SUMMARY
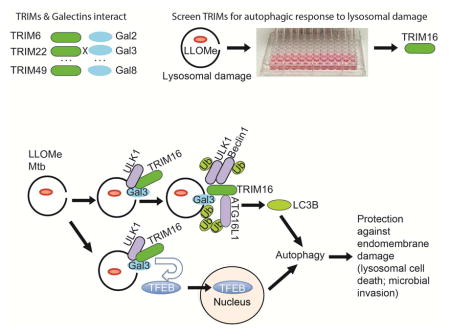
Selective autophagy performs an array of tasks to maintain intracellular homeostasis, sterility, and organellar and cellular functionality. The fidelity of these processes depends on precise target recognition and limited activation of the autophagy apparatus in a localized fashion. Here we describe cooperation in such processes between the TRIM family and Galectin family of proteins. TRIMs, which are E3 ubiquitin ligases, displayed propensity to associate with Galectins. One specific TRIM, TRIM16, interacted with Galectin-3 in an ULK1-dependent manner. TRIM16, through integration of Galectin- and ubiquitin-based processes, coordinated recognition of membrane damage with mobilization of the core autophagy regulators ATG16L1, ULK1, and Beclin 1 in response to damaged endomembranes. TRIM16 affected mTOR, interacted with TFEB and influenced TFEB’s nuclear translocation. The cooperation between TRIM16 and Galectin-3 in targeting and activation of selective autophagy protects cells from lysosomal damage and Mycobacterium tuberculosis invasion.
eTOC BLURB
Selective authophagy contributes to intracellular homeostasis. Chauhan, Kumar, Jain, Ponpuak et al. show that TRIM family proteins, via interactions with Galectin proteins, recognize membrane damage and direct autophagic homeostasis of lysosomal and phagosomal organelles. They function by assembling core autophagy factors and influencing mTOR and TFEB activity.
INTRODUCTION
Autophagy maintains nutrient, energy and organellar homeostasis, participates in intracellular quality control and ensures functionality and sterility of the eukaryotic cell (Mizushima et al., 2011). The most commonly studied form of autophagy, referred to as macroautophagy, is a pathway dependent on highly conserved ATG factors whereby cytoplasmic cargo is captured into autophagosomes, endomembranous organelles decorated with Atg8 in yeast, or in mammalian cells with mammalian Atg8 homologs (mAtg8s) (Mizushima et al., 2011). Autophagy is co-regulated transcriptionally with lysosomal biogenesis via TFEB and other MiT/TEF factors (Napolitano and Ballabio, 2016).
The core autophagy machinery in mammals includes several interconnected components: (i) a phosphatidylinositol 3-kinase VPS34 complex containing ATG14L (Sun et al., 2008), and Beclin 1 (Liang et al., 1999), which upon activation initiates autophagy through production of phosphatidylinositol 3-phosphate (PI3P); (ii) a key upstream protein Ser/Thr kinase Atg1/ULK1 (Mizushima et al., 2011), which phosphorylates and activates Beclin 1 (Russell et al., 2013); (iii) the six mAtg8 homologs (LC3A, B and C, GABARAP, GABARAPL1, and GABARAPL2) undergoing ATG5-ATG12/ATG16L1 E3 ligase-sponsored C-terminal lipidation with phosphatidylethanolamine (PE), a modification engendering their membrane association (Kabeya et al., 2004), and playing distinct roles in autophagosome membrane biogenesis. (iv) A PI3P-binding ATG factor known as WIPI2, connecting PI3P production, ULK1, and the PE conjugation system; WIPI2 recognizes PI3P-modified membranes and interacts with ATG16L1 (Dooley et al., 2014), whereas in turn ATG16L1 is both a localizer of the E3 ligase-like system leading to mAtg8 PE-lipidation (Fujita et al., 2008) and a binding partner for FIP200, a component of the ULK1 complex systems (Fujita et al., 2013). (v) The above systems are set in motion by various signals transduced by upstream signaling systems including Ser/Thr protein kinases mTOR, AMPK, and MK2/MK3 (Kim et al., 2011; Wei et al., 2015).
Less is known about how selective autophagy connects with the upstream regulatory systems, albeit the homing and fidelity of selective autophagy is an area of intense study. The best-understood processes underlying selective autophagy in mammalian systems involve a group of ubiquitin-binding receptors termed Sequestosome 1/p62-like receptors (SLRs) (Birgisdottir et al., 2013; Deretic et al., 2013). The unifying property for SLRs is their ability to bind ubiquitin and associate with LC3. Individual or distinct subsets of SLRs recognize ubiquitin or phosphorylated ubiquitin on targets, delivering them to autophagosomes (Khaminets et al., 2016). Other recognition tags, provided by cytosolic lectins termed Galectins have been implicated in selective autophagy (Thurston et al., 2012). Galectins bind to β-galactoside glycoconjugates (normally sequestered from the cytosol by being located within the lumen-oriented endofacial membrane leaflets) when they become exposed to the cytosol upon endomembrane damage, such as within phagosomes harboring bacteria with membrane penetrating properties (Thurston et al., 2012), or lysosomes (Maejima et al., 2013) and phagosomes (Fujita et al., 2013) perforated by damaging inanimate objects. While intriguing, Galectin-based selective autophagy has thus far been linked to only one of the SLRs, NDP52 (Thurston et al., 2012). Furthermore, how recognition of autophagic targets tagged by ubiquitin or Galectins is integrated with the localized activation of the core autophagic apparatus is not well understood.
The TRIM proteins (Reymond et al., 2001) play a dual role as receptors and regulators of autophagy (Kimura et al., 2015; Kimura et al., 2016; Mandell et al., 2014). TRIMs recognize targets and assemble autophagy regulators ULK1 and Beclin 1 in their activated forms (Kimura et al., 2015; Mandell et al., 2014). The majority of TRIMs contain E3 ligase domains (Kimura et al., 2016; Reymond et al., 2001). Here we report that TRIMs possess another surprising feature, i.e. that they broadly interact with Galectins. Using lysosomal and phagosomal damage models, we show that one TRIM, TRIM16, recognizes endomembrane damage through interactions with Galectin-3 in an ULK1-dependent manner, with ULK1 playing a structural role in enhancing TRIM16-Galectin-3 interactions. TRIM16 binds ATG16L1 and associates with key autophagy regulators ULK1 and Beclin 1. Furthermore, TRIM16 influences TFEB and mTOR activation states.
RESULTS
TRIMs and Galectins interact
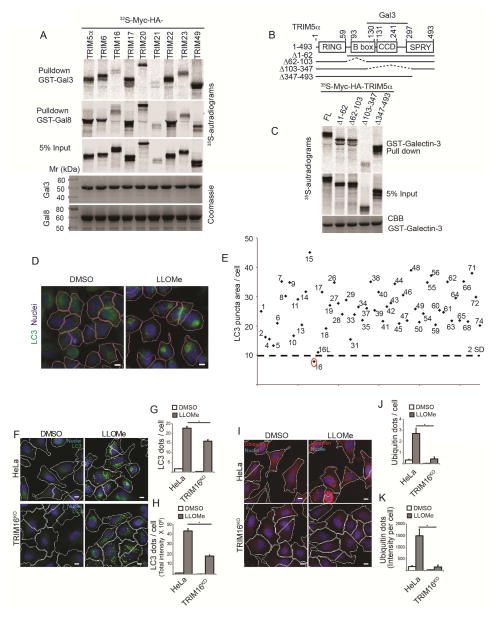
During the screens uncovering a broad role of TRIMs in autophagy (Kimura et al., 2015; Mandell et al., 2014), we observed an unanticipated propensity of TRIMs to bind Galectins. Of the TRIMs tested, TRIM5α, TRIM6, TRIM17, TRIM20, TRIM22, TRIM23 and TRIM49 bound both Galectin-3 and Galectin-8, whereas TRIM16, TRIM21, TRIM55 and TRIM56 did not (Figures 1A and S1A). Galectin-8 bound NDP52 as expected (Thurston et al., 2012) and its close homolog TAX1BP1 but not Optineurin and other controls (Figure S1B). For specificity, we mapped the Galectin-3-binding region in TRIM5α and found that a region spanning TRIM5α’s CCD, but not its B-Box or SPRY domains, was required for Galectin-3 (Figure 1B and Figure S1C). Thus, TRIMs, through specific regions as delimited within TRIM5α, have a propensity to associate with Galectins.
