Abstract
The present study aimed to investigate the expression of Golgi phosphoprotein-3 (GOLPH3) protein in colon cancer tissues and the association with the prognosis of patients. In total, 98 patients with colon cancer admitted to The First Affiliated Hospital of Henan University of Science and Technology for surgery between June 2011 and June 2013 were taken as the observation group. In addition, 15 healthy individuals, determined by enteroscopy, were taken as the control group. The expressions of GOLPH3 mRNA and protein were detected by reverse transcription-polymerase chain reaction and immunohistochemistry, respectively. The patients were divided into GOLPH3-positive and GOLPH3-negative groups according to the expression of GOLPH3. The expression of GOLPH3 in colon cancer and its association with the prognosis of patients was analyzed. The expression of GOLPH3 mRNA and protein in colon cancer tissues was significantly increased compared with normal colon mucosa (P<0.05); among the tissues, GOLPH3 was not expressed in 29 patients and positively expressed in 69 patients. The expression of GOLPH3 was negatively associated with the tumor differentiation degree, and positively associated with tumor invasion depth, lymph node metastasis and clinical stages in GOLPH3-positive patients. The cumulative recurrence rates at 1, 2 and 3 years were significantly lower in GOLPH3-negative patients (P<0.05). The survival rates at 1, 2 and 3 years in the GOLPH3-positive group were significantly higher than that of the GOLPH3-negative patients (P<0.05). In conclusion, the positive expression of GOLPH3 mRNA and protein in colon cancer tissue was significantly increased compared with the control group. GOLPH3 expression was closely associated with the pathological features, consisting of tissue typing, clinical stage, degree of tumor invasion and lymph node metastasis, and GOLPH3 expression. Patients with GOLPH3 overexpression also had a poor prognosis.
Keywords: colon cancer, GOLPH3, recurrence rate, survival rate, prognosis
Introduction
Colon cancer is the third most common type of malignant tumor of the gastrointestinal tract, and it has the highest mortality rate (1). Clinical data showed that with an improvement in living standards and a change of diet, the incidence rate of colon cancer showed a rapidly rising trend year by year and tended to be diagnosed at younger ages (2,3). The incidence rate of colon cancer rose from 6.1/100,000 in 1972–1974 to 11.2/100,000 in 1987–1989 worldwide (4). This rise was an 84.6% increase and the average annual increase was 4.2% (4). The incidence rate of colon cancer ranks 4th in the world, subsequent to stomach, lung and liver cancer, seriously affecting the social labor force, social production and quality of life for numerous families (5,6). For the treatment of colon cancer, radical treatment is surgery, and the auxiliary treatments consist of radiotherapy and chemotherapy, reducing tumor burden and the number of tumor cells to achieve a therapeutic effect. For the majority of colon cancer patients at the progressive stage it is too late for successful treatment of the malignancy, and thus, chemotherapy and palliative care may be the remaining therapeutic strategies. Recurrence and metastasis are the main causes of treatment failure (7,8). Therefore, clarification of the pathogenesis of colon cancer may play an important role in the early diagnosis, early treatment and prognosis of colon cancer. The Golgi phosphoprotein-3 (GOLPH3) gene has been identified as an oncogene, localized in the network structure on the reverse side of the Golgi apparatus (9). GOLPH3 is a phosphorylated Golgi membrane protein that is involved in the protein sorting of the Golgi apparatus (10,11). GOLPH3 is involved in the occurrence, development, apoptosis, proliferation and differentiation of human cancer, regulating the sensitivity of cancer cells to anti-tumor drugs (12). There are few studies that have researched the mechanisms of GOLPH3 in colon cancer (13). Based on this, the expression of GOLPH3 in colon cancer and its clinical significance were analyzed in the present study to provide guidance for the clinical treatment of colon cancer.
