Abstract
The present study aimed to investigate the interaction between miR-196 and its target gene homeobox B8 (HoxB8) in colorectal cancer (CRC) cells, and the sensitivity of miR-196 and HoxB8 to fluorouracil, leucovorin and oxaliplatin (FOLFOX4) chemotherapy (1,200 mg/m2 fluorouracil, 200 mg/m2 leucovorin and 85 mg/m2 oxaliplatin). In total, 80 tissue samples were collected in the present study. In total, 50 patients undergoing preoperative chemotherapy completed at least 3 cycles (2 weeks per cycle) of 85 mg/m2 oxaliplatin (day 1) combined with a 2 h injection of 200 mg/m2 leucovorin (days 1 and 2), a bolus injection of 400 mg/m2 and 44 h continuous intravenous infusion of 1,200 mg/m2 fluorouracil. Complete response and partial response were included in the chemotherapy sensitive group (25 patients), and stable disease and progressive disease were included in the chemotherapy resistant group (25 patients). In addition, 30 patients without preoperative chemotherapy were examined for mRNA and protein expression of miR-196 and HoxB8. The expression of the mRNA and protein of miR-196 and HoxB8 was analyzed in 30 CRC and normal mucosa tissue samples. In addition, the expression of the mRNA and protein of miR-196 and HoxB8 was measured in 50 tissue samples obtained from patients that had received FOLFOX4 neoadjuvant chemotherapy. The expression levels of miR-196 and HoxB8 mRNA in CRC tissues were significantly increased compared with the corresponding normal mucosa tissue (P<0.05). The miR-196 mRNA was significantly correlated with lymph node metastasis, tumor stage and distant metastasis (P<0.05). miR-196 was indicated to be negatively correlated with HoxB8 mRNA expression (r=−0.458; P<0.05). The relative amount of miR-196 in the chemotherapy-sensitive group of patients was 0.949±0.691, which was increased compared with the chemotherapy-resistant group (0.345±0.536; P<0.01). The relative level of HoxB8 mRNA in the chemotherapy-sensitive group was 0.490±0.372, which was decreaesd compared with the chemotherapy-resistant group (0.725±0.438; P<0.05). HoxB8 protein expression level in the chemotherapy-sensitive group was decreased compared with the chemotherapy-resistant group (Z=−2.396; P=0.017). Overall, miR-196 was correlated with metastasis and prognosis, and HoxB8 was highly expressed in CRC tissues. The difference in the gene expression of miR-196 and HoxB8 may be associated with the sensitivity to FOLFOX4 for CRC patients. In addition, the highly expressed miR-196 increased the sensitivity of CRC cells to chemotherapy with FOLFOX4 by inhibiting HoxB8.
Keywords: miR-196, homeobox B8, colorectal cancer, neoadjuvant chemotherapy, FOLFOX4
Introduction
Colorectal cancer (CRC) is one of the most common malignant neoplasms worldwide with the third highest morbidity and mortality rates among all neoplasms (1). In the early stages of disease, numerous patients diagnosed with CRC may be successfully treated by surgery, although post-surgery chemotherapy is necessary for patients with locally advanced cancer and distant metastases. At present, fluorouracil, leucovorin and oxaliplatin (FOLFOX4) chemotherapy is one of the more commonly used treatments in patients with locally advanced CRC.
MicroRNAs (miRNAs) are a class of small non-coding RNAs containing ~22 nucleotides, which suppress gene expression post-transcriptionally by base pairing with the 3′-untranslated regions (UTR) of target mRNA (2). Over 700 human miRNAs have been indicated to participate in almost every cellular process investigated, including diverse biological functions and processes, such as development, differentiation, metabolism, growth, proliferation and apoptosis (3). The abnormal expressions of miRNAs are hypothesized to be associated with various human pathologies, including malignant disease, heart and neurodegenerative diseases (4). miRNA-196 (miR-196) is one of the human miRNAs encoded at three paralogous locations in the A, B and C mammalian homeobox (Hox) clusters and has extensive, evolutionarily conserved complementarity to messages of HoxB8, HoxC8 and HoxD8 (5). Previous studies have shown that miR-196 is overexpressed in several tumor tissue samples, including CRC, and has an active role in cancer progression and metastasis, exhibiting tumorigenic (6) or inhibitory effects (7). In addition, increasing numbers of studies indicate that miR-196 is important in the development of the immune system through targeted genes, including the Hox gene family (7–10). The present study aimed to investigate the association between the expression levels of miR-196 and HoxB8 and the clinicopathological features of CRC. In addition, the effect of miR-196 on the target gene HoxB8 at the RNA and protein level was explored. Finally, the difference in the expression levels of miR-196b and HoxB8 between the chemotherapy-sensitive and chemotherapy-resistant groups was measured.
