Abstract
Previous studies have demonstrated that microRNA-21 (miR-21) is an oncogene and is significantly upregulated in tumor tissue. However, its association with T-cell acute lymphoblastic lymphoma/leukemia (T-ALL) remains poorly understood. The aim of the present study was to investigate the effects of miR-21 on T-ALL cells by constructing Jurkat cells infected with recombinant adenovirus adv-miR-21 or adv-anti-miR-21. In addition, the target gene of miR-21 was identified by reverse transcription-quantitative polymerase chain reaction and western blotting. The results demonstrated that miR-21 expression in Jurkat cells was markedly upregulated. Furthermore, upregulating miR-21 expression in Jurkat cells promoted cell proliferation and invasion and decreased the apoptosis rate. By contrast, knockdown of miR-21 in Jurkat cells suppressed proliferation and invasion and increased the apoptosis rate. Furthermore, the results indicated that signal transducer and activator of transcription (STAT) 3 was targeted by miR-21, and that miR-21 inhibited STAT3 expression at the protein level rather than at the messenger RNA level. In conclusion, targeting the inhibition of miR-21 may be a novel therapeutic strategy for patients with T-ALL.
Keywords: T-cell acute lymphoblastic lymphoma/leukemia, microRNA-21, signal transducer and activator of transcription 3, target gene, feedback
Introduction
Acute lymphoblastic leukemia (ALL) is an aggressive hematological tumor characterized by the overproduction of lymphoblasts (immature lymphoid cells) in the peripheral blood, bone marrow and other organs, including the lymph nodes and spleen (1). T-cell ALL (T-ALL) is a subtype of ALL based on immunophenotype (1,2). It accounts for ~12% of cases of ALL in children, whereas it constitutes ~25% of cases in adult patients with ALL (2,3). Generally speaking, patients with T-ALL have a poorer prognosis than other ALL patients, with an increased risk of experiencing early relapse, induction failure and isolated central nervous system relapse (4). Therefore, it is essential to identify efficient markers and novel clinical treatment targets of ALL to enhance the efficacy of existing regimens while reducing their toxic side effects in order to improve the prognosis of patients with ALL.
MicroRNAs (miRNAs or miRs) are single-stranded noncoding regulatory RNA molecules (5) that regulate gene expression by binding to the 3′-untranslated regions (UTR) of messenger RNA (mRNA) transcripts and repressing the translation or degrading the target mRNA (6). miRNAs regulate cell differentiation, proliferation and apoptosis, and in this way they influence the occurrence and development of inflammation, endocrine diseases and tumors (5). It has been demonstrated that specific miRNAs act as oncogenes or anti-oncogenes by regulating cellular pathways (7). Furthermore, previous studies have suggested that miR-21 is an oncogenic miRNA that is significantly upregulated in tumor tissue, peripheral blood and particular body fluids (such as the cerebrospinal fluid) of patients with malignancies, including lung, hepatic and breast cancer, as well as B-cell and central nervous system lymphoma (8–11). miR-21 binds to the 3′-UTR region of target genes, including programmed cell death 4 (PDCD4), phosphatase and tensin homolog (PTEN) and reversion inducing cysteine rich protein with kazal motifs (RECK), thus promoting proliferation and invasion whilst inhibiting apoptosis (12–14). However, the expression and biological function of miR-21 in T-ALL remains poorly understood. In the present study, levels of miR-21 expression were measured in a T-ALL cell line (Jurkat) to investigate the mechanism of miR-21 oncogenic function. Jurkat cells were infected with recombinant adenovirus adv-miR-21 or adv-anti-miR-21 and the effects of miR-21 expression and knockout on cell proliferation, invasion and apoptosis were explored.
Signal transducer and activator of transcription (STAT) 3 is involved in a number of cellular functions. It regulates genes associated with cell growth, movement and apoptosis, and is overactive in various types of cancer, including breast, pancreatic and prostate cancer, as well as leukemia and lymphoma (15). Evidence suggests that miRNAs are closely associated with the STAT3 signaling pathway. It has been demonstrated that miR-21 is involved in STAT3-mediated tumorigenesis, and that the STAT3 protein regulates the expression of miR-21 by binding to the STAT3-binding site within the promoter of the miR-21 gene on chromosome 17q23.2 (16,17). Furthermore, Fujita et al (18) identified a feedback mechanism between nuclear factor (NF)-IB and miR-21. Previous studies using TargetScan and PicTar (16,17) suggested that STAT3 may be a target of miR-21. Therefore, the present study investigated whether a feedback loop between STAT3 and miR-21 exists. To verify this hypothesis, a luciferase reporter assay, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting were performed.
Materials and methods
Cell culture
Cells from the human Jurkat cell line (obtained from the Committee on Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China) were cultured in RPMI 1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Biological Industries, Beit-Haemek, Israel). Human kidney (HEK) 293A and 293T cell lines (both obtained from the Committee on Type Culture Collection of the Chinese Academy of Sciences) were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) containing 10% FBS. Cells from the Jurkat, HEK 293A and 293T cell lines were kept in the laboratory and incubated in cell culture incubators (Sanyo Electric, Co., Ltd., Osaka, Japan) at 37°C with 5% CO2.