Patients and methods
General data
In total, 98 cases of colon cancer patients randomly selected for surgery in The First Affiliated Hospital of Henan University of Science and Technology (Luoyang, China) between June 2011 and June 2013 were taken as the observation group. The patients did not receive radiotherapy or other associated anti-tumor treatments prior to the surgery. The colon cancer tissue was resected and confirmed as cancer tissue by pathology. In total, 61 patients were male and 37 patients were female. The ages ranged between 30 and 80 years old and the average age was 56.78±10.67 years (mean ± standard deviation). The Dukes' clinical stages consisted of 42 cases of stage II and 56 cases of stage III colon cancer (14). The tissue typing, performed using the World Health Organization histological classification system, consisted of 53 patients with poorly differentiated and 45 patients with highly differentiated colon cancer (15). The infiltration degree consisted of 40 patients with T1-T2 and 58 patients with T3-T4. In addition, normal biopsy samples collected from 15 healthy individuals following routine colonoscopy were used as the control group. Of these, 9 patients were male and 6 patients were females. The patient ages ranged between 42 and 78 years and the average age was 63.26±11.32 years. The present study was conducted in accordance with the Declaration of Helsinki and was conducted with approval from the Ethics Committee of the First Affiliated Hospital of Henan University of Science and Technology (Luoyang, China). Written informed consent was obtained from all participants.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tumor tissues of the observation group and biopsy tissues of the control group, according to the RNA extraction kit protocol (Takara Biotechnology Co., Ltd., Dalian, China). In total, 25 µl RNase-free water was added to the precipitate and total RNA was obtained. RNA was transcribed to complementary DNA (cDNA) using a Moloney Murine Leukemia Virus Reverse Transcription kit (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. This cDNA was then used as the template for PCR following concentration determination. The primers were designed by Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA) according to the GOLPH3 mRNA sequence provided by GeneBank (ID number, NM_022130.3): GOLPH3 forward, 5′-AAGCCGTTCTTGACAAATGG-3′ and reverse, 5′-ATTGGTGTTGGCCTTCAGAC-3′; and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward, 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse, 5′-AGGGGCCATCCACAGTCTTC-3′. The primer was diluted in double distilled water (10 µmol/l−1). The PCR reaction system was prepared for RT-qPCR subsequent to the optimization of primer specificity and annealing temperature. The PCR reaction system comprised of: 10 µl SYBR Green qPCR Master Mix (Bio-Rad Laboratories, Inc., Hercules, CA, USA), 1 µl cDNA, 1 µl upstream and downstream primer, respectively. The PCR reaction conditions were as follows: Pre-degeneration, 94°C for 2 min; degeneration, 94°C for 30 sec; annealing, 60°C for 30 sec; and extension, 72°C for 30 sec, for a total of 35 cycles. PCR was performed using DNA polymerase from Takara Biotechnology Co., Ltd and SYBR green (Bio-Rad Laboratories, Inc.). To determine the specificity of the product, a solubility curve was constructed. Finally, the data were read directly from the fluorescence qPCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) using the 2−ΔΔCq method (16).
Immunohistochemical detection
The tissue specimens in the observation and control groups were embedded in paraffin, then sectioned into 4-mm slices and adhered onto poly-lysine acid-treated slides. All slides were subjected to the same protocol, as follows: Placed in a 67°C oven and baked for 2 h; immersed in xylene 3 times, 5 min/immersion; immersed in absolute ethyl alcohol for 5 min; immersed in 95% ethanol for 5 min; immersed in 75% ethanol for 5 min, washed in phosphate buffered saline (PBS) 3 times (3 min/wash); and then placed in 10 mM pH 6.0 citrate sodium buffer solution and heated for 10 min prior to being cooled for 30 min. The steps were repeated 2 times. Subsequently, the slides were washed with PBS 3 times (3 min/wash) and incubated in 3% H2O2 solution at room temperature for 10 min to block the activity of endogenous peroxide hydrogenase. The slides were then washed with PBS 3 times and incubated in rabbit anti-human GOLPH3 polyclonal antibody (catalog no., ab82377; dilution, 1:100; Abcam, Cambridge, UK) at 4°C overnight. Subsequent to incubation the slides were washed with PBS with Tween 20 (PBST) 3 times (3 min/wash). The sheep anti-rabbit IgG secondary antibody (catalog no., A0208; dilution, 1:500; Beyotime Institue of Biotechnology, Shanghai, China) was then added, the slides were incubated for 1 h at room temperature and washed with PBST 3 times (3 min/wash). The substrate 3,3′-diaminobenzidine was used for staining, hematoxylin was used as a counterstain and 0.1% hydrochloric acid was used for differentiation, followed by washing in PBS to return the color to blue. The slides were flushed with unfiltered water, redyed, dehydrated, hyalinized and sealed.