Figure 1. TRIMs and Galectins interact.
(A) GST pulldowns of in vitro translated and radiolabeled 35S-Myc-HA-TRIMs (as indicted) with GST-Galectin-3 and GST-Galectin-8. (B) Mapping of Galectin-3 interaction domain within TRIM5α. Deletion constructs of TRIM5a used in C. (C) TRIM5α constructs depicted in B were radiolabeled as in A and subjected to GST-pulldowns. with GST-Galectin-3. (D–E) siRNA screen of human TRIMs (identified by TRIM numbers) for effects on LLOMe-induced autophagosome formation by high content microscopy and automated image acquisition and quantification (HC). Panels in D, program-assigned masks superimposed on epifluorescent images of HeLa cells treated with LLOMe or DMSO vehicle. Pink mask, automatically defined cell boundaries (primary objects). Blue, nuclei stained with Hoechst 3342 (10 μg/ml). Red mask, machine identified endogenous LC3 puncta (target objects). Scale bar, 10 μm. Graph in E, Autophagosome abundance (total area of endogenous LC3 puncta/cell; staining with 1:500 antibody against LC3B; PM036 from MBL) in cells subjected to TRIM knockdowns in 96 well plates and treated with 0.5 mM LLOMe for 2 h (E). Dashed line, 2 SD below the mean (cumulative, LC3 puncta area/cell) for all TRIMs tested. N≥500 cells imaged per well. (F–H) Autophagic response (HC, LC3 puncta) to LLOMe (0.5 mM, 2 h) in HeLa cells and their CRISPR TRIM16KO mutant derivative A9. (I–K) Ubiqutination response, revealed with FK2 mouse monoclonal antibody (1:500 dilution; MBL D058-3) and quantified by HC (ubiquitin puncta, yellow mask) in HeLa vs. TRIM16KO HeLa mutant A9. Same conditions as in F–H. Data: means (n>3); t-test *, p < 0.05.
Screen for TRIMs involved in autophagic response to lysosomal damage
Galectins participate in autophagic response to endomembrane perforations caused by lysosomal damaging agents and by bacteria (Fujita et al., 2013; Maejima et al., 2013; Thurston et al., 2012). Based on interactions between Galectins and TRIMs detected above, we hypothesized that TRIMs might play a role in autophagic response to endomembrane damage. The lysosomal damaging agent Leu-Leu-O-Me (LLOMe), which is condensed into a membranolytic polymer via the transpeptidase action of cathepsin C within lysosomes (Fujita et al., 2013; Maejima et al., 2013), elicited a consistent LC3 dose response with dynamic range suitable for a screen. We employed high content microscopy with automated image acquisition and quantification (HC) (Kimura et al., 2015; Mandell et al., 2014) of endogenous LC3B puncta (Figure 1D) and screened the human TRIM family by siRNA knockdowns in HeLa cells for effects on LC3 response to LLOMe treatment (Figure 1E). TRIM16 showed the strongest effect (Figure 1E).
TRIM16 is necessary for autophagic response and ubiquitination upon lysosomal damage
The screening data were confirmed by follow-up siRNA knockdowns (Figure S1C) and by generating a TRIM16 CRISPR knockout in HeLa cells (TRIM16KO). Two independent CRISPR mutants, HeLa TRIM16KO A9 (Figure S1D,E) and C2 (Figure S1F) were generated. TRIM16 knockdowns diminished LC3B response (Figure S1G,H). HC imaging analysis in TRIM16 knockout cell line A9 demonstrated that TRIM16 was required for a full autophagic response to lysosomal injury caused by LLOMe (Figure 1F–H). This was confirmed in a different TRIM16 CRISPR clone, C2 (Figure S1I).
In addition to LC3+ autophagosomes (Maejima et al., 2013), LLOMe-induced lysosomal damage elicits ubiquitin puncta formation on lysosomes (Maejima et al., 2013), in a dose response fashion (Figure S1J). HC screen using ubiquitin puncta as a lysosomal-damage response readout (Maejima et al., 2013) again indicated a requirement for TRIM16 (Figure S1K,L). TRIM16 was needed for optimal ubiquitin response to LLOMe when tested by siRNA knockdowns (Figure S1M,N), and was absolutely required for ubiquitin dots formation when tested in TRIM16 CRISPR knockout HeLa A9 cells (Figure 1I–K). This was confirmed in the TRIM16 CRISPR knockout clone C2 (Figure S1O).
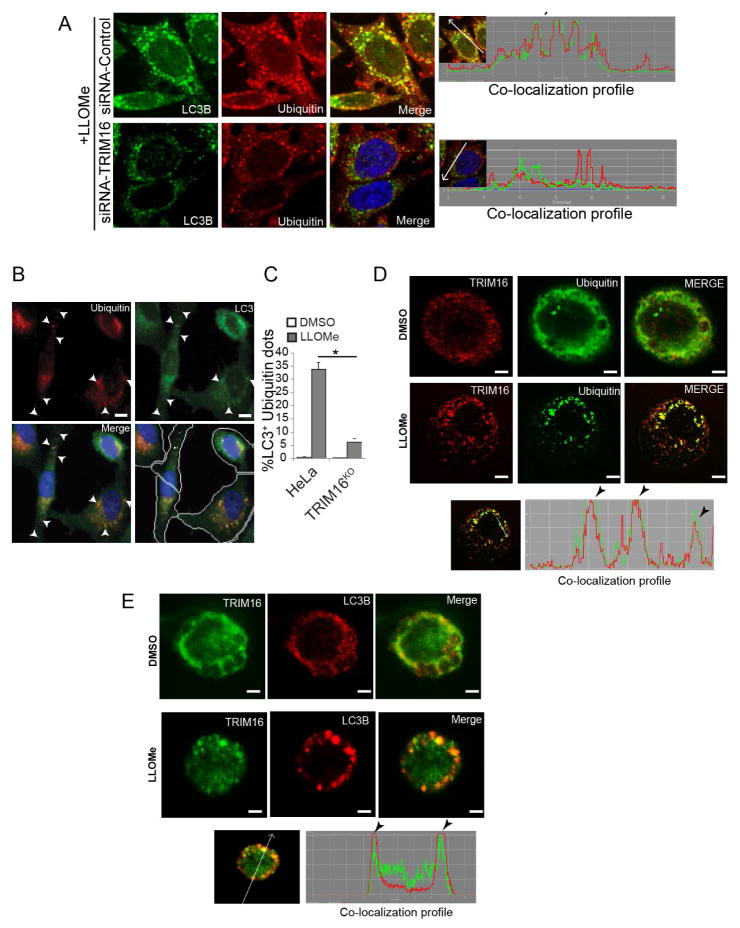
TRIM16 was required for colocalization between LC3B and ubiquitin puncta in response to LLOMe (Figure 2A and S2A) as quantified by HC imaging (Figure 2B,C). TRIM16 puncta were morphologically prominent in response to LLOMe in leukocytes (Figure 2D,E) but not in HeLa cells, most likely due to differential cathepsin C levels. TRIM16 puncta and ubiquitin profiles (Figure 2D) and TRIM16 puncta and LC3B puncta (Figure 2E) colocalized in THP-1 cells exposed to LLOMe. Thus, TRIM16 is required for optimal ubiqutination and autophagic response in response to lysosomal damage.
Figure 2. Colocalization between TRIM16, ubiquitin and LC3B.
(A) Confocal microscopy analysis of endogenous LC3B puncta and endogenous ubiquitin profiles localization in cells knocked down for TRIM16. Right, line tracers showing fluorescence peaks. (B–C) HC analysis of LC3B and ubiquitin profiles colocalization (%) in HeLa cells and CRISPR TRIM16KO mutant (A9) exposed to 0.5 mM LLOMe for 2 h. Yellow mask, ubiquitin puncta. Scale bar, 10 μm. (D,E) Confocal image analysis of THP1 cells untreated or treated with 0.5 mM LLOMe and immunostained for endogenous TRIM16, ubiquitin or LC3B. Bottom panels, line tracings representing fluorescence peaks. Scale bar, 10 μm.