Construction of recombinant adenovirus
High expression of miRNA-21 by recombinant vector pAV-MCMV-EGFP-3FLAG-miRNA-21 (Shanghai Sunbio Technology Co., Ltd, Shanghai, China) and knockdown of miR-21 by recombinant vector pAVsiRNA1.1-anti-miR-21 (Shanghai Sunbio Technology Co., Ltd.) were successfully completed. The adenovirus expression plasmid and the adenovirus backbone plasmid underwent co-transfection in 293A cells using Trans-EZ (Shanghai Sunbio Technology Co., Ltd.) to package adv-miR-21, adv-miR-21-control, adv-anti-miR-21 and adv-anti-miR-21-control. Virus titer was determined by Titer-EZ adenoviral titer detection reagent (Shanghai Sunbio Technology Co., Ltd.).
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
To compare the expression of miR-21 in Jurkat cells with normal lymphocytes, peripheral blood samples were obtained from patients at the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China) from July 2013 to October 2013, once written informed consent was obtained, in accordance with the Declaration of Helsinki. Experiments conformed to regulatory standards and were approved by the Zhengzhou University Life Science Ethics Review Committee (Zhengzhou, China). Lymphocytes from normal human peripheral blood were isolated using the Ficoll-Paque® Plus medium (GE Healthcare Life Sciences, Chalfont, UK). Jurkat cells were infected with recombinant adenovirus, adv-miR-21, adv-miR-21-control, adv-anti-miR-21 or adv-anti-miR-21-control, and collected 48 h after infection. Total RNA was extracted from Jurkat cells and normal lymphocytes using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), and RT-qPCR was performed as described previously (8). The sequences of the primers (synthesized by Genewiz, Inc., South Plainfield, NJ, USA) are presented in Table I. U6 (with a stem-loop primer) was used as reference gene. Each sample was analyzed in triplicate. The ΔΔCq values were calculated following normalization to reference U6 RNA and quantified using the 2−ΔΔCq method (9).
Table I.
Primers for miR-21.
| Primers | Sequence (5′→3′) |
|---|---|
| miR21-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAACATC |
| miR21-qPCR | Forward: TAGCTTATCAGACTGA |
| Reverse: TGGCAGGGTCCGAGGT | |
| U6-RT | TCGTATCCATGGCAGGGTCCGAGGTATTCGCCATGGATACGACACAAAAATATGGAACGCTT |
| U6-qPCR | Forward: GTGCTCGCTTCGGCAGCACA |
| Reverse: TGGCAGGGTCCGAGGT |
miR-21, microRNA-21; RT, reverse transcription; qPCR, quantitative polymerase chain reaction.
Cell proliferation assay
Cell proliferation in vitro was measured using a CCK-8 Cell Counting kit (Vazyme, Piscataway, NJ, USA). Jurkat cells were infected with recombinant adenovirus for 24, 48, 72 and 96 h in a 96-well plate. A total of 10 µl CCK-8 was added to each well, and samples were incubated for a further 4 h in a cell culture incubator. Optical density (OD) values were measured using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) at 450 nm. Each experiment was performed in triplicate. Cell viability was calculated by the following formula: Cell viability = (dosing OD-blank OD / control cells OD-blank OD) ×100%.
In vitro invasion assay
For the in vitro invasion assay, Jurkat cells were infected with recombinant adenovirus. Cells were seeded on a Matrigel™-coated membrane matrix (BD Biosciences, Franklin Lakes, NJ, USA) in the insert of a 24-well Transwell culture plate fitted with polycarbonate filters (8-µm pore size; Costar; Corning Incorporated, Corning, NY, USA) 72 h following infection. Cells in the upper chamber were cultured in RPMI 1640 medium without FBS, while 10% FBS was added to the lower chamber. After 24 h, migrating cells located in the lower chamber were counted directly under a microscope (Olympus Corporation, Tokyo, Japan). The invasive ability of Jurkat cells was determined by the number of cells observed migrating into the lower chamber.
Flow cytometric analysis
A PE Annexin V Apoptosis Detection kit I (BD Biosciences) was used to detect the apoptosis rate of Jurkat cells. Jurkat cells were infected with recombinant adenovirus for 96 h. Following the collection and resuspension of 1×105 cells using 100 µl 1X Binding Buffer, 5 µl PE Annexin V and 5 µl 7-aminoactinomycin D were added to each sample for 15 min at room temperature and a further 400 µl of 1X Binding Buffer was added to each system. Stained samples were analyzed within 1 h by flow cytometry (LSR II; BD Biosciences).
Prediction of target genes of miR-21
TargetScan (www.targetscan.org) and PicTar (pictar.mdc-berlin.de) were used to identify possible target genes of miR-21. Among the same target genes that the two prediction software identified, the target genes of different scores were filtered according to the conservation and the integrated score of the base distribution of the 3′-UTR of the target genes. Finally, STAT3 was selected as the target gene of miR-21 that was associated with T-ALL (Fig. 1).
