Abstract
Nanodiamond (ND) is a renowned material in nonviral small interfering RNA (siRNA) carrier field due to its unique physical, chemical, and biological properties. In our previous work, it was proven that ND could deliver siRNA into cells efficiently and downregulate the expression of desired protein. However, synthesizing a high-efficient tumor-targeting carrier using ND is still a challenge. In this study, a novel carrier, NDCONH(CH2)2NH-VDGR, was synthesized for siRNA delivery, and its properties were characterized with methods including Fourier transform infrared spectrometry, transmission electron microscopy, scanning electron microscopy, gel retardation assay, differential scanning calorimetry, confocal microscopy, releasing test, real-time polymerase chain reaction (PCR) assay, enzyme-linked immunosorbent assay (ELISA), flow cytometry, cytotoxicity assay, and gene-silencing efficacy assay in vitro and in vivo. The mechanism of NDCONH(CH2)2NH-VDGR/survivin-siRNA-induced tumor apoptosis was evaluated via flow cytometer assay using Annexin V–fluorescein isothiocyanate/propidium iodide staining method. The NDCONH(CH2)2NH-VDGR/survivin-siRNA nanoparticle with 60–110 nm diameter and 35.65±3.90 mV zeta potential was prepared. For real-time PCR assay, the results showed that the expression of survivin mRNA was reduced to 46.77%±6.3%. The expression of survivin protein was downregulated to 48.49%±2.25%, as evaluated by ELISA assay. MTT assay showed that NDCONH(CH2)2NH-VDGR/survivin-siRNA had an inhibitory effect on MCF-7 cell proliferation. According to these results, the survivin-siRNA could be delivered, transported, and released stably, which benefits in increasing the gene-silencing effect. Therefore, as an siRNA carrier, NDCONH(CH2)2NH-VDGR was suggested to be used in siRNA delivery system and in cancer treatments.
Keywords: NDCONH(CH2)2NH-VDGR, siRNA carrier, RNAi, survivin
Introduction
Cancer is one of the greatest threats to human health, and the means of inhibiting tumor cell growth have been proved to be effective methods in cancer treatment; there are various methods of inhibiting tumor cell growth, such as RNA interference (RNAi).1 RNAi has been receiving attention in the field of tumor therapy for its suppressing effect on gene expression.2,3 RNAi transforms the double-stranded RNA into small RNAs and then guides a protein nuclease to destroy their targeted mRNA. The process is defined as post-transcriptional gene silencing.4 Small interfering RNA (siRNA) is a kind of small RNA with 20–25 base pairs, which could integrate into the RNA-induced silencing complex and lead to a downregulated expression of a targeted gene.5,6 As a precise and efficient method, siRNA-mediated therapy is considered to be promising in cancer treatment. However, the naked siRNA has low delivery rate when crossing the cell membrane, due to disadvantages including negative-charged surface, hydrophilic functional groups, and large molecular size.7,8 In addition, the naked siRNA can be degraded by ribonuclease quite rapidly in cells or body, and hence, it is unstable.9,10 Thus, an efficient carrier system to overcome poor intracellular uptake and instability problems is required.
Survivin is an apoptosis inhibitor and a member of apoptosis protein (IAP) family, and plays an important role in inhibiting apoptosis.11–13 According to the published literatures, survivin cannot be detected in tissues except the testis, thymus, and placenta. Yet it can be overexpressed in several kinds of cancer cells including breast cancer and gastric cancer cells.14,15 Therefore, survivin is used as a drug target for novel cancer therapies. Inhibiting the expression of survivin protein can increase the apoptotic rate and inhibit tumor growth.16,17 However, naked survivin-siRNA has difficulty crossing the cell membrane and is easily decomposed, and hence, significant efforts have been focused on searching for a low-toxic, highly-efficient, and highly-biocompatible carrier for survivin delivery.18,19
Nanodiamond (ND) has been an siRNA delivery carrier due to its unique physical, chemical, and biological properties.19–21 In our previous work, it was proven that it could deliver VEGF-siRNA into HeLa cells efficiently and downregulate the expression of VEGF protein.22 The surface of ND is covered with a large amount of functional groups such as carboxyl and hydroxyl groups, and it can also be further decorated to improve the performance and transfection ability.20–22 For example, lysine-functionalized ND was synthesized as a gene carrier, and the modification resulted in a good dispersibility.23 However, synthesizing a high-efficient targeting carrier using ND is still a challenge.24 RGD peptides, ligands for αvβ3-intergrin, are known as an effective tool for targeting tumor cells.25–27 In this study, we prepared a novel targeted gene delivery system, NDCONH(CH2)2NH-VDGR, containing ND (gene delivery vehicle), H-Arg-Gly-Asp-Val-OH (targeting agent), and EDA (joint arm), and the targeting and transfection efficiency was studied.
Materials and methods
Materials and instruments
ND, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), penicillin, and streptomycin were purchased from Sigma-Aldrich (St Louis, MO, USA). Boc-Arg(Tos)-OH, H-Gly-OBzl⋅HCl, Boc-Asp(OMe)-OH, H-Val-OBzl⋅HCl, dicyclohexylcarbodiimide, N1-((ethylimino) methylene)-N3,N3-dimethylpropane-1,3-diamine (EDC), and hydroxybenzotriazole (HOBt) were purchased from GL Biochem Ltd (Shanghai, People’s Republic of China). Boc-HN-(CH2)2-NH2 was purchased from GL Biochem Ltd. Anti-survivin siRNA and fluorescein-labeled survivin-siRNA (FAM-survivin-siRNA) were purchased from GenePharma Co., Ltd (Shanghai, People’s Republic of China). The sequences of sense and antisense (forward sequence, 5′-GCATGGGTCCCCCGACGTTG-3′; reverse sequence, 5′-GCTCCGGCCAGAGGCCTCAA-3′) RNA were synthesized and purified by GenePharma Co., Ltd. The scramble sequence of siRNA, which served as normal control (NC) of siRNA, was purchased from GenePharma Co., Ltd. RPMI-1640 medium, fetal bovine serum (FBS), and trypsin were purchased from Hyclone Laboratories Inc. (Logan, UT, USA). Dimethylsulfoxide (DMSO) was purchased from AppliChem GmbH Company (Darmstadt, Germany). Radioimmunoprecipitation assay (RIPA) kit and protease inhibitor cocktail (1%, Cat No: 539134) were purchased from Life Scientific (Dublin, Ireland). Bicinchoninic acid (BCA) protein kit was purchased from Pierce Biotechology (Waltham, MA, USA). Human Survivin Quantikine ELISA Kit was purchased from R&D Systems Company (Minneapolis, MN, USA). High Capacity cDNA Reverse Transcription Kit, TaqMan Gene Expression Master Mix, TaqMan Gene Expression Assay Kit (survivin assay, GAPDH assay), propidium iodide (PI), RNase, and FITC Annexin V/Dead Cell Apoptosis Kit were purchased from Life Invitrogen. Trizol was obtained from Invitrogen. Other chemicals and reagents were of chemical grade unless otherwise specified. Eppendorf tubes were used (Eppendorf, Hamburg, Germany).
Zetasizer Nano ZS (Malvern Instruments, Worcestershire, UK), zeta potential analyzer (ZetaPlus; Brookhaven Instruments Corporation, New York, USA), differential scanning calorimeter (204F1; Netzsch, Bavaria, Germany), transmission electron microscope (JEOL, Tokyo, Japan), scanning electron microscope (S-4800; Hitachi, Tokyo, Japan), Victor X5 plate reader (PerkinElmer, Waltham, MA, USA), real-time polymerase chain reaction system (Applied Biosystems; Thermo Fisher Scientific, Waltham, MA, USA), confocal laser scanning microscope (Leica, Munich, Germany), centrifuge (5810R; Eppendorf), and atomic force microscope (Veeco Instruments, Inc., Town of Oyster Bay, NY, USA) were obtained and used.
Preparation of NDCONH(CH2)2NH-VDGR/survivin-siRNA
Synthesis of carboxylated ND
Carboxylic acid-functionalized ND (ND-COOH) was synthesized using oxidation method described in Figure 1 (step i). Firstly, 300 mg of ND power was added to 20 mL of concentrated H2SO4 and HNO3 (v:v =3:1) mixture and stirred at 40°C for 24 h. Then, the suspension was centrifuged at 5,000 g for 5 min, the supernatant was discarded, and then 10 mL of 0.1 M NaOH was added. The mixture was stirred at 90°C for 1 h, and centrifuged at 5,000 g for 15 min, and the sediment was dispersed in 10 mL of 0.1 M hydrochloric acid. The mixture was refluxed at 90°C for 1 h. After 1 h, the suspension was centrifuged and washed three times with distilled water. The sediment was freeze dried, and carboxylated ND was obtained.20,24,25
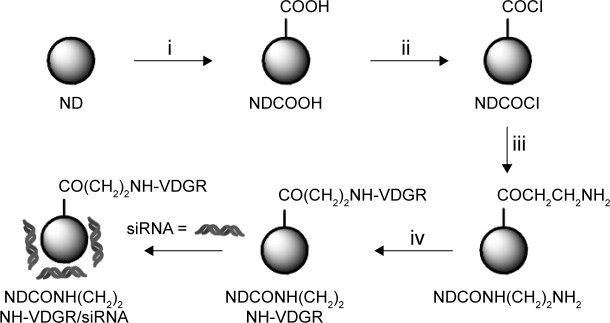
Figure 1.
Synthesis of NDCONH(CH2)2NH-VDGR/survivin-siRNA. Conditions in each step: (i) H2SO4/HNO3; (ii) SOCl2; (iii) NH2(CH2)2NH-Boc/THF and HCl/EA; and (iv) EDC/Boc-Arg(Tos)-Gly-Asp(OMe)-Val-OH, NaOH/MeOH, and TFA/TfOH.