TRIM16 is in protein complexes with Galectin-3 in cells
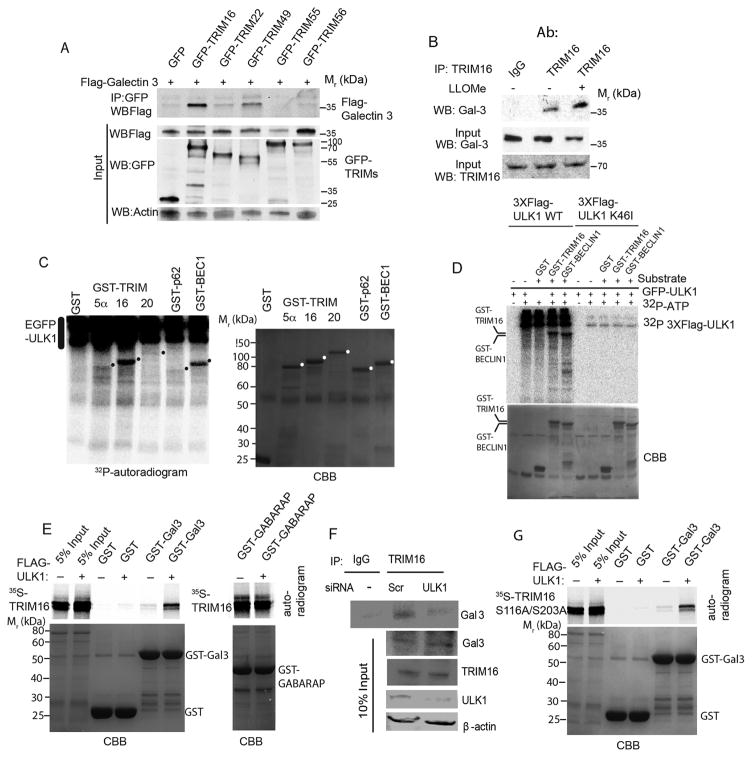
Galectin-3 is a marker for damaged lysosomes (Aits et al., 2015). Consistent with the prior reports using GFP-Galectin-3 (Maejima et al., 2013) we found endogenous Galectin-3 colocalizing with or juxtaposed to a number of LC3 profiles induced in response to LLOMe in different cell types (Figure S2B). Although the initial GST-Galectin pull-downs (Figure 1A) indicated that TRIM16 did not bind Galectin-3 in vitro, the finding that TRIM16 was a stand-out in our screens, and the acknowledged pivotal role of Galectins, and specifically that of Galectin-3, in recognition of lysosomal damage (Aits et al., 2015; Maejima et al., 2013), prompted us to re-asses the apparent absence of TRIM16-Galectin-3 interactions. TRIM16 was found in co-IPs with Galectin-2 and Galectin-3 in cellular extracts (Figure S3A). We focused on Galectin-3 as an accepted marker for damaged lysosomes (Aits et al., 2015). A reverse tag co-IP (Figure 3A) confirmed TRIM16-Galectin-3 association. TRIM55 and TRIM56 were not found in co-IPs with Galectin-3 (Figure 3A) indicating selectivity of Galectin-3-TRIM16 associations. Endogenous protein co-IPs detected TRIM16 and Galectin-3 in common complexes (Figure 3B). The association between endogenous TRIM16 and Galectin-3 increased in cells exposed to LLOMe (Figure 3B).
Figure 3. TRIM16 is phosphorylated by ULK1 and interacts with Galectin-3 in the presence of ULK1 as a platform.
(A) Co-immunoprecipitation (Co-IP) analysis of interaction between Galectin-3 and a panel of TRIMs in HEK293T cell lysates expressing GFP or GFP-TRIM fusions and Flag-Galectin-3. (B) Co-IP analysis of endogenous TRIM16 and endogenous Galectin-3 in the absence and presence of LLOMe. (C) Indicated GST-fusion proteins were incubated in a phosphorylation reaction (with γ32P ATP) with EGFP-ULK1 enriched from HEK293T cells and products separated by PAGE. Left, autoradiogram; right, coommassie brilliant blue (CBB). Dots, position of GST-fusion protein bands on the CBB gel. (D) Flag-ULK1 wt or K46I catalytic ULK1 mutant were incubated with GST fusion protein substrates and processed as in C. (E) In vitro translated and radiolabeled [35S] myc-HA-TRIM16 wild type incubated with potential interactors in the presence (+) or absence (−) of Flag-ULK1 and cold ATP, GST pulldowns performed and amount of [35S] radiolabeled Myc-HA-TRIM16 determined by PAGE and autoradiography. Amounts of GST fusion proteins are shown in coommassie brilliant blue (CBB)-stained gels (F) Co-IP analysis of interactions between endogenous TRIM16 and Gal3 proteins in HeLa cells knocked down for ULK1 by siRNA. (G) GST pulldown analysis as in E, using [35S] myc-HA-TRIM16-S116A/S203A mutant instead of wild type TRIM16. Note that both wt (in E) and the ULK1-non-phosphorylatable TRIM16-S116A/S203A (in G) mutant promote association between TRIM16 and Galectin-3.
TRIM16 interacts with Galectin-3 in an ULK1-dependent manner
We investigated the basis for the association between Galectin-3 and TRIM16 detected in cells. TRIM16 turned out to be a substrate for phosphorylation (Figure 3) by ULK1 (Figure S3B) similarly to the known ULK1 substrate Beclin 1 (Russell et al., 2013) (Figure 3C). Using a different preparations of ULK1 (Figure S3C), and comparing active vs. kinase dead ULK1 we established that TRIM16 was a substrate for ULK1 phosphorylation (Figure 3D). We next tested whether ULK1 modulated TRIM16 and Galectin-3 association observed in cells. GST pull-downs (initially negative for TRIM16-Galectin-3 interactions; Figure 1A), were repeated in the presence of ULK1. We observed enhancement of interactions between GST-Galectin-3 and TRIM16 (in vitro translated and radiolabeled) when ULK1 was added (Figure 3E), contrasting with no effect on the binding (Mandell et al., 2014) between TRIM16 and GABARAP (Figure 3E). In keeping with the above, a knockdown of ULK1 reduced the levels of Galectin-3 found in complexes with TRIM16 (Figure 3F).
TRIM16 was phosphorylated at S116 and S203 in an ULK1-dependent manner as determined by mass spectrometry (Figure S3D,F). When these TRIM16 residues were doubly mutated into a non-phosphorylatable form (S116A and S203A), TRIM16 nevertheless retained its capacity to promote TRIM16-Galectin-3 interactions (Figure 3G). Thus, ULK1 serves primarily as a platform for formation of TRIM16-Galectin-3 complexes. This played a function in response to LLOMe, as determined by using ULK1/2 knockout MEFs. Wild type MEFs showed LC3 (Figure S3G) and ubiquitin (Figure S3H) response to LLOMe, whereas ULK1/2 MEFs could not mount that response. Since as shown previously (Fujita et al., 2013), ubiquitin dots precede LC3 membrane formation, these findings also indicate that ULKs are required for ubiquitination events preceding LC3 response.