Figure 1.
miR-21 targets STAT3 by binding to the 3′-UTR region of STAT3. miR-21, microRNA-21; STAT3, signal transducer and activator of transcription; UTR, untranslated region; F, forward; R, reverse; hsa, Homo sapiens.
Dual luciferase reporter assay
A dual luciferase reporter assay was performed to validate that STAT3 was the target gene of miR-21. The 3′-UTR fragment, which contained the binding site of miR-21 to STAT3 mRNA, was cloned, as well as a mutant fragment that lacked the binding site. Then, either the 3′-UTR fragment or the mutant fragment was recombined with the pmir-GLO Vector (Promega Corporation, Madison, WI, USA), consisting of the Renilla luciferase (hRluc-neo) gene and a firefly luciferase gene in the XhoI site, using the ClonExpress® MultiS One Step Cloning kit (Vazyme). For the reporter assays, HEK 293T cells were seeded onto 24-well plates. The recombinant pmir-GLO-3′-UTR vector or the pmir-GLO-mutant-3′-UTR vector were transfected into HEK 293T cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, 6 h following transfection, either adv-miR-21 or adv-miR-21-control were added to the HEK 293T cells. Following 48 h of cultivation, firefly and Renilla luciferase activities were detected using the Dual-Glo®Luciferase Assay System (Promega Corporation) and a MicroLumatPlus LB96V luminometer (Berthold Technologies GmbH & Co. KG, Bad Wildbad, Germany). Firefly luciferase values were normalized to Renilla luciferase activity, and the ratio of firefly/Renilla luciferase activity was presented. Primers for STAT3-3′-UTR-wild type or mutant fragment are presented in Table II.
Table II.
Primers for STAT3-3′-UTR-wild type or mutant fragments.
| Primer | Sequence (5′→3′) |
|---|---|
| STAT3-3′-UTR fragment | F: TTGGCTGGCTAGCTCGCCTCT |
| R: CTTATAGGTAGGTAAGCAACC | |
| STAT3-3′-UTR-wild type | F: GTTGTTTAAACGAGCTCGTGGCAACTCAAAACCACC |
| R: GGTCGACTCTAGACTCGAGGGGCTCAGCTCCTCTCAG | |
| STAT3-3′-UTR-mutation | |
| Fragment 1 | F: GTTGTTTAAACGAGCTCGTGGCAACTCAAAACCACC |
| R: TTCTGTTTATCAGTTATATGTACTGAAGAGT | |
| Fragment 2 | F: ACTCTTCAGTACATATAACTGATAAACAGAA |
| R: GGTCGACTCTAGACTCGAGGGGCTCAGCTCCTCTCAG |
F, forward; R, reverse; UTR, untranslated region; STAT3, signal transducer and activator of transcription.
STAT3 mRNA quantification by RT-qPCR
Normal lymphocytes, Jurkat cells and Jurkat cells infected with adv-miR-21-control or adv-miR-21 were collected 48 h following infection. Total RNA was extracted with TRIzol and subjected to RT. In the RT step, TransScript First-Strand cDNA Synthesis Supermix (Beijing Transgen Biotech Co., Ltd., Beijing, China) was used to synthesize the complementary DNA of STAT3, and the cycling conditions were as follows: 5 min at 25°C, 60 min at 42°C, 5 min at 85°C, and hold at 4°C. STAT3 mRNA qPCR was performed as described previously (19). The expression of STAT3 mRNA was normalized to the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA. The sequences of the primers (synthesized by Genewiz, Inc.) are presented in Table III.
Table III.
Primers for STAT3 messenger RNA quantification by qPCR.
| Primers | Sequence (5′→3′) |
|---|---|
| STAT3-qPCR | F: GGAGGCATTCGGAAAGTATTGTCG |
| R: ATGGTATTGCTGCAGGTCGTTGGT | |
| GAPDH-qPCR | F: AGAAGGCTGGGGCTCATTTG |
| R: AGGGGCCATCCACAGTCTTC |
qPCR, quantitative polymerase chain reaction; F, forward; R, reverse; STAT3, signal transducer and activator of transcription; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Western blot assay
Firstly, Jurkat cells were harvested following infection with adv-miR-21-control or adv-miR-21 for 72 h. Jurkat cells and normal lymphocytes were subsequently lysed with radioimmunoprecipitation assay lysis buffer (Beijing Dingguo Changsheng, Biotechnology Co., Ltd., Beijing, China), and total protein was extracted. Protein concentrations were measured by a BCA Protein Assay kit (Beijing Dingguo Changsheng, Biotechnology Co., Ltd.), according to a standardized curve. Equal protein amounts were separated on 10% Tris-glycine sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). Membranes were blocked with 5% nonfat dried milk for 2 h at room temperature and incubated with antibodies against STAT3 (1:2,000; catalogue number 12640; Cell Signaling Technology, Inc., Danvers, MA, USA) and GAPDH (1:3,000; catalogue number sc-47724; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. The quantification of western blotting was performed using image processing and analysis software (Fusion SL; Vilber Lourmat Deutschland GmbH, Eberhardzell, Germany), and the relative expression of STAT3 protein was normalized to GAPDH.