Materials and methods
Patients and chemotherapy
The CRC tissue specimens of 80 patients were collected from patients with CRC at the Department of Colorectal Surgery in the Union Hospital of Fujian Medical University (Fuzhou, China) between November 2008 and March 2012. In addition, normal-appearing mucosae were identified to be directly adjacent to the focal lesions, 2 samples from which were collected for the extraction of RNA and protein. The excised tissues were quickly washed in saline and then immediately placed in liquid nitrogen and stored at −80°C until use. All patients were diagnosed by pathological examination, and the clinical and pathological data are listed in Table I.
Table I.
Patient characteristics.
| Gender, n | Tumor location, n | Stage of disease, n | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Group | Total, n | Age range, years (mean) | Male | Female | Rectum | Colon | II | III | IV |
| Chemotherapy | 50 | 25–82 (58) | 30 | 20 | 20 | 30 | 14 | 7 | 29 |
| Sensitive | 25 | 30–82 (59) | 15 | 10 | 11 | 14 | 6 | 3 | 16 |
| Resistant | 25 | 25–81 (57) | 15 | 10 | 9 | 16 | 8 | 4 | 13 |
| P-value | >0.05a | >0.05b | >0.05c | >0.05d | |||||
Sensitive vs. resistant
Male vs. female
rectum vs. colon
II vs. III vs. IV.
In total, 30 patients did not undergo preoperative chemotherapy. Among these 30 patients, the median age was 64.5 years (20–90 years) and there were 19 men and 11 women. One-half of these 30 patients were aged <60 years, and 10 of the patients had a tumor diameter of >4 cm and 20 patients had tumors <4 cm in diameter. In total, 16 and 14 patients were diagnosed with colon and rectal cancer, respectively, and 18, 8 and 4 of the tumors were classified as ulcerative, protrusive and infiltrating, respectively. Among these 30 tumors, 1 was graded as well-differentiated, one was identified as poorly-differentiated and the other 28 were classified as moderately differentiated tumors. According to the American Joint Committee on Cancer/Union for International Cancer Control tumor-node-metastasis (TNM) staging system in CRC (7th edition) (11), 3 cases of stage I, 7 cases of stage II, 17 cases of stage III and 17 cases of stage IV disease were identified based on the TNM staging. Also, 3 cases of T1 stage, 5 cases of T2 stage, 21 cases of T3 stage and 1 case of T4 stage disease were categorized based on the invasion depth. In addition, 18 cases demonstrated lymph node metastasis and 12 cases were without lymph node metastasis, 3 cases showed distant metastases to the liver and the remaining 27 did not demonstrate distant metastasis.
The remaining 50 specimens obtained from patients that underwent neoadjuvant chemotherapy were quickly placed in 10% formalin fixative and sent to the Department of Pathology for paraffin-embedding. All patients signed the informed consent form and agreed to undergo the FOLFOX4 chemotherapy program. The 50 patients completed at least 3 cycles (2 weeks per cycle) of 85 mg/m2 oxaliplatin (day 1) combined with a 2-h injection of 200 mg/m2 leucovorin (days 1 and 2), a bolus injection of 400 mg/m2 and 44-h continuous intravenous infusion of 1,200 mg/m2 fluorouracil. Routine abdominal and pelvic computed tomography (SOMATOM Sensation 40; Siemens AG, Munich, Germany) or magnetic resonance scans (Magnetom Trio 3.0T; Siemens AG) were performed prior to and subsequent to chemotherapy, and the Response Evaluation Criteria In Solid Tumors was used to assess the response to chemotherapy (12). Complete response and partial response were included in the chemotherapy-sensitive group (25 patients), and stable disease and progressive disease (PD) were included in the chemotherapy-resistant group (25 patients). The two groups have similar demographics, as shown in Table I.
Methods
Reagents and instruments
The kits for extracting total RNA (miRNeasy Mini kit), reverse transcription-polymerase chain reaction (RT-PCR; miScriptReverse Transcription kit) and quantitative fluorescence PCR (miScript SYBR GreenPCR kit) were purchased from Qiagen GmbH (Hilden, Germany). The miR-196, HoxB8 and U6 primers were synthesized and obtained from Shanghai Biotechnology Corporation (Shanghai, China), as shown in Table II. The ProteoPrep® Total Extraction Sample kit and Bicinchoninic Acid (BCA) kit were purchased from GE Healthcare Life Sciences (Chalfont, UK) and Biyuntian Biotech Co., Ltd., (Nanjing, China), respectively. Mouse anti-human HoxB8 polyclonal antibody (#ab55244; dilution, 1:1,000) and mouse anti-β-actin monoclonal antibody (#ab8226; dilution, 1:2,000) were purchased from Abcam (Cambridge, UK). The microplate reader (DU-600; Beckman Coulter, Inc., Brea, CA, USA), real-time PCR instrument (ABI-7500) and image scanner were purchased from Beckman Coulter, Inc. (Brea, CA, USA), Ambion (Thermo Fisher Scientific Inc., Waltham, MA, USA) and GE Healthcare Life Sciences, respectively.
Table II.