Abbreviations: siRNA, small interfering RNA; THF, tetrahydrofuran; EA, ethyl alcohol; EDC, N1-((ethylimino)methylene)-N3,N3-dimethylpropane-1,3-diamine; TFA, trifluoroacetic acid; TfOH, trifluoromethanesulfonic acid; ND, nanodiamond.
Synthesis of NDCONH(CH2)2NH2
Carboxylated ND (200 mg) was added to 10 mL of thionyl chloride, the reaction was carried out at 70°C for 12 h, and NDCOCl was obtained (Figure 1, step ii). NH2CH2CH2NH-Boc (50 mg) and tetrahydrofuran were then added, and the suspension was stirred at 60°C for 6 h. After the reaction, the mixture was centrifuged at 5,000 g for 15 min and washed three times with distilled water. After freeze drying, 200 mg of NDCONH-CH2CH2NH-Boc was obtained. The NDCONH-CH2CH2NH-Boc was then added into HCl/ethyl alcohol in an ice-bath for 4 h, and NDCONH(CH2)2NH2 was synthesized (Figure 1, step iii).
Preparation of NDCONH(CH2)2NH-VDGR
NDCONH(CH2)2NH2 (50 mg), 20 mg of EDC, 20 mg of HOBt, and 25 mg of Boc-Arg(Tos)-Gly-Asp(OMe)-Val-OH were dispersed in 20 mL of DMF. The suspension was stirred at room temperature for 24 h and then centrifuged at 5,000 g for 15 min. The sediment was washed three times with distilled water, and after freeze drying, 50 mg of NDCONH(CH2)2NH-Val-Asp(OMe)-Gly-Arg(Tos)-Boc was obtained. NDCONH(CH2)2NH-Val-Asp(OMe)-Gly-Arg(Tos)-Boc was added to 20 mL of methanol, and pH value of the mixture was adjusted to 12 using 2 mol/L NaOH. NDCONH(CH2)2NH-Val-Asp-Gly-Arg(Tos)-Boc was obtained after 2 h. NDCONH(CH2)2NH-Val-Asp-Gly-Arg(Tos)-Boc (30 mg) was added into 10 mL of trifluoroacetic acid/trifluoromethanesulfonic acid (v/v, 4:1) and left for 1 h. Then, the suspension was centrifuged at 5,000 g for 5 min, and the sediment was washed three times with distilled water. After the sediment was freeze dried, NDCONH(CH2)2NH-Val-Asp-Gly-Arg-H (NDCONH(CH2)2NH-VDGR) was obtained (Figure 1, step iv).
Calculation of the linked amount of Boc-Arg(Tos)-Gly-Asp(OMe)-Val-OH
NDCONH(CH2)2NH-VDGR was obtained as a gray precipitate. In order to calculate the amount of Boc-Arg(Tos)-Gly-Asp(OMe)-Val-OH linked to NDCONH(CH2)2NH2, the ultraviolet (UV) absorbance of Boc-Arg(Tos)-Gly-Asp(OMe)-Val-OH in N,N-dimethylformamide (DMF) was measured using UV and visible spectrophotometer. The standard absorption curve of Boc-Arg(Tos)-Gly-Asp(OMe)-Val-OH was established under a UV detection wavelength of 268 nm. Three groups of 10 mg of NDCONH(CH2)2NH2 were dispersed in DMF, and 5 mg of Boc-Arg(Tos)-Gly-Asp(OMe)-Val-OH was added into DMF. After the reactions were completed, the suspension was centrifuged, and the supernatant was diluted 20-fold to measure the absorbance at 268 nm; the value was recorded as A1. Then, the precipitates were washed with DMF, and the absorbance of the supernatant was measured; the value was recorded as A2. These experiments were carried out repeatedly until all the absorbance values of supernatants reached zero (An =0).
Preparation of NDCONH(CH2)2NH-VDGR/survivin-siRNA nanoparticles
The NDCONH(CH2)2NH-VDGR/survivin-siRNA nanoparticles were prepared by adding NDCONH(CH2)2NH-VDGR to survivin-siRNA solution. The mixture was gently shaken and incubated for 30 min at room temperature. All complexes used in this experiment were prepared using fresh diethylpyrocarbonate (DEPC) water.
Characteristics of NDCONH(CH2)2NH-VDGR
The size and shape of ND and NDCONH(CH2)2NH-VDGR were evaluated by scanning electron microscopy (SEM), transmission electron microscopy (TEM), and atomic force microscopy (AFM), and the samples were dispersed in mouse plasma. The structure of the nanoparticle was determined by Fourier transform infrared (FT-IR) spectra. Zeta potentials of NDCONH(CH2)2NH-VDGR/survivin-siRNA were determined using zeta potential analyzer.
Gel retardation assay
Gel electrophoresis was carried out using 1% agarose gel to evaluate the loading capacity of NDCONH(CH2)2NH-VDGR. Similar to the method mentioned for the preparation of NDCONH(CH2)2NH-VDGR/survivin-siRNA, nanoparticles were prepared at different weight ratios (NDCONH(CH2)2NH-VDGR:survivin-siRNA =10:1, 20:1, 25:1, 30:1, 40:1). Then, the samples were loaded into the agarose gel, respectively, and the experiment was carried out in Tris–ethylene diamine tetraacetic acid (TE) buffer at a constant voltage of 120 V for 20 min. The result was obtained using UV gel image system.
Calorimetric analysis of NDCONH(CH2)2 NH-VDGR-absorbing survivin-siRNA
Calorimetric analyses were carried out with differential scanning calorimeter to confirm the loading of survivin-siRNA onto the nanoparticles. In this study, 20 nM NDCONH(CH2)2NH-VDGR suspension and 20, 40, 60, and 80 nM NDCONH(CH2)2NH-VDGR/survivin-siRNA were used. Differential scanning calorimetry (DSC) curves were obtained in a pierced aluminum pan with the temperature increasing from 20°C to 200°C (20°C min−1).
In vitro release of survivin-siRNA from NDCONH(CH2)2NH-VDGR/survivin-siRNA
Naked survivin-siRNA (20 nM), ND/survivin-siRNA, and NDCONH(CH2)2NH-VDGR/survivin-siRNA were dissolved in 500 μL DEPC water and sealed in a dialysis bag. The dialysis bag was suspended in 5 mL of TE buffer (10 mL of Tris–HCl and 1 mM EDTA, pH 8.0) at 37°C±0.5°C at a rotation speed of 100 g. TE buffer was refreshed at 0, 1, 2, and 4 h. Solution outside the bag was collected, centrifuged, and measured using a plate reader. The excitation and emission wavelengths were set at 492 and 520 nm, respectively. The amount of released survivin-siRNA was calculated according to the standard curve of survivin-siRNA. All survivin-siRNA was FAM labeled.
Cell culture
MCF-7 cells (human breast cancer cells; Institute of Basic Medical Sciences Chinese Academy of Medical Sciences, People’s Republic of China) were cultured with RPMI-1640 medium containing 10% FBS at 37°C in humidified atmosphere containing 5% CO2. MCF-7 cells were cultured to 70%–80% confluence for further use.
Cell transfection
MCF-7 cells (density of 2×105 cells/well) were seeded in six-well dishes and incubated overnight at 37°C under 5% CO2. Before transfection, medium was discarded, and cells were rinsed with phosphate-buffered saline (PBS) solution. Then, cells were treated with 100 nM survivin-siRNA, NDCONH(CH2)2NH-VDGR/NC, Lipo/NC, NDCONH(CH2)2NH-VDGR/survivin-siRNA, and Lipo/survivin-siRNA, respectively. Survivin-siRNA, NDCONH(CH2)2NH-VDGR/NC, and Lipo/NC were used as negative control, while Lipo/survivin-siRNA was used as positive control. After 4 h of incubation, the medium in each well was replaced with medium containing serum, and the cells were cultured for another 44 h.
Confocal microscopy analysis of NDCONH(CH2)2NH-VDGR/survivin-siRNA
The relocation of survivin-siRNA was determined by confocal laser scanning microscopy. Firstly, MCF-7 cells were seeded in 35 mm glass bottom culture dishes at a density of 1×106 cells/well in 2 mL medium and were incubated overnight. After the incubation, cells were attached onto the dishes and were transfected by 100 nM NDCONH(CH2)2 NH-VDGR/survivin-siRNA and survivin-siRNA for 48 h. Same volume of medium was added to the control. After 48 h, 1 mL of 4 μg/mL Hoechst 33342 was added to stain the cell nuclei for 30 min. Then, the liquid was abandoned, and cells were washed with PBS solution. Intracellular location of survivin-siRNA was observed by confocal microscope, and the samples were excited at 350 and 492 nm and emitted at 460 and 518 nm for Hoechst 33342 and FAM, respectively. All survivin-siRNA used in the experiment was FAM labeled.
Cytotoxicity assay
MTT assay was used to evaluate the cytotoxicity of NDCONH(CH2)2NH-VDGR against MCF-7 cells. MCF-7 cells were seeded into 96-well plate at the density of 5×105 cells/well and were cultured overnight for cell attachment. Then, 10–200 μg/mL of NDCONH(CH2)2NH-VDGR in 20 μL medium was added into experimental wells, and 20 μL of fresh medium was added to control wells. Six replicates were included in each group. After 48 h incubation, 25 μL of 5 mg/mL MTT solution was added into each well. The cells were incubated at 37°C for another 4 h. Then, the medium in each well was removed carefully, 200 μL of DMSO was added into each well, and the 96-well plates were shaken for 10 min. Finally, the optical density (OD) was measured at 490 nm. All experiments were repeated three times.