Statistical analysis
All experiments were performed three times. Data were processed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA) and presented as the mean ± standard deviation. Statistical analysis was conducted using Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-21 in Jurkat cells, normal lymphocytes and Jurkat cells infected with recombinant adenovirus
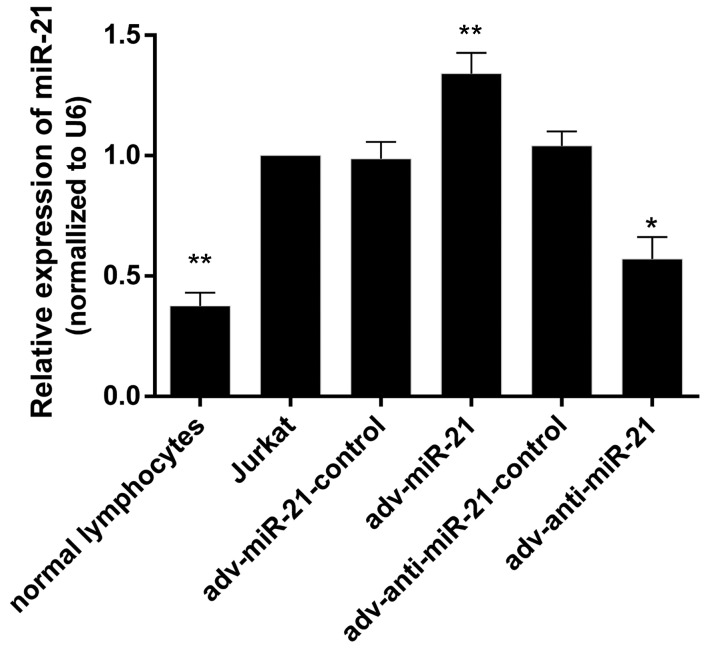
RT-qPCR was performed to detect miR-21 levels in normal lymphocytes, Jurkat cells, and Jurkat cells infected with adv-miR-21-control, adv-miR-21, adv-anti-miR-21-control or adv-anti-miR-21. The results indicated that the expression levels of miR-21 were 0.37±0.03, 1.00±0.00, 0.98±0.07, 1.34±0.86, 1.04±0.06 and 0.57±0.09 in normal lymphocytes, Jurkat cells, and Jurkat cells infected with adv-miR-21-control, adv-miR-21, adv-anti-miR-21-control and adv-anti-miR-21, respectively. The expression of miR-21 in Jurkat cells was significantly higher than in normal lymphocytes (P<0.01), and miR-21 levels were significantly upregulated or downregulated in Jurkat cells infected with adv-miR-21 (P<0.01) or adv-anti-miR-21 (P<0.05), respectively, compared with normal Jurkat cells (Fig. 2). These results indicated that high expression of miR-21 and knockdown of miR-21 by recombinant adenovirus successfully occurred.
Figure 2.
Relative expression of miR-21 in Jurkat cells, normal lymphocytes and Jurkat cells infected with recombinant adenovirus. The expression of miR-21 in Jurkat cells was significantly higher (**P<0.01) than in normal lymphocytes. miR-21 levels in the adv-miR-21 group were significantly upregulated (**P<0.01) compared with the adv-miR-21-control group, while miR-21 levels in the adv-anti-miR-21 group were significantly downregulated (*P<0.05) compared with the adv-anti-miR-21-control group. miR-21, microRNA-21; adv, adenovirus.
Upregulation of miR-21 in Jurkat cells promotes proliferation, while knockdown of miR-21 in Jurkat cells inhibits proliferation
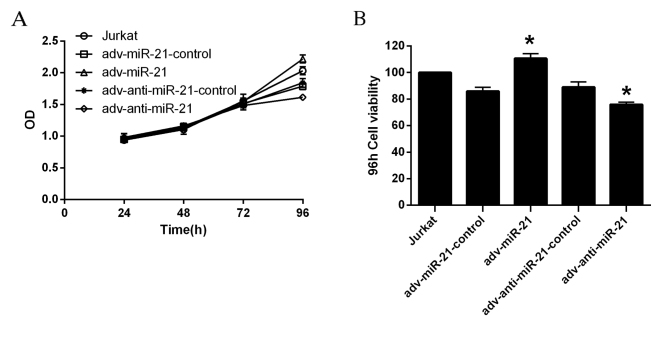
According to the OD values obtained, the proliferation of Jurkat cells infected with adv-miR-21 or adv-anti-miR-21 was significantly different to that observed in the relevant control group 96 h after infection, while the differences were not significant at 24, 48 or 72 h (Fig. 3A). The viability of Jurkat cells was calculated by the formula: Cell viability=(dosing OD-blank OD/control cells OD-blank OD)x100%. As demonstrated in Fig. 3B, the viability of Jurkat cells infected with adv-miR-21 significantly increased to 128.80% after 96 h compared with ~90% viability in the adv-miR-21 control. However, the viability of Jurkat cells infected with adv-anti-miR-21 was significantly inhibited to 85.33% compared with ~90% viability in adv-anti-miR-21-control after 96 h. Therefore, upregulation of miR-21 in Jurkat cells promoted proliferation, while knockdown of miR-21 in Jurkat cells inhibited proliferation.
