Abstract
Objective
Chromosomal aberrations are frequently associated with birth defects and pregnancy losses. Trisomy13, Trisomy 18 and Trisomy 21 are the most common, clinically relevant fetal aneusomies. This study used a transcriptomics approach to identify the molecular signatures at the maternal-fetal interface in each aneuploidy.
Methods
We profiled placental gene expression (13–22 wks) in T13 (n = 4), T18 (n = 4), and T21 (n = 8), and in euploid pregnancies (n = 4).
Results
We found differentially expressed (DE) transcripts (≥ 2-fold) in T21 (n = 160), T18 (n = 80), and T13 (n=125). The majority were upregulated and most of the misexpressed genes were not located on the relevant trisomic chromosome, suggesting genome-wide dysregulation. A smaller number of the DE transcripts were encoded on the trisomic chromosome, suggesting gene dosage. In T21, <10% of the genes were transcribed from the Down syndrome critical region (21q21-22), which contributes to the clinical phenotype. In T13, 15% of the upregulated genes were on the affected chromosome (13q11-14) and in T18 the percentage increased to 24% (18q11-22 region).
Conclusion
The trisomic placental (and possibly fetal) phenotypes are driven by the combined effects of genome-wide phenomena and increased gene dosage from the trisomic chromosome.
INTRODUCTION
T13, T18 and T21 are clinically significant aneusomies that impact pregnancy outcome. In the United States, T21 with an incidence of 1/697 affects approximately 350,000 families and an estimated 6,000 children with this syndrome are born each year, representing the leading cause of mental retardation among live births1. Concurrently, the mortality rate associated with Down syndrome (DS) is decreasing. As a result, the number of affected individuals is increasing and predicted to double in the next 10 years2. T18, which represents the second most common autosomal trisomy in pregnancies that are carried to term, occurs in ~1 in 3,000 live births3. The numbers increase significantly when pregnancy losses are included. As compared to T21, the developmental issues and birth defects associated with T18 are more serious3–5. Fifty percent of T18 infants carried to term will be stillborn, and abnormalities in placentation such as abruption, intrauterine growth restriction, preterm delivery and preeclampsia are significantly represented5, 6. T13 is associated with the most adverse outcomes. The incidence varies between 1 in 10–15,000 live births7. The result is less than a 1% live birth rate and pregnancies that reach viability have the highest risk of maternal and fetal complications7, 8. Given the increased number of affected individuals and the pregnancy complications that are associated with their birth, there is an urgent need to gain more knowledge about the molecular and genomic bases of common aneuploidies.
Although the genetic causes of these trisomies have been known since the mid-20th century9, the mechanisms by which three copies of chromosomes 13, 18 or 21 disrupt normal development are not well understood. In T21, two major theories have been proposed for the DS phenotype. According to the “gene dosage effect” hypothesis, imbalances in the genetic load of a small number of critical genes on the affected chromosome could drive the DS phenotype10–12. Alternatively, in the “developmental instability” hypothesis, a global disruption of developmental homeostasis due to a chromosomal imbalance causes the phenotypic profile of DS, and could also explain the similarities observed among the common trisomies13–18. With regard to trisomies 13 and 18, molecular knowledge about the origins of the phenotypic alterations that are associated with these conditions is limited.
In this study, we profiled gene expression at the maternal-fetal interface in T13, T18 and T21. T21 was associated with the highest number of dysregulated genes. However, pathway analysis of the differentially expressed transcripts suggested that T13 and T18 had the greatest impact on crucial signaling pathways, which could explain why these aneuploidies are associated with poorer pregnancy outcomes than DS. The chromosomal locations of the differentially-expressed genes highlighted the importance of genome-wide effects and trisomic dosage. We theorize that the aberrations in gene expression that were identified give insights into the etiology of pregnancy complications that are associated with these trisomies. For example, several dysregulated molecules were involved in apoptosis and cell cycle regulation, processes that go awry in placental defects19. sFLT1, which circulates at higher levels in pregnancy complications20, 21, was up regulated in T13. These data enable a better understanding of how the consequences of a trisomic genome ultimately play out at a cellular level and point to potential therapeutic targets for improving pregnancy outcomes. It will be interesting to learn whether the general principles that emerged from this study of the extraembryonic transcriptome hold true for other cell types of the affected fetuses.
MATERIALS AND METHODS
Tissue Collection
All tissues were collected with informed consent under a protocol approved by The University of California San Francisco (UCSF), Committee on Human Research. Trisomy 13 (13–19 wks), trisomy 18 (14–20 wks), trisomy 21 (18–22 wks) and normal control (12–20 wks) placentas were collected immediately following voluntary interruption of pregnancy. The basal plate was dissected from the placenta proper, rinsed in PBS, and diced into approximately 3 × 3-mm pieces, which were snap frozen in liquid nitrogen and stored at −80°C, as previously published 22. For immunolocalization, biopsy samples of the basal plates were fixed in 3% formaldehyde in PBS, passed through a sucrose gradient (5–15% in PBS), and frozen in optimal cutting temperature (OCT) medium.
Cytogenetics
The affected pregnancies were diagnosed prenatally by chorionic villus sampling or amniocentesis, by using a standard karyotype method 23, 24. Fluorescence in situ hybridization confirmed the karyotypes25. Only cases with <5% mosaicism were included in the analysis.
Total RNA Extraction
RNA was isolated from snap-frozen basal plate specimens by using RNeasy Mini kits (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA concentration/quality was initially assessed on a Nanodrop spectrophotometer (Thermo Scientific) followed by evaluation using an Agilent Bioanalyzer 2100 System. Only samples with a RNA integrity number (RIN) of ≥9 were used in the microarray experiments.
Microarray analyses
These experiments were performed using the GeneChip Human Genome U133 Plus 2.0 array (Affymetrix). Sample processing and hybridization were accomplished by using the protocols devised by the UCSF Gladstone (NHLBI) Genomics Core Facility as previously described22. The microarrays were normalized together by using Robust Multi-array Average. Significant differential expression among groups (control vs. T13, T18 or T21) was determined using the Bioconductor (v.3) package limma (v.3) 26, 27 within the software environment R (v.3). Differentially expressed genes were determined by statistical analysis of B-H corrected p scores of ≤0.05 and absolute fold changes of ≥2. Microarray data were deposited in NCBI’s Gene Expression Omnibus (accession no. GSE70102).
PCA analyses
The methods that we used have been published28. Specifically, we used FactoMineR, an R software package for conventional and multivariate PCA.
Pathway Analysis
To determine whether there was a significant overrepresentation of differentially expressed genes in particular functional pathways or physiologic processes, the data were analyzed by using Ingenuity Pathway Analysis software (http://www.ingenuity.com). The results were displayed by using Circos29.
qRT-PCR
Reverse transcription of basal plate (total) RNA was carried out using the TaqMan Gold RT-PCR kit (Applied Biosystems) as described by the manufacturer. qRT-PCR was performed in triplicate by using standard protocols on an Applied Biosystems 7900HT Sequence Detection System. All templates were amplified by using commercially available primer/probe sets. Results were reported as the relative mRNA levels ± SD for each group. Differences among target expression levels were estimated by the ΔΔCT method with normalization to GAPDH/18S. Differences between means were assessed using a 2-tailed Student’s t test (p < 0.05), assuming unequal variance. The values shown are the mean ± SD. The list of primers with their sequence annotations is shown in Table S2.