TRIM16 interacts with key autophagy regulators ULK1 and Beclin 1
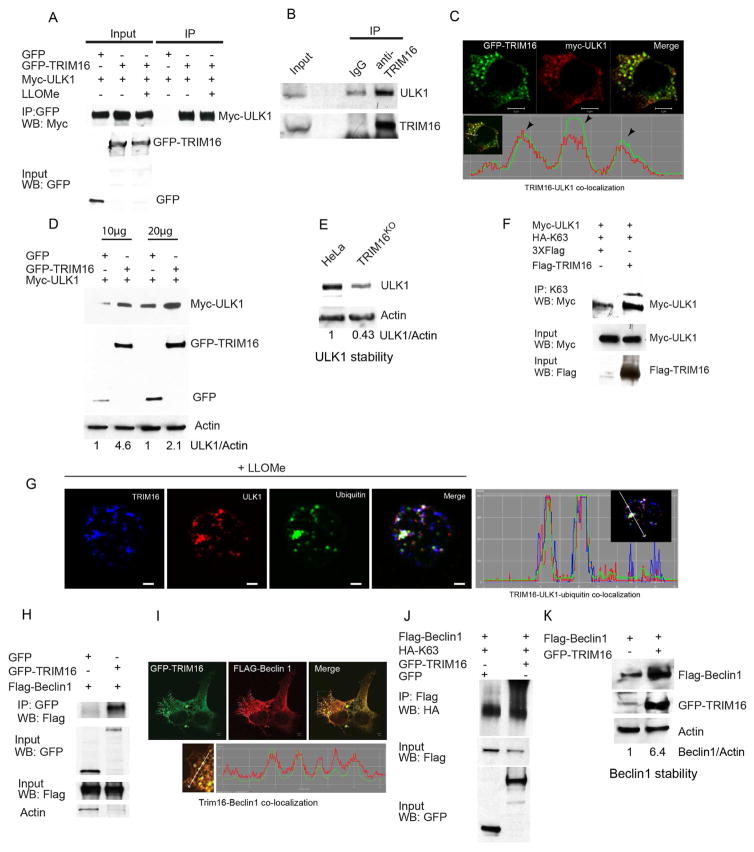
TRIM16 showed a capacity to associate with ULK1 (Figure 4A,B) and colocalized (Figure 4C) in cells. Co-expression of GFP-TRIM16 increased levels of myc-ULK1 (Figure 4D), whereas a TRIM16 knockdown diminished endogenous ULK1 (Figure 4E). TRIM16 promoted ULK1 K63-linked ubiquitination (Figure 4F), a modification stabilizing autophagy regulators (Nazio et al., 2013). This action of TRIM16 could be either direct, since it has an E3 ubiquitin ligase activity (Bell et al., 2012) (confirmed in Figure S4A), or indirect by recruiting/activating other E3 ligases controlling activity of ULK1 (Nazio et al., 2013). The latter possibility was underscored by the co-immunoprecipitation of TRIM16 with cullin ubiquitin ligase components (Cullin 4A; Figure S4B) implicated in regulation of autophagy (Antonioli et al., 2014). LLOMe-elicited TRIM16 puncta colocalized with ubiquitin and ULK1 (Figures 4G and S4C). GFP-TRIM16 co-IPed (Figure 4H) and co-localized (Figure 4I) with Flag-Beclin 1. Co-expression of GFP-TRIM16 with Flag-Beclin 1 stimulated K63-linked ubiquitination of Beclin 1 (Figure 4J), which could be direct or through other E3 ligases described previously (Shi and Kehrl, 2010; Xia et al., 2013), and increased cellular levels of Flag-Beclin 1 (Figure 4K).
Figure 4. TRIM16 interacts, co-localizes with, and stabilizes ULK1 and Beclin 1.
(A) Co-IP analysis of TRIM16 and ULK1 interactions in HEK293T lysates from cells expressing GFP or GFP-TRIM16 and Myc-ULK1. (B) Co-IP analysis of interactions between endogenous TRIM16 and endogenous ULK1 (HeLa lysates). (C) Confocal microscopy images of HEK293T cells transiently expressing GFP-TRIM16 and Myc-ULK1, and co-localization trace profiles. (D) HEK293T cell lysates co-expressing Myc-ULK1 and either GFP or GFP-TRIM16 were subjected to immunoblotting analysis. (E) Western blot analysis of relative abundance of endogenous ULK1 in wild type HeLa and CRISPR TRIM16KO HeLa derivative A9. (F) Analysis of ULK1 ubiquitination in cells co-expressing Myc-ULK1 and HA-tagged ubiquitin C mutated for all lysines except Lysine 63 (HA-K63) in the absence and presence of Flag-TRIM16. Immunoprecipitation was performed with K63 antibody followed by the Western blotting (WB) with indicated antibodies. (G) Confocal images of THP1 cells (treated with LLOMe); immunofluorescence with TRIM16, ULK1 and ubiquitin antibodies. Right, co-localization tracer profile along the line indicated in the inset. (H) Co-IP analysis of interactions between TRIM16 and Beclin1 in HEK293T lysates of cells expressing GFP or GFP-TRIM16 and Flag-Beclin1. (I) Confocal images of HEK293T cells transiently expressing GFP-TRIM16 and Flag-Beclin 1. Co-localization profile tracer along line indicated in the enlarged region. (J) Analysis of Beclin 1 ubiquitination in cells co-expressing Flag-Beclin 1 and HA-tagged Ubiquitin C (HA-K63) and either GFP or GFP-TRIM16. IP, immunoprecipitation performed with Flag antibody, followed by WB (Western blotting) with indicated antibodies. (K) HEK293T cell lysates expressing Flag-Beclin 1 in absence and presence GFP-TRIM16 were subjected to immunoblotting with antibodies as indicated.
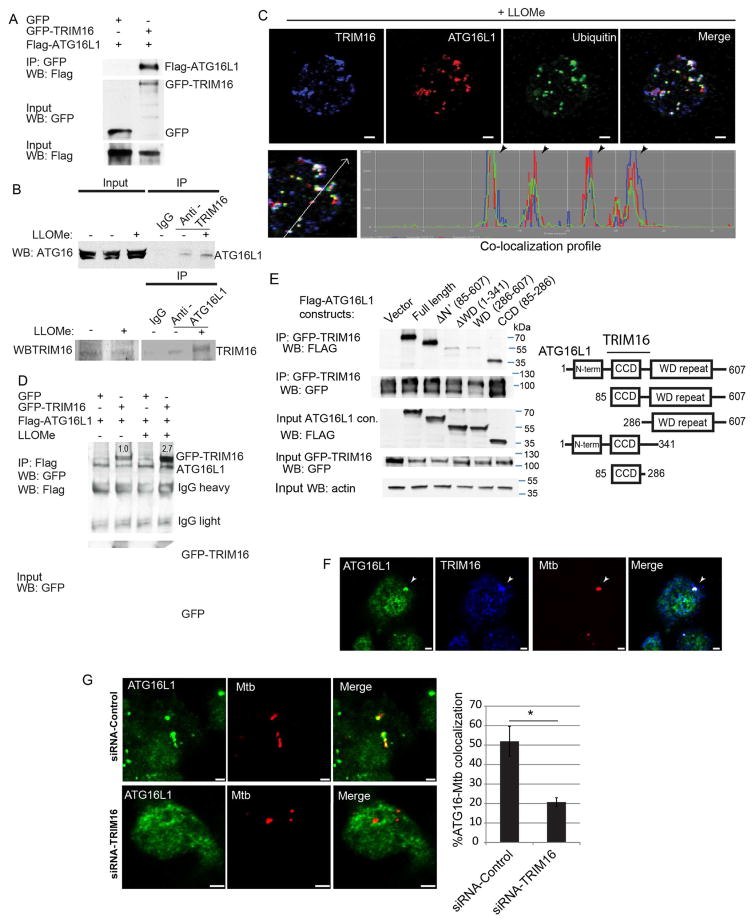
ATG16L1 associates with TRIM16 upon endomembrane damage
ATG16L1 is a core autophagy factor implicated in response to lysosomal damage (Fujita et al., 2013), through a convergence of at least three different association events: FIP200, binding to upstream residues within ATG16L1, ubiquitin recognized by ATG16L1’s C-terminal WD domain, and an unidentified factor that required the middle section of ATG16L1 (Fujita et al., 2013). Thus, we tested whether TRIM16 interacted with ATG16L1. GFP-TRIM16 and Flag-ATG16L1 co-IP-ed (Figure 5A) and colocalized in cells (Figure S5A). Endogenous ATG16L1 and TRIM16 coimmunoprecipitated (Figure 5B) and colocalized in THP1 cells treated with LLOMe (Figure 5C). Endogenous TRIM16 and ATG16L1 colocalized with ubiquitin puncta in THP1 macrophages exposed to LLOMe (Figure 5C).
Figure 5. TRIM16 interacts with and recruits ATG16L1 in response to endomembrane damage.