Figure 3.
Effect of miR-21 on the proliferation of Jurkat cells. (A) OD values and (B) viability of Jurkat cells at 96 h after infection with recombinant adenovirus. The viability of Jurkat cells infected with adv-miR-21 increased to 128.80% at 96 h, which was significantly higher than the adv-miR-21-control (*P<0.05); the viability of Jurkat cells infected with adv-anti-miR-21 decreased to 85.33% at 96 h, which was significantly lower compared with the adv-anti-miR-21-control (*P<0.05). miR-21, microRNA-21; adv, adenovirus; OD, optical density.
In vitro invasion assay
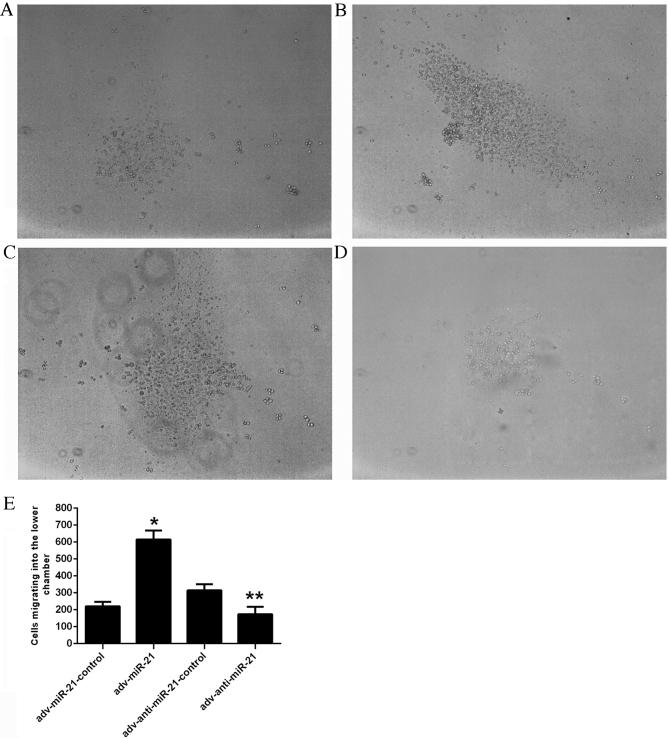
An in vitro Matrigel Transwell analysis was applied to determine the influence of miR-21 on the invasive ability of Jurkat cells. Due to the characteristics of suspension cells, Jurkat cells migrating into the lower chamber floated in the culture medium rather than attaching to the membrane of the Transwell. The number of cells migrating into the lower chamber was 220.11±26.78 in the adv-miR-21-control group, 615.33±52.20 in the adv-miR-21 group, 314.00±36.70 in the adv-anti-miR-21-control group and 173.89±43.50 in the adv-anti-miR-21 group (Fig. 4E). A higher proportion of Jurkat cells infected with adv-miR-21 migrated into the lower chamber than the adv-miR-21-control group (P<0.001), while fewer Jurkat cells infected with adv-anti-miR-21 migrated into the lower chamber compared with the adv-anti-miR-21-control group (P<0.01; Fig. 4). These results indicated that upregulating miR-21 expression in Jurkat cells significantly promoted invasive ability, whereas downregulating miR-21 expression significantly inhibited invasive ability.
Figure 4.
Effect of miR-21 on the invasive ability of Jurkat cells. Jurkat cells infected with different recombinant adenoviruses (A) adv-miR-21-control, (B) adv-miR-21, (C) adv-anti-miR-21-control and (D) adv-anti-miR-21 migrated into the lower chamber. (E) Jurkat cells infected with adv-miR-21 exhibited an increased invasive ability compared with the adv-miR-21-control group (*P<0.05), whereas the invasive ability of Jurkat cells infected with adv-anti-miR-21 was significantly inhibited compared with the adv-anti-miR-21-control group (**P<0.01). miR-21, microRNA-21; adv, adenovirus.
miR-21 influences the apoptosis of Jurkat cells
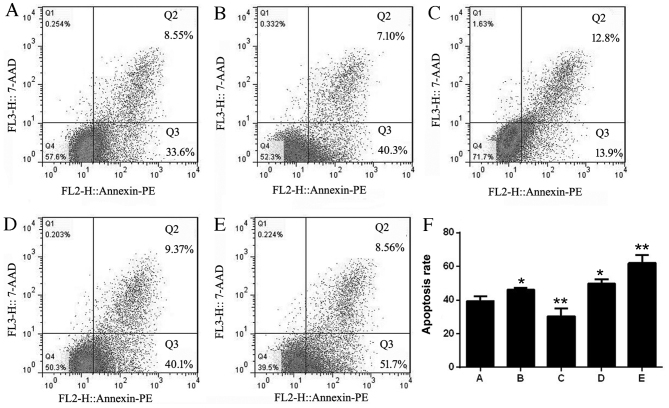
The apoptosis rates of each group were 39.42±2.90 (non-infected Jurkat cells), 46.20±1.18 (adv-miR-21-control), 30.36±4.71 (adv-miR-21), 49.62±2.77 (adv-anti-miR-21-control) and 61.96±4.75 (adv-anti-miR-21). The results of flow cytometry analysis indicated that the apoptosis rates of Jurkat cells infected with adv-miR-21-control or adv-anti-miR-21-control were upregulated compared with control Jurkat cells, indicating that the adenovirus affected the apoptosis of Jurkat cells. However, compared with the adv-miR-21-control group, there was a significant decrease in the apoptosis rate of Jurkat cells infected with adv-miR-21. In addition, a significant increased rate of apoptosis was also detected in Jurkat cells infected with adv-anti-miR-21 compared with the adv-anti-miR-21-control group (Fig. 5). These results demonstrated that upregulating miR-21 expression in Jurkat cells inhibited apoptosis, while downregulating miR-21 promoted apoptosis.