Immunohistochemistry and Immunoblotting
Sources of primary antibodies used in these studies are listed in Table S2. Five μm frozen sections cut from optimal cutting temperature-embedded tissues were washed in PBS three times for 5 min each, then incubated in blocking buffer (3% BSA/0.1% Tween-20) for 30 min. Next, the sections were incubated for 1h with a primary antibody diluted 1:100 in blocking buffer. These antibodies included anti-Rac1 (Millipore, cat. #05-389), anti-S100A7 (Imgenex cat. # IMG-409E), anti-SSFA2 (Sigma, cat. #SAB1100788), anti-IGFBP5 (R&D systems, cat. #AF875), anti-USP16 (Abcam, cat. #ab121650), and an anti-cytokeratin (CK7) developed in the Fisher lab 30. Tissue sections were washed and incubated with species-specific secondary antibodies for 30 min. Secondary antibodies (Jackson Immunoresearch) included TRITC-conjugated donkey anti-rat (prod. #712-026-153), FITC-conjugated donkey anti-mouse (Jackson Immunoresearch cat. #715-095-151), FITC-conjugated donkey anti-goat (cat. # 705-096-147), and FITC-conjugated donkey anti-rabbit (cat. #711-096-152). Then the tissue sections were washed, rinsed in dH2O, dried, and mounted using Vectashield Mounting medium with DAPI (cat. #H-1200). Immunoreactivity was imaged using a Leica DM 5000B fluorescence microscope equipped with a Leica DFC 350FX digital camera (Leica Instruments). Immunoblotting was performed as previously described31.
RESULTS
Placental Gene Expression in the Setting of Trisomy Suggested Genome-Wide Dysregulation Dominated Gene Dosage Effects
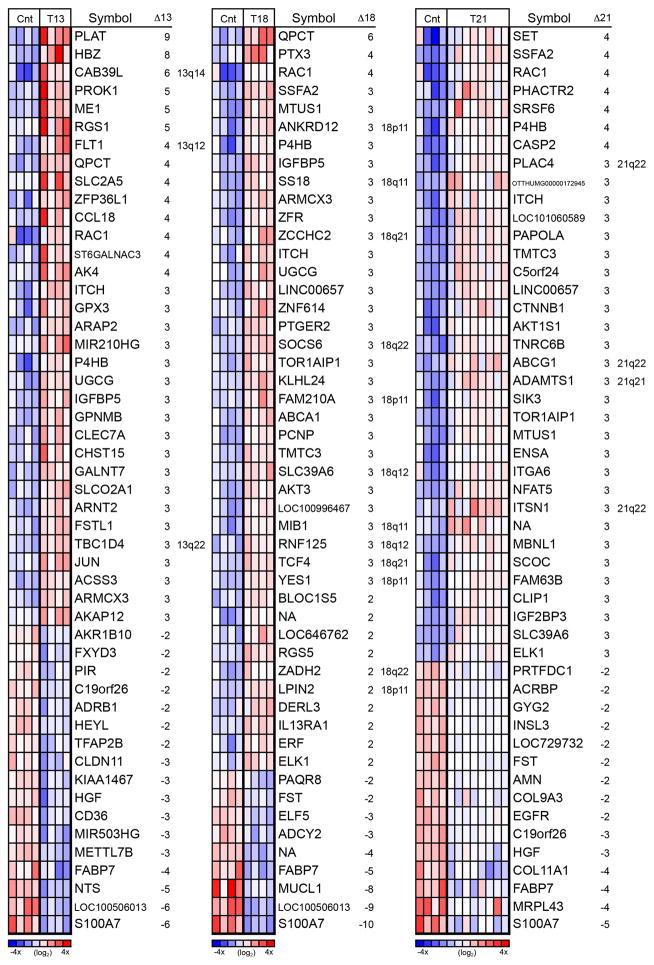
First, we determined the gene expression profiles of 16 aneuploid samples, including 4 cases of T13, 4 cases of T18 and 8 cases of T21. The samples were collected between the gestational ages of 13 and 22 wks. Four gestational age-matched, euploid control samples were also analyzed in parallel. Maternal age, gravidity, parity, gestational age and karyotype for each specimen are summarized in Table S1. Using a microarray approach, we identified, among all the aneuploid samples of the maternal-fetal interface, a total of 281 differentially expressed (DE) annotated genes (≥ 2 fold). By trisomy, the results were as follows: T13 (n = 125; 108 upregulated and 17 downregulated), T18 (n = 80; 71 upregulated and 9 downregulated) and T21 (n = 160; 145 upregulated and 15 downregulated). Figure 1 shows the 50 most highly DE genes for each trisomy. Heat maps of the entire data sets are shown in Figure S1, A–D.
Figure 1.
Heatmap of the 50 most highly differentially expressed (DE) genes for each trisomy revealed unique and overlapping patterns of genome-wide dysregulation. The majority of DE genes in placentas affected by T13 (left), T18 (middle), and T21 (right) were not on the trisomic chromosome. Heatmaps are relative to euploid controls (Cnt) and based on the normalized log2 intensity, centered to median values and colored with a range of −4 to +4. Red denotes up regulated, white denotes intermediate and blue denotes down regulated expression levels. Each column represents data from one sample and each row represents one probe set. Misregulated transcripts that originated from the trisomic chromosome are indicated by inclusion of the chromosomal region in the right hand column of each panel. The majority of DE genes did not reside on the affected chromosome.
A minority of the overexpressed genes was located on the relevant trisomic chromosome (Figure S1). For instance, in the case of T21, <10% of the genes were transcribed from that chromosome (Figure S1D). They included, ADAMTS1 and ATP-binding cassette, subfamily G, member 132, 33, which were increased 3-fold as compared to controls. In the brain, these genes play functional roles in neurocognitive pathways 34, 35. For T13 and T18, a higher proportion of the differentially expressed genes were located on the trisomic chromosome (Figure S1B and C, respectively). In T13, 19/108 of the up regulated genes were on the affected chromosome. All were located on the long arm, 13q11-14. In T18, 19/80 of the up regulated genes were on the affected chromosome; 15 were located on the long arm (18q11-22). These data suggested that T13 and T18, like T21, are associated with altered transcription from a critical region of the trisomic chromosome. However, in all the syndromes, many of the genes on the aneuploid chromosome were not up regulated perhaps due to a compensating repressive mechanism.