Primer sequence of real-time PCR.
| Primer name | Primer sequence |
|---|---|
| miR-196 | TAGGTAGTTTCCTGTTGTTGGG |
| HoxB8-F | ACGTGCTTCTTTGTAATGACCA |
| HoxB8-R | TGTAACAATTGCCCACAGCG |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
Upstream primer of miR-196 was the miScript universal primer of the miScript SYBR Green PCR kit. PCR, polymerase chain reaction; miR, micro RNA; Hox, homeobox; F, forward; R, reverse.
Total RNA extraction
Total RNA, including messenger RNA (mRNA) and miRNA, was extracted from fresh and pathological paraffin tissues following the protocol of the miRNeasy Mini kit. The concentration and purity of RNA were measured at an absorbance (A) value of 280 and 260 nm using a microplate reader (DU-600; Beckman Coulter, Inc.), and the specimens with A260:A280 ratios of 1:1.8 and 1:2.1 were used for subsequent experiments.
RT-PCR
Prior to RT-PCR, 16 µl DNase Booster Buffer (Qiagen GmbH) was added to total RNA extracts, followed by adding 10 µl DNaseI liquid (Qiagen GmbH). Based on the protocol of the miScript Reverse Transcription kit (Takara Biotechnology Co., Ltd., Dalian, China), the RT reaction mixture was prepared (2X miRNA RT buffer, 10 µl; 0.1% bovine serum albumin, 2 µl; miRNA reverse transcriptase mixture, 2 µl; RNA quantification, 0.5 µg; plus RNase-free water up to a 20-µl total volume of reaction solution) with the reaction occurring at 37°C for 60 min, then 85°C for 5 sec. The synthesized complementary DNA (cDNA) were stored at −20°C. RT-PCR was performed on the Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific Inc.). RNAse-free water and DNase Booster Buffer and DNaseI liquid + U6 served as the negative and RT controls, respectively.
Quantitative PCR
According to the quantitative PCR kit (Takara Biotechnology Co., Ltd.), the miR-196 and HoxB8 reaction mixture was prepared using 10 µl SYBR Premix ExTaq II, 0.8 µl F-Primer and 0.8 µl R-Primer, and correction dye was made with 0.4 µl ROX Reference Dye II, 6 µl RNase-free water and 2 µl cDNA in the condition of pre-denaturation at 95°C for 30 sec. Quantitative PCR was performed on the Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific Inc.). The PCR reaction conditions consisted of 95°C for 3 sec and then at 60°C for 30 sec. There was a total of 40 cycles of PCR, with U6 as an internal reference. The average quantification cycle (Cq) was obtained from triplicate using 2−ΔΔCq (13).
HoxB8 protein expression analysis
Fresh tissue samples, including cancer and normal mucosa tissue from 30 patients, were analyzed using western blot analysis. Each sample weighed 50 mg and was ground into powder in liquid nitrogen. Radio-immunoprecipitation assay lysate buffer (1 ml) was added into the powder and the supernatant was transferred to a 1.5 ml Eppendorf tube for 30-min lysis on ice. The protein was obtained from centrifuged supernatant at 4°C for 30 min at 15,000 × g and the concentration was determined with a BCA kit (GE Healthcare Life Sciences). A total of 25 µg of the protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, which was electrotransferred onto hydrophobic polyvinylidene membranes with a pore size of 0.45 µm (EMD Millipore, Billerica, MA, USA), and underwent conventional blocking, primary antibody incubation with mouse monoclonal anti-HoxB8 (dilution, 1:1,000; catalog no. ab55244; Abcam) and mouse monoclonal anti-β-actin (dilution, 1:2,000; catalog no. ab8226; Abcam) antibodies for 30 min at room temperature, secondary goat anti-mouse IgG horseradish peroxidase-conjugated antibody (dilution, 1:500; catalog no., SA101-102; Tiangen Biotechnology Co., Ltd, Beijing, China) incubation for 30 min at room temperature, enhanced chemiluminescence (ECL) color exposure and internal reference to β-actin. The film was scanned by the image scanner (XHCV-3D20; Amersham; GE Healthcare Life Sciences) and pixel intensity value was obtained by Quantity One software (version 4.62; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The expression of HoxB8 protein was shown as the ratio of HoxB8 to β-actin.
The concentration of HoxB8 protein in the 50 samples embedded in paraffin wax was measured using the immunohistochemical streptavidin-biotin complex method (14). Phosphate-buffered saline was used for the negative control and image of the positive control was from GE Healthcare Life Sciences. The following standards were used.