In vitro antiproliferation assay
MTT assay was used to evaluate the tumor growth after treatment with synthesized compounds. Firstly, MCF-7 cells were seeded into 96-well plates at the density of 5×105 cells/well and were cultured overnight. The cells were transfected with NDCONH(CH2)2NH-VDGR, survivin-siRNA, NDCONH(CH2)2NH-VDGR/NC, NDCONH(CH2)2NH-VDGR/survivin-siRNA, Lipo/NC, and Lipo/survivin-siRNA, respectively, at the concentration of 30, 60, 90, and 120 nM, and all test groups were incubated for 48 h. In order to investigate the relationship between the concentration of ND-b-VDGR and tumor growth inhibition, MCF-7 cells were divided into different groups containing 5–200 nM for 48 h. After 48 h, 25 μL of 5 mg/mL sterile MTT solution was added into each well, and the cells were incubated at 37°C for 4 h. Then, the medium in each well was removed carefully. DMSO (200 μL) was added into each well, and the 96-well plates were shaken for 10 min. The OD was measured at 490 nm. All experiments were repeated for three times.
Analysis of apoptosis by flow cytometry (Annexin V–FITC/PI staining)
MCF-7 cells (2×105 cells/well) in six-well dishes were transfected by 100 nM survivin-siRNA, NDCONH(CH2)2NH-VDGR/survivin-siRNA, and NDCONH(CH2)2NH-VDGR for 4 h, and the untransfected cells served as blank control. After the transfection, the medium in each well was replaced with serum medium, and the cells were cultured for another 44 h. After 44 h, MCF-7 cells were harvested and washed by PBS solution. Then, the harvested cells were dispersed in 100 μL of binding buffer (1×) containing 5 μL of Annexin V–fluorescein isothiocyanate (FITC) and 1 μL of PI (100 μg/mL), respectively. Cells were stained for 15 min at room temperature in dark. After 15 min, 400 μL of binding buffer (1×) was added to each tube, and the cells were analyzed by flow cytometry. MCF-7 cells stained only by Annexin-V or PI served as control.
Analysis of cell cycle by flow cytometry
MCF-7 cells were transfected at a density of 2×105 cells/well in six-well dishes with 100 nM survivin-siRNA, NDCONH(CH2)2NH-VDGR/survivin-siRNA, and Lipo/survivin-siRNA for 4h. The untransfected cells served as blank control. Then, the medium in each well was replaced with serum medium, and the cells were incubated at 37°C continuously. MCF-7 cells were harvested at 12, 24, and 48 h, and the harvested MCF-7 cells were dispersed into 10 mL of 80% alcohol and were stored at −20°C. Then, the cells were centrifuged and washed by cold PBS solution for three times, and dispersed in 500 μL PBS solution containing 5 μL RNase and PI (50 μg/mL). The mixture was left at room temperature for 30 min and analyzed by flow cytometry.
Real-time PCR
Real-time PCR, a variant of PCR, is a technique commonly used in molecular biology to detect RNA expression. In this study, real-time PCR was used to evaluate the expression of survivin-mRNA. MCF-7 cells (2×105 cells/well) in six-well dishes were transfected by 100 nM survivin-siRNA, NDCONH(CH2)2NH-VDGR/NC, NDCONH(CH2)2NH-VDGR/survivin-siRNA, Lipo/NC, and Lipo/survivin-siRNA for 48 h. After 48 h, Trizol reagent was used to extract the total cell RNA, and all operations were carried out on ice. The concentration of the RNA was measured by Nanodrop-1000. RNA (2 μg) was reverse-transcribed into cDNA using PCR amplification instrument, and the concentration of cDNA was measured by Nanodrop-1000. cDNA (80 ng) was subjected to real-time PCR according to standard instructions. Primer pairs for survivin (forward sequence, 5′-GCATGGGTCCCCCGACGTTG-3′; reverse sequence, 5′-GCTCCGGCCAGAGGCCTCAA-3′) and GAPDH (forward sequence, 5′-CAAATTCCATGGCACCGTCA-3′; reverse sequence, 5′-GGAGTGGGTGTCGCTGTTGA-3′) were synthesized and purified by GenePharma Co., Ltd. The mRNA expression of survivin was evaluated against GAPDH mRNA. The Ct value was calculated using delta-delta Ct (2−ΔΔCt) method. All experiments were repeated for three times.
Enzyme-linked immunosorbent assay
MCF-7 cells (2×105 cells/well) were seeded in six-well plates. After the cells reached 70%–80% confluence, they were transfected with 100 nM survivin-siRNA, NDCONH(CH2)2NH-VDGR/NC, NDCONH(CH2)2NH-VDGR/survivin-siRNA, Lipo/NC, and Lipo/survivin-siRNA for 48 h. Then, the whole-cell protein was extracted, and all operations were carried out on ice. Firstly, the medium in six-well plates was discarded, and each well was washed by PBS solution for three times. Then, 1 mL of PBS solution was added into each well, and the cells were collected by cell scraper into 1.5 mL centrifuge tubes, which were centrifuged at 4,000 g for 5 min subsequently. The supernatant was abandoned, and 100 μL of protein lysis buffer (RIPA:cocktail:200 mM phenylmethane sulfonyl fluoride, 100:1:1, v/v/v) was added into each tube. After 30 min, tubes were centrifuged at 4,000 g for 10 min, and the supernatant was collected. Then, BCA Protein Quantification Kit was used to measure the protein concentration of the supernatant. Survivin enzyme-linked immunosorbent assay (ELISA) kit was used to measure the amount of survivin protein according to the instructions. All experiments were repeated for three times.
In vivo antitumor assay
Male ICR mice (6-week-old, 18–22 g) were obtained from the Animal Department of Capital Medical University (Beijing Laboratory Animal Center, Beijing, People’s Republic of China). All animal work was performed according to the Health Guidelines of the Capital Medical University, and protocols were approved by the Institutional Animal Ethics Committee of Capital Medical University. Tumor xenograft was made by inoculating 1×107 S180 cells into all mice. A dial caliper was used to monitor tumor volumes (tumor volume = length × width2/2). After the volume of the tumor reached 150 mm3 approximately, all mice were randomly divided into four groups, each containing 12 mice. Four groups of mice were treated with NDCONH(CH2)2NH-VDGR/survivin-siRNA (0.3 mg/kg survivin-siRNA and 7.5 mg/kg NDCONH(CH2)2NH-VDGR), naked survivin-siRNA (0.3 mg/kg), doxorubicin (DOX; 2 μmol/kg), and saline (NS) every other day, respectively, for a total of five times via tail injections. After 10 days, mice were sacrificed to harvest the brain, liver, spleen, lung, heart, kidney, and tumor. Deionized water was added and mixed with viscera, and the organs and tumor tissues from NDCONH(CH2)2NH-VDGR/survivin-siRNA group were homogenized on ice to extract RGDV. The homogenized liquid was centrifuged at 12,000 g for 10 min. The supernatant was collected, and 200 μL of supernatant and 400 μL of methanol were added, respectively, into 1.5 mL centrifuge tubes to precipitate protein. The suspension was centrifuged at 12,000 g for 10 min. The supernatant was collected and concentrated by rotary evaporator, and 200 μL chromatographic pure methanol was used to resolve the residue. Magnetic sector mass analyzer was used to analyze the samples.
Statistical analysis
All experiments were repeated at least three times. Quantitative data were expressed as the mean ± standard deviation. Statistical analysis was performed using SPSS V13.0 software. Student’s t-test was used to evaluate the differences between groups. A P-value <0.05 was considered to be statistically significant, and a P-value <0.01 was considered to be very statistically significant.
Results and discussion
Calculation of the amount of effectively linked NDCONH(CH2)2NH2RGDV
Boc-Arg(Tos)-Gly-Asp(OMe)-Val-OH was linked on the surface of ND by the NDCONH(CH2)2NH-VDGR preparation method mentioned earlier. The RGDV concentration in supernatant was used to calculate the amount of successfully linked RGDV. The results indicated that 1.28±0.12 mg of RGDV was successfully linked on 10 mg of NDCONH(CH2)2NH2.
SEM, TEM, AFM, particle size, zeta potential, and FT-IR
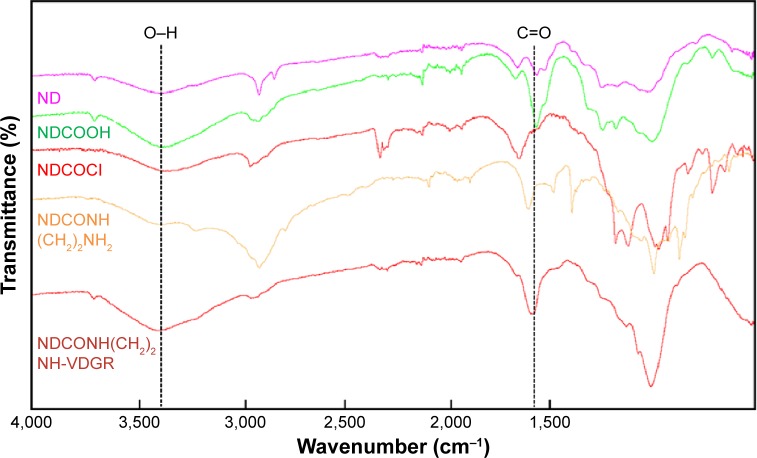
The FT-IR spectra of ND, NDCO2H, NDCOCl, NDCONH (CH2)2NH2, and NDCONH(CH2)2NH-VDGR are shown in Figure 2. The appearance of characteristic absorption peaks at 3,200–3,500 and 1,700 cm−1 indicated the presence of carboxyl group. After the reaction of acylating chlorination, the peak at 3,200–3,500 cm−1 was weakened. The appearance of absorption peaks at 2,900 and 1,500 cm−1 showed that ethyldiamine was successfully linked to ND-COOH. The spectra of NDCONH(CH2)2NH-VDGR showed that the peak of carboxyl group appeared at 3,200–3,500 (O–H stretching vibration) and 1,700 cm−1 (C=O stretching vibration). Meanwhile, the appearance of the peak at 1,670 cm−1 (amide I) indicated that H-Arg-Gly-Asp-Val-OH was covalently linked to NDCONH(CH2)2NH2.