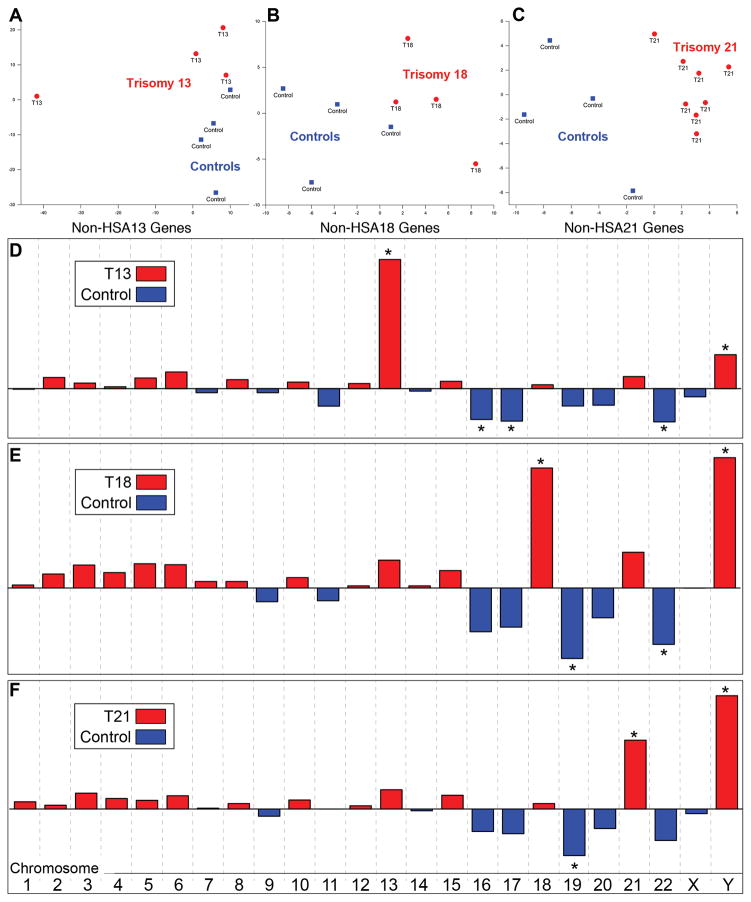
Next, we were interested in whether expression of genes from the non trisomic chromosomes had a distinguishing pattern. A principal component analysis (PCA) was used to answer this question. The results are shown in Figure 2, A–C. The genes expressed from the non trisomic chromosomes separated each aneuploid condition (red) from the euploid samples (blue), suggesting genome-wide effects. In the latter study, PCA showed that control and trisomy samples separated based on gene expression patterns from the affected chromosome, but gene expression from the non trisomic chromosomes failed to distinguish the two groups.
Figure 2.
PCA analyses highlighted dysregulation of genes encoded on the nontrisomic chromosomes. (A–C) The genes expressed from the non trisomic chromosomes (non-HSA) separated each aneuploid condition (red) from the euploid samples (blue), suggesting genome-wide effects. (D–F) A categorical PCA revealed the involvement of gene dysregulation on specific non trisomic chromosomes for each syndrome and overlapping patterns. In all cases, the duplicated chromosome was highlighted. Expression of genes on the Y-chromosome identified pregnancies in which the fetus was male. * denotes p ≤ 0.05 as determined by FactoMineR.
A categorical PCA was used to identify the chromosomes whose gene expression drove the patterns observed in Figure 2, A–C. The results are shown in Figure 2, D–F. In all cases, genes on the affected chromosome were statistically significant contributors that separated the trisomic (red) and control (blue) samples. Additionally, genes on the Y chromosome identified pregnancies in which the offspring was male. In T13, genes on chromosomes 16, 17 and 22 enabled discrimination. For T18, the results were driven by genes on chromosomes 19 and 22. A similar pattern was observed for T21, although the chromosome 22 effects did not reach statistical significance. Overall, the pattern of gene dysregulation across the non trisomic chromosomes was very similar for T13, T18 and T21, again suggestive of genome-wide effects.
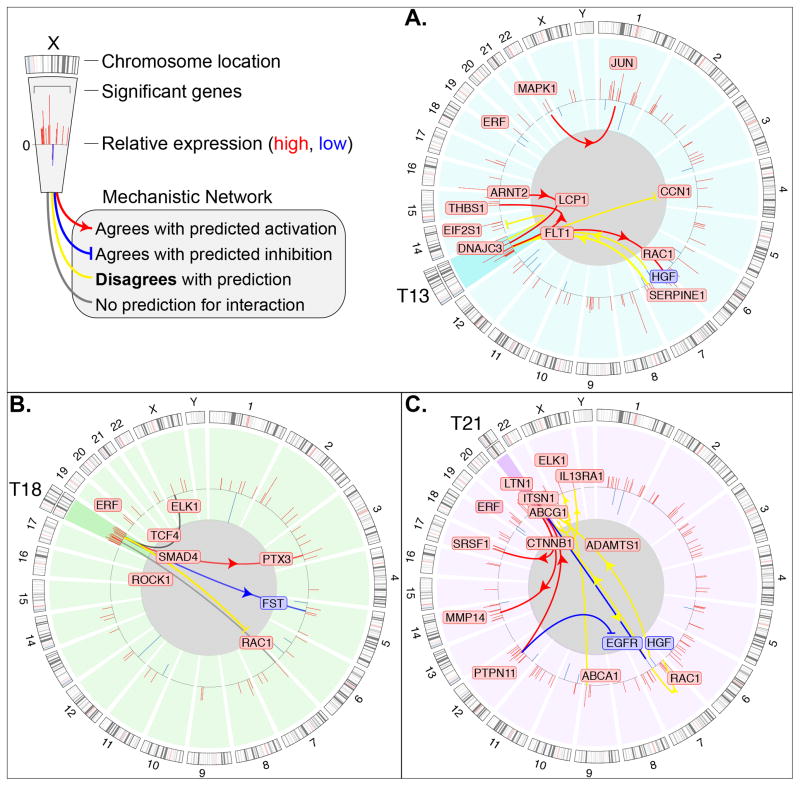
We further investigated this possibility by using IPA mechanistic network analysis. Circos plots of the DE genes in trisomies 13, 18 and 21 are shown Figure 3. From this graphical representation, it was again evident that only a portion of the DE genes was encoded by the duplicated chromosome. Examples included DNAJC3, FLT1 and LCP1 (chromosome 13); TCF4, SMAD4 and ROCK 1 (Chromosome 18); and ABCG1, CTNNB1 and ITSN1 (chromosome 21). Other DE genes were not located on the aneuploid chromosome. These included ARNT2, JUN and HGF (in T13); ELK1, PTX3 and FST (in T18); and ELK1, EGFR, and PTPN11 (in T21). In addition, a set of up and down regulated genes was common to all the trisomies, for example, RAC1 and ERF (Figure S2).
Figure 3.
A Circos plot of DE mRNAs and their mechanistic networks for each trisomy suggested the important roles of genes that were encoded on the duplicated and non duplicated chromosomes. Individual diagrams depict the molecular signature for each trisomy (A, T13; B, T18; C, T21). The outer track is the human chromosome map; the inner track is the loci of the DE genes. Red and blue boxes indicate up and down regulated transcripts, respectively; the height of the red and blue bars is proportional to the fold change values (legend, upper left corner). The trisomic chromosome in each panel is highlighted in a darker tone. The lines drawn between DE genes indicate mechanistic associations as determined by IPA. Some of the associations between dysregulated genes agreed with the predictions (red lines = activation; blue lines = inhibition) and others did not (yellow lines).
Individual diagrams depict the molecular signature for each trisomy. The outer track is the human chromosome map; the inner track is the loci of the DE genes. Red and blue boxes indicate up and down regulated transcripts, respectively; the height of the red and blue bars is proportional to the fold change values (legend, upper left corner). The trisomic chromosome in each panel is highlighted in a darker tone. The lines drawn between DE genes indicate mechanistic associations involving the trisomic chromosome as determined by IPA.