Observation index
GOLPH3 was located in the network structure on the opposite side of the Golgi apparatus, so brown yellow or brown particles in the cytoplasm were considered to be positively stained cells following immunohistochemical detection. The results were analyzed according to the cell staining intensity and percentage of GOLPH3-positive cells. In total, 5 high-power fields were randomly selected for every section and 100 target cells were counted under high power fields. The average percentage of 5 high-power fields was used for result determination. For evaluation the cell staining criteria were set as: 0% staining was 0; positive cell percentage ≤10% was 1; 11–50% was 2; 51–75% was 3; and >75% was 4. The intensity of the dye was also classified into numerical criteria: No dyeing was 0; faint yellow was 1; deep yellow was 2; and brown yellow was 3. The two scores were multiplied to calculate the final score. A total score of 4 was the demarcation point. Therefore, a total score of <4 was defined as GOLPH3-negative and ≥4 was GOLPH3-positive. The prognosis of the patients was evaluated by the postoperative recurrence rate and survival rate at 1, 2 and 3 years.
Statistical analysis
All data were analyzed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). The measurement data are shown as the mean ± standard deviation. The comparison among the multi-group measurement data was performed using analysis of variance. The comparison between the two groups was shown using Fisher's least significant difference test. The cumulative incidence of cancer development among the groups was performed using Kaplan-Meier curves. The comparison of the survival rate and recurrence rate among the groups was performed using Log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
Comparison of GOLPH3 mRNA expression in 2 groups
The results of GOLPH3 primer analysis showed that the primer amplification curve was accurate and the repeatability was high (Fig. 1A). The primer specificity was good and there were no miscellaneous peaks subsequent to amplification (Fig. 1B). GOLPH3 mRNA level in the 2 groups by RT-qPCR method showed that the average level of GOLPH3 mRNA was significantly higher than that of the control group (P<0.001) (Fig. 1C).
Figure 1.
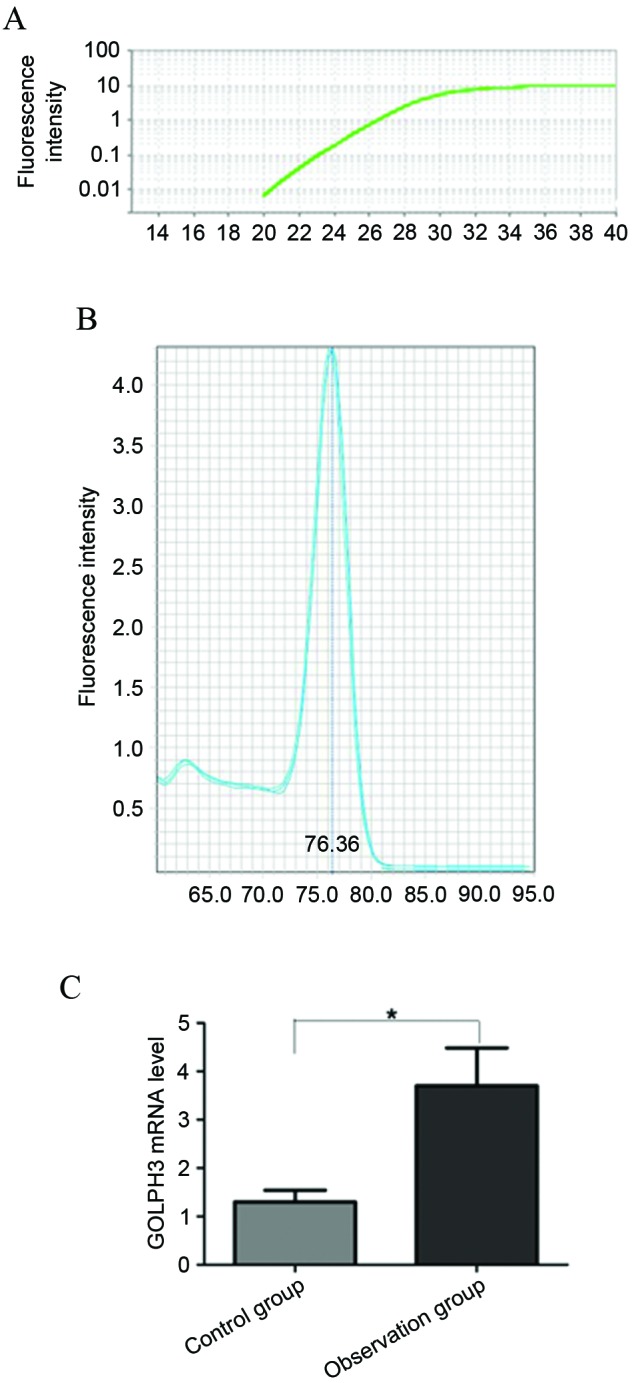
Reverse transcription-quantitative PCR analysis of the GOLPH3 mRNA levels in the 2 groups. (A) PCR amplification curve. (B) The melting curve of PCR primers. (C) Comparison of GOLPH3 mRNA level in observation group and control group. *P<0.05. PCR, polymerase chain reaction; GOLPH3, Golgi phosphoprotein-3.
