. A structural basis for 14-3-3 sigma functional specificity. J. Biol.Chem. 2005;280:18891–18898. doi: 10.1074/jbc.M500982200. [DOI] [PubMed] [Google Scholar]
- Wu K, Lu G, Sehnke P, Ferl RJ. The heterologous interactions among plant 14-3-3 proteins and identification of regions that are important for dimerization. Arch. Biochem. Biophys. 1997;339:2–8. doi: 10.1006/abbi.1996.9841. [DOI] [PubMed] [Google Scholar]
- Wu L, Liu D, Wu J, Zhang R, Qin Z, Liu D, Li A, Fu D, Zhai W, Mao L. Regulation of Flowering Locus T by a microRNA in Brachypodium distachyon. Plant Cell. 2013;25:4363–4377. doi: 10.1105/tpc.113.118620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W, Liu C, Hou X, Yu H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell. 2010;22:1733–1748. doi: 10.1105/tpc.109.073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Chung KS, Jung SH, Yoo SY, Lee JS, Ahn JH. BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. 2010;63:241–253. doi: 10.1111/j.1365-313X.2010.04234.x. [DOI] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF. A decimal code for the growth stages of cereals. Weed Research. 1974;14:415–21. [Google Scholar]
- Zeevaart JAD. Florigen coming of age after 70 years. Plant Cell, 2006;18:1783–1789. doi: 10.1105/tpc.106.043513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZT, Zhou Y, Li Y, Shao SQ, Li BY, Shi HY, Li XB. Interactome analysis of the six cotton 14-3-3s that are preferentially expressed in fibers and involved in cell elongation. J. Exp. Bot. 2010;61:3331–3344. doi: 10.1093/jxb/erq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.