With regard to the mechanistic network analyses (Figure 3), approximately half the associations between dysregulated genes on the trisomic chromosome and those in other genome locations agreed with the predictions (red lines = activation; blue lines = inhibition). Oftentimes disagreement was observed (yellow lines). Additionally, mechanistic associations between DE genes located on non trisomic chromosomes were also identified for T13 and T21. Taken together, the data presented in Figures 1–3 suggested that genome-wide dysregulation and critical regions on the duplicated chromosome play important roles in the placental pathologies that are associated with the aneuploidies that we studied. Additionally, these results raised the possibility that the same mechanisms may contribute to the phenotypes of the offspring. It was also clear that the DE genes included molecules with central functions in many processes that are dysregulated in T13, T18 and T21.
We used Ingenuity Pathway Analysis (IPA) software to gain additional insights into these observations. For the trisomy datasets as a whole, IPA showed that many molecular functions and physiological processes were perturbed. These results, summarized by functional categories, are shown in Figure S3. Overall, they highlighted the important role of aberrations in cell signaling as a large majority of DE genes were involved in these processes. This result suggested that for all the syndromes we studied, global signaling defects are major drivers of the phenotypes. Furthermore, different signaling pathways were associated with different aneusomies. The affected pathways were involved in stem cell maintenance, neuronal development, immune functions, cardiovascular biology, growth and cancer. The results resonated with other work from our group. For example, we showed dysregulation of semaphorin expression in preeclampsia 31 and SEMA3F signals were upregulated in T21. Thus, many of the dysregulated pathways played important roles in processes that regulate functions that are fundamental to the phenotypic manifestation—both embryonic and extraembryonic—of the trisomies on which we focused.
Confirmation of the Microarray Data
For validation, we used two approaches: qRT-PCR to assess relative mRNA levels and antibody-based methods to confirm differential expression at the protein level. The qRT-PCR analyses focused on genes that exhibited the greatest change and those that encoded proteins with potentially important functions. The sequences of the primer/probe sets are shown in Table S2. All showed the expected expression patterns that were predicted based on the microarray data (Figure S4 bar graphs vs. insets; P < 0.05).
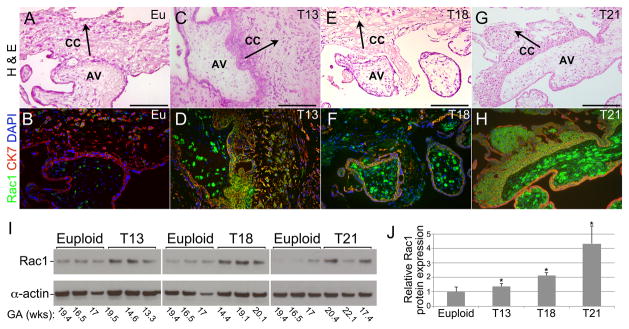
Additionally, we used an immunolocalization approach to confirm misexpression, at the protein level, of several differentially expressed mRNAs. This strategy also enabled identification of the cell types that were involved. Initially, we focused on DE genes that were common to the three trisomies. This included Rac1, a small G protein that is involved in a wide variety of signaling pathways and in the migration of extravillous trophoblasts 36. At the protein level, low expression was detected in euploid samples (Figure 4B). In the setting of trisomy, strong immunoreactivity was observed in association with the stromal cores of chorionic villi (Figure 4D, F, H). Rac1 expression, which was also upregulated in the various trophoblast populations, was particularly prominent in the columns that contained cytotrophoblasts that are destined to invade the uterus. These findings were confirmed by immunoblotting (Figure 4; I and J). Of note, in T21, inter-sample variations in expression were observed. This was in accord with our previous work, which showed variable phenotypes in this trisomy despite a confirmed T21 genotype and the absence of mosaicism 19.
Figure 4.
Expression of RAC1, a GTPase with diverse functions, was up regulated at the protein level in the three trisomies. (A–H) As compared to euploid placentas (A, B; 18wks), RAC1 immunoreactivity (B, D, F, H) was increased in the stromal cores of anchoring villi (AV) and cell columns (CC) in T13 (C, D; 19 wks), T18 (E, F; 23 wks), and T21 (G, H; 17 wks). Staining of invasive cytotrophoblasts was variably detected and most pronounced in T21. (I) Immunoblotting confirmed aneuploidy-associated upregulated expression of RAC1. (J) Quantitation of expression normalized to actin and relative to control euploid samples showed statistically significant (*) increases in the 3 trisomies (p=0.02). Size bars = 100 μm. Eu, euploid; CK7, cytokeratin 7; GA, gestational age; arrows indicate direction of cytotrophoblast invasion.
With regard to downregulation, decreased levels of the mRNA encoding S100 calcium-binding protein A7 (S100A7), also known as psoriasin, were observed in all the trisomies. This molecule regulates angiogenesis in a VEGF-dependent manner 37, 38. Immunolocalization showed that, in euploid placentas, this molecule was highly expressed in the villous cores (Figure 5A). Immunostaining was significantly downregulated in the T13, T18 and T21 samples (Figure 5B, C and D). This result was confirmed by immunoblotting (Figure 5E and F).
Figure 5.
Expression of S100 calcium-binding protein A7 (S100A7) was down regulated in the three trisomies. (A) In euploid samples (e.g., 21 wks), strong S100A7 immunoreactivity was detected in the stromal cores of anchoring villi (AV); the cell columns (CC) of emigrating cytotrophoblasts stained variably and weakly. (B–D) (B) Little to no staining was observed in comparable regions from T13 (18 wks), (C) T18 (20 wks) and (D) T21 (22 wks) placentas. When present, antibody binding was detected only in association with the villous stromal compartments. (E) Immunoblotting showed S100A7 expression in normal pregnancy and uniform downregulation in T13, T18 and T21. (F) Quantitation of expression normalized to actin and relative to control euploid samples showed statistically significant (*) decreases in the 3 trisomies (p= 0.02). Size bars = 100 μm; Eu, euploid; CK7, cytokeratin 7; GA, gestational age; arrows indicate direction of cytotrophoblast invasion.
We also used an immunolocalization approach to validate upregulation, at the protein level, of genes that were DE in a subset of the syndromes. This category included insulin-like growth factor binding protein 5 (IGFBP5) expression, which was upregulated in T13 and T18. In accord with the microarray data, immunoreactivity was largely absent in chorionic villi from normal pregnancies (Figure S5A), although some regions of brighter staining were detected. In T13 and T18, strong signals were evident in association with cytotrophoblasts that emigrated from the villi to form columns (Figure S5B and C). Sperm-specific antigen 2 (SSFA2) mRNA was highly upregulated in T18 and T21 as compared to euploid controls. Immunostaining confirmed this result at the protein level. Very little signal was detected in samples from normal pregnancies (Figure S5D). In contrast, the stroma and some cytotrophoblasts were strongly immunoreactive in T18 and T21 (Figure S5E and F). Finally, USP16 mRNA upregulation was confined to T21. This molecule, which resides in the 21q21-22 region, has been associated with accelerated senescence in the DS murine model 39. At the protein level, no USP16 immunoreactivity was observed in euploid samples (Figure S5G, H and I). In T21, strong staining was detected in association with villous cores and variably weaker signals were associated with invasive cytotrophoblasts (Figure S5J, K and L). Together, these results showed that, for the molecules we studied, the observed mRNA differences were paralleled by changes at the protein level. Frequently, antigen expression was dysregulated in both the stromal and trophoblast compartments.