Comparison of GOLPH3 protein expression in 2 groups
The immunohistochemical results of GOLPH3 protein expression analysis in the specimens of the observation and control groups showed that GOLPH3 was mainly expressed in the cytoplasm and showed low to no expression in normal colon tissue. The expression of GOLPH3 in colon cancer tissues was increased significantly (P=0.012; Fig. 2). According to the expression of GOLPH3 in colon cancer tissues in the observation group, 29 patients were identified as the GOLPH3-negative group and 69 patients were identified as the GOLPH3-positive group.
Figure 2.
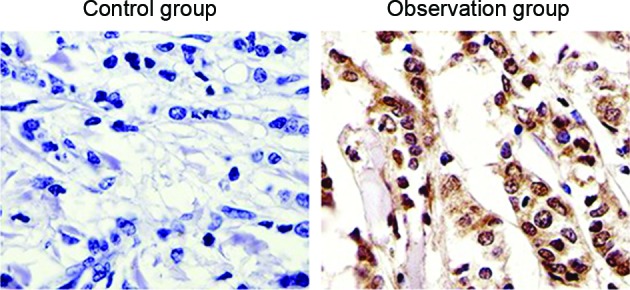
GOLPH3 protein levels in the 2 groups by immunohistochemical analysis (magnification, ×40). The immunohistochemical results of GOLPH3 protein expression analysis showed that GOLPH3 was mainly expressed in the cytoplasm. The expression of GOLPH3 in colon cancer tissues was increased significantly. GOLPH3, Golgi phosphoprotein-3.
Association between GOLPH3 expression and clinical features
As shown in Table I, GOLPH3 protein expression was associated with the clinical stage, histological differentiation degree, lymph node metastasis and infiltration degree (P<0.05; Table I), but GOLPH3 expression was unassociated with gender (P>0.05).
Table I.
Association between clinicopathological features and Golgi phosphoprotein-3 expression level in the observation group.
| Gender | Clinical stages | Tissue types | Lymphatic metastasis | Infiltration degree | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pathological features | Male | Female | II | III | Poorly differentiated | Highly differentiated | Yes | No | T1-T2 | T3-T4 |
| Cases, n | 61 | 37 | 42 | 56 | 53 | 45 | 55 | 43 | 40 | 58 |
| Positive cases, n | 43 | 26 | 23 | 45 | 42 | 27 | 43 | 26 | 22 | 46 |
| Percentage, % | 70.5 | 70.3 | 54.8 | 80.4 | 79.2 | 60 | 78.2 | 60.5 | 55 | 79.3 |
| χ2 | 0.238 | 7.275 | 4.356 | 5.021 | 6.254 | |||||
| P-value | 0.598 | 0.012 | 0.057 | 0.036 | 0.011 | |||||
Comparison of postoperative recurrence and survival rate of patients with different levels of GOLPH3 protein
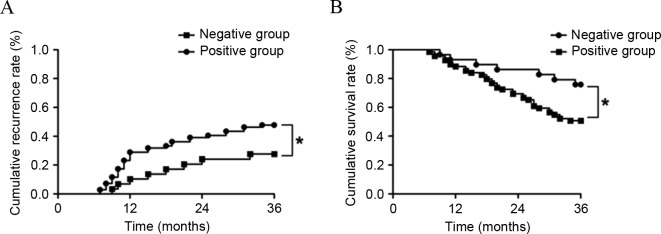
As shown in Fig. 3, the patients were followed up for >36 months subsequent to being treated for 3 months. In total, 8 cases in the GOLPH3-negative group and 34 cases in the GOLPH3-positive group relapsed. Kaplan-Meier curve showed that the cumulative recurrence rates at 1, 2 and 3 years were 10.34 (3/29 patients), 24.14 (7/29 patients) and 27.59% (8/29 patients) in the GOLPH3-negative group. The cumulative recurrence rates at 1, 2 and 3 years were 28.99 (20/69 patients), 39.13 (27/69 patients) and 49.28% (34/69 patients) in the GOLPH3-positive group. Log-rank test showed that the cumulative recurrence rate in the GOLPH3-negative group was significantly lower that of the GOLPH3-positive group (P=0.005). The statistical results of survival rate showed the cumulative survival rates at 1, 2 and 3 years were 93.10 (27 patients), 86.21 (25 patients) and 72.41% (21 patients) in the GOLPH3-negative patients. The cumulative survival rates at 1, 2 and 3 years were 88.41 (61 patients), 69.57 (48 patients) and 50.72% (35 patients) in the GOLPH3-positive patients. The cumulative survival rates at 1, 2 and 3 years in the GOLPH3-negative patients were significantly increased compared with those of GOLPH3-positive patients (P=0.012).