Figure 2.
FT-IR spectra of ND, NDCO2H, NDCOCl, NDCONH(CH2)2NH2, and NDCONH(CH2)2NH-VDGR.
Abbreviations: FT-IR, Fourier transform infrared; ND, nanodiamond.
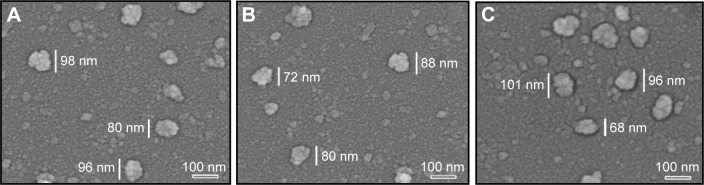
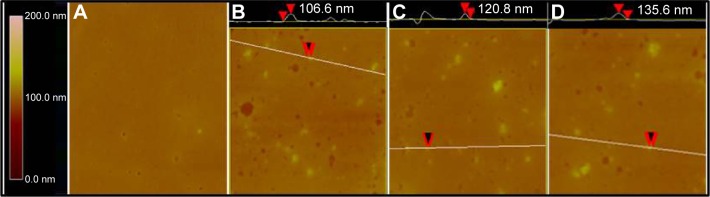
TEM images showed that ND, NDCONH(CH2)2NH-VDGR, and NDCONH(CH2)2NH-VDGR/survivin-siRNA nanoparticles, which comprised smaller NDs (around 10 nm in diameter, marked by the red circles), were approximately 30–90 nm in diameter (Figure 3). In Figure 4, SEM images show that the ND, NDCONH(CH2)2NH-VDGR, and NDCONH(CH2)2NH-VDGR/survivin-siRNA formed irregular nano-globes from 60 to 110 nm in diameter, which resulted from the aggregation of smaller nanoparticles. These smaller nanoparticles with high surface energy, less than 10 nm in diameter, can assemble themselves into clusters in order to minimize their surface energy.28 The nano-structures were then characterized with the AFM images as shown in Figure 5. The results showed that ND, NDCONH(CH2)2NH-VDGR, and NDCONH(CH2)2NH-VDGR/survivin-siRNA formed nanoparticles which were 106.6, 120.8, and 135.6 nm in diameter, respectively. The results suggested that the size and shape of nanoparticles would stay stable as nano-globes and could be stable after delivery into blood circulation.
Figure 3.
TEM images of ND (A), NDCONH(CH2)2NH-VDGR (B), and NDCONH(CH2)2NH-VDGR/survivin-siRNA (C).
Abbreviations: TEM, transmission electron microscopy; ND, nanodiamond; siRNA, small interfering RNA.
Figure 4.
SEM images of ND (A), NDCONH(CH2)2NH-VDGR (B), and NDCONH(CH2)2NH-VDGR/survivin-siRNA (C).
Abbreviations: SEM, scanning electron microscopy; ND, nanodiamond; siRNA, small interfering RNA.
Figure 5.
AFM images of mouse plasma alone (A), ND in mouse plasma (B), NDCONH(CH2)2NH-VDGR in mouse plasma (C), and NDCONH(CH2)2NH-VDGR/survivin-siRNA in mouse plasma (D).
Abbreviations: AFM, atomic force microscopy; ND, nanodiamond; siRNA, small interfering RNA.
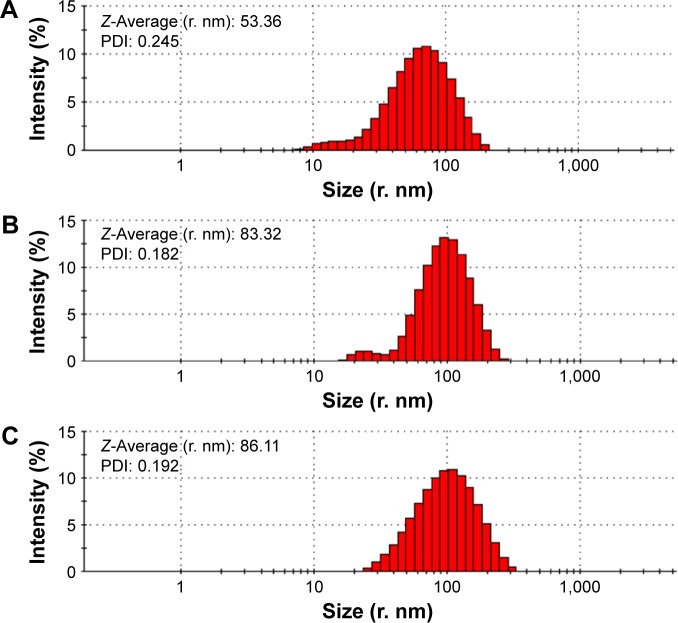
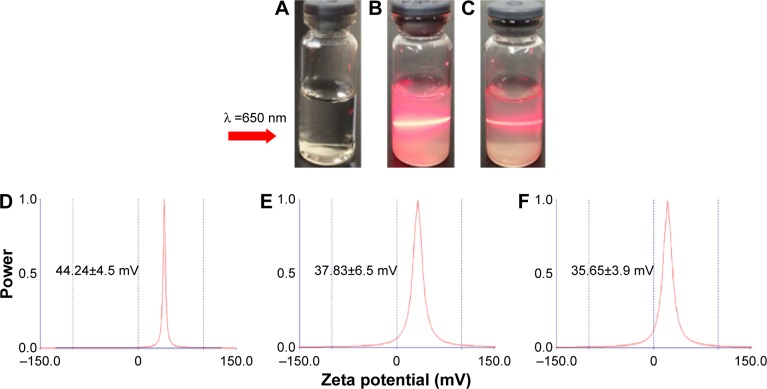
Figure 6 displays that the average hydrodynamic radius of ND, NDCONH(CH2)2NH-VDGR, and NDCONH(CH2)2NH-VDGR/survivin-siRNA nanoparticles was 53.36, 83.32, and 86.11 nm, respectively. The hydrodynamic radius of nanoparticles was determined by dynamic light scattering method, a technique that measures the size of nanoparticles in a hydrated state with the coating material and the solvent layer attached to the nanoparticles;29–31 therefore, the values were much larger than that measured by TEM and SEM in a dried state. In addition, the polydispersity indexes of NDCONH(CH2)2NH-VDGR or NDCONH(CH2)2NH-VDGR/survivin-siRNA were less than 0.2, which indicated that the nanoparticles were well dispersed and uniform. Figure 7 displays that the zeta potentials of ND, NDCONH(CH2)2NH-VDGR, and NDCONH(CH2)2NH-VDGR/survivin-siRNA were 44.24±4.5, 37.83±6.5, and 35.65±3.85, respectively, which benefits the cellular uptake and indicated that the dispersion systems had good stability.32 As shown in Figure 7B and C, the Tyndall phenomenon was observed clearly, which suggested that ND and NDCONH(CH2)2NH-VDGR could form a nano-level suspension.25,33
Figure 6.
Particle size and PDI of ND (A) and NDCONH(CH2)2NH-VDGR (B), and NDCONH(CH2)2NH-VDGR/survivin-siRNA (C).
Abbreviations: PDI, polydispersity index; ND, nanodiamond; siRNA, small interfering RNA.
Figure 7.
Tyndall effect of ND (B) and NDCONH(CH2)2NH-VDGR (C). Water (A) served as control. The zeta potential of ND (D), NDCONH(CH2)2NH-VDGR (E), and NDCONH(CH2)2NH-VDGR/survivin-siRNA (F).
Abbreviations: ND, nanodiamond; siRNA, small interfering RNA.
Gel retardation assay
According to Dolmatov’s research,34 ND was synthesized using detonation synthesis method, which resulted in multiple positively charged surfaces. It is reported that ND has the ability to absorb drugs by means of electrostatic interactions and then deliver them into cells.23,35,36 The gel retardation assay was used to confirm the loading capacity of survivin-siRNA onto NDCONH(CH2)2NH-VDGR. The retardation results of NDCONH(CH2)2NH-VDGR and survivin-siRNA (w/w) are listed in Figure 8.
Figure 8.
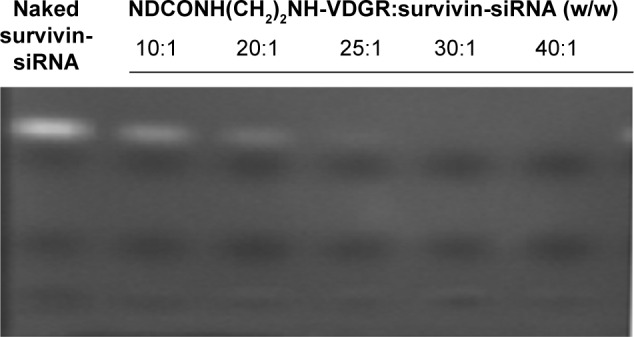
Agarose gel retardation of naked survivin-siRNA and NDCONH(CH2)2NH-VDGR/survivin-siRNA at different N/P ratios.
Abbreviations: siRNA, small interfering RNA; N, negative; P, positive.
Naked survivin-siRNA served as control, and the free siRNA was visualized as a bright band on the gel. The results showed that, when the ratio of NDCONH(CH2)2NH-VDGR and survivin-siRNA reached 25:1 (w/w), the bright band disappeared, and the results suggested that survivin-siRNA could be absorbed onto NDCONH(CH2)2NH-VDGR completely.37 According to the results, NDCONH(CH2)2NH-VDGR/survivin-siRNA nanoparticles were prepared at the ratio of 25:1 (w/w).