DISCUSSION
Inadequate formation and function of the placenta are associated with adverse pregnancy outcomes 22, 30, 40–43. In the early stages, aberrations may result in miscarriages, and later on, the great obstetrical syndromes 19, 44, 45. The latter may lead to iatrogenic preterm delivery with the goal of preventing fetal death and/or maternal complications. In the present study, we investigated the effects of commonly occurring aneuploidies on placental gene expression at the maternal-fetal interface. Our goal was to gain insights into how T13, T18 and T21 impact placental development in this critical region. Since these aneuploidies are associated with an increased risk of pregnancy complications, we were also interested in learning whether the results of our analysis might add to the understanding of the etiology of syndromes such as preeclampsia and preterm birth 8, 20, 31, 46–52. Additionally, we wanted to better understand the details of gene dysregulation 19, 22, 31, 50, 53–55.
It was clear from our data that gene dosage effects are only partially responsible for the phenotypic alterations we observed in the aneuploid placentas. The majority of misexpressed mRNAs were not encoded on the relevant trisomic chromosome (Figure 1 and S1). A few genes were upregulated in all three trisomies (Figure S2) and the magnitude of up and down regulation was generally greater than the 1.5 fold that would be predicted based on dosage effects alone. Together, these findings suggested the existence of more complicated mechanisms of dysregulation, involving genome-wide phenomena that are largely not understood. In this regard, it will be important to determine if alterations at the epigenetic level play a role. Thus, we are very interested in profiling methylation patterns together with histone modifications in the setting of the aneuploidies we studied. It is also possible that the up regulated expression of transcription factors and/or other DNA binding proteins on the trisomic chromosome leads to altered transcription of targets elsewhere. For example, SAP18, a major HDAC1/2 co-repressor 56 that is encoded on chromosome 13, was up regulated by 2-fold in Patau syndrome samples. Also, BRWD1, a major epigenetic regular on chromosome 21 57, was up regulated in Down syndrome. Thus, the enhanced expression of these molecules could impact the placental epigenome. TCF4, located on chromosome 18 and overexpressed (3-fold) in Edwards syndrome placentas, interacts with other bHLH transcription factors, creating networks that regulate differentiation of diverse cell types; mutations lead to a variety of disease phenotypes 58. The expression of SMAD4, also encoded on chromosome 18, was up regulated (2-fold) in T18, suggesting disruptions in TGF beta-mediated processes 59, 60. Thus, our data suggested logical hypotheses regarding mechanisms of genome-wide dysregulation in the trisomies we studied.
In terms of the obstetric clinical course, trisomic cases often have poor outcomes, for example, preeclampsia and intrauterine growth restriction 8, 20, 31, 46–52, 61. These conditions, which can be associated with all the aneuploidies that we studied, are most common in T13 and T18 pregnancies 8, 48, 49, 62, 63. In contrast, T21-affected pregnancies have a more favorable outcome, not only over the course of pregnancy, but also during the postnatal period 64. In fact, Down syndrome sometimes goes undiagnosed, being discovered only at birth 65. Interestingly, pathway analyses revealed that aberrant gene expression in the three trisomies converged on a set of signaling pathways involving key functions mediated, for example, by integrins, HGF, CXCR4 and Rho family members. Conversely, other pathways were unique to individual trisomies such as the association of NF-kappa B, NO and cAMP signaling with T13. In this regard, dysregulated PEDF signaling was unique to T18. T21 impacted the most number of pathways. In addition to commonalities with T13 and T18, numerous other signals were dysregulated, including those involving IL-2, IL-6, CREB, NANOG, RhoA and EGF. Together these results suggested that the pathway level dysregulations that were associated with T13 and T18 have a more profound negative impact on pregnancy outcome. T21 had more pleiotropic effects, but the sum total was more likely to result in a live birth. Thus, the transcriptomic data from the maternal-fetal interface help explain, in molecular terms, the disparate pregnancy outcomes in the various trisomies and, perhaps, the fetal manifestations of these aneuploidies.
We noted with interest that many of the misexpressed genes and the pathways that they regulate play important roles in placental development (Figure 1). For example, HIF family members have oxygen dependent and oxygen independent functions in trophoblast differentiation 66. In this regard, ARNT2 was overexpressed in T13 as was FLT1, which has been implicated in the pathogenesis of preeclampsia 31. Conversely, HGF, which plays an important role in trophoblast differentiation/invasion 67, was down regulated in this syndrome. T18 was associated with a down regulation of ELF5, which defines an epigenetically regulated stem cell compartment in the human placenta 68. Along with HGF, EGFR was downregulated in T21. Given that EGF signals enhance invasion, the net effect could be faulty invasion in Down syndrome, 69, which we observed here and in another study 19. Thus, we found the misexpression of many genes that could play important roles in the placental pathologies that are associated with the aneuploidies that we studied.
Our immunolocalization studies confirmed and extended the mRNA-level data. Differentially expressed gene products were frequently associated with cells of the villous cores and trophoblasts, particularly the cytotrophoblast subpopulation that forms the cell columns that attach the placenta to the uterine wall. In general, the functions of the molecules we chose for further analysis are consistent with their misexpression driving abnormal placental development in the setting of the aneuploidies we studied. For instance, overexpression of Rac1 in the villous stroma could dysregulate cell growth and, in cytotrophoblasts, may affect migration 36. S100A7 was strongly expressed in the villous stroma of euploid placentas and downregulated in all three trisomies. Given its role in regulating angiogenesis through VEGF-mediated mechanisms 38, 70, it is possible that misexpression could be associated with abnormal vascularization of the villous tree. Creating an abnormal trophoblast niche is another possibility. SSFA2 (also known as KRAP), which interacts with actin 71, could affect adhesion of the stromal cells and trophoblast populations in the setting of T18 and T21. Finally, IGFBP5 was significantly upregulated in several compartments of T13- and T18-affected placentas. In general, its proteolysis liberates IGF 72. Pregnancy-associated plasma protein-A, which is expressed at high levels by the placenta, cleaves IGFBP5, increasing IGF bioavailability. USP16 is a deubiquitinase that controls a key epigenetic switch that regulates stem self-renewal vs. senescence in Down syndrome 39. Collectively, at the protein level, we observed dysregulated expression of molecules with known or implied important functions in placental development.
In summary, our study showed that gene expression at the maternal-fetal interface in the setting of the common trisomies is associated with a unique gene signature that is related to the chromosome of origin as well as misregulated gene expression at the genome level. Our findings also led us to conclude that the mechanisms of gene dysregulation were due to a combination of genome-wide phenomena and dosage effects. Future studies will determine the extent to which the newly described chromosomal locations contribute to gene dysregulation of the offspring in Patau and Edwards syndromes. This is particularly important given the small sample size of the current study. We are also interested in pursuing these findings in terms of individual cell types, focusing on syncytiotrophoblasts, likely an important source of cell free fetal DNA in maternal blood, and trophoblast progenitors, which play a fundamental role in placental development. Improving our knowledge of the molecular consequences of the most common aneuploidies has the potential to yield novel therapeutic approaches for mitigating the effects of chromosome duplication on key developmental processes.
Supplementary Material
Figure S1. Complete DE gene lists annotated with the gene symbol/name, chromosomal location, and fold change. (A) Combined microarray data from T13, T18, and T21 placentas as compared with euploid controls. (B, C, D) Transcripts differentially expressed in each trisomy compared to euploid controls (T13, T18, and T21 respectively).
Figure S2. Common misregulated transcripts across all trisomic placentas. Eleven genes were DE in all the trisomies studied. Nine transcripts were up-regulated and two transcripts were down regulated.