Chemiluminescence detection was performed using an ECL western blotting detection kit (Western Bright ECL kit; Advansta Inc., Menlo Park, CA, USA) and quantified by scanning densitometry. β-actin was used as control reference, and the ratio of HoxB8 to β-actin indicated the HoxB8 expression level. The following criteria were used: The percentage of positive cells for each of the sections and colored shades were scored with semi-quantitative analysis, and the fields were randomly selected from five directions, consisting of up, center, down, left and right, under a high magnification (×400). The color was determined based on intensity score, as follows: 0, no staining; 1, light yellow staining; 2, brown staining; and 3, deep brown staining. The percentage of positive cells was also scored, as follows: 0, <5% stained cells; 1, 5–25% stained cells; 2, 25–50% stained cells; 3, 50–75% stained cells; and 4, >75% stained cells. The mean value was calculated for each case with the aforementioned scoring methods and the final score was obtained by multiplying these two scores. The expression of HoxB8 was qualitatively determined by the final score: 0, for negative (−); 1–3, for weakly positive (+); 4–7, for positive (++); 8–12, for strongly positive (+++). All analyses were performed with the double-blind method (15).
Statistical analysis
Statistical analyses were conducted using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). The data were expressed as the mean ± standard deviation. The paired t-test, Kruskal-Wallis H test and χ2 test were employed to compare the miR-196 mRNA and HoxB8 mRNA expression level in CRC tissues and corresponding normal mucosa tissues. The Mann-Whitney U test was used to analyze the association between the mRNA expression of miR-196 and HoxB8 and the clinicopathological features of CRC and sensitivity to chemotherapy. The correlation was examined by Spearman's rank correlation coefficient analysis. All statistics were based on two-sample paired tests. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression levels of miR-196 mRNA and HoxB8 mRNA
The relative expression levels of miR-196 mRNA and HoxB8 mRNA were 0.873±1.020 and 1.265±1.040 in CRC tissues, respectively, and 0.513±0.262 and 0.889±0.548 in the corresponding normal mucosa tissues, respectively. The expression levels of miR-196 mRNA and HoxB8 mRNA in CRC tissues were significantly increased compared with the normal mucosa tissues (P=0.001), with a negative correlation (r=−0.458; P=0.011). The relative miR-196 expression levels in the chemotherapy-sensitive group and chemotherapy-resistant group were 0.949±0.691 and 0.345±0.536, respectively. miR-196 expression in the chemotherapy-sensitive group was significantly increased compared with the expression in the chemotherapy-resistant group, and the Mann-Whitney U test demonstrated a statistically significant difference between the two groups (Z=−3.172; P=0.002). The relative expression level of HoxB8 mRNA in the chemotherapy-sensitive group (0.490±0.372) was increased compared with the chemotherapy-resistant group (0.725±0.438). In addition, there was a significant difference between the two groups, supported by Mann-Whitney U test (Z=−2.222; P=0.026). In addition, a negative correlation was identified (r=−0.595; P=0.001).
HoxB8 protein expression level
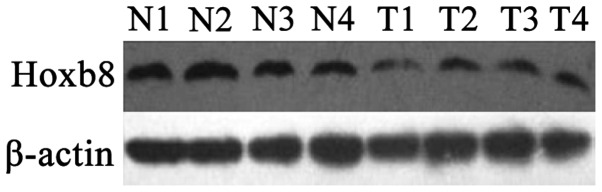
HoxB8 protein was decreased in CRC tissues (0.532±0.07) and highly expressed in the corresponding normal mucosa tissues (0.647±0.04). There was a statistically significant difference between these two groups (P=0.001; Fig. 1). Fig. 2 shows the results of the western blot analysis for the four typical CRC tissue samples and their corresponding normal mucosa tissue samples. There was no significant association between HoxB8 protein and mRNA expression (r=−0.236; P=0.210). HoxB8 protein in CRC tissues was mainly expressed in the cytoplasm (Fig. 3). The Mann-Whitney U test demonstrated that the difference was statistically significant (Z=−2.396; P=0.017) and the positive rate of HoxB8 expression in the chemotherapy-sensitive group was decreased compared with the chemotherapy-resistant group (Table III).
Figure 1.
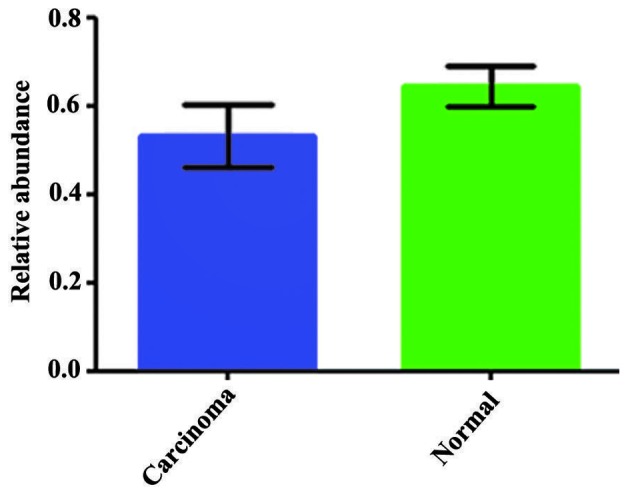
Polymerase chain reaction analysis of homeobox B8 protein expression in colorectal carcinoma and the normal mucosa tissues.
Figure 2.
Western blot analysis of HoxB8 protein expression in colorectal carcinoma and normal mucosa tissues. Hox, homeobox; N, normal mucosa; T, colorectal carcinoma.