Figure 3.
Comparison of cumulative recurrence and survival rates at 1, 2 and 3 years in GOLPH3-negative and GOLPH3-positive groups. (A) Curve of cumulative recurrence rate. (B) Curve of cumulative survival rate. *P<0.05. GOLPH3, Golgi phosphoprotein-3.
Discussion
The preferential site for colon cancer to develop is at the junction of the rectum and sigmoid colon. Colon cancer is one of the most common types of malignant tumor of the gastrointestinal tract, although the incidence and mortality rates of colon cancer are lower than those of gastric, esophageal and lung cancers. However, with the development of the economy and living style, including a change of dietary structure, colon cancer has become one of the fastest growing types of malignant tumor in China, which has a detrimental effect upon morbidity and mortality rates (17,18). In recent years, with the development of clinical medicine, the treatment success of colon cancer has improved; however, the 5-year survival rate has not been significantly improved for advanced stage colon cancer (19). The mortality rate of colon cancer ranks second for malignant tumors (20). Therefore, additional studies investigating the prognosis and therapies of colon cancer is important in order to improve the survival rate.
A previous study showed that GOLPH3 belonged to the family of Golgi apparatus matrix proteins, and was the first identified example of an oncogene having powerful trafficking ability at the reverse side of Golgi network within yeast and humans (21). Scott et al (22) conducted a genomic analysis on a variety of solid tumors; it was found that GOLPH3 was highly expressed in solid tumors, including melanoma, brain glioma, prostate cancer and non-small cell cancer. The positive rate of GOLPH3 was 56% in non-small cell lung cancer. GOLPH3 was confirmed as an oncogene subsequent to the knockout of RNA interference. GOLPH3 was attached to the N-terminal of glycosyl-acyl transferase, and these proteases were fixed on the Golgi apparatus, to regulate the processing and modification of glycoproteins. GOLPH3 was identified to move between the Golgi apparatus and cell membrane, which enables its involvement in a variety of cellular processes. GOLPH3 overexpression can regulate the signal transduction of various cellular pathways, particularly the protein glycosylation pathway, which may eventually lead to tumorigenesis. Studies have confirmed that the carcinogenic mechanism of GOLPH3 is involved in tumor occurrence and development via mechanistic target of rapamycin complex 1 (mTORC1) and mTORC2 signal transductions (23,24). Therefore, GOLPH3 can be taken as a critical reference factor of tumor prognosis and may be used in new drug screening (25,26). Feng et al (27) studied the expression of GOLPH3 in ovarian carcinoma tissue in 40 cases of ovarian cancer patients. The results showed that the overall expression of GOLPH3 was significantly higher than that of para-carcinoma tissue. In addition, the expression was associated with the patients' surgical pathological stages, histological differentiation, therapeutic response and survival. Yu et al (28) detected the expressions of GOLPH3, caspase-3 and apoptosis index in 62 cases of colon cancer patients. The results showed the positive expression rate of caspase-3 and the apoptosis index in colorectal cancer tissues with high expression of GOLPH3 were significantly decreased compared with colorectal cancer tissues with low expression of GOLPH3, showing that the expression of GOLPH3 in colorectal carcinoma was negatively associated with cell apoptosis.