DSC of NDCONH(CH2)2NH-VDGR/survivin-siRNA
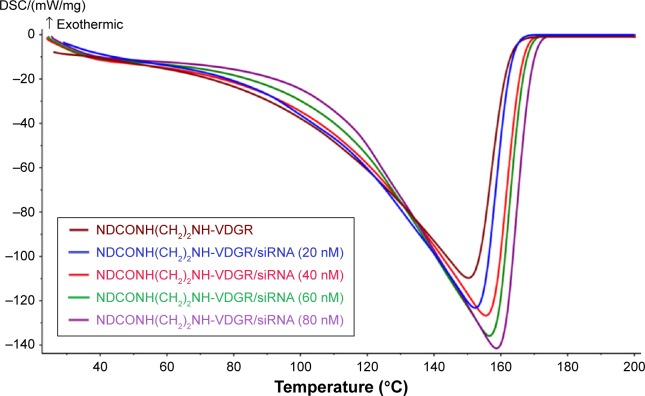
DSC is an auxiliary tool to prove the connection of survivin-siRNA and NDCONH(CH2)2NH-VDGR in Figure 9.25 The result showed that with the increasing amount of loaded survivin-siRNA, the endothermic peaks shifted from 152°C to 159°C. The changes in the DSC curves resulted from the interactions between NDCONH(CH2)2NH-VDGR and survivin-siRNA,30,38 which also suggested that survivin-siRNA was successfully absorbed onto the surface of NDCONH(CH2)2NH-VDGR.
Figure 9.
The DSC curves of NDCONH(CH2)2NH-VDGR and different concentrations of NDCONH(CH2)2NH-VDGR/survivin-siRNA.
Abbreviations: DSC, differential scanning calorimetry; siRNA, small interfering RNA.
Release of survivin-siRNA from NDCONH(CH2)2NH-VDGR/survivin-siRNA
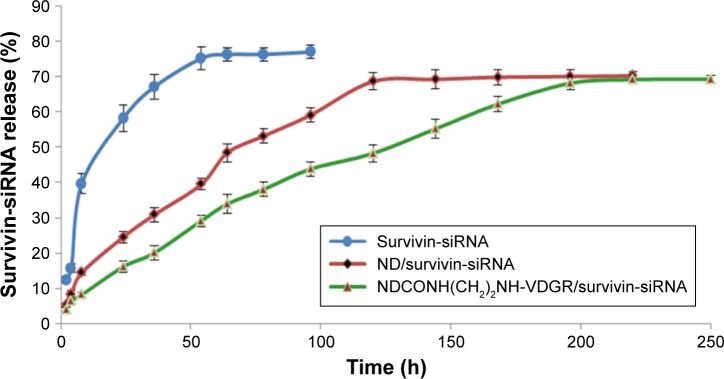
The release curves of survivin-siRNA in TE buffer are shown in Figure 10. Naked survivin-siRNA was released quickly, and the cumulative releasing amount reached 70% within 54 h. But the release of survivin-siRNA from NDCONH(CH2)2NH-VDGR/survivin-siRNA was slow, and only 30% of survivin-siRNA was released from NDCONH(CH2)2NH-VDGR/survivin-siRNA in the first 54 h. The accumulated releasing amount increased to approximately 60% in 200 h gradually, which was attributed to the electrostatic interaction between the carrier and the drug.39 The results indicated that compared to the ND/survivin-siRNA group, the release of survivin-siRNA from NDCONH(CH2)2NH-VDGR/survivin-siRNA lasted longer, and this sustained-release feature of survivin-siRNA from NDCONH(CH2)2NH-VDGR could benefit its gene-silencing effect.
Figure 10.
The cumulative releasing percentage of survivin-siRNA from NDCONH(CH2)2NH-VDGR/survivin-siRNA, ND/survivin-siRNA, and naked survivin-siRNA in TE buffer (n=3).
Abbreviations: siRNA, small interfering RNA; ND, nanodiamond; TE, Tris–ethylene diamine tetraacetic acid.
Cellular uptake of NDCONH(CH2)2NH-VDGR/survivin-siRNA
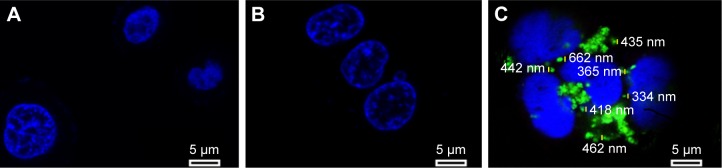
To test the internalization of siRNA delivery, MCF-7 cells were transfected for 6 h and observed under confocal microscope; the result is shown in Figure 11. The MCF-7 cells were treated with PBS solution (blank control), naked survivin-siRNA, and NDCONH(CH2)2NH-VDGR/survivin-siRNA at a dose of 100 nM, respectively. Green fluorescence (FAM-survivin-siRNA) was observed around the nucleus (blue fluorescence) of MCF-7 cells in NDCONH(CH2)2NH-VDGR/FAM-survivin-siRNA treated group, compared to naked survivin-siRNA group. Therefore, it could be concluded that survivin-siRNA can be transferred into the cytoplasm of MCF-7 cells by NDCONH(CH2)2NH-VDGR, and NDCONH(CH2)2NH-VDGR/survivin-siRNA exhibited much higher transfection efficiency than naked survivin-siRNA. The results suggested that NDCONH(CH2)2NH-VDGR has good transfection ability which might benefit its gene-silencing effect.
Figure 11.
Confocal images of control (A), cells treated with naked FAM-survivin-siRNA (B), and cells treated with NDCONH(CH2)2NH-VDGR/FAM-survivin-siRNA (C).
Abbreviation: siRNA, small interfering RNA.
NDCONH(CH2)2NH-VDGR has no cytotoxicity
Cytotoxicity of gene carrier is one of the main disadvantages and needs to be considered. ND has been proven to have high transfection efficiency with low cytotoxicity.40,41 The interaction of integrin with the RGDV sequence has been widely used in drug delivery systems.42,43 Arg-Gly-Asp-Val-modified ND should be bio-safe and nontoxic. MTT assay was used to demonstrate the above-mentioned hypothesis, and MCF-7 cell line was used. The concentrations of NDCONH(CH2)2NH-VDGR used were from 10 to 100 μg/mL. As shown in Figure 12, NDCONH(CH2)2NH-VDGR in all groups had no significant inhibition effect on proliferation of cells, and the cell viability in each group was above 85%. Therefore, NDCONH(CH2)2NH-VDGR could be a genetic drug carrier with no cytotoxicity.
Figure 12.
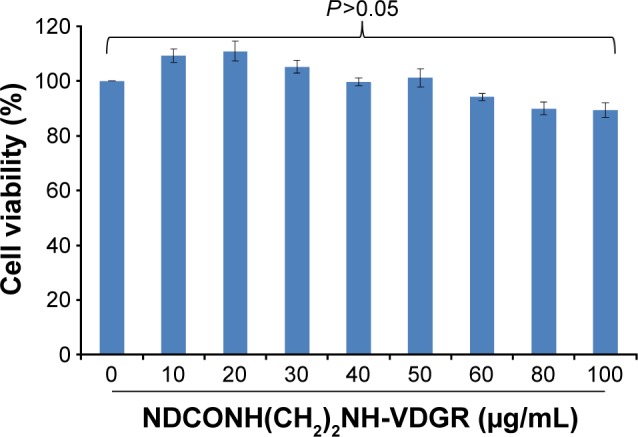
Cytotoxicity assay using MTT method. Data are presented as the average ± SD (n=3).
Abbreviations: MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; SD, standard deviation.
Cell proliferation-inhibitory activity in vitro
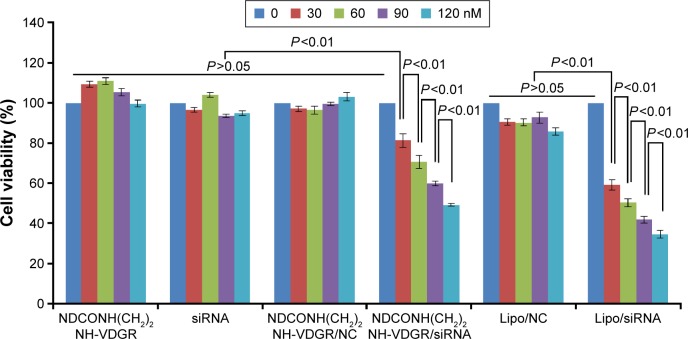
The most important standard for the stabilized drug–carrier complex is to measure the pharmacological activity of drug.44,45 In this study, survivin-siRNA gene expression was expected to be downregulated in order to inhibit the tumor growth. MCF-7 cells were treated with NDCONH(CH2)2 NH-VDGR, survivin-siRNA, NDCONH(CH2)2NH-VDGR/NC, NDCONH(CH2)2NH-VDGR/survivin-siRNA, Lipo/NC, and Lipo/survivin-siRNA at concentration of 30, 60, 90, and 120 nM, respectively, and the results of selected concentration of compounds against the cell viability are shown in Figure 13. NDCONH(CH2)2NH-VDGR, survivin-siRNA, NDCONH(CH2)2NH-VDGR/NC, and Lipo/NC served as negative controls, and Lipo/survivin-siRNA served as positive control. There was no difference between the cell viability of NDCONH(CH2)2NH-VDGR, NDCONH(CH2)2NH-VDGR/NC, naked survivin-siRNA, and Lipo/NC groups, which indicated no inhibitory activity. In NDCONH(CH2)2NH-VDGR/survivin-siRNA group, the cell viability was very significantly lower than that of NDCONH(CH2)2NH-VDGR/NC group (P<0.01), while in Lipo/survivin-siRNA group, values of all the concentration groups were similar. The results suggested that survivin-siRNA was successfully delivered into MCF-7 cells and could effectively suppress cell proliferation. Moreover, with the increase in the amount of NDCONH(CH2)2NH-VDGR/survivin-siRNA, the cell viability was reduced from 80% to less than 50% gradually. This decreased pattern indicated that the cell proliferation-inhibitory activity of NDCONH(CH2)2NH-VDGR/survivin-siRNA had a concentration-dependent trend.