(A) Co-IP analysis of interaction between TRIM16 and ATG16L1 in HEK293T lysates from cells co-expressing GFP or GFP-TRIM16 with Flag-ATG16L1. (B) Co-IP and reverse Co-IP analyses of interaction between endogenous TRIM16 and endogenous ATG16L1 in resting and LLOMe-treated HeLa cells. (C) Confocal images of THP1 cells treated with LLOMe and processed for immunofluorescence microscopy analysis with TRIM16, ATG16L1 and ubiquitin antibodies. Bottom, co-localization profile measurement along straight line using LSM510 software. (D) Co-IP analysis of interactions between TRIM16 and ATG16L1 from LLOMe treated and untreated HEK293T cell lysates co-expressing GFP or GFP-TRIM16 with Flag-ATG16L1. (E) Mapping of ATG16L1 regions interacting with TRIM16. Lysates of HEK293T cells co-expressing GFP-TRIM16 and Flag-ATG16L1 variants (see scheme, right panel) were subjected to immunoprecipitation with anti-GFP and blots were probed as indicated. (F) Confocal images of RAW264.7 macrophage cell line infected with Alexa-568-labeled M. tuberculosis Erdman and immunostained for ATG16L1 and TRIM16. (G) RAW264.7 cells were transfected with siRNAs against TRIM16 or control siRNAs for 48 h. Cells were then infected with Alexa-568-labeled M. tuberculosis Erdman for 4 h followed by immunofluorescence staining for ATG16L1. Data, means ± SEM; n=3 (at least 100 phagosomes per condition); *, p<0.05 (t-test). Bar, 2 μm.
LLOMe treatment increased TRIM16 and ATG16L1 association (Figure 5D). ATG16L1 K63 ubiquitination was enhanced by TRIM16 (Figure S5B) but TRIM16 had no effect on levels of ATG16L1 (Figure S5C). TRIM16 association was delimited to ATG16L1 residues 85-286 (Figures 5E and S5D) encompassing residues in ATG16L1 implicated in binding of an unknown factor initiating autophagic response to lysosomal damage (Fujita et al., 2013).
We next employed another model of endomembrane damage, based on phagosome damage caused by M. tuberculosis, which permeabilizes phagosomal membranes eliciting autophagic response including LC3B and ubiquitin (Watson et al., 2012). For these experiments we used murine RAW264.7 macrophages, and detected colocalization between ATG16L1, TRIM16 and M. tuberculosis (Figure 5F). ATG16L1 and M. tuberculosis colocalization was reduced upon knocking down TRIM16 in RAW264.7 cells (Figure 5G and S5E). Thus, TRIM16 and ATG16L1 cooperate in autophagic responses to different types of endomembrane damage.
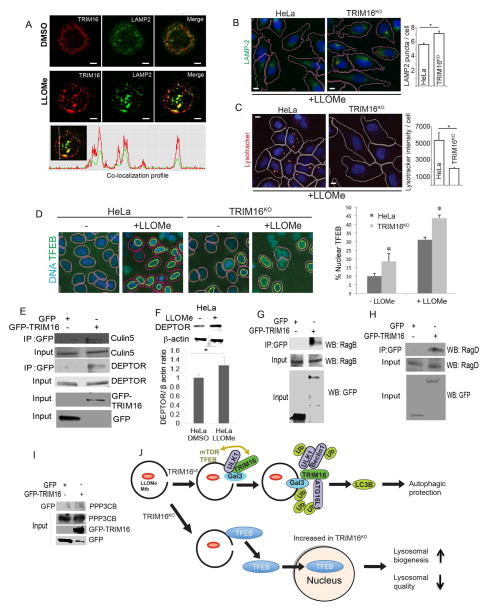
TRIM16 affects lysosomal quality and quantity
Following LLOMe exposure, TRIM16 translocated to LAMP2-positive lysosomal profiles in THP1 cells (Figure 6A), a cell type highly susceptible to LLOMe-caused lysosomal damage. We next used TRIM16KO HeLa cells to assess the status of lysosomes upon LLOMe treatment (Figure 6B,C). Although the number of lysosomes increased (Figure 6B), the quality of these lysosomes was compromised, as evidenced by lessened acidification quantified by LysoTracker Red (Figure 6C).
Figure 6. TRIM16 is required for lysosomal quality control and function.
(A) Confocal immunofluorescence microscopy images of THP1 cells untreated or treated with 0.5 mM LLOMe and stained for endogenous TRIM16 and LAMP2. (B) HC analysis of lysosome abundance (green, anti-LAMP2 staining; blue, Hoeschst 3342 nuclear stain) in HeLa cells or their CRISPR TRIM16KO derivative A9 following 1 mM LLOMe treatment for 2 h. (C) HC analysis of lysosomal acidification (Lysotracker Red) in HeLa cells or their CRISPR TRIM16KO derivative A9 following 1 mM LLOMe treatment for 2 h. (D) HC analysis of nuclear partition of TFEB in resting HeLa cells vs. their CRISPR TRIM16KO derivative cells (A9), and translocation of TFEB to the nucleus upon LLOMe treatment. HC masks: pink, nuclei (blue, Hoechst stain), yellow, nuclear TFEB (green fluorescence, Alexa-488). Data: means (n>3); t-test *, p < 0.05. All HC experiments were carried out in 96 well plates, with >12 wells/condition with >500 valid objects/well. (E) Co-IP analysis of interaction between GFP-TRIM16 and endogenous DEPTOR and Cullin-5 in HEK293T lysates from cells expressing GFP or GFP-TRIM16. (F) Levels of DEPTOR in extracts of HeLa cells subjected to LLOMe-induced lysosomal damage (1 mM LLOMe, 2_h) (G,H) Co-IP analysis of interactions between GFP-TRIM16 and endogenous RagB (G) or RagD (H) in HEK293T lysates from cells expressing GFP or GFP-TRIM16. (I) Co-IP analysis of interactions between GFP-TRIM16 and endogenous calcineurin catalytic subunit isoform β (PPP3CB) as in E, G and H. (J) Model of TRM16 action and consequences of its absence. Top, pictorial summary of relationships in wild type TRIM16 cells. Bottom, a depiction of what happens in the absence of TRIM16-dependent homeostatic repair of lysosomal membranes.
Lysosomal and autophagosomal systems are co-activated by the transcriptional factor TFEB (Settembre et al., 2011). TFEB is phosphorylated by Ser/Thr protein kinases, notably by mTOR, whereupon it resides in inactive cytoplasmic complexes, but translocates to the nucleus upon starvation to activate transcription (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012). TRIM16KO HeLa had increased TFEB presence in the nucleus under basal conditions cells (Figure 6D). TFEB translocated to the nucleus in response to lysosomal damage caused by LLOMe (Figures 6D and S6A), and mTOR activity was inhibited upon LLOMe treatment (Figure S6B). The LLOMe-induced levels of nuclear TFEB were higher in TRIM16KO cells relative to wt HeLa (Figure 6D). Thus, TRIM16 controls quantity and quality of lysosomes and affects localization of TFEB.
TRIM16 is in complexes with regulators of mTOR, and with calcineurin and TFEB
The increased TFEB in the nucleus of TRIM16KO may be secondary to defective lysosomes in cells lacking TRIM16 quality control. Nevertheless, we found TRIM16 in protein complexes regulating mTOR and TFEB (Napolitano and Ballabio, 2016). TRIM16 immunoprecipitates from cells expressing GFP-TRIM16 contained endogenous DEPTOR (Figure 6E), an mTOR inhibitor (Peterson et al., 2009) whose stability is regulated by Cullin-5 (Antonioli et al., 2014). Cellular DEPTOR increased upon LLOMe treatment (Figure 6F) and TRIM16-DEPTOR complexes contained Cullin-5 (Figure 6E). GFP-TRIM16 protein immunoprecipitates contained endogenous RagB and RagD (Figure 6G,H) factors recruiting mTOR to lysosomes where it is activated (Bar-Peled and Sabatini, 2014). GFP-TRIM16 coimmunoprecipitated with Flag-TFEB (Figure S6C). GFP-TRIM16 coimmunoprecipitated with endogenous calcineurin catalytic subunit isoform β (PPP3CB) (Figure 6I) known to dephosphorylate TFEB and facilitate its translocation to the nucleus (Medina et al., 2015). Flag-TRIM16 coimmunoprecipitated with endogenous PPP3CB (Figure S6D). The TRIM16 region required for PPP3CB association was delimited to the 166-373 segment of TRIM16 (Figure S6E,F). Thus, under homeostatic conditions when all components are present, TRIM16 coordinates its activities with mTOR, calcineurin and TFEB. When TRIM16 is absent, as in TRIM16KO cells, basal levels of nuclear TFEB increase due to perturbed interactions and/or indirectly due to compromised lysosomal quality (Figure 6J).