Figure 5.
Effect of miR-21 on the apoptosis of Jurkat cells. Flow cytometry analysis of (A) Jurkat cells, (B) adv-miR-21-control, (C) adv-miR-21, (D) adv-anti-miR-21-control, and (E) adv-anti-miR-21. (F) The apoptosis rate of Jurkat cells increased in cells infected with adv-miR-21-control and adv-anti-miR-21-control compared with non infected Jurkat cells (*P<0.05). The apoptosis rate of the adv-miR-21 group was significantly inhibited compared with the adv-miR-21-control (**P<0.01) and the adv-anti-miR-21-control groups, indicating that downregulating miR-21 in Jurkat cells significantly increased apoptosis (**P<0.01). miR-21, microRNA 21; adv, adenovirus; PE, phycoerythrin; 7-AAD, 7-aminoactinomycin D.
STAT3 is a direct target gene of miR-21
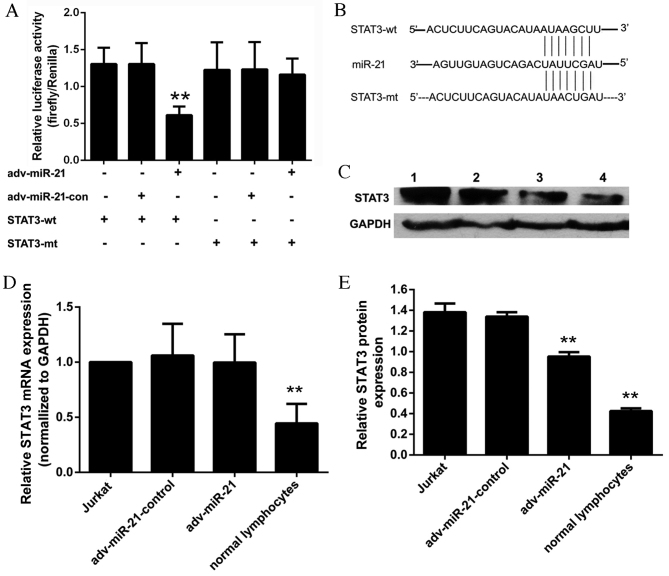
STAT3 was selected as the predicted target gene of miR-21 associated with T-ALL. The results of the dual luciferase reporter assay (Fig. 6A) indicated that the relative luciferase activity of the pmir-GLO-STAT3-3′-UTR-wild-type (STAT3-wt) reporter was significantly inhibited in the adv-miR-21 group (0.61±0.12) compared with the adv-miR-21-control group (1.31±0.28). However, the relative luciferase activity of the pmir-GLO-STAT3-3′-UTR-mutant (STAT3-mt) reporter (1.22±0.37) was almost the same as the STAT3-mt-adv-miR-21-control group (1.23±0.37) and the STAT3-mt-adv-miR-21 group (1.16±0.22). These results suggested that miR-21 targets STAT3 by binding to a specific region of the 3′-UTR.
Figure 6.
Effect of miR-21 on STAT3 in Jurkat cells. (A) The relative luciferase activity of the STAT3-wt reporter was significantly inhibited in the adv-miR-21 group compared with the control group (**P<0.01). However, the relative luciferase activity of the STAT3-mt reporter was almost the same as the control group (P=0.690). (B) Sequence of the 3′-UTR fragment that contained the binding site to miR-21 of STAT3 mRNA and a mutant 3′-UTR fragment that lacked the binding site to miR-21. (C) Lanes 1–4 represent the results from a western blot analysis of Jurkat cells, adv-miR-21-control, adv-miR-21 and normal lymphocytes, respectively. The relative STAT3 protein expression normalized to GAPDH protein in normal lymphocytes was significantly lower than that in Jurkat cells (**P<0.01). (D) The relative STAT3 mRNA expression normalized to GAPDH mRNA in normal lymphocytes was significantly lower than that in Jurkat cells (**P<0.01); however, there was no significant difference in relative STAT3 mRNA levels between the adv-miR-21 group and the adv-miR-21-control group (P=0.79). (E) The relative STAT3 protein expression in Jurkat cells infected with adv-miR-21 was significantly lower than that in the adv-miR-21-control group (**P<0.01). Wt, wild type; mt, mutant; miR-21, microRNA-21; con, control; adv, adenovirus; STAT3, signal transducer and activator of transcription; UTR, untranslated region; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; mRNA, messenger RNA.
