Abstract
Treponema pallidum subspecies pallidum is the causative agent of syphilis, a chronic, multistage, systemic infection that remains a major global health concern. The molecular mechanisms underlying T. pallidum pathogenesis are incompletely understood, partially due to the phylogenetic divergence of T. pallidum. One aspect of T. pallidum that differentiates it from conventional Gram-negative bacteria, and is believed to play an important role in pathogenesis, is its unusual cell envelope ultrastructure; in particular, the T. pallidum peptidoglycan layer is chemically distinct, thinner and more distal to the outer membrane. Established functional roles for peptidoglycan include contributing to the structural integrity of the cell envelope and stabilization of the flagellar motor complex, which are typically mediated by the OmpA domain-containing family of proteins. To gain insight into the molecular mechanisms that govern peptidoglycan binding and cell envelope biogenesis in T. pallidum we report here the structural characterization of the putative OmpA-like domain-containing protein, Tp0624. Analysis of the 1.70 Å resolution Tp0624 crystal structure reveals a multi-modular architecture comprised of three distinct domains including a C-terminal divergent OmpA-like domain, which we show is unable to bind the conventional peptidoglycan component diaminopimelic acid, and a previously uncharacterized tandem domain unit. Intriguingly, bioinformatic analysis indicates that the three domains together are found in all orthologs from pathogenic treponemes, but are not observed together in genera outside Treponema. These findings provide the first structural insight into a multi-modular treponemal protein containing an OmpA-like domain and its potential role in peptidoglycan coordination and stabilization of the T. pallidum cell envelope.
Introduction
Treponema pallidum subspecies pallidum (hereafter T. pallidum) is a medically important spirochete bacterium and the causative agent of syphilis, a chronic, multistage systemic disease transmitted sexually, through direct contact with an infectious lesion, or in utero. It is estimated that the current global burden is 36 million cases with 11 million new infections each year [1]. Congenital syphilis is also a major global health concern with approximately 1.4 million pregnant women infected annually [2]. Furthermore, symptomatic syphilis infections increase the risk of HIV transmission and acquisition 2- to 5-fold [3].
Eighteen years have passed since the publication of the first complete T. pallidum genome sequence [4], yet the molecular basis of T. pallidum pathogenesis remains enigmatic. Research efforts are complicated by the fact that T. pallidum is an obligate human pathogen that cannot be cultured in vitro or genetically modified. Moreover, compared to conventional Gram-negative bacteria, T. pallidum has an inherently fragile cell envelope [5, 6] composed of a chemically distinct peptidoglycan layer that is thinner and more spatially separated from the outer membrane, as well as periplasmic flagella (endoflagella) that are located between the outer membrane and the peptidoglycan layer [5, 6]. These ultrastructural differences suggest that treponemes such as T. pallidum may harbor divergent molecular strategies for linking the peptidoglycan layer with the inner and outer membranes to stabilize the bacterial cell envelope.
In Gram-negative bacteria, cell envelope stabilization is partially achieved through Outer Membrane Protein A (OmpA)-like domain-containing proteins that bind non-covalently to peptidoglycan. This domain is commonly found in bacterial proteins that support outer membrane integrity and indirectly facilitate host-pathogen interactions by acting as a structural bridge between the peptidoglycan layer and the outer membrane [7–10]. In conventional Gram-negative bacteria the stabilization of the outer membrane via OmpA domain-containing proteins occurs via one of three mechanisms: insertion of an N-terminal beta-barrel domain [11] or an N-terminal lipid anchor distal to the C-terminal OmpA-domain into the outer membrane [12], or association of the OmpA-domain with bacterial outer membrane porins [7–10]. OmpA-like domains that contain peptidoglycan binding sites are also found in bacterial flagellar motor complex proteins, including motility protein B (MotB) [13–15]. MotB is an essential component of the torque-generating stator ring in the flagellar motor complex [16–18] where its N-terminus is anchored in the cytoplasmic membrane via a membrane-spanning helix and its C-terminal OmpA-like domain directed into the periplasm where it coordinates the peptidoglycan layer [13, 15]. It appears that the peptidoglycan-binding OmpA-like domain was acquired by outer membrane-associated proteins and inner membrane-associated flagellar motor complex proteins from a common ancestor long before their protein functions diverged [19]. Accordingly, the OmpA-like domains of these otherwise functionally divergent proteins exhibit significant sequence similarity and share a common structural fold [13, 15, 19].