Figure 3.
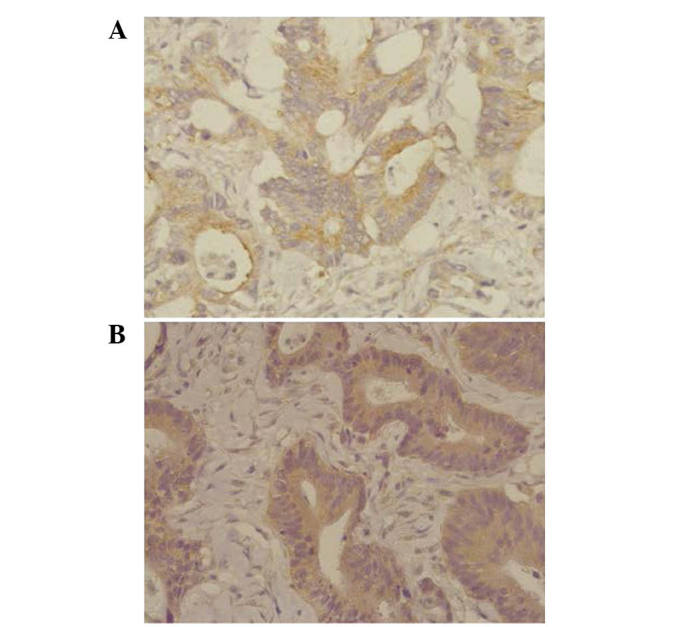
Expression of homeobox B8 protein in colorectal carcinoma tissues (streptavidin-biotin complex method; magnification, ×400). (A) Chemotherapy-sensitive group. (B) Chemotherapy-resistant group.
Table III.
Results of positive rates of HoxB8 protein between the chemotherapy-sensitive and chemotherapy-resistant groups.
| HoxB8 expression, n (%) | |||||||
|---|---|---|---|---|---|---|---|
| Group | Cases, n | − | + | ++ | +++ | Z-value | P-value |
| Sensitive | 25 | 3 (12) | 16 (64) | 4 (16) | 2 (8) | −2.396 | 0.017 |
| Non-sensitive | 25 | 2 (8) | 7 (28) | 13 (52) | 3 (12) | ||
HoxB8, homeobox B8; -, negative expression; +, weakly positive expression; ++ positive expression; +++, strongly positive expression.
Association between miR-196 mRNA and HoxB8 mRNA expression and the clinical pathological features of CRC
As shown in Table IV, miR-196 expression was significantly associated with CRC lymph node metastasis (P=0.003), tumor stage (I+II and III+IV; P=0.003) and distant metastasis (P=0.005), but was not associated with the tumor site (P=0.901), tumor size (P=0.692), gross type (P=0.290), depth of invasion (P=0.542), tissue differentiation (P=0.615), age (P=0.178) or gender (P=0.333). HoxB8 mRNA expression was not associated with any of the aforementioned clinicopathological features.
Table IV.
Association between the mRNA expression of miR-196 and HoxB8 and the clinicopathological features of colorectal cancer.
| Clinicopathological characteristics | n | Expression of miR-196 mRNA | Z-value | P-value | Expression of HoxB8 mRNA | Z-value | P-value |
|---|---|---|---|---|---|---|---|
| Gender | −0.968 | 0.333 | −0.667 | 0.505 | |||
| Male | 19 | 1.551±1.100 | 1.767±1.237 | ||||
| Female | 11 | 2.090±1.311 | 1.403±0.733 | ||||
| Age | −1.348 | 0.178 | −1.597 | 0.110 | |||
| >60 years | 15 | 1.412±1.000 | 1.879±1.205 | ||||
| ≤60 years | 15 | 2.085±1.298 | 1.388±0.914 | ||||
| Tumor diameter, cm | −0.396 | 0.692 | −0.352 | 0.725 | |||
| >4 | 10 | 1.889±1.195 | 1.810±1.206 | ||||
| ≤4 | 20 | 1.678±1.210 | 1.545±1.033 | ||||
| Macroscopic type of tumora | 2.479b | 0.290 | 2.516b | 0.284 | |||
| Ulcerative | 18 | 1.658±1.183 | 1.782±1.175 | ||||
| Protrusive | 8 | 1.489±1.151 | 1.658±1.036 | ||||
| Infiltrative | 4 | 2.634±1.167 | 0.915±0.347 | ||||
| Tissue differentiationa | 0.973b | 0.615 | 0.410b | 0.815 | |||
| High | 1 | 0.704±0.000 | 1.202±0.000 | ||||
| Moderate | 28 | 1.792±1.214 | 1.674±1.107 | ||||
| Low | 1 | 1.595±0.000 | 0.942±0.000 | ||||
| Tumor staging | −2.948 | 0.003 | −1.276 | 0.202 | |||
| I+II | 10 | 0.847±0.349 | 1.854±1.071 | ||||
| III+IV | 20 | 2.199±1.207 | 1.523±1.095 | ||||
| Depth of invasion | −0.610 | 0.542 | −0.516 | 0.606 | |||
| T1+T2 | 8 | 1.635±1.391 | 1.454±0.914 | ||||
| T3+T4 | 22 | 1.790±1.140 | 1.699±1.147 | ||||
| Tumor location | −0.125 | 0.901 | −0.333 | 0.739 | |||
| Rectum | 16 | 1.696±1.133 | 1.609±0.882 | ||||
| Colon | 14 | 1.809±1.290 | 1.661±1.305 | ||||
| Lymph node metastasis | −2.921 | 0.003 | −1.482 | 0.138 | |||
| Yes | 18 | 2.245±1.153 | 1.504±1.132 | ||||
| No | 12 | 1.005±0.816 | 1.827±1.012 | ||||
| Distant metastasis | −2.800 | 0.005 | −0.726 | 0.468 | |||
| Yes | 3 | 4.146±0.758 | 1.220±0.840 | ||||
| No | 27 | 1.482±0.892 | 1.679±1.107 |
Kruskal Wallis H test
χ2. Data are expressed as the mean ± standard deviation. miR, micro RNA; mRNA, messenger RNA; Hox, homeobox; miR, microRNA.