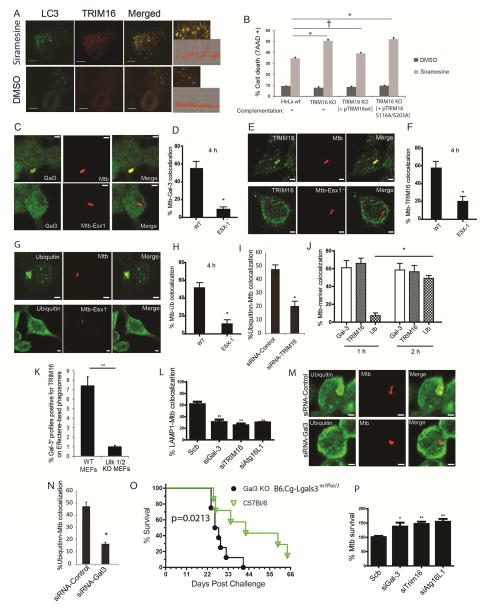
TRIM16 protects cells against consequences of endomembrane damage
Although LLOMe can cause limited lysosomal damage in HeLa, it cannot cause significant lysosomal cell death in these cells. However, other lysosome damaging agents can (Petersen et al., 2013). We thus used lysosome-destabilizing experimental anticancer lysosomotropic agent siramesine (Ostenfeld et al., 2008), because it can promote cell death in HeLa cells (Petersen et al., 2013). Siramesine, unlike LLOMe, caused in HeLa cells the formation of TRIM16 puncta that colocalized with LC3B (Figure 7A). Absence of TRIM16 in TRIM16KO HeLa cells increased sensitivity to cell death caused by siramesine (Figure S7A,B). Wild type TRIM16 but not mutant TRIM16S116A/S203A rescued this phenotype (Figure 7B). This indicates that whereas S116 and S203 (the residues phosphorylated by ULK1) are not critical for Galectin-3-TRIM16 interactions they are required for protection against lysosomal damage-induced cell death.
Figure 7. TRIM16-Galectin-3 system protects against endomembrane damage associated with lysosomal dysfunction and pathogen-mediated phagosomal perforation.
(A) Confocal images of HeLa cells treated with DMSO or siramesine; immunofluorescence, endogenous LC3B and TRIM16 revealed with corresponding antibodies. Graphs on the right, line tracing colocalization analyses. (B) HeLa, CRISPR TRIM16KO HeLa cells A9, or TRIM16KO HeLa cells A9 transfected with expression plasmids for wt TRIM16 or TRIM16S116A/S203A mutant were incubated with siramesine and cell death was measured using 7AAD nuclear staining. Data, means; n>3; †, p ≥ 0.05 *, p < 0.05 (ANOVA). (C,D) RAW264.7 macrophages were infected with Alexa-568-labeled wild-type M. tuberculosis Erdman or its ESX-1 mutant at MOI=10 for 4 h and then processed for confocal microscopy analysis of M. tuberculosis (Mtb) colocalization with Galectin-3. Data, means ± SEM (n>3; at least 100 Mtb phagosomes per condition were quantified); *, p<0.05 (t-test). Bar 2 μm. (E,F) RAW264.7 macrophages were infected with Alexa-568-labeled wild-type Mtb Erdman (WT) or its ESX-1 mutant at MOI=10 for 4 h and then processed for confocal microscopy analysis for the colocalization of Mtb with TRIM16. Data and statistics as in E. (G,H) RAW264.7 cells were infected with Alexa-568-labeled Mtb wild-type Erdman or its ESX-1 mutant for 4 h and then processed for immunofluorescence staining with anti-ubiquitin. Data and statistics as in D. (I) RAW264.7 macrophages were transfected with siRNAs against TRIM16 or control siRNAs for 48 h. Cells were then infected with Alexa-568-labeled wild-type Mtb Erdman for 4 h and processed for confocal microscopy analysis for the colocalization of Mtb with ubiquitin. Data and statistics as in D. (J) Time course of marker appearance on Mtb phagosomes; RAW264.7 macrophages were infected with Alexa-568-labeled wild-type M. tuberculosis Erdman at MOI=10 for 1 and 2 h and processed and data analyzed as in panels C–H. (K) Effectene-coated beads were phagocytosed by MEFs in 96 well plates, incubated for up to 24 h (> 3h), stained with antibodies, and processed for HC microscopy (see images in Figure S7D). Data: means (n>3); t-test *, p < 0.05. Ninety six-well plates, with >12 wells/condition with >500 valid objects/well. (L) RAW264.7 cells were transfected with siRNAs against TRIM16, Galectin-3, or ATG16L1 or control siRNAs for 48 h. Cells were then infected with Alexa-568-labeled wild-type Mtb Erdman for 4 h followed by immunofluorescence staining for LAMP1 and % colocalization of Mtb phagosomes with LAMP1 determined (for representative images see Figure S7G). Data, means ± SEM (n=3; at least 100 Mtb phagosomes per condition were quantified). (M,N) RAW264.7 cells were transfected with siRNAs against Galectin-3 or control siRNAs for 48 h. Cells were then infected with Alexa-568-labeled wild-type Mtb Erdman for 4 h followed by immunofluorescence staining for ubiquitin. Data and statistics as in L. (O) Survival of wt C57BL and Galectin-3 C57BL knockout mice in a short-term acute infection model (high dose; 1-3xe3 CFU) with M. tuberculosis Erdman aerosols. For data from a chronic model of infection with lower doses of M. tuberculosis, see Figure S7J,K). (P) RAW264.7 cells were transfected with siRNAs against TRIM16, Galectin-3, or ATG16L1 or control siRNAs for 48 h. Cells were then infected with wild-type Mtb Erdman for 1 h (t=0) at MOI=10. Cells were then washed three times with complete medium to remove uninternalized mycobacteria and then continued to grow in complete medium for another 24 h (t=24). Cells were then harvested for CFU analysis of Mtb intracellular survival. Data, means ± SEM of CFU at t=24 normalized to CFU at t=0 (n>3 independent experiments). *p < 0.05 and **p < 0.01, t-test relative to the scrambled (control) siRNA set at 100%.
Galectin-3, TRIM16, and ATG16L1 protect against M. tuberculosis infection
M. tuberculosis ESX-1 secretion substrates cause phagosomal damage (Manzanillo et al., 2013; Watson et al., 2012). We examined whether TRIM16 and its interactors contribute to autophagic control of M. tuberculosis. Galectin-3 and TRIM16 localized to phagosomes when macrophages were infected with M. tuberculosis wild type strain Erdman and not when its ESX-1 mutant was used (Figure 7C–F). Ubiqutin dots were present on wild type phagosomes but not on phagosomes with the ESX-1 mutant (Figure 7G,H). TRIM16 was needed for ubiquitin colocalization on M. tuberculosis phagosomes (Figure 7I and S7C), and ubiquitin appeared after the recruitment of Galectin-3 and TRIM16 (Figure 7J).
A question arose whether ULK1 affected colocalization of Galectin 3 and TRIM16 on phagosomes. We used MEFs and phagosomes with membrane-damaging beads coated with Effectene (Fujita et al., 2013). The Ulk1KO/Ulk2KO MEFs had diminished numbers of and reduced colocalization between TRIM16 and Galectin-3 puncta on phagosomes relative to wt MEFs (Fig. 7K and S7D–F).