The T. pallidum protein Tp0624 is annotated as belonging to the OmpA-OmpF porin (OOP) family [20]. The C-terminal region of Tp0624 is predicted to adopt an OmpA domain architecture, but it is unclear what role a canonical peptidoglycan binding domain might possess in an organism that contains an unusual form of peptidoglycan and has a divergent cell envelope ultrastructure. Further complicating the functional prediction of Tp0624 is the observation that the N-terminal two thirds of the protein does not show significant sequence homology to any functionally- or structurally-characterized protein. The lack of sequence identity is consistent with T. pallidum being a phylogenetically distinct bacterium with no known orthologs nor assigned functions for almost 30% of its predicted protein-coding genes [4]. Moreover, the extreme genomic reduction in obligate pathogens such as T. pallidum often translates to the expression of multi-modular proteins with increased functional complexity [21, 22].
Treponema pallidum transcriptomic [23] and proteomic [24] studies have shown that Tp0624 is expressed during infection, yet its precise biological role is unknown. The presence of an OmpA-like domain suggests a potential role for Tp0624 in peptidoglycan coordination and cell envelope stabilization, which could be facilitated by the uncharacterized N-terminal region. To gain insight into the function of Tp0624, we first determined its three-dimensional structure, which revealed a trimodular architecture incorporating a highly divergent OmpA-like domain and a previously uncharacterized tandem domain with intriguing implications for peptidoglycan coordination. Collectively, these data provide insight into how the unusual cell envelope structure may be stabilized in the human pathogen T. pallidum.
Materials and Methods
Ethics statement
All animal studies were approved by the local institutional review board at the University of Victoria and were conducted in strict accordance with standard accepted principles as set forth by the Canadian Council on Animal Care (CCAC) and National Institutes of Health in a facility accredited by the American Association for the Accreditation of Laboratory Animal Care.
Bacterial growth and genomic DNA extraction
T. pallidum subsp. pallidum (Nichols strain) was propagated in, and extracted from, New Zealand White rabbits as described elsewhere [25]. Genomic DNA was purified from treponemes using the DNeasy Tissue kit (Qiagen). All DNA purifications were conducted according to the manufacturer’s instructions for Gram-negative bacteria.
Cloning, protein production and purification
Eight signal sequence prediction servers were used to predict the presence and position of a potential cleavable signal peptidase type I (SPI) signal sequence within the N-terminus of Tp0624: PrediSi (http://www.predisi.de/index.html; cleavage at T61), Signal BLAST (http://sigpep.services.came.sbg.ac.at/signalblast.html; cleavage at G58), Signal-CF (http://www.csbio.sjtu.edu.cn/bioinf/Signal-CF/; cleavage at A59), Spoctopus (http://octopus.cbr.su.se/; cleavage at G58), SPEPLip (http://gpcr.biocomp.unibo.it/cgi/predictors/spep/pred_spepcgi.cgi; cleavage at A59), Signal3L (http://www.csbio.sjtu.edu.cn/bioinf/Signal-3L/; cleavage at A30), EMBOSS sigcleave (http://www.bioinformatics.nl/cgi-bin/emboss/sigcleave; cleavage at A32), and SignalP (http://www.cbs.dtu.dk/services/SignalP/; SPI signal sequence not fully predicted, but graphical analysis shows a below cut-off cleavage prediction at A59). For expression of the predicted mature full-length protein, a DNA fragment encoding residues T61-D476 of Tp0624 was PCR-amplified from T. pallidum (Nichols Strain) genomic DNA using the following primers: 5´-CAG ACT GCT AGC ACC CAG GAT GCC GTA CAC ACC G (sense primer with NheI restriction site [underlined]) and 5´-CTG TCT G GC GGC CGC ATC GCG CAA GAT GGT GAT CTC C (antisense primer with NotI restriction site [underlined]). The PCR amplicon was then cloned into a modified pET28a (VWR International) vector with a TEV protease cleavable N-terminal hexa-histidine tag. The construct was confirmed by DNA sequencing. The construct was transformed into the E. coli expression strain BL21 Star (DE3) (Invitrogen). Transformants were cultured at 37°C in LB broth containing 100 μg/mL ampicillin until the A600 reached 0.4. Cultures were induced with 0.4 mM IPTG for 16 h at 25°C and harvested by centrifugation at 4°C. Cell pellets were resuspended in buffer (20 mM HEPES pH 7.5, 500 mM NaCl, 20 mM imidazole) and flash frozen in liquid nitrogen and stored at -80°C.