Figure S3. Ingenuity Pathway Analyses (IPA) of the DE genes associated with each trisomy displayed as a Circos plot. Additional analyses of the DE genes in T13, T18 and T21 placentas as compared to euploid controls showed the profound impact of these aneuploidies at the pathway level. The Circos plot shows DE genes for each chromosome (right side) linked to the IPA pathways (left side). Overall, this analysis highlighted the important role of aberrations in cell signaling as a large majority of DE genes were involved in these processes.
Figure S4. RT-qPCR confirmed dysregulation of a subset of DE genes in the trisomic syndromes. (A) Targets up regulated in T13. (B) Targets up and down regulated in T18. (C) Targets up and down regulated in T21. Genes with relevant biological functions and/or those that were dysregulated in 2 or more trisomies were studied. With regard to the latter, IGFBP5 was up regulated in trisomy 13 and trisomy 18. SSFA2 was up regulated in trisomy 18 and trisomy 21. Insets show the corresponding microarray data for each gene. Insets in each panel show log2 microarray intensity comparisons between trisomy and euploid samples (p<0.05).
Figure S5. Immunolocalization of DE gene products in trisomic placentas confirmed dysregulation at the protein level. A euploid (eu) placental biopsy (19 wks) (A) showed minimal expression of insulin-like growth factor binding protein 5 (IGFBP5) as compared with up regulation in (B) T13 (19 weeks) and (C) T18 (19 weeks). These findings were in accord with this molecule’s role in placentation and decidualization. Very little sperm-specific antigen 2 (SSFA2) immunoreactivity was detected in euploid placentas (D). In contrast, the stroma and some cytotrophoblasts were strongly immunoreactive in T18 (E) and T21 (F). No USP16 immunoreactivity was observed in euploid samples (G, H and I). In T21, strong staining was detected in association with villous cores and variably weaker signals were associated with invasive cytotrophoblasts (J, K and L). Size bars = 100 μm; AV, Anchoring villi; CK, cytokeratin; arrows indicate direction of trophoblast invasion.
Clinical characteristics of the placental donors in the study.
PCR Primers used to validate the microarray results.
Antibodies used to validate the microarray results via immunolocalization and immunoblotting.
There are limited data about trisomy 13, trisomy 18 and trisomy 21 molecular signatures in terms of placental gene expression.
We identified the dysregulation of several genes that are involved in extraembryonic development, which might be associated with a substantially increased rate of complications in trisomic pregnancies.
Our data suggested that the trisomic placental (and possibly fetal) phenotypes are driven by the combined effects of genome-wide phenomena and critical region gene dosage from the trisomic chromosome.
Acknowledgments
Funding: Dr. Katherine Bianco was supported by a Reproductive Scientist Development Program Award (K12HD000849) and a Clinical Investigator Award (K08HD069518-01). Placental tissue samples were provided by the NIH Placental Bank at UCSF, funded under NIH U54 HD055764.
The authors thank the UCSF Clinical Cytogenetics Laboratory and Dr. Jing Way for assistance with the karyotyping and FISH analyses, and Mr. Gabriel Goldfien for assistance with tissue collection.
Footnotes
Conflict of interest: All authors declare that there is no conflict of interest.
References
- 1.Parker SE, Mai CT, Canfield MA, et al. Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res A Clin Mol Teratol. 2010;88:1008–16. doi: 10.1002/bdra.20735. [DOI] [PubMed] [Google Scholar]
- 2.CDC. Morbidity and Mortality Weekly Report. Atlanta: Center for Disease Control; Jan 6, 2006. Report No.: 51 & 52. [Google Scholar]
- 3.Edwards JH, Harnden DG, Cameron AH, et al. A new trisomic syndrome. Lancet. 1960;1:787–90. doi: 10.1016/s0140-6736(60)90675-9. [DOI] [PubMed] [Google Scholar]
- 4.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–91. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 5.Irving C, Richmond S, Wren C, et al. Changes in fetal prevalence and outcome for trisomies 13 and 18: a population-based study over 23 years. J Matern Fetal Neonatal Med. 2010 doi: 10.3109/14767051003758879. [DOI] [PubMed] [Google Scholar]
- 6.Fisher JM, Harvey JF, Lindenbaum RH, et al. Molecular studies of trisomy 18. Am J Hum Genet. 1993;52:1139–44. [PMC free article] [PubMed] [Google Scholar]
- 7.Baty BJ, Blackburn BL, Carey JC. Natural history of trisomy 18 and trisomy 13: I. Growth, physical assessment, medical histories, survival, and recurrence risk. American journal of medical genetics. 1994;49:175–88. doi: 10.1002/ajmg.1320490204. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg RF, Kliman HJ, Cohen AW. Preeclampsia, trisomy 13, and the placental bed. Obstetrics and gynecology. 1991;78:505–8. [PubMed] [Google Scholar]
- 9.Jacobs PA, Baikie AG, Court Brown WM, et al. The somatic chromosomes in mongolism. Lancet. 1959;1:710. doi: 10.1016/s0140-6736(59)91892-6. [DOI] [PubMed] [Google Scholar]
- 10.Mao R, Zielke CL, Zielke HR, et al. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics. 2003;81:457–67. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 11.Antonarakis SE. Chromosome 21: from sequence to applications. Curr Opin Genet Dev. 2001;11:241–6. doi: 10.1016/s0959-437x(00)00185-4. [DOI] [PubMed] [Google Scholar]
- 12.Rozovski U, Jonish-Grossman A, Bar-Shira A, et al. Genome-wide expression analysis of cultured trophoblast with trisomy 21 karyotype. Hum Reprod. 2007;22:2538–45. doi: 10.1093/humrep/dem214. [DOI] [PubMed] [Google Scholar]
- 13.FitzPatrick DR, Ramsay J, McGill NI, et al. Transcriptome analysis of human autosomal trisomy. Human molecular genetics. 2002;11:3249–56. doi: 10.1093/hmg/11.26.3249. [DOI] [PubMed] [Google Scholar]
- 14.Letourneau A, Santoni FA, Bonilla X, et al. Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature. 2014;508:345–50. doi: 10.1038/nature13200. [DOI] [PubMed] [Google Scholar]
- 15.Korenberg JR, Kawashima H, Pulst SM, et al. Molecular definition of a region of chromosome 21 that causes features of the Down syndrome phenotype. Am J Hum Genet. 1990;47:236–46. [PMC free article] [PubMed] [Google Scholar]
- 16.Korbel JO, Tirosh-Wagner T, Urban AE, et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12031–6. doi: 10.1073/pnas.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korenberg JR, Chen XN, Schipper R, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shapiro BL. Whither Down syndrome critical regions? Hum Genet. 1997;99:421–3. doi: 10.1007/s004390050383. [DOI] [PubMed] [Google Scholar]
- 19.Wright A, Zhou Y, Weier JF, et al. Trisomy 21 is associated with variable defects in cytotrophoblast differentiation along the invasive pathway. Am J Med Genet A. 2004;130:354–64. doi: 10.1002/ajmg.a.30254. [DOI] [PubMed] [Google Scholar]
- 20.Herraiz I, Droge LA, Gomez-Montes E, et al. Characterization of the soluble fms-like tyrosine kinase-1 to placental growth factor ratio in pregnancies complicated by fetal growth restriction. Obstetrics and gynecology. 2014;124:265–73. doi: 10.1097/AOG.0000000000000367. [DOI] [PubMed] [Google Scholar]