Figure 13.
Viability of MCF-7 cells after treatment with different concentrations of NDCONH(CH2)2NH-VDGR, survivin-siRNA, NDCONH(CH2)2NH-VDGR/NC, NDCONH(CH2)2NH-VDGR/survivin-siRNA, Lipo/NC, and Lipo/survivin-siRNA for 48 h. Data are presented as the average ± SD (n=3).
Abbreviations: siRNA, small interfering RNA; NC, normal control; SD, standard deviation.
The MCF-7 cells were treated with NDCONH(CH2)2NH-VDGR/survivin-siRNA at the concentration of 5–200 nM. The results showed that with the increasing concentration of NDCONH(CH2)2NH-VDGR/survivin-siRNA, cell viability decreased gradually as shown in Figure 14. The IC50 of NDCONH(CH2)2NH-VDGR/survivin-siRNA was calculated at 110.6 nM. According to the results, NDCONH(CH2)2NH-VDGR is a highly efficient carrier to deliver survivin-siRNA.
Figure 14.
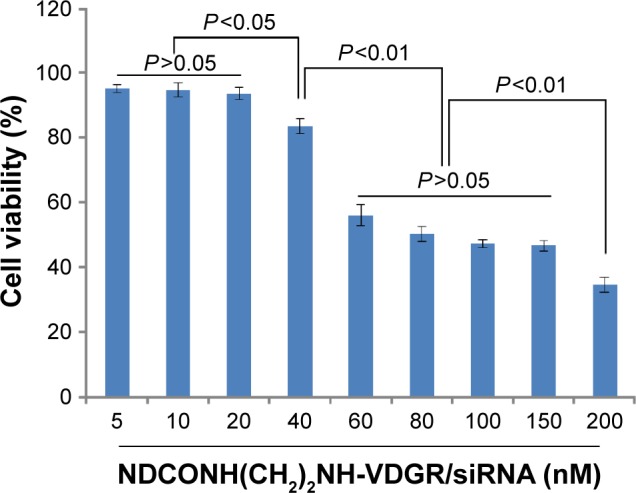
Antiproliferation effect of NDCONH(CH2)2NH-VDGR/survivin-siRNA against MCF-7 cells at different concentrations. Data are presented as the average ± SD (n=3).
Abbreviations: siRNA, small interfering RNA; SD, standard deviation.
Induction of cell cycle arrest and apoptosis
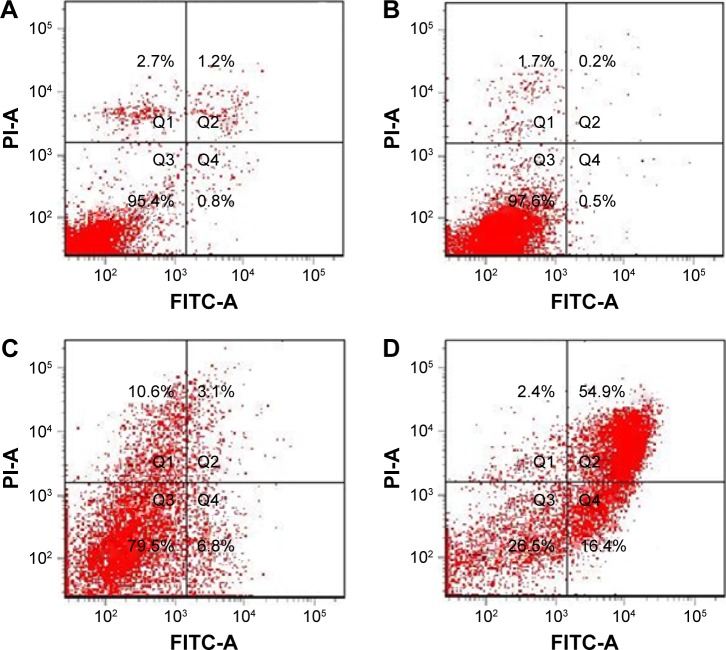
The results of apoptosis analysis (Figure 15) showed that the apoptosis rate of the NDCONH(CH2)2NH-VDGR/survivin-siRNA group (71.3%) was remarkably higher than that of naked siRNA (0.7%), NDCONH(CH2)2NH-VDGR (9.9%), and blank control group (2.0%), which confirmed that survivin-siRNA had been efficiently delivered into MCF-7 cells by the NDCONH(CH2)2NH-VDGR nanoparticles to promote apoptosis. The results indicated that the proliferation-inhibitory activity of NDCONH(CH2)2NH-VDGR/survivin-siRNA might be caused by induction of apoptosis.46
Figure 15.
The apoptosis of MCF-7 cells induced by survivin-siRNA (B), NDCONH(CH2)2-NH-VDGR (C), and NDCONH(CH2)2NH-VDGR/survivin-siRNA (D). Cells treated with PBS solution served as control (A) (n=3).
Abbreviations: siRNA, small interfering RNA; PBS, phosphate-buffered saline; FITC-A, fluorescein isothiocyanate–Annexin V; PI-A, propidium iodide–Annexin V.
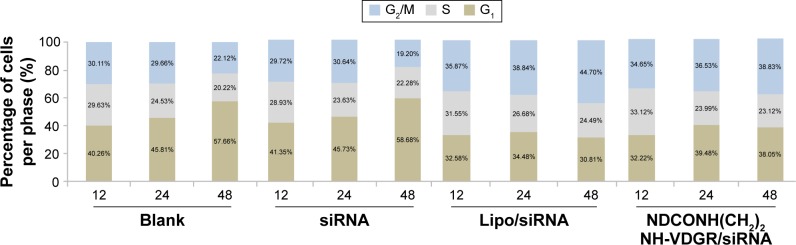
To confirm the apoptotic ability of NDCONH(CH2)2NH-VDGR/survivin-siRNA, the cell cycle study was performed. As shown in Figure 16, cell cycle arrest at G2/M was induced by NDCONH(CH2)2NH-VDGR/survivin-siRNA at 48 h (38.83%), while the cell cycle profile of MCF-7 cells did not change significantly (19.20%) when treated with naked survivin-siRNA compared to blank group (22.12%). The result suggested that the inhibitory effect of NDCONH(CH2)2NH-VDGR/survivin-siRNA resulted from the apoptosis by cell cycle arrest at G2/M.
Figure 16.
The cell percentage of blank, naked survivin-siRNA, NDCONH(CH2)2NH-VDGR/survivin-siRNA, and Lipo/survivin-siRNA group in each period of cell cycle (n=3).
Abbreviation: siRNA, small interfering RNA.
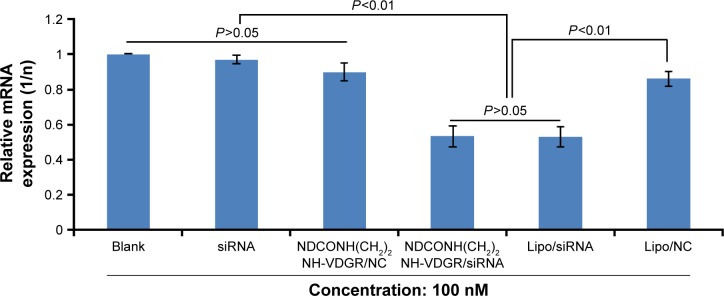
Real-time PCR
The expression of survivin mRNA was determined by real-time PCR in MCF-7 cells, and the results are shown in Figure 17. NDCONH(CH2)2NH-VDGR/NC served as negative control and Lipo/survivin-siRNA served as positive control. The results indicated that the expression of survivin mRNA in NDCONH(CH2)2NH-VDGR/survivin-siRNA group (0.5323±0.063) was significantly decreased compared with negative control group (0.9056±0.048) and equal to the positive control group (0.5306±0.061). The results could further prove that NDCONH(CH2)2NH-VDGR/survivin-siRNA could successfully downregulate the expression of survivin mRNA.
Figure 17.
Survivin-mRNA expression of MCF-7 cells treated with NDCONH(CH2)2NH-VDGR/survivin-siRNA. Data are presented as the average ± SD (n=3).
Abbreviations: siRNA, small interfering RNA; SD, standard deviation; NC, normal control.
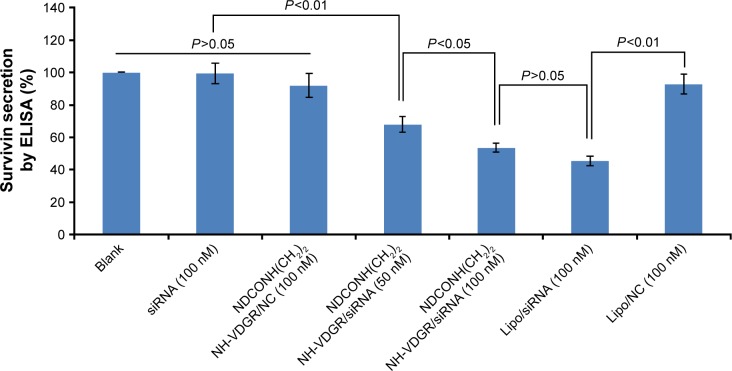
Enzyme-linked immunosorbent assay
ELISA was used to evaluate the protein expression in NDCONH(CH2)2NH-VDGR/survivin-siRNA-treated MCF-7 cells. Cells treated with PBS solution and Lipo/survivin-siRNA served as blank and positive control, respectively. The result of ELISA is shown in Figure 18; it indicated that NDCONH(CH2)2NH-VDGR/NC, naked survivin-siRNA, and Lipo/NC group had no obvious gene-silencing effect, while NDCONH(CH2)2NH-VDGR/survivin-siRNA group (100 nM) showed significantly decreased expression of survivin, compared with Lipo/survivin-siRNA (P>0.05). The results indicated that the expression of survivin protein was significantly downregulated by NDCONH(CH2)2NH-VDGR/survivin-siRNA.