Purification of recombinant protein was performed using immobilized nickel affinity and size exclusion chromatography (SEC). Briefly, recombinant His-tagged protein was loaded onto 1 ml HisTrap FF affinity columns (GE Healthcare) pre-packed with pre-charged Nickel Sepharose 6 Fast Flow resin. Columns were then washed with buffer (20 mM HEPES pH 7.5, 500 mM NaCl, 20 mM imidazole) and bound recombinant protein was eluted in a buffer consisting of 20 mM HEPES pH 7.5, 500 mM NaCl, and 500 mM imidazole. Tp0624 was then further purified by SEC on a HiLoad™ 16/60 Superdex™ 75 column (GE Healthcare) pre-equilibrated in HEPES-buffered saline (HBS: 20 mM HEPES pH 7.5, 150 mM NaCl). Notably, Tp0624 eluted from the SEC as a monomer (S1 Fig). The N-terminal His tag was removed by cleavage with TEV protease at 16°C for 16 h. Cleaved protein was then purified by SEC in HBS as described above, and concentrated to 10 mg/mL.
Crystallization and data collection
Crystals of Tp0624 were originally identified in the MCSG-1 screen (Microlytic) using sitting drops at 18°C. The final, refined drops were set in hanging drops and consisted of 3.6 μL Tp0624 at 10 mg/mL in HBS with 1.8 μL of reservoir solution (0.1 M HEPES pH 6.8, 0.25 M MgCl2, 14% PEG3350) and were equilibrated against 300 μL of reservoir solution. Crystals appeared within 24 h and grew to a final size within 3 days. Crystals were cryoprotected in reservoir solution supplemented with 12.5% glycerol and flash cooled in liquid nitrogen. Diffraction data were collected on beamline 08ID-1 at the Canadian Light Source (CLS). For phase determination, Tp0624 was crystallized using the same technique as for the native crystals but the reservoir solution consisted of 0.1 M HEPES pH 6.8, 0.45 M MgBr2, 14% PEG3350. Diffraction data were collected on beamline 08ID-1 at CLS using an optimized wavelength of 0.9201 Å for the f” bromine edge.
Data processing, structure solution and refinement
Diffraction data were processed to 1.70 Å (Tp0624) or 1.92 Å (Tp0624-Br) resolution using Imosflm [26] and Aimless [27]. The structure of Tp0624 was solved by Bromide single wavelength anomalous dispersion. A total of 53 high confidence Br sites were identified using ShelxC/D [28] and refined using Phenix.phaser-ep [29]. High quality phases enabled building and registering of approximately 75% of the backbone using Phenix.autobuild [30]. The Tp0624 native structure was solved by molecular replacement using a single Tp0624 chain from the Br-phased model in Phaser [31]. COOT [32] was used for model building and selection of solvent atoms, and the model was refined in Phenix.refine [33]. Complete structural validation was performed with Molprobity [34], including analysis of the Ramachandran plots, which showed greater than 98% of residues in the most favored conformations. Five percent of reflections were set aside for Rfree calculation. Data collection and refinement statistics are presented in Table 1. The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 5JIR.
Table 1. Data collection and refinement statistics.