In the present study, the detection result of GOLPH3 mRNA and protein in colorectal cancer patients showed that the expressions of GOLPH3 mRNA and protein were significantly increased in colon tissue of colon cancer patients, indicating that GOLPH3 gene signal transduction pathway in the colorectal cancer tissue was in an active state, resulting in the increase of GOLPH3 protein. In addition, the current study also revealed that the expression of GOLPH3 in colorectal carcinoma tissue was positively associated with: Clinical stages of disease; histological differentiation degree; lymph node metastasis; and infiltration degree. The cumulative recurrence rates at 1, 2 and 3 years in GOLPH3-positive patients were significantly higher than those of the GOLPH3-negative patients, and the cumulative survival rates were significantly lower than those of the GOLPH3-negative patients. Overall, these results confirmed that GOLPH3 was involved in the occurrence, development, invasion, metastasis and recurrence of colon cancer.
In conclusion, GOLPH3 was overexpressed in colon cancer tissue and was closely associated to clinical pathological features and prognosis, which may be used as a reference for clinical diagnosis and treatment.
References
- 1.Kolligs FT. Diagnostics and Epidemiology of colorectal cancer. Visc Med. 2016;32:158–164. doi: 10.1159/000446488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slattery ML, Pellatt DF, Wolff RK, Lundgreen A. Genes, environment and gene expression in colon tissue: A pathway approach to determining functionality. Int J Mol Epidemiol Genet. 2016;7:45–57. [PMC free article] [PubMed] [Google Scholar]
- 3.Hou TY, Davidson LA, Kim E, Fan YY, Fuentes NR, Triff K, Chapkin RS. Nutrient-Gene interaction in colon cancer, from the membrane to cellular physiology. Annu Rev Nutr. 2016;36:543–570. doi: 10.1146/annurev-nutr-071715-051039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin F, Devesa SS, Chow WH, Zheng W, Ji BT, Fraumeni JF, Jr, Gao YT. Cancer incidence trends in urban shanghai, 1972–1994: An update. Int J Cancer. 1999;83:435–440. doi: 10.1002/(SICI)1097-0215(19991112)83:4<435::AID-IJC1>3.3.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 5.Aoyagi T, Terracina KP, Raza A, Takabe K. Current treatment options for colon cancer peritoneal carcinomatosis. World J Gastroenterol. 2014;20:12493–12500. doi: 10.3748/wjg.v20.i35.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramalingam S, Subramaniam D, Anant S. Manipulating miRNA Expression: A novel approach for colon cancer prevention and chemotherapy. Curr Pharmacol Rep. 2015;1:141–153. doi: 10.1007/s40495-015-0020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia X, Wu W, Zhang K, Cen G, Jiang T, Cao J, Huang K, Huang C, Qiu Z. Prognostic significance of complications after laparoscopic colectomy for colon cancer. PLoS One. 2014;9:e108348. doi: 10.1371/journal.pone.0108348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim CW, Kim WR, Kim HY, Kang J, Hur H, Min BS, Baik SH, Lee KY, Kim NK. Learning curve for single-incision laparoscopic anterior resection for sigmoid colon cancer. J Am Coll Surg. 2015;221:397–403. doi: 10.1016/j.jamcollsurg.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Guo F, Gao X, Wu Y. Golgi phosphoprotein 3 expression predicts poor prognosis in patients with prostate cancer undergoing radical prostatectomy. Mol Med Rep. 2015;12:1298–1304. doi: 10.3892/mmr.2015.3455. [DOI] [PubMed] [Google Scholar]
- 10.Ng MM, Dippold HC, Buschman MD, Noakes CJ, Field SJ. GOLPH3L antagonizes GOLPH3 to determine Golgi morphology. Mol Biol Cell. 2013;24:796–808. doi: 10.1091/mbc.E12-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu L, Chen L, Banfield DK. A conserved N-terminal arginine-motif in GOLPH3-family proteins mediates binding to coatomer. Traffic. 2012;13:1496–1507. doi: 10.1111/j.1600-0854.2012.01403.x. [DOI] [PubMed] [Google Scholar]