Discussion
miRNA is a class of endogenous gene encoded by ~21–25 nucleotides. miRNAs are small non-coding single stranded RNA molecules involved in a variety of biological processes, including early development, apoptosis, proliferation and tumor occurrence (16,17). For example, miRNA may be combined with a target gene, through complete or partial binding, by forming the RNA-induced silencing complex to induce lysis of the target mRNA or inhibit protein translation (18,19). In addition, miRNA may be involved in the occurrence and evolution of malignancies as oncogenes or tumor suppressor genes (20,21).
miR-196 is located in the upstream of the Hox gene family and has a negative effect on expression of the majority of Hox gene family members, in which the most common target gene is HoxB8 (5,22–24). Previous studies have demonstrated that miR-196 is highly expressed in gastric cancer (25,26) and malignant glioma (27). Schimanski et al (22) indicated that 29% of 7 CRC tissue samples overexpressed miR-196a with quantitative RT-PCR compared with normal colon mucosa. The present study also indicated that miR-196 was upregulated in the cancer tissue samples, indicating the possible participation of miR-196 in the occurrence of CRC. In addition, the expression level of miR-196 was associated with lymph node metastases, staging and distant metastasis, but was not associated with the location, size, gross type, depth of invasion, differentiation, age and gender.
Lymph node metastasis and tumor stage are indicators of cancer invasion, suggesting that miR-196 is involved in cancer metastasis and prognosis. According to the 2014 NCCN treatment guidelines for CRC (28), the patients with distant metastases are recommended to be administered with chemotherapy prior to surgery. However, in the present study, 3 patients with distant metastases, all of which were liver metastases, directly underwent surgery without chemotherapy due to limited budgets. With these 3 samples, the expression level of miR-196 was indicated to be significantly correlated with distant metastasis; this suggests that the overexpression of miR-196 may be associated with a poor prognosis, and that miR-196 may be a prognosis marker for CRC. Certain chemotherapy drugs, including 5-fluorouracil and oxaliplatin, may alter the expression level of miRNAs in CRC. Meng et al (29) indicated that the expression profiles of miRNA changed with the intervention of chemotherapy drugs, and proposed to use miRNA as a predictor for evaluating the chemotherapy drugs. Schimanski et al (22) also identified that the increased concentration of miR-196 may enhance the sensitivity of CRC cell to platinum drugs. The results of the present study showed that the expression levels of miR-196 in patients with CRC decreased following chemotherapy with FOLFOX4. In addition, the expression level of miR-196 in the chemotherapy-sensitivity patient group was increased compared with the chemotherapy-resistant patient group, suggesting an increased sensitivity to FOLFOX4, which was consistent with results of the study by Schimanski et al (22).
HoxB8 is one of the homologous gene family members encoding the DNA-binding domain of the nucleoprotein, which is located in human chromosome 17 (30). HoxB8 is a sequence-specific transcription factor that is important for cell differentiation, growth and organ formation (31). Rawat et al (32) indicated that the deletion of the N-terminal domain in the caudal type homeobox 2 led to the suppression of the Hox gene, which caused abnormal differentiation of hematopoietic stem cells and the development of acute myeloid leukemia in mice.