Figure 18.
Survivin protein expression of MCF-7 cells treated with NDCONH(CH2)2NH-VDGR/survivin-siRNA. Data are presented as the average ± SD (n=3).
Abbreviations: siRNA, small interfering RNA; SD, standard deviation; ELISA, enzyme-linked immunosorbent assay; NC, normal control.
Antitumor activity of NDCONH(CH2)2 NH-VDGR/survivin-siRNA in vivo
The results from animal experiments were important standard for assessing the availability of gene carriers.47,48 Therefore, in this study, ICR mice were used to evaluate the activity of NDCONH(CH2)2NH-VDGR/survivin-siRNA in vivo. The average tumor weight of NS (control), survivin-siRNA, NDCONH(CH2)2NH-VDGR/survivin-siRNA, and DOX group is shown in Figure 19.
Figure 19.
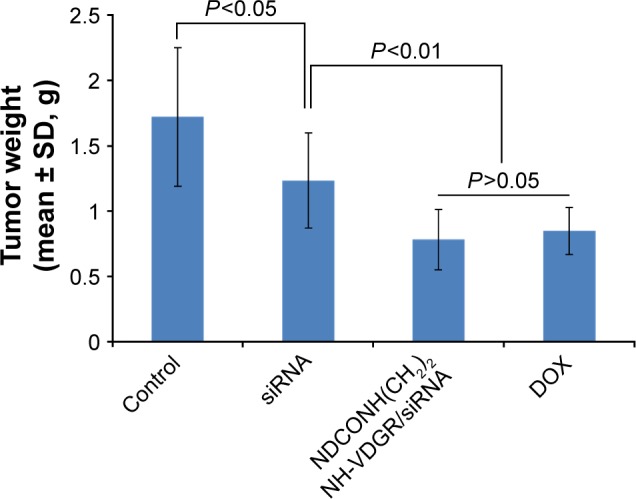
Tumor weights of the normal control (NS), survivin-siRNA group, NDCONH(CH2)2NH-VDGR/survivin-siRNA group, and DOX group (n=10).
Abbreviations: siRNA, small interfering RNA; DOX, doxorubicin; SD, standard deviation.
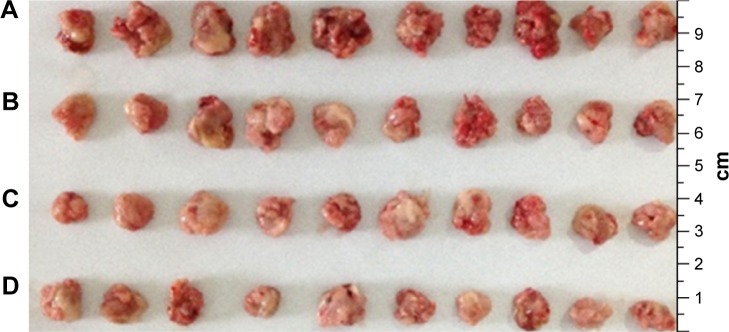
The tumor weights were not significantly different (P>0.05) between the naked survivin-siRNA group and NS group, but the tumor weights of both the groups were significantly higher than the NDCONH(CH2)2NH-VDGR/survivin-siRNA group (P<0.01), which indicated that the naked survivin-siRNA could not inhibit the tumor growth. And the tumor weight of NDCONH(CH2)2NH-VDGR/survivin-siRNA group was equal to that of DOX group (P>0.05), which suggested that NDCONH(CH2)2NH-VDGR/survivin-siRNA exerted strong inhibition on tumor growth in vivo. It could be further concluded that survivin-siRNA was successfully transported into cells by NDCONH(CH2)2NH-VDGR and had inhibited the growth of tumor in vivo. The similar results can be obtained from Figure 20; the tumor size in the NDCONH(CH2)2NH-VDGR/survivin-siRNA group was smaller than the NS group, which indicated that the tumor growth was effectively suppressed by NDCONH(CH2)2NH-VDGR/survivin-siRNA.
Figure 20.
Tumor tissues from normal control (NS) group (A), survivin-siRNA group (B), NDCONH(CH2)2NH-VDGR/survivin-siRNA group (C) and DOX group (D).
Abbreviations: siRNA, small interfering RNA; DOX, doxorubicin.
Targeting efficiency of NDCONH(CH2)2 NH-VDGR/survivin-siRNA in vivo
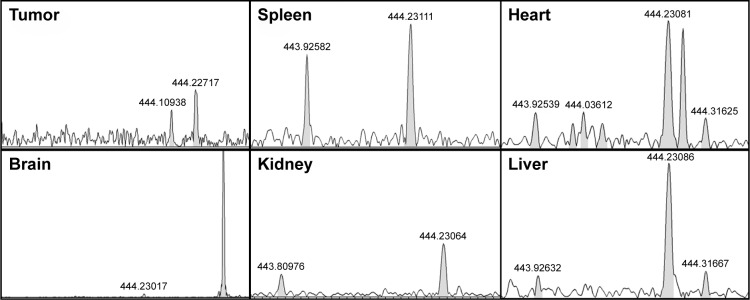
Gene-silencing selectivity of NDCONH(CH2)2NH-VDGR/survivin-siRNA depends on the presence of RGD-targeting peptide,30 and the magnetic sector mass was applied for RGDV detection.28 The mass-to-charge ratio (m/z) of RGDV was 444.22012 [M-H]−. The spectra are shown in Figure 21.
Figure 21.
MS spectra of organ homogenate in NDCONH(CH2)2NH-VDGR/survivin-siRNA group. RGDV (m/z): 444.22012 [M-H]−.
Abbreviations: MS, mass spectrometry; siRNA, small interfering RNA.
The mass spectrometry results showed that RGDV was detected in tumors of NDCONH(CH2)2NH-VDGR/survivin-siRNA treatment group, which confirmed that NDCONH(CH2)2NH-VDGR/survivin-siRNA has tumor-targeting ability as an siRNA carrier. It was also suggested that RGDV-modified ND could be a potential tumor-targeting carrier.
Conclusion
In this study, a novel targeted gene delivery system, NDCONH(CH2)2NH-VDGR, containing ND (gene delivery vehicle), H-Arg-Gly-Asp-Val-OH (targeting agent), and EDA (joint arm) was synthesized. The results suggested that not only could survivin-siRNA be loaded onto the surface of NDCONH(CH2)2NH-VDGR and successfully delivered into MCF-7 cells but it could also downregulate the expression of survivin protein and result in tumor apoptosis. Most important of all, the nanoparticles could effectively target the tumor cells, and the survivin-siRNA could be delivered into tumor cells by NDCONH(CH2)2NH-VDGR, which showed a strong inhibitory activity on tumor growth in vivo and in vitro. All the results confirmed that NDCONH(CH2)2NH-VDGR was an efficient targeted gene carrier. Modified ND was a potential gene carrier, and further studies are required.
Acknowledgments
This work was supported by the National Natural Science Foundation (81502688), the Basic Clinical Key Research Grant (16JL72) from the Capital Medical University, the Importation and Development of High-Caliber Talents Project of Beijing Municipal Institutions (2013–2015), the Natural Science Foundation of Capital Medical University (2015ZR14), and China’s 55th postdoctoral scientific research funds (2014M550768). The authors gratefully acknowledge the support from Beijing area Major Laboratory of Peptide and Small Molecular Drugs, Engineering Research Center of Endogenous Prophylactic of Ministry of Education of China, and Beijing Laboratory of Biomedical Materials.