| Tp0624-Br SAD | Tp0624 native | |
|---|---|---|
| Data collection statistics | ||
| Spacegroup | P212121 | P212121 |
| a, b, c (Å) | 62.78, 104.3, 141.3 | 63.02, 103.50, 140.68 |
| α, β, γ (deg.) | 90, 90, 90 | 90, 90, 90 |
| Wavelength | 0.9201 | 0.9794 |
| Resolution range (Å) | 47.11–1.95 (2.00–1.95) | 63.02–1.70 (1.73–1.70) |
| Measured reflections | 879019 (58876) | 592930 (29342) |
| Unique reflections | 68488 (4574) | 101666 (4969) |
| Redundancy | 12.8 (12.9) | 5.8 (5.9) |
| Completeness (%) | 100.0 (100.0) | 99.9 (100.0) |
| I/σ(I) | 12.4 (4.0) | 9.4 (2.2) |
| Rmerge | 0.172 (0.745) | 0.089 (0.684) |
| Refinement statistics | ||
| Resolution (Å) | 51.77–1.70 | |
| Rwork / Rfree | 0.189/0.215 | |
| No. of atoms | ||
| Protein (A/B) | 3233/3189 | |
| Glycerol/Chloride | 24/8 | |
| Solvent | 346 | |
| B-values (Å2) | ||
| Protein (A/B) | 28.1/28.2 | |
| Glycerol/Chloride | 33.5/24.6 | |
| Solvent | 29.1 | |
| r.m.s. deviation from ideality | ||
| Bond lengths (Å) | 0.008 | |
| Bond angles (deg.) | 1.10 | |
| Ramachandran statistics (%) | ||
| Most favoured | 98.4 | |
| Allowed | 1.6 | |
| Disallowed | 0 |
Values in parentheses are for the highest resolution shell.
5% of reflections were set aside for calculation of Rfree.
Bioinformatic identification of Tp0624 orthologs
Orthologs of Tp0624 were identified using BLASTp (PSI-BLAST algorithm), the T. pallidum query sequence (Tp0624; NCBI accession number WP_010882070) and the NCBI non-redundant database. Tp0624 residues A64-F177, Y198-C317, and D332–D476 were used for the identification of orthologous proteins containing domains with significant homology to Tp0624 domains 1, 2, and 3, respectively. All identified orthologs had an e-value cutoff of 0.00026.
Results and Discussion
A non-canonical signal peptide suggests that Tp0624 is exported from the cytoplasm
To assess the potential for Tp0624 to adopt a subcellular localization consistent with canonical OmpA-containing proteins (i.e. exported from the cytoplasm), we first sought to establish the presence of a signal peptide. Of the eight signal sequence servers used, seven predicted a cleavable signal peptidase type I (SPI) signal sequence within the N-terminus of Tp0624. The majority of these programs predict cleavage to occur near residue 60 indicating an unusually long signal peptide. Notably, however, atypically long signal sequences that exhibit diversity in the length of the extreme N-terminal region of the signal peptide have been reported [35]. The assignment of a signal sequence was further supported based on the analysis of the region immediately prior to the predicted cleavage site, which contains the hallmarks of SPI-cleaved signal peptides including a positively charged N-terminal region, a central hydrophobic region, and a neutral/polar C-terminal region [36]. Moreover, an alignment of eighteen Tp0624 homologs (S2 Fig) revealed that significant sequence homology was observed only after the predicted signal peptide indicating that the N-terminus is structurally independent of the protein core. Based on these observations, we revisited our recent proteomic analysis of T. pallidum isolated from an infected rabbit. Somewhat unexpectedly, we identified a peptide that mapped to the predicted signal sequence region of Tp0624 [24]. While this peptide may have originated from the pre-protein before signal peptidase cleavage, it also may reflect that the signal peptide is not cleaved following export from the cytoplasm resulting in Tp0624 being anchored in the inner membrane via an N-terminal hydrophobic transmembrane segment with its head group directed into the periplasm. Consistent with this possibility is the fact that the predicted signal peptide central region (F18-F52) is very hydrophobic (GRAVY [grand average of hydropathy] value = 1.166) compared to the mature protein (GRAVY value = -0.220). While we are unable to definitively assign a subcellular location for Tp0624, there is strong bioinformatic evidence that Tp0624, like other OmpA-containing proteins, is exported from the cytoplasm.