Vider et al (33) supported that HoxB6, HoxB8, HoxC8 and HoxC9 were abnormally expressed in the various stages of CRC and that HoxB8 is important in the progression of cancer (34). However, Vider et al (33) did not investigate the association with clinicopathological features. In the present study, HoxB8 was also indicated to be highly expressed in the CRC tissues, which is consistent with the findings of Vider et al (33). Furthermore, the present study explored the association between HoxB8 and the clinicopathological features of CRC, and indicated that the expression level of HoxB8 was not associated with lymph node metastasis, staging, distant metastasis, tumor gross type, depth of invasion, differentiation, tumor size, tumor location, age or gender. The preliminary results of the present study demonstrated that HoxB8 was downregulated in the FOLFOX4-sensitivity patient group. Using the expression of HoxB8 to predict the sensitivity to FOLFOX4, the specificity, sensitivity and accuracy of FOLFOX4 were 76.9, 82.4 and 79.7%, respectively. HoxB8 may be used to predict the sensitivity to FOLFOX4 in patients with CRC (35). Furthermore, the present study indicated that the expression level of HoxB8 decreased with FOLFOX4 chemotherapy and that HoxB8 expression was decreased in the FOLFOX4-sensitive patient group, suggesting that an increased level of HoxB8 may decrease the sensitivity to FOLFOX4.
miRNA lyses target mRNA or inhibits the translation of target mRNA by complete or partial binding to the 3′-UTR in the target mRNA, and then downregulates the target mRNA. Kawasaki et al (36) showed that miR-196 inhibited the expression of HoxB8 in HL60 cells by directly lysing the target HoxB8 gene. The present study demonstrated that HoxB8 was highly transcripted in CRC but lowly expressed in the CRC tissues, and that miR-196 was negatively correlated with HoxB8 mRNA. However, miR-196 expression was not associated with the concentration of HoxB8 protein, which suggests that miR-196 regulates its target gene, HoxB8, in the transcription stage. Numerous studies have indicated that the expression pattern of miRNAs has a certain time-sequence and phasic manner, meaning varied expression levels in various tissues and at various development stages (20). A proportional association between miR-196 and HoxB8 may therefore exist. When the ratio between miR-196 and HoxB8 does not reach the threshold, miR-196 completely or partially inhibits the expression of HoxB8 or invokes the development of a tumor (22). Overexpressed miR-196 may cause the development of a tumor as an oncogene, while downregulated miR-196 may suppress the development of tumor by inhibiting its target oncogene, HoxB8.
Due to the varied expression of miR-196 and HoxB8 between the chemotherapy-sensitive and chemotherapy-resistant groups, miR-196 and HoxB8 are hypothesized to aid prediction of the response of the patient to FOLFOX4 chemotherapy for the CRC. In addition, miR-196 and HoxB8 may be used to predict the toxicity of the FOLFOX4 chemotherapy. By measuring the expression of miR-196 and HoxB8, patients that may benefit from FOLFOX4 chemotherapy may be selected and moved towards to personalized treatment. In addition, miR-196 and HoxB8 may be used as targets for drugs, either by inhibiting the expression of HoxB8 to increase the sensitivity to drugs or by indirectly changing the sensitivity through the regulation of upstream miR-196. All these findings lay the foundation of developing novel drugs to treat CRC.
Acknowledgements
The present study was funded by the Natural Science Foundation of China (grant no., 81472777), the Young and Middle-aged Talent Training Project of the health ministry of Fujian Province (grant no., 2013-ZQN-ZD-11), the Ministry of Health National Clinic Key Special Department Constructive Project (grant no., 2012 649), the Natural Science Foundation of Fujian Province (grant no. 2016J01465) and the Youth scientific research fundation of Fujian Health Department (grant no. 2011-1-9).
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson LJ, Ansel KM. MicroRNA regulation of lymphocyte tolerance and autoimmunity. J Clin Invest. 2015;125:2242–2249. doi: 10.1172/JCI78090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 6.Braig S, Mueller DW, Rothhammer T, Bosserhoff AK. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell Mol Life Sci. 2010;67:3535–3548. doi: 10.1007/s00018-010-0394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Zhang M, Chen H, Dong Z, Ganapathy V, Thangaraju M, Huang S. Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer Res. 2010;70:7894–7904. doi: 10.1158/0008-5472.CAN-10-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang G, Han D, Chen X, et al. MiR-196a exerts its oncogenic effect in glioblastoma multiforme by inhibition of IκBα both in vitro and in vivo. Neuro Oncol. 2014;16:652–661. doi: 10.1093/neuonc/not307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang F, Tang J, Zhuang X, Zhuang Y, Cheng W, Chen W, Yao H, Zhang S. MiR-196a promotes pancreatic cancer progression by targeting nuclear factor kappa-B-inhibitor alpha. PLoS One. 2014;9:e87897. doi: 10.1371/journal.pone.0087897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng T, Wang J, Chen X, Liu L. Role of microRNA in anticancer drug resistance. Int J Cancer. 2010;126:2–10. doi: 10.1002/ijc.24782. [DOI] [PubMed] [Google Scholar]