- 12.Kunigou O, Nagao H, Kawabata N, Ishidou Y, Nagano S, Maeda S, Komiya S, Setoguchi T. Role of GOLPH3 and GOLPH3L in the proliferation of human rhabdomyosarcoma. Oncol Rep. 2011;26:1337–1342. doi: 10.3892/or.2011.1413. [DOI] [PubMed] [Google Scholar]
- 13.Qiu CZ, Wang MZ, Yu WS, Guo YT, Wang CX, Yang XF. Correlation of GOLPH3 gene with Wnt signaling pathway in human colon cancer cells. J Cancer. 2016;7:928–934. doi: 10.7150/jca.13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan DS. Clinical Oncology. Third. Beijing: Science Press; 1999. p. 215. (In Chinese) [Google Scholar]
- 15.Ionilă M, Mărgăritescu C, Pirici D, Mogoantă SS. Mucinous adenocarcinoma of the colon-a histochemical study. Rom J Morphol Embryol. 2011;52:783–790. [PubMed] [Google Scholar]
- 16.Reuther S, Reiter M, Raabe A, Dikomey E. Effect of irradiation on the expression of DNA repair genes studied in human fibroblasts by real-time qPCR using three methods of reference gene validation. Radiat Environ Biophys. 2013;52:463–469. doi: 10.1007/s00411-013-0482-9. [DOI] [PubMed] [Google Scholar]
- 17.Lavotshkin S, Jalas JR, Torisu-Itakura H, Ozao-Choy J, Lee JH, Sim MS, Stojadinovic A, Wainberg Z, Bifulco CB, Fox BA, Bilchik AJ. Immunoprofiling for prognostic assessment of colon cancer: A novel complement to ultrastaging. J Gastrointest Surg. 2015;19:999–1006. doi: 10.1007/s11605-015-2759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill S. Adjuvant therapy for resected high-risk colon cancer: Current standards and controversies. Indian J Med Paediatr Oncol. 2014;35:197–202. doi: 10.4103/0971-5851.142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zakaria F, El-Mashad N. Inadequate lymph node sampling as a risk factor in stage II colon cancer. Gulf J Oncolog. 2014;1:46–55. [PubMed] [Google Scholar]
- 20.Uddin S, Bavi PP, Hussain AR, Alsbeih G, Al-Sanea N, Abduljabbar A, Ashari LH, Alhomoud S, Al-Dayel F, Ahmed M, Al-Kuraya KS. Leptin receptor expression in Middle Eastern colorectal cancer and its potential clinical implication. Carcinogenesis. 2009;30:1832–1840. doi: 10.1093/carcin/bgp145. [DOI] [PubMed] [Google Scholar]
- 21.Abraham RT. GOLPH3 links the Golgi network to mTOR signaling and human cancer. Pigment Cell Melanoma Res. 2009;22:378–379. doi: 10.1111/j.1755-148X.2009.00596.x. [DOI] [PubMed] [Google Scholar]
- 22.Scott KL, Kabbarall O, Liang MC, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv XL, Tian H, Lu M, Yue WM, Li SH, Gao C, Si LB. The expression of Golgi phosphoprotein 3 (GOLPH3) in non-small cell lung cancer and its clinical significance. Chin J Cancer Biother. 2014;21:554–558. [Google Scholar]
- 24.Isaji T, Im S, Gu W, Wang Y, Hang Q, Lu J, Fukuda T, Hashii N, Takakura D, Kawasaki N, et al. An oncogenic protein Golgi phosphoprotein 3 up-regulates cell migration via sialylation. J Biol Chem. 2014;289:20694–20705. doi: 10.1074/jbc.M113.542688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sechi S, Frappaolo A, Belloni G, Giansanti MG. The roles of the oncoprotein GOLPH3 in contractile ring assembly and membrane trafficking during cytokinesis. Biochem Soc Trans. 2015;43:117–121. doi: 10.1042/BST20140264. [DOI] [PubMed] [Google Scholar]
- 26.Buschman MD, Rahajeng J, Field SJ. GOLPH3 links the Golgi, DNA damage, and cancer. Cancer Res. 2015;75:624–627. doi: 10.1158/0008-5472.CAN-14-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng LP, Su WM. Relationship between Golph3 and prognosis of epithelial ovarian cancer. J Prac Med. 2013;29:243–245. (In Chinese) [Google Scholar]
- 28.Yu WS, Qiu CZ, Wang CX, Wu YY, Huang ZX. Relationship between Golph3 expression and apoptosis in colorectal carcinoma tissues. Chin J Clin Oncol. 2013;40:1094–1097. (In Chinese) [Google Scholar]