Tp0624 adopts an ordered alpha/beta structure
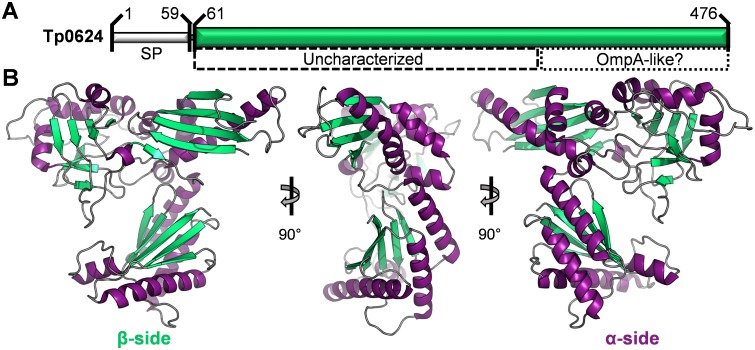
To establish the optimal start site for the recombinant Tp0624, we integrated secondary structure predictions with the sequence alignment of the eighteen Tp0624 homologs (S2 Fig). Based on these analyses, a construct incorporating T61 to D476 was cloned for expression in E. coli (Fig 1A). The recombinant hexa-histidine fusion protein was purified by nickel affinity and size exclusion chromatography (SEC), which showed an elution profile consistent with a Tp0624 monomer (S1 Fig). Crystallization trials were set with purified Tp0624 and diffraction data on optimized crystals were collected to a resolution of 1.70 Å. In the absence of a suitable molecular replacement model, crystals of Tp0624 were soaked in bromide, data collected at the optimized wavelength for the f” bromine edge, and the structure phased by single wavelength anomalous dispersion (Table 1). The structure revealed that Tp0624 crystallized with two molecules in the asymmetric unit of the primitive orthorhombic unit cell with each monomer related by a root mean square deviation (rmsd) of 0.70 Å over 393 Cα atoms. A minimal interface between the two protomers is consistent with the SEC data indicating that Tp0624 is a functional monomer. Of the two molecules, chain A was the most completely modeled and was used for subsequent analyses unless otherwise stated. The overall architecture of Tp0624 is that of a thin triangle with approximate dimensions of 73 x 63 x 33 Å. Notably, the secondary structure elements are highly polarized with one side of the triangle comprised primarily of three beta-sheets and the opposite side dominated by ten alpha-helices (Fig 1B).
Fig 1. Overall structure of Tp0624.
(A) Sequence features of Tp0624. SP—signal peptide (approximately residues 1 to 60). Green bar—construct used for structural studies (T61-D476). Dashed box—region with predicted secondary structure elements but no sequence similarity to any characterized protein. Dotted box—putative OmpA-like C-terminal domain. (B) Orthogonal views of Tp0624 tertiary structure shown as a cartoon with beta-strands in green, alpha helices in purple, and connecting coil in grey. Note the slightly flattened, triangle-like architecture with two distinct sides defined by either beta-strands (left) or alpha-helices (right).
Tp0624 adopts a highly modular architecture
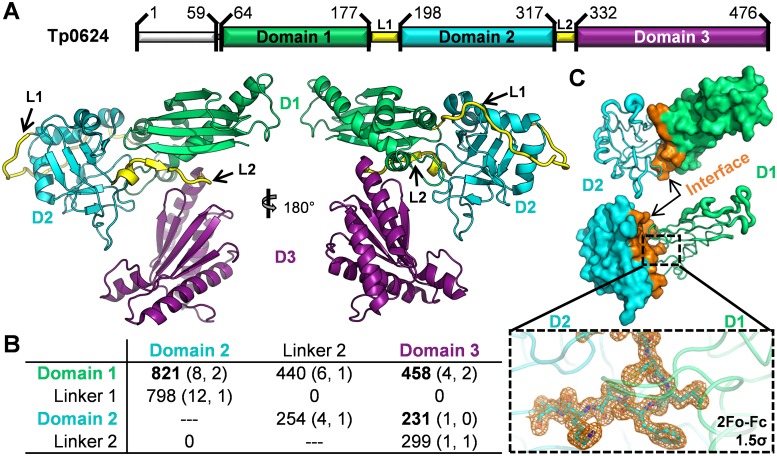
Structural analysis reveals that Tp0624 is comprised of three distinct domains connected by two interdomain linkers: domain 1 (D1, A64-F177) is linked via a 20 residue linker (L1) to domain 2 (D2, Y198-C317), which is in turn linked via a 14 residue linker (L2) to domain 3 (D3, D332-D476) (Fig 2A). Linker 1 stretches across the back and around the end of D2 (Fig 2A), burying nearly 800 Å2 at the interface and forming 12 hydrogen bonds with D2 (Fig 2B) [37]. The N-terminal portion of L1 is well ordered while the C-terminal portion that wraps around the end of D2 displays significant flexibility. Despite the observed flexibility in L1, the buried interface and ordered residues between D1 and D2 suggest that these two domains function as an integrated unit (Fig 2B and 2C). While L2 connects D2 and D3, it also forms the terminal strand in the D1 beta-sheet highlighting the overall integrated nature of the Tp0624 modular architecture.