- 21.Jardim LL, Rios DR, Perucci LO, et al. Is the imbalance between pro-angiogenic and anti-angiogenic factors associated with preeclampsia? Clin Chim Acta. 2015;447:34–8. doi: 10.1016/j.cca.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Winn VD, Haimov-Kochman R, Paquet AC, et al. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology. 2007;148:1059–79. doi: 10.1210/en.2006-0683. [DOI] [PubMed] [Google Scholar]
- 23.Caspersson T, Farber S, Foley GE, et al. Chemical differentiation along metaphase chromosomes. Exp Cell Res. 1968;49:219–22. doi: 10.1016/0014-4827(68)90538-7. [DOI] [PubMed] [Google Scholar]
- 24.Kottler MJ. From 48 to 46: cytological technique, preconception, and the counting of human chromosomes. Bull Hist Med. 1974;48:465–502. [PubMed] [Google Scholar]
- 25.Trask BJ. Fluorescence in situ hybridization: applications in cytogenetics and gene mapping. Trends Genet. 1991;7:149–54. doi: 10.1016/0168-9525(91)90378-4. [DOI] [PubMed] [Google Scholar]
- 26.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 28.Le S, Josse J, Husson F. FactoMineR: An R package for multivariate analysis. J Stat Softw. 2008;25:1–18. [Google Scholar]
- 29.Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damsky C, Schick SF, Klimanskaya I, et al. Adhesive interactions in peri-implantation morphogenesis and placentation. Reprod Toxicol. 1997;11:367–75. doi: 10.1016/s0890-6238(96)00149-9. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Gormley MJ, Hunkapiller NM, et al. Reversal of gene dysregulation in cultured cytotrophoblasts reveals possible causes of preeclampsia. The Journal of clinical investigation. 2013;123:2862–72. doi: 10.1172/JCI66966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson PF, Kannangara TS, Eadie BD, et al. Cognition, learning behaviour and hippocampal synaptic plasticity are not disrupted in mice over-expressing the cholesterol transporter ABCG1. Lipids in health and disease. 2009;8:5. doi: 10.1186/1476-511X-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pereira PL, Magnol L, Sahun I, et al. A new mouse model for the trisomy of the Abcg1-U2af1 region reveals the complexity of the combinatorial genetic code of down syndrome. Human molecular genetics. 2009;18:4756–69. doi: 10.1093/hmg/ddp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds LE, Watson AR, Baker M, et al. Tumour angiogenesis is reduced in the Tc1 mouse model of Down’s syndrome. Nature. 2010;465:813–7. doi: 10.1038/nature09106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miguel RF, Pollak A, Lubec G. Metalloproteinase ADAMTS-1 but not ADAMTS-5 is manifold overexpressed in neurodegenerative disorders as Down syndrome, Alzheimer’s and Pick’s disease. Brain Res Mol Brain Res. 2005;133:1–5. doi: 10.1016/j.molbrainres.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Grewal S, Carver JG, Ridley AJ, et al. Implantation of the human embryo requires Rac1-dependent endometrial stromal cell migration. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16189–94. doi: 10.1073/pnas.0806219105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moog-Lutz C, Bouillet P, Regnier CH, et al. Comparative expression of the psoriasin (S100A7) and S100C genes in breast carcinoma and co-localization to human chromosome 1q21-q22. Int J Cancer. 1995;63:297–303. doi: 10.1002/ijc.2910630225. [DOI] [PubMed] [Google Scholar]
- 38.Emberley ED, Murphy LC, Watson PH. S100A7 and the progression of breast cancer. Breast cancer research: BCR. 2004;6:153–9. doi: 10.1186/bcr816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adorno M, Sikandar S, Mitra SS, et al. Usp16 contributes to somatic stem-cell defects in Down’s syndrome. Nature. 2013;501:380–4. doi: 10.1038/nature12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benirschke K, Kaufmann P. Pathology of the human placenta. 3. New York: Springer-Verlag; 1995. p. xviii.p. 871. [Google Scholar]
- 41.Fisher SJ, Cui TY, Zhang L, et al. Adhesive and degradative properties of human placental cytotrophoblast cells in vitro. J Cell Biol. 1989;109:891–902. doi: 10.1083/jcb.109.2.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maltepe E, Bakardjiev AI, Fisher SJ. The placenta: transcriptional, epigenetic, and physiological integration during development. The Journal of clinical investigation. 2010;120:1016–25. doi: 10.1172/JCI41211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Red-Horse K, Rivera J, Schanz A, et al. Cytotrophoblast induction of arterial apoptosis and lymphangiogenesis in an in vivo model of human placentation. The Journal of clinical investigation. 2006;116:2643–52. doi: 10.1172/JCI27306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bianco K, Caughey AB, Shaffer BL, et al. History of miscarriage and increased incidence of fetal aneuploidy in subsequent pregnancy. Obstetrics and gynecology. 2006;107:1098–102. doi: 10.1097/01.AOG.0000215560.86673.22. [DOI] [PubMed] [Google Scholar]
- 45.Houlihan OA, O’Donoghue K. The natural history of pregnancies with a diagnosis of trisomy 18 or trisomy 13; a retrospective case series. BMC pregnancy and childbirth. 2013;13:209. doi: 10.1186/1471-2393-13-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chelbi ST, Vaiman D. Genetic and epigenetic factors contribute to the onset of preeclampsia. Mol Cell Endocrinol. 2008;282:120–9. doi: 10.1016/j.mce.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 47.Enquobahrie DA, Abetew DF, Sorensen TK, et al. Placental microRNA expression in pregnancies complicated by preeclampsia. American journal of obstetrics and gynecology. 2011;204:178, e12–21. doi: 10.1016/j.ajog.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson WP, Penaherrera MS, Jiang R, et al. Assessing the role of placental trisomy in preeclampsia and intrauterine growth restriction. Prenat Diagn. 2010;30:1–8. doi: 10.1002/pd.2409. [DOI] [PubMed] [Google Scholar]
- 49.Silasi M, Rana S, Powe C, et al. Placental expression of angiogenic factors in Trisomy 13. American journal of obstetrics and gynecology. 2011;204:546, e1–4. doi: 10.1016/j.ajog.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 50.Winn VD, Gormley M, Paquet AC, et al. Severe preeclampsia-related changes in gene expression at the maternal-fetal interface include sialic acid-binding immunoglobulin-like lectin-6 and pappalysin-2. Endocrinology. 2009;150:452–62. doi: 10.1210/en.2008-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Bianco K, Huang L, et al. Comparative analysis of maternal-fetal interface in preeclampsia and preterm labor. Cell Tissue Res. 2007;329:559–69. doi: 10.1007/s00441-007-0428-0. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Y, Damsky CH, Chiu K, et al. Preeclampsia is associated with abnormal expression of adhesion molecules by invasive cytotrophoblasts. The Journal of clinical investigation. 1993;91:950–60. doi: 10.1172/JCI116316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aronow BJ, Richardson BD, Handwerger S. Microarray analysis of trophoblast differentiation: gene expression reprogramming in key gene function categories. Physiol Genomics. 2001;6:105–16. doi: 10.1152/physiolgenomics.2001.6.2.105. [DOI] [PubMed] [Google Scholar]