TRIM16 was required for translocation of M. tuberculosis to LAMP1+ compartments (Figure 7L and Figure S7G) and TRIM16 localized on LAMP1+ autophagolysosomes (Figure S7H). Like TRIM16, Galectin-3 was required for translocation of M. tuberculosis to LAMP1+ compartments (Figure 7L and Figure S7G). Galectin-3 was required for ubiquitin deposits on M. tuberculosis phagosomes (Figure 7M,N). Galectin-3 and TRIM16 colocalized on M. tuberculosis phagosomes (Figure S7I). Finally, Galectin-3 protected mice in models of acute (Figure 7O) and chronic (Figure S7J,K) infection following aerosol exposure to M. tuberculosis. ATG16L1 was required for translocation of M. tuberculosis to autophagolysosomal compartments (Figure 7L and Figure S7G). Finally, all three components, Galectin-3, TRIM16, and ATG16L1 were required for control of intracellular M. tuberculosis (Figure 7P). Thus, TRIM16 along with Galectin-3 and ATG16L1, protects cells from the invading mycobacteria. In summary, the Galectin-3-TRIM16-ATG16L1 axis affords autophagic protection against lysosomal and phagosomal damage in diverse physiological contexts.
DISCUSSION
This work shows that TRIMs (Reymond et al., 2001) and Galectins (Arthur et al., 2015; Blidner et al., 2015; de Waard et al., 1976; Nabi et al., 2015) interact and that the TRIM16-Galectin-3 system organizes autophagic response to endomembrane damage. TRIM16 controls ubiquitination of damaged compartments, and regulates the core autophagy regulators ULK1, Beclin 1, and ATG16L1, which confer localized autophagic sequestration of damaged lysosomes (Maejima et al., 2013). TRIM16 also affects TFEB activation and nuclear translocation. Thus, TRIM16, in cooperation with Galectin-3, organizes core autophagy factors and orchestrates sequential stages of autophagic responses to lysosomal and phagosomal damage. These relationships are depicted in Figure 6J.
Our understanding of the role of TRIMs (Kimura et al., 2015; Kimura et al., 2016; Mandell et al., 2014) and Galectins (Aits et al., 2015; Chen et al., 2014; Fujita et al., 2013; Hung et al., 2013; Maejima et al., 2013; Thurston et al., 2012) in autophagy is growing. The present study underscores the significance of both of these families of proteins and reports the key finding that these two systems interact. This work furthermore expands the number of autophagic receptors and regulators of autophagy that utilize Galectins as cofactors in recognition of autophagic targets (e.g. damaged membranes).
The interaction of TRIM16 with ATG16L1 is of particular interest. ATG16L1, occupies a unique place among core autophagy factors by acting as a hub that brings together the principal parts of the autophagic apparatus: (i) ATG16L1 is a component of the LC3-PE conjugation system (Mizushima et al., 2011); (ii) ATG16L1 interacts with the WIPI2 (Dooley et al., 2014), which in turn binds to PI3P produced by Beclin1-VPS34; and (iii) ATG16L1 associates with FIP200, a component of the ULK1 complex systems (Dooley et al., 2014; Fujita et al., 2013). Although ATG16L1 has intrinsic affinity for ubiquitin it requires an additional (hitherto unidentified) factor in order to be recruited to the correct membranes (Fujita et al., 2013). TRIM16 fits the properties of this missing link as it binds to the region of ATG16L1 spanning the critical residues in ATG16L1 postulated (Fujita et al., 2013) to interact with the putative factor that homes ATG16L1 to damaged lysosomal membranes. Thus, TRIM16, by interacting with ATG16L1 and Galectin-3, guides the placement of ATG16L1 on damaged membranes.
TRIM16 and its interacting partner Galectin-3 protect cells from lysosomal cell death or microbial invasion. Galectin-3 has been previously observed on mycobacterial phagosomes and implicated in control of mycobacteria in a short term infection of Galectin-3 knockout mice (Beatty et al., 2002). These studies are congruent with the results of murine survival studies in the aerosol M. tuberculosis infection model reported here. Given the connections to autophagic machinery via TRIM16, the role of Galectin-3 in control of bacteria or pathology associated with mycobacterial infection can now be mechanistically linked to the TRIM-driven process of precision autophagy (Kimura et al., 2016), which differs from bulk autophagy. TRIM16 is also known as estrogen-responsive B box protein, and its role in cancer (a general property of TRIMs (Hatakeyama, 2011)) has been linked to specific effects on immune signaling (Sutton et al., 2014), cell survival (Kim et al., 2013), cell migration and metastasis (Marshall et al., 2010; Sutton et al., 2014) through various mechanisms, including measures of membrane repair (Cheung et al., 2012; Marshall et al., 2010), with the latter potentially overlapping with the processes described in this work.
TRIM16 absence elevates TFEB’s partitioning to the nucleus. Both of these proteins can localize to lysosomes as their station for exerting regulatory and effector functions. TFEB is coupled to the mTOR system, with mTOR phosphorylating TFEB to keep it locked in the cytosol, which can be reversed upon starvation that inactivates mTOR, further coupled with Ca2+ efflux from the lysosomes thus activating calcineurin to dephosphorylate TFEB and allow its nuclear translocation (Medina et al., 2015; Settembre et al., 2011; Settembre et al., 2012). These processes are modulated by TRIM16 action as suggested through interactions of TRIM16 shown here for DEPTOR, Rag GTPases, calcineurin, and TFEB itself. Absence of these interactions may lead to elevated TFEB in the nucleus. In summary, the relationships demonstrated here show convergence of precision autophagy systems in control of autophagic responses to endomembrane damage of significance in cancer and infectious diseases.
EXPERIMENTAL PROCEDURES
Cells and cell lines
RAW264.7, 293T, THP-1 and HeLa cells were obtained directly from ATCC and maintained in ATCC recommended media. Ulk1/2 double knockout MEFs and matching wild type MEFs were from Sharon Tooze, The Francis Crick Institute.
Cell culture and biochemical methods
Cells and cell lines, GST pull-downs, ULK1 phosphorylation assay, mass spectrometry, antibodies source and dilutions, immunoblotting, coimmunoprecipitation, plasmids, siRNA, and transfection are described in Supplementary Experimental Procedures.
CRISPR knockout cell lines and their complementation
To generate CRISPR knock out cell lines, HeLa cells were transfected with a PX458 (Addgene plasmid # 48138) (Ran et al., 2013) encoding the U6 promoter, human TRIM16 target sequence located within the first exon (AGTTGGATCTAATGGCTCCA, with the 5′ nucleotide A changed into a G; this sequence is followed on the chromosome by a protospacer adjacent motif GGG and was selected using crispr.mit.edu/guides site) fused to a chimeric guide RNA, S. pyogenes Cas9, and GFP. Transfected cells (green fluorescence) were sorted by flow cytometry and single cell clones analyzed by immunoblotting for a loss of TRIM16 band. Positive clones were subjected to next generation sequencing (Illumina; Massachusetts General Hospital core) to characterize the mutation (Figure S1F). For complementation, TRIM16KO HeLa cells were transfected with empty vector, WT TRIM 16 or phosphorylation mutated TRIM16 with > 60% transfection efficiency. After 24 h of transfection cells were subjected to response assays.
Bacterial strains and procedures
M. tuberculosis wild-type Erdman and ESX-1 mutant (Manzanillo et al., 2012) were cultured in Middlebrook 7H9 broth supplemented with 0.05% Tween 80, 0.2% glycerol, and 10% oleic acid, albumin, dextrose, and catalase (OADC; BD Biosciences) at 37°C and homogenized to generate single-cell suspension for macrophage infection studies. For acute (short-term) aerosol infection, C57BL mice or their Galectin-3 knockout derivative B6.Cg-Lgals3m1Poi/J (Jackson Laboratory) were exposed to high dose M. tuberculosis Erdman aerosols (1-4 x e3 CFU deposition) as previously described (Castillo et al., 2012), with a modification of using a GlasCol apparatus for aerosol delivery, and survival monitored for 2.5 months post-infection. For chronic (long-term) infection with lower doses of M. tuberculosis, mice were exposed in a GlasCol apparatus to medium dose (6 x e2 CFU initial lung deposition) and low dose (2 x e2 CFU initial lung deposition) of M. tuberculosis Erdman aerosols as previously described (Manzanillo et al., 2012), (Castillo et al., 2012)(Castillo et al., 2012)and survival monitored for up to 200 days post-infection. For intracellular mycobacterial survival assays and fluorescence microscopy see Supplementary Experimental Procedures.