Fig 2. Tp0624 is organized into two units consisting of D1-D2 and D3.
(A) Top: structure-based assignment of Tp0624 domains (D1, green; D2, cyan; D3, purple) and linkers (L1 and L2, yellow). Bottom: cartoon of Tp0624 tertiary structure colored by domain as in (top). (B) Table of inter-domain/linker interfaces of Tp0624 listed as “Interface Area in Å2 (# Hydrogen Bonds, # Salt Bridges)” as defined by PISA [37]. (C) Interface of D1-D2. Top: D2 (cyan) shown as B-factor defined coil with increased mobility indicated by thicker coil. D1 shown as green surface, with residues contacting D2 colored orange. Middle: Inverse display of (top). Inset: Sigma-A weighted electron density mesh (orange) contoured at 1.5 sigma for a loop of D2 (cyan) that protrudes into D1 (green).
The C-terminal domain, D3, which is classified based on sequence to be part of the OmpA-OmpF porin (OOP) family [20], is the largest of the three domains and is formed by a five-stranded beta-sheet of mixed parallel and anti-parallel strands in the pattern 1-2-5-3-4 flanked on one face by three alpha-helices (Fig 2A). The core of the central domain, D2, adopts a pseudo beta-sandwich fold, with a top sheet of four parallel strands, and a bottom twisted sheet of four mixed strands; the two sheets are in parallel planes but orthogonally packed (Fig 2A). The N-terminal domain, D1, of Tp0624 is comprised primarily of a four-stranded anti-parallel beta-sheet (1-2-4-3 connectivity), supplemented by a short fifth strand derived from L2, with an extended helix that packs diagonally against the full length of one face of the beta-sheet. This domain core is supplemented by three short helices packed at the base of the domain, with the first interfacing with D3, the second bracing against the D1 primary helix, and the third forming a substantial portion of the interface with D2 (Fig 2A).
Tp0624 D3 adopts a divergent OmpA-like domain that is unable to bind the diaminopimelic acid of conventional peptidoglycan
To gain additional insight into potential functions of the three Tp0624 domains, we searched for related structures using the DALI server [38]. We first analyzed D3, which showed strong structural homology to the C-terminal domain of E. coli Outer Membrane Protein A (OmpA), which contributes to cell envelope stability by providing a structural link between the peptidoglycan layer and outer membrane [7–10, 12]. The closest structural homolog of Tp0624 D3 is the OmpA-like domain from Vibrio PomB, an inner membrane-anchored MotB ortholog and component of the flagellar motor that anchors the stator to the peptidoglycan layer (PDB ID 3WPW, Z-score 17.0 with scores < 2 being spurious, rmsd of 2.4 Å over 135 positions). The function of all OmpA-like domains that have been characterized to date is centered on their coordination of peptidoglycan via a pocket at the base of the domain [11]. A structural overlay on an OmpA-like domain with a bound peptidoglycan fragment (Fig 3A; Acinetobacter baumannii OmpA-like domain, PDB ID 3TD5) reveals key differences between Tp0624 D3 and canonical OmpA-like domains. For example, a loop that forms the outer border of the peptidoglycan binding site in other OmpA-like domains is displaced by nearly 15 Å in Tp0624 (Fig 3A, asterisk). In addition, while the base of the binding site tends to be lined by small residues (e.g. Thr-Gly in PDB ID 3TD5), a Leu-His pair presents a possible steric hindrance to binding in the analogous region of Tp0624 (Fig 3A, insets). Sequence and structural analysis shows that binding of peptidoglycan in the pocket of OmpA-like domains depends on the motif TD(x)7NxxLSxxRA, with Asp2, Asn10, and Arg17 being the most critical and highly conserved binding residues [11]. In contrast, Tp0624 D3 homologs from spirochetes present a conserved TAD(x)6E/QxxLSxxRA motif (Fig 3A). Most notably, the Asp residue that coordinates the head group of the peptidoglycan-specific diaminopimelic acid (DAP) is mutated to Ala in Tp0624 D3 and this Ala is completely conserved