Footnotes
Disclosure
The authors confirm that the content of the paper entails no conflicts of interest.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Hannon GJ. RNA interference. Nature. 2002;418(6894):244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 4.Hutvágner G, Zamore PD. RNAi: nature abhors a double-strand. Curr Opin Genet Dev. 2002;12(2):225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Li Z, Han Y, Liang LH, Ji A. Nanoparticle-based delivery system for application of siRNA in vivo. Curr Drug Metab. 2010;11(2):182–196. doi: 10.2174/138920010791110863. [DOI] [PubMed] [Google Scholar]
- 6.Aliabadi HM, Landry B, Sun C, Tang T, Uludağ H. Supramolecular assemblies in functional siRNA delivery: where do we stand? Biomaterials. 2012;33(8):2546–2569. doi: 10.1016/j.biomaterials.2011.11.079. [DOI] [PubMed] [Google Scholar]
- 7.Lo Muzio L, Pannone G, Leonardi R, et al. Survivin, a potential early predictor of tumor progression in the oral mucosa. J Dent Res. 2003;82(11):923–928. doi: 10.1177/154405910308201115. [DOI] [PubMed] [Google Scholar]
- 8.Abe T, Goda K, Futami K, Furuichi Y. Detection of siRNA administered to cells and animals by using a fluorescence intensity distribution analysis polarization system. Nucleic Acids Res. 2009;37(7):e56. doi: 10.1093/nar/gkp131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thakur A, Fitzpatrick S, Zaman A, et al. Strategies for ocular siRNA delivery: potential and limitations of non-viral nanocarriers. J Biol Eng. 2012;6:7. doi: 10.1186/1754-1611-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conti DS, Brewer D, Grashik J, Avasarala S, da Rocha SR. Poly(amidoamine) dendrimer nanocarriers and their aerosol formulations for siRNA delivery to the lung epithelium. Mol Pharm. 2014;11(6):1808–1822. doi: 10.1021/mp4006358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trabulo S, Cardoso AM, Santos-Ferreira T, Cardoso AL, Simões S, Pedroso de Lima MC. Survivin silencing as a promising strategy to enhance the sensitivity of cancer cells to chemotherapeutic agents. Mol Pharm. 2011;8(4):1120–1131. doi: 10.1021/mp100426e. [DOI] [PubMed] [Google Scholar]
- 12.Altieri DC. Survivin – the inconvenient IAP. Semin Cell Dev Biol. 2015;39:91–96. doi: 10.1016/j.semcdb.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamm I, Wang Y, Sausville E, et al. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58(23):5315–5320. [PubMed] [Google Scholar]
- 14.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3(8):917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 15.Wang TT, Qian XP, Liu BR. Survivin: potential role in diagnosis, prognosis and targeted therapy of gastric cancer. World J Gastroenterol. 2007;13(20):2784–2790. doi: 10.3748/wjg.v13.i20.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smolewski P, Robak T. Inhibitors of apoptosis proteins (IAPs) as potential molecular targets for therapy of hematological malignancies. Curr Mol Med. 2011;11(8):633–649. doi: 10.2174/156652411797536723. [DOI] [PubMed] [Google Scholar]
- 17.Mobahat M, Narendran A, Riabowol K. Survivin as a preferential target for cancer therapy. Int J Mol Sci. 2014;15(2):2494–2516. doi: 10.3390/ijms15022494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim NJ, Jiang D, Jacobi AM, et al. Synthesis and characterization of mannosylated pegylated polyethylenimine as a carrier for siRNA. Int J Pharm. 2012;427(1):123–133. doi: 10.1016/j.ijpharm.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo X, Huang L. Recent advances in nonviral vectors for gene delivery. Acc Chem Res. 2012;45(7):971–979. doi: 10.1021/ar200151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mochalin VN, Shenderova O, Ho D, Gogotsi Y. The properties and applications of nanodiamonds. Nat Nanotechnol. 2011;7(1):11–23. doi: 10.1038/nnano.2011.209. [DOI] [PubMed] [Google Scholar]
- 21.Mansoorianfar M, Shokrgozar MA, Mehrjoo M, Tamjid E, Simchi A. Nanodiamonds for surface engineering of orthopedic implants: enhanced biocompatibility in human osteosarcoma cell culture. Diam Relat Mater. 2013;40:107–114. [Google Scholar]
- 22.Cui C, Wang Y, Yang K, et al. Preparation and characterization of RGDS/nanodiamond as a vector for VEGF-siRNA delivery. J Biomed Nanotechnol. 2015;11(1):70–80. doi: 10.1166/jbn.2015.2005. [DOI] [PubMed] [Google Scholar]
- 23.Kaur R, Chitanda JM, Michel D, et al. Lysine-functionalized nanodiamonds: synthesis, physiochemical characterization, and nucleic acid binding studies. Int J Nanomedicine. 2012;7:3851–3866. doi: 10.2147/IJN.S32877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Look J, Wilhelm N, Briesen HV, et al. Ligand-modified human serum albumin nanoparticles for enhanced gene delivery. Mol Pharm. 2015;12(9):3202–3213. doi: 10.1021/acs.molpharmaceut.5b00153. [DOI] [PubMed] [Google Scholar]
- 25.Jin S, Wang Y, Zhu H, et al. Nanosized aspirin-Arg-Gly-Asp-Val: delivery of aspirin to thrombus by the target carrier Arg-Gly-Asp-Val tetrapeptide. ACS Nano. 2013;7(9):7664–7673. doi: 10.1021/nn402171v. [DOI] [PubMed] [Google Scholar]
- 26.Nazli C, Ergenc TI, Yar Y, Acar HA, Kizilel S. RGDS-functionalized polyethylene glycol hydrogel-coated magnetic iron oxide nanoparticles enhance specific intracellular uptake by HeLa cells. Int J Nanomedicine. 2011;7:1903–1920. doi: 10.2147/IJN.S29442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kai T, Schiffelers RM, Molema G, Kok RJ. RGD-based strategies for selective delivery of therapeutics and imaging agents to the tumor vasculature. Drug Resist Updates. 2005;8(6):381–402. doi: 10.1016/j.drup.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 28.El-Say K. Nanodiamond as a drug delivery system: applications and prospective. J Appl Pharm Sci. 2011;1(6):29–39. [Google Scholar]
- 29.She W, Luo K, Zhang C, et al. The potential of self-assembled, pH-responsive nanoparticles of mPEGylated peptide dendron–doxorubicin conjugates for cancer therapy. Biomaterials. 2012;34(5):1613–1623. doi: 10.1016/j.biomaterials.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Olariu CI, Yiu HHP, Bouffier L, et al. Multifunctional Fe3O4 nanoparticles for targeted bi-modal imaging of pancreatic cancer. J Mater Chem. 2011;21:12650–12659. [Google Scholar]
- 31.Palma RD, Peeters S, Bael MJV, et al. Silane ligand exchange to make hydrophobic superparamagnetic nanoparticles water-dispersible. Chem Mater. 2007;19(7):1821–1831. [Google Scholar]
- 32.Xu R, Wu C, Xu H. Particle size and zeta potential of carbon black in liquid media. Carbon. 2007;45(14):2806–2809. [Google Scholar]
- 33.Thomas E. Laser pointer and the Tyndall effect. J Chem Educ. 1996;73(5):470. [Google Scholar]
- 34.Dolmatov VY. Detonation-synthesis nanodiamonds: synthesis, structure, properties and applications. Cheminform. 2007;38:35. [Google Scholar]
- 35.Shimkunas RA, Robinson E, Lam R, et al. Nanodiamond–insulin complexes as pH-dependent protein delivery vehicles. Biomaterials. 2009;30(29):5720–5728. doi: 10.1016/j.biomaterials.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Zhang XQ, Chen M, Lam R, Xu X, Osawa E, Ho D. Polymer-functionalized nanodiamond platforms as vehicles for gene delivery. ACS Nano. 2009;3(9):2609–2616. doi: 10.1021/nn900865g. [DOI] [PubMed] [Google Scholar]
- 37.Yang F, Huang W, Li Y, et al. Anti-tumor effects in mice induced by survivin-targeted siRNA delivered through polysaccharide nanoparticles. Biomaterials. 2013;34(22):5689–5699. doi: 10.1016/j.biomaterials.2013.03.047. [DOI] [PubMed] [Google Scholar]
- 38.Ficarra R, Ficarra P, Di Bella MR, et al. Study of beta-blockers/beta-cyclodextrins inclusion complex by NMR, DSC, X-ray and SEM investigation. J Pharm Biomed Anal. 2000;23(1):33–40. doi: 10.1016/s0731-7085(00)00261-2. [DOI] [PubMed] [Google Scholar]
- 39.Smith AH, Robinson EM, Zhang XQ, et al. Triggered release of therapeutic antibodies from nanodiamond complexes. Nanoscale. 2011;3(7):2844–2848. doi: 10.1039/c1nr10278h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaur R, Badea I. Nanodiamonds as novel nanomaterials for biomedical applications: drug delivery and imaging systems. Int J Nanomedicine. 2013;8:203–220. doi: 10.2147/IJN.S37348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrand AM, Huang H, Carlson C, et al. Are diamond nanoparticles cytotoxic? J Phys Chem B. 2007;111(1):2–7. doi: 10.1021/jp066387v. [DOI] [PubMed] [Google Scholar]
- 42.Gilad Y, Noy E, Senderowitz H, Albeck A, Firer MA, Gellerman G. Dual-drug RGD conjugates provide enhanced cytotoxicity to melanoma and non-small lung cancer cells. Peptide Sci. 2016;106(2):160–171. doi: 10.1002/bip.22800. [DOI] [PubMed] [Google Scholar]
- 43.Garate A, Santos E, Pedraz JL, Hernández RM, Orive G. Evaluation of different RGD ligand densities in the development of cell-based drug delivery systems. J Drug Target. 2015;23(9):806–812. doi: 10.3109/1061186X.2015.1020428. [DOI] [PubMed] [Google Scholar]
- 44.Maver Uroš, Godec A, Bele M, et al. Novel hybrid silica xerogels for stabilization and controlled release of drug. Int J Pharm. 2007;330(1–2):164–174. doi: 10.1016/j.ijpharm.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 45.Wang W, Fang C, Wang X, et al. Modifying mesoporous silica nanoparticles to avoid the metabolic deactivation of 6-mercaptopurine and methotrexate in combinatorial chemotherapy. Nanoscale. 2013;5(14):6249–6253. doi: 10.1039/c3nr00227f. [DOI] [PubMed] [Google Scholar]
- 46.Liu S, Huang W, Jin MJ, Fan B, Xia GM, Gao ZG. Inhibition of murine breast cancer growth and metastasis by survivin-targeted siRNA using disulfide cross-linked linear PEI. Eur J Pharm Sci. 2016;82:171–182. doi: 10.1016/j.ejps.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 47.Beloor J, Choi CS, Nam HY, et al. Arginine-engrafted biodegradable polymer for the systemic delivery of therapeutic siRNA. Biomaterials. 2012;33(5):1640–1650. doi: 10.1016/j.biomaterials.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 48.Xie Y, Qiao H, Su Z, Chen M, Ping Q, Sun M. PEGylated carboxymethyl chitosan/calcium phosphate hybrid anionic nanoparticles mediated hTERT siRNA delivery for anticancer therapy. Biomaterials. 2014;35(27):7978–7991. doi: 10.1016/j.biomaterials.2014.05.068. [DOI] [PubMed] [Google Scholar]