High content microscopy, confocal microscopy, flow cytometry, cell survival
High content microscopy with automated image acquisition and quantification was carried out using a Cellomics HCS scanner and iDEV software (Thermo) in 96-well plates (Mandell et al., 2014). Immunofluorescence confocal microscopy was performed using an LSM510 confocal microscope and Zeiss software package. For quantification of puncta or total cell fluorescence, image J was used as described previously (Chauhan et al., 2015). For quantifying cell death, HeLa and derivative cells treated with 15 μM siramesine for 12 h were incubated with 7AAD (BD Pharmingen™; cat# 559925) for 30 min and fluorescence of 7AAD bound to nuclear DNA measured in a flow cytometer.
Supplementary Material
HIGHLGHTS.
TRIM proteins and Galectins interact
TRIM16 works with Galectin-3 in recognition of damaged lysosomes and phagosomes
TRIM16 controls autophagy to contain lysosomal damage and M. tuberculosis invasion
TRIM16 affects mTOR and TFEB and controls lysosomal quality and quantity
Acknowledgments
We thank Drs. J. Cox for M. tuberculosis strains, F. Randow for YFP-Galectins s, S. Tooze for Ulk1/2 knockout MEFs, A. Ballabio for information on TFEB, and C. Cleyrat for CRISPR consultations. This work was supported by grants AI042999 and AI111935 from NIH. T. Johansen was supported by grants 196898 and 71043-PR-2006-0320.
Footnotes
Author contributions. SK, SC, AJ, MP designed and carried out experiments. MHM, TK, SWC, RP, MAM, JAB carried out experiments. TJ designed experiments. VD conceived the project, designed and carried out experiments, and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aits S, Kricker J, Liu B, Ellegaard AM, Hamalisto S, Tvingsholm S, Corcelle-Termeau E, Hogh S, Farkas T, Holm Jonassen A, et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy. 2015;11:1408–1424. doi: 10.1080/15548627.2015.1063871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonioli M, Albiero F, Nazio F, Vescovo T, Perdomo AB, Corazzari M, Marsella C, Piselli P, Gretzmeier C, Dengjel J, et al. AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev Cell. 2014;31:734–746. doi: 10.1016/j.devcel.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Arthur CM, Baruffi MD, Cummings RD, Stowell SR. Evolving mechanistic insights into galectin functions. Methods Mol Biol. 2015;1207:1–35. doi: 10.1007/978-1-4939-1396-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–406. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty WL, Rhoades ER, Hsu DK, Liu FT, Russell DG. Association of a macrophage galactoside-binding protein with Mycobacterium-containing phagosomes. Cell Microbiol. 2002;4:167–176. doi: 10.1046/j.1462-5822.2002.00183.x. [DOI] [PubMed] [Google Scholar]
- Bell JL, Malyukova A, Holien JK, Koach J, Parker MW, Kavallaris M, Marshall GM, Cheung BB. TRIM16 acts as an E3 ubiquitin ligase and can heterodimerize with other TRIM family members. PloS one. 2012;7:e37470. doi: 10.1371/journal.pone.0037470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir AB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. Journal of cell science. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- Blidner AG, Mendez-Huergo SP, Cagnoni AJ, Rabinovich GA. Re-wiring regulatory cell networks in immunity by galectin-glycan interactions. FEBS Lett. 2015;589:3407–3418. doi: 10.1016/j.febslet.2015.08.037. [DOI] [PubMed] [Google Scholar]
- Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S, Delgado-Vargas M, Timmins GS, Bhattacharya D, Yang H, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3168–3176. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Mandell MA, Deretic V. IRGM Governs the Core Autophagy Machinery to Conduct Antimicrobial Defense. Mol Cell. 2015;58:507–521. doi: 10.1016/j.molcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Khambu B, Zhang H, Gao W, Li M, Chen X, Yoshimori T, Yin XM. Autophagy induced by calcium phosphate precipitates targets damaged endosomes. The Journal of biological chemistry. 2014;289:11162–11174. doi: 10.1074/jbc.M113.531855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung BB, Koach J, Tan O, Kim P, Bell JL, D’Andreti C, Sutton S, Malyukova A, Sekyere E, Norris M, et al. The retinoid signalling molecule, TRIM16, is repressed during squamous cell carcinoma skin carcinogenesis in vivo and reduces skin cancer cell migration in vitro. J Pathol. 2012;226:451–462. doi: 10.1002/path.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waard A, Hickman S, Kornfeld S. Isolation and properties of beta-galactoside binding lectins of calf heart and lung. The Journal of biological chemistry. 1976;251:7581–7587. [PubMed] [Google Scholar]
- Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation, and immunity. Nature Reviews Immunology. 2013 doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 Links LC3 Conjugation with PI3P, Autophagosome Formation, and Pathogen Clearance by Recruiting Atg12-5-16L1. Mol Cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, Umemoto T, Saitoh T, Nakatogawa H, Kobayashi S, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- Hung YH, Chen LM, Yang JY, Yang WY. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nature communications. 2013;4:2111. doi: 10.1038/ncomms3111. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Khaminets A, Behl C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PY, Rahmanto AS, Tan O, Norris MD, Haber M, Marshall GM, Cheung BB. TRIM16 overexpression induces apoptosis through activation of caspase-2 in cancer cells. Apoptosis. 2013;18:639–651. doi: 10.1007/s10495-013-0813-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, Deretic V. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol. 2015;210:973–989. doi: 10.1083/jcb.201503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Mandell M, Deretic V. Precision Autophagy directed by Receptor-Regulators. Journal of Cell Science. 2016 doi: 10.1242/jcs.163758. In press. [DOI] [PMC free article] [PubMed]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013;32:2336–2347. doi: 10.1038/emboj.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, Silvestri G, Munch J, Kirchhoff F, Simonsen A, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell. 2014;30:394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium Tuberculosis Activates the DNA-Dependent Cytosolic Surveillance Pathway within Macrophages. Cell host & microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall GM, Bell JL, Koach J, Tan O, Kim P, Malyukova A, Thomas W, Sekyere EO, Liu T, Cunningham AM, et al. TRIM16 acts as a tumour suppressor by inhibitory effects on cytoplasmic vimentin and nuclear E2F1 in neuroblastoma cells. Oncogene. 2010;29:6172–6183. doi: 10.1038/onc.2010.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of atg proteins in autophagosome formation. Annual review of cell and developmental biology. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Nabi IR, Shankar J, Dennis JW. The galectin lattice at a glance. J Cell Sci. 2015;128:2213–2219. doi: 10.1242/jcs.151159. [DOI] [PubMed] [Google Scholar]
- Napolitano G, Ballabio A. TFEB at a glance. J Cell Sci. 2016 doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nature cell biology. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- Ostenfeld MS, Hoyer-Hansen M, Bastholm L, Fehrenbacher N, Olsen OD, Groth-Pedersen L, Puustinen P, Kirkegaard-Sorensen T, Nylandsted J, Farkas T, et al. Anti-cancer agent siramesine is a lysosomotropic detergent that induces cytoprotective autophagosome accumulation. Autophagy. 2008;4:487–499. doi: 10.4161/auto.5774. [DOI] [PubMed] [Google Scholar]
- Petersen NH, Olsen OD, Groth-Pedersen L, Ellegaard AM, Bilgin M, Redmer S, Ostenfeld MS, Ulanet D, Dovmark TH, Lonborg A, et al. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell. 2013;24:379–393. doi: 10.1016/j.ccr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. The EMBO journal. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton SK, Koach J, Tan O, Liu B, Carter DR, Wilmott JS, Yosufi B, Haydu LE, Mann GJ, Thompson JF, et al. TRIM16 inhibits proliferation and migration through regulation of interferon beta 1 in melanoma cells. Oncotarget. 2014;5:10127–10139. doi: 10.18632/oncotarget.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA Targets Bacteria for Autophagy by Activating the Host DNA-Sensing Pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, An Z, Zou Z, Sumpter R, Su M, Zang X, Sinha S, Gaestel M, Levine B. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. eLife. 2015;4 doi: 10.7554/eLife.05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, Huang G, Ye B, Li C, Dai Z, et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 2013;32:2685–2696. doi: 10.1038/emboj.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.