- 54.Ben-Tabou de-Leon S, Davidson EH. Modeling the dynamics of transcriptional gene regulatory networks for animal development. Developmental biology. 2009;325:317–28. doi: 10.1016/j.ydbio.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rawn SM, Cross JC. The evolution, regulation, and function of placenta-specific genes. Annu Rev Cell Dev Biol. 2008;24:159–81. doi: 10.1146/annurev.cellbio.24.110707.175418. [DOI] [PubMed] [Google Scholar]
- 56.Heideman MR, Lancini C, Proost N, et al. Sin3a-associated Hdac1 and Hdac2 are essential for hematopoietic stem cell homeostasis and contribute differentially to hematopoiesis. Haematologica. 2014;99:1292–303. doi: 10.3324/haematol.2013.092643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z, Huang Y, Li H, et al. Excess of rare variants in genes that are key epigenetic regulators of spermatogenesis in the patients with non-obstructive azoospermia. Sci Rep. 2015;5:8785. doi: 10.1038/srep08785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forrest MP, Waite AJ, Martin-Rendon E, et al. Knockdown of human TCF4 affects multiple signaling pathways involved in cell survival, epithelial to mesenchymal transition and neuronal differentiation. PLoS One. 2013;8:e73169. doi: 10.1371/journal.pone.0073169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heldin CH, Moustakas A. Role of Smads in TGFbeta signaling. Cell Tissue Res. 2012;347:21–36. doi: 10.1007/s00441-011-1190-x. [DOI] [PubMed] [Google Scholar]
- 60.Moustakas A, Souchelnytskyi S, Heldin CH. Smad regulation in TGF-beta signal transduction. J Cell Sci. 2001;114:4359–69. doi: 10.1242/jcs.114.24.4359. [DOI] [PubMed] [Google Scholar]
- 61.Chen DB, Wang W. Human placental microRNAs and preeclampsia. Biology of reproduction. 2013;88:130. doi: 10.1095/biolreprod.113.107805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boyd PA, Lindenbaum RH, Redman C. Pre-eclampsia and trisomy 13: a possible association. Lancet. 1987;2:425–7. doi: 10.1016/s0140-6736(87)90960-3. [DOI] [PubMed] [Google Scholar]
- 63.Tuohy JF, James DK. Pre-eclampsia and trisomy 13. Br J Obstet Gynaecol. 1992;99:891–4. doi: 10.1111/j.1471-0528.1992.tb14436.x. [DOI] [PubMed] [Google Scholar]
- 64.Koduri P, Giraldo M, Shlossman P, et al. Intrapartum fetal heart rate characteristics of nonanomalous fetuses with trisomy 21. American journal of perinatology. 2012;29:415–8. doi: 10.1055/s-0032-1304821. [DOI] [PubMed] [Google Scholar]
- 65.de Groot-van der Mooren MD, Gemke RJ, Cornel MC, et al. Neonatal diagnosis of Down syndrome in The Netherlands: suspicion and communication with parents. J Intellect Disabil Res. 2014;58:953–61. doi: 10.1111/jir.12125. [DOI] [PubMed] [Google Scholar]
- 66.Maltepe E, Krampitz GW, Okazaki KM, et al. Hypoxia-inducible factor-dependent histone deacetylase activity determines stem cell fate in the placenta. Development. 2005;132:3393–403. doi: 10.1242/dev.01923. [DOI] [PubMed] [Google Scholar]
- 67.Nasu K, Zhou Y, McMaster MT, et al. Upregulation of human cytotrophoblast invasion by hepatocyte growth factor. J Reprod Fertil Suppl. 2000;55:73–80. [PubMed] [Google Scholar]
- 68.Hemberger M, Udayashankar R, Tesar P, et al. ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Human molecular genetics. 2010;19:2456–67. doi: 10.1093/hmg/ddq128. [DOI] [PubMed] [Google Scholar]
- 69.Bass KE, Morrish D, Roth I, et al. Human cytotrophoblast invasion is up-regulated by epidermal growth factor: evidence that paracrine factors modify this process. Developmental biology. 1994;164:550–61. doi: 10.1006/dbio.1994.1223. [DOI] [PubMed] [Google Scholar]
- 70.Krop I, Marz A, Carlsson H, et al. A putative role for psoriasin in breast tumor progression. Cancer research. 2005;65:11326–34. doi: 10.1158/0008-5472.CAN-05-1523. [DOI] [PubMed] [Google Scholar]
- 71.Fujimoto T, Shirasawa S. KRAS-induced actin-interacting protein: a potent target for obesity, diabetes and cancer. Anticancer Res. 2011;31:2413–7. [PubMed] [Google Scholar]
- 72.Beattie J, Allan GJ, Lochrie JD, et al. Insulin-like growth factor-binding protein-5 (IGFBP-5): a critical member of the IGF axis. Biochem J. 2006;395:1–19. doi: 10.1042/BJ20060086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Complete DE gene lists annotated with the gene symbol/name, chromosomal location, and fold change. (A) Combined microarray data from T13, T18, and T21 placentas as compared with euploid controls. (B, C, D) Transcripts differentially expressed in each trisomy compared to euploid controls (T13, T18, and T21 respectively).
Figure S2. Common misregulated transcripts across all trisomic placentas. Eleven genes were DE in all the trisomies studied. Nine transcripts were up-regulated and two transcripts were down regulated.
Figure S3. Ingenuity Pathway Analyses (IPA) of the DE genes associated with each trisomy displayed as a Circos plot. Additional analyses of the DE genes in T13, T18 and T21 placentas as compared to euploid controls showed the profound impact of these aneuploidies at the pathway level. The Circos plot shows DE genes for each chromosome (right side) linked to the IPA pathways (left side). Overall, this analysis highlighted the important role of aberrations in cell signaling as a large majority of DE genes were involved in these processes.
Figure S4. RT-qPCR confirmed dysregulation of a subset of DE genes in the trisomic syndromes. (A) Targets up regulated in T13. (B) Targets up and down regulated in T18. (C) Targets up and down regulated in T21. Genes with relevant biological functions and/or those that were dysregulated in 2 or more trisomies were studied. With regard to the latter, IGFBP5 was up regulated in trisomy 13 and trisomy 18. SSFA2 was up regulated in trisomy 18 and trisomy 21. Insets show the corresponding microarray data for each gene. Insets in each panel show log2 microarray intensity comparisons between trisomy and euploid samples (p<0.05).
Figure S5. Immunolocalization of DE gene products in trisomic placentas confirmed dysregulation at the protein level. A euploid (eu) placental biopsy (19 wks) (A) showed minimal expression of insulin-like growth factor binding protein 5 (IGFBP5) as compared with up regulation in (B) T13 (19 weeks) and (C) T18 (19 weeks). These findings were in accord with this molecule’s role in placentation and decidualization. Very little sperm-specific antigen 2 (SSFA2) immunoreactivity was detected in euploid placentas (D). In contrast, the stroma and some cytotrophoblasts were strongly immunoreactive in T18 (E) and T21 (F). No USP16 immunoreactivity was observed in euploid samples (G, H and I). In T21, strong staining was detected in association with villous cores and variably weaker signals were associated with invasive cytotrophoblasts (J, K and L). Size bars = 100 μm; AV, Anchoring villi; CK, cytokeratin; arrows indicate direction of trophoblast invasion.
Clinical characteristics of the placental donors in the study.
PCR Primers used to validate the microarray results.
Antibodies used to validate the microarray results via immunolocalization and immunoblotting.