- 11.Edge SB, Compton C. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:143–164. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 12.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Kojima A, Uchida I, Sekizaki T, Sasaki Y, Ogikubo Y, Tamura Y. Rapid detection and identification of Clostridium chauvoei by PCR based on flagellin gene sequence. Vet Microbiol. 2001;78:363–371. doi: 10.1016/S0378-1135(00)00339-4. [DOI] [PubMed] [Google Scholar]
- 14.Qiu JY, Liu P, Shi C, Han B. Low-grade myofibroblastic sarcomas of the maxilla. Oncol Lett. 2015;9:619–625. doi: 10.3892/ol.2014.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salmanidis M, Brumatti G, Narayan N, Green BD, van den Bergen JA, Sandow JJ, Bert AG, Silke N, Sladic R, Puthalakath H, et al. Hoxb8 regulates expression of microRNAs to control cell death and differentiation. Cell Death Differ. 2013;20:1370–1380. doi: 10.1038/cdd.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blenkiron C, Miska EA. miRNAs in cancer: Approaches, aetiology, diagnostics and therapy. Hum Mol Genet. 2007;16:R106–R113. doi: 10.1093/hmg/ddm056. [DOI] [PubMed] [Google Scholar]
- 17.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 18.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 19.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 20.Akao Y, Nakagawa Y, Naoe T. Let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 21.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 22.Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, Moehler M, Gockel I. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089–2096. doi: 10.3748/wjg.15.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chopra VS, Mishra RK. ‘Mir’acles in hox gene regulation. Bioessays. 2006;28:445–448. doi: 10.1002/bies.20401. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Z, Boyle TJ, Liu Z, Murray JI, Wood WB, Waterston RH. A negative regulatory loop between microRNA and Hox gene controls posterior identities in Caenorhabditis elegans. PLoS Genet. 2010;6:e1001089. doi: 10.1371/journal.pgen.1001089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai KW, Hu LY, Wu CW, Li SC, Lai CH, Kao HW, Fang WL, Lin WC. Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosomes Cancer. 2010;49:969–980. doi: 10.1002/gcc.20804. [DOI] [PubMed] [Google Scholar]
- 26.Liao YL, Hu LY, Tsai KW, Wu CW, Chan WC, Li SC, Lai CH, Ho MR, Fang WL, Huang KH, Lin WC. Transcriptional regulation of miR-196b by ETS2 in gastric cancer cells. Carcinogenesis. 2012;33:760–769. doi: 10.1093/carcin/bgs023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan Y, Mizoguchi M, Yoshimoto K, Hata N, Shono T, Suzuki SO, Araki Y, Kuga D, Nakamizo A, Amano T, et al. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin Cancer Res. 2010;16:4289–4297. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 28.National Comprehensive Cancer Network, corp. http://www.nccn.org/professionals/physician_gls/f_guidelines.asp#colon. [Aug 2;2014 ];Clinical Practice Guidelines in Oncology.
- 29.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 30.Yue Y, Farcas R, Thiel G, Bommer C, Grossmann B, Galetzka D, Kelbova C, Küpferling P, Daser A, Zechner U, Haaf T. De novo t(12;17) (p13.3;q21.3) translocation with a breakpoint near the 5′end of the HOXB gene cluster in a patient with developmental delay and skeletal malformations. Eur J Hum Genet. 2007;15:570–577. doi: 10.1038/sj.ejhg.5201795. [DOI] [PubMed] [Google Scholar]
- 31.Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 32.Rawat VP, Thoene S, Naidu VM, Arseni N, Heilmeier B, Metzeler K, Petropoulos K, Deshpande A, Quintanilla-Martinez L, Bohlander SK, et al. Overexpression of CDX2 perturbs HOX gene expression in murine progenitors depending on its N-terminal domain and is closely correlated with deregulated HOX gene expression in human acute myeloid leukemia. Blood. 2008;111:309–319. doi: 10.1182/blood-2007-04-085407. [DOI] [PubMed] [Google Scholar]
- 33.Vider BZ, Zimber A, Hirsch D, Estlein D, Chastre E, Prevot S, Gespach C, Yaniv A, Gazit A. Human colorectal carcinogenesis is associated with deregulation of homeobox gene expression. Biochem Biophys Res Commun. 1997;232:742–748. doi: 10.1006/bbrc.1997.6364. [DOI] [PubMed] [Google Scholar]
- 34.Vider BZ, Zimber A, Chastre E, Gespach C, Halperin M, Mashiah P, Yaniv A, Gazit A. Deregulated expression of homeobox-containing genes, HOXB6, B8, C8, C9 and Cdx-1, in human colon cancer cell lines. Biochem Biophys Res Commun. 2000;272:513–518. doi: 10.1006/bbrc.2000.2804. [DOI] [PubMed] [Google Scholar]
- 35.Lu X, Pan J, Li S, Shen S, Chi P, Lin H, Huang Y, Xu Z, Huang S. Establishment of a predictive genetic model for estimating chemotherapy sensitivity of colorectal cancer with synchronous liver metastasis. Cancer Biother Radiopharm. 2013;28:552–558. doi: 10.1089/cbr.2012.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawasaki H, Taira K. MicroRNA-196 inhibits HOXB8 expression in myeloid differentiation of HL60 cells. Nucleic Acids Symposium. 2004;48:211–212. doi: 10.1093/nass/48.1.211. [DOI] [PubMed] [Google Scholar]