miR-21 regulates STAT3 by inhibiting translation
Western blot analysis and qPCR were applied to evaluate the effects of miR-21 on STAT3. The relative STAT3 mRNA expression of the Jurkat cell, adv-miR-21-control, adv-miR-21 and normal lymphocyte groups were 1.00±0.00, 1.06±0.29, 0.99±0.27 and 0.44±0.18, respectively (Fig. 6D). The results of western blot analysis demonstrated that the relative grayscale rates of the Jurkat cells, adv-miR-21-control, adv-miR-21 and normal lymphocytes groups were 1.38±0.08, 1.33±0.04, 0.95±0.04 and 0.42±0.02, respectively (Fig. 6E). Furthermore, the results indicated that the levels of STAT3 mRNA (Fig. 6D) and STAT3 protein (Fig. 6C and E) in normal lymphocytes were significantly lower than those in Jurkat cells. Following infection with adv-miR-21, endogenous STAT3 protein levels were significantly decreased compared with the control group (Fig. 6E), however STAT3 mRNA levels were approximately the same as the control group (Fig. 6D). These results suggest that miR-21 targets STAT3 by inhibiting translation rather than degrading the target mRNA.
Discussion
Aberrant expression of miR-21 has been observed in various types of malignancies, and it serves an important role in tumorigenesis and other oncogenic processes by controlling the expression of target genes (8–11). The current study demonstrated that miR-21 expression in Jurkat cells was significantly upregulated compared with normal lymphocytes, indicating that miR-21 may be a novel diagnostic marker for T-ALL.
To further investigate the effects of miR-21 on T-ALL, Jurkat cells were infected with miR-21 recombinant adenovirus, which induced either increased expression or knockdown of miR-21. The present study identified that upregulating miR-21 increased the proliferation and invasive ability of Jurkat cells and decreased their rate of apoptosis. Conversely, knockdown of miR-21 suppressed the proliferation and invasive ability of Jurkat cells and promoted their apoptosis. These results suggest that miR-21 may serve an oncogenic role in T-ALL by regulating multiple biological functions, including proliferation, apoptosis or invasion of T-ALL cells.
Previous studies have demonstrated that miR-21 may be oncogenic, as it regulates target genes, including PTEN, PDCD4 and B-cell lymphoma (Bcl)-2, which are associated with cell proliferation, apoptosis and invasion (10,12,13). The effects of miR-21 on a specific target are context and sequence specific (20). A perfect match of ≥6 nucleotides (nucleotides 2–7) is required to bind miR-21 to the 3′-UTR of the target gene and form the RNA induced silencing complex, which is responsible for regulating the translation of target genes (21,22). Degradation of miR-21 target mRNA may be the predominant mechanism in plants, whereas inefficient translation of the target gene may be a more common regulation mechanism in animals (22). TargetScan and PicTar are examples of common computational target prediction softwares used to predict miRNA target sites conserved among orthologous 3′-UTRs of protein-coding genes (23). In the present study, TargetScan and PicTar were used to predict the target genes of miR-21 and the mechanism of action of miR-21 in Jurkat cells. Among all the predicted target genes, PDCD4, PTEN, Bcl-2 and RECK had previously been demonstrated to be target genes of miR-21 (12–14). STAT3 was another predicted target gene of miR-21, and previous studies have determined that the STAT3 protein is able to regulate miR-21 expression by binding to the miR-21 gene promoter (16,17,24). Therefore, it was hypothesized that a feedback loop may exist between miR-21 and STAT3.
STAT3 is a member of the STATs family, and serves an important role in malignant transformation and oncogenesis by regulating cell proliferation, differentiation and invasion (25). Members of the miR-17 cluster family regulate STAT3, which in turn regulates certain cancer-associated miRNAs, including miR-155 and miR-21, by binding to the promoters of these genes (26). It has previously been demonstrated the existence of regulatory feedback loops between the STAT3 pathway and miRNAs in different cancer contexts, including interleukin-6 (IL-6) receptor-STAT3-NF-κB-Lin-28-let-7a and IL-6-STAT3-miR-24/miR-629-hepatocyte nuclear factor 4α-miR-124 (27).
The present study established that STAT3 expression in Jurkat cells was significantly higher than in normal lymphocytes. The results of the luciferase reporter assay demonstrated that miR-21 could bind to a specific region of the STAT3 3′-UTR. Furthermore, the results of western blot assay indicated that upregulating the expression of miR-21 in Jurkat cells resulted in decreased levels of STAT3 protein, whereas there were no significant differences between the levels of STAT3 mRNA among any of the groups. Therefore, the current study determined that STAT3 was a target gene of miR-21 and, as STAT3 protein is able to regulate miR-21 expression by binding to the miR-21 gene promoter, the existence of a regulatory feedback loop between miR-21 and STAT3 was suggested. This feedback loop may balance miR-21 and STAT3 expression in Jurkat cells, in order to precisely regulate the biological function of Jurkat cells.
In conclusion, miR-21 affects T-ALL by promoting the proliferation and invasion and inhibiting the apoptosis of Jurkat cells. In addition, miR-21 targets STAT3 by inhibiting translation rather than degrading the target mRNA. The regulatory mechanism of miR-21 in Jurkat cells is subtle and complicated. Future investigations are necessary to improve the understanding of the interactions between miR-21 and STAT3 and to facilitate the development of novel therapeutic strategies to treat T-ALL, for which the current study may provide valuable evidence.
Acknowledgements
The present study was supported by grants from the Youth Foundation of the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China; grant no. 13Y0165) the Key Science and Technology Research Project of the Education Department of Henan province (Zhengzhou, China; grant no. 13A320413) and the General Program of the National Natural Science Foundation of China (Beijing, China; grant no. 81470364).
References
- 1.Jabbour EJ, Faderl S, Kantarjian HM. Adult acute lymphoblastic leukemia. Mayo Clin Proc. 2005;80:1517–1527. doi: 10.4065/80.11.1517. [DOI] [PubMed] [Google Scholar]
- 2.Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, Cheng C, Su X, Rubnitz JE, Basso G, et al. Early T-cell precursor leukaemia: A subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23:6306–6315. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L, Cohen HJ, Sallan SE, Asselin BL. Childhood T-cell acute lymphoblastic leukemia: The Dana-Farber cancer institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21:3616–3622. doi: 10.1200/JCO.2003.10.116. [DOI] [PubMed] [Google Scholar]
- 5.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 6.Yendamuri S, Calin GA. The role of microRNA in human leukemia: A review. Leukemia. 2009;23:1257–1263. doi: 10.1038/leu.2008.382. [DOI] [PubMed] [Google Scholar]
- 7.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 8.Baraniskin A, Kuhnhenn J, Schlegel U, Chan A, Deckert M, Gold R, Maghnouj A, Zöllner H, Reinacher-Schick A, Schmiegel W, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117:3140–3146. doi: 10.1182/blood-2010-09-308684. [DOI] [PubMed] [Google Scholar]
- 9.Gu L, Song G, Chen L, Nie Z, He B, Pan Y, Xu Y, Li R, Gao T, Cho WC, Wang S. Inhibition of miR-21 induces biological and behavioral alterations in diffuse large B-cell lymphoma. Acta Haematol. 2013;130:87–94. doi: 10.1159/000346441. [DOI] [PubMed] [Google Scholar]
- 10.Liu C, Yu J, Yu S, Lavker RM, Cai L, Liu W, Yang K, He X, Chen S. MicroRNA-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J Hepatol. 2010;53:98–107. doi: 10.1016/j.jhep.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 11.Wu R, Jiang Y, Wu Q, Li Q, Cheng D, Xu L, Zhang C, Zhang M, Ye L. Diagnostic value of microRNA-21 in the diagnosis of lung cancer: Evidence from a meta-analysis involving 11 studies. Tumour Biol. 2014;35:8829–8836. doi: 10.1007/s13277-014-2106-7. [DOI] [PubMed] [Google Scholar]
- 12.Xu LF, Wu ZP, Chen Y, Zhu QS, Hamidi S, Navab R. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and Bcl-2 in lung squamous carcinoma, Gejiu City, China. PLoS One. 2014;9:e103698. doi: 10.1371/journal.pone.0103698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 14.Wu MF, Yang J, Xiang T, Shi YY, Liu LJ. miR-21 targets Fas ligand-mediated apoptosis in breast cancer cell line MCF-7. J Huazhong Univ Sci Technolog Med Sci. 2014;34:190–194. doi: 10.1007/s11596-014-1257-5. [DOI] [PubMed] [Google Scholar]
- 15.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Löffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermüller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 17.van der Fits L, van Kester MS, Qin Y, Out-Luiting JJ, Smit F, Zoutman WH, Willemze R, Tensen CP, Vermeer MH. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sezary syndrome. J Invest Dermatol. 2011;131:762–768. doi: 10.1038/jid.2010.349. [DOI] [PubMed] [Google Scholar]
- 18.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Ou H, Li Y, Kang M. Activation of miR-21 by STAT3 Induces Proliferation and Suppresses apoptosis in nasopharyngeal carcinoma by targeting PTEN gene. PLoS One. 2014;9:e109929. doi: 10.1371/journal.pone.0109929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 24.Bourguignon LY, Earle C, Wong G, Spevak CC, Krueger K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31:149–160. doi: 10.1038/onc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohanbash G, Okada H. MicroRNAs and STAT interplay. Semin Cancer Biol. 2012;22:70–75. doi: 10.1016/j.semcancer.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, Young BM, De Langhe SP, Bellusci S, Shi W, Parnigotto PP, Warburton D. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol. 2009;333:238–250. doi: 10.1016/j.ydbio.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Q, Li YY, He WF, Zhang ZZ, Zhou Q, Liu X, Shen Y, Huang TT. Interplay between microRNAs and the STAT3 signaling pathway in human cancers. Physiol Genomics. 2013;45:1206–1214. doi: 10.1152/physiolgenomics.00122.2013. [DOI] [PubMed] [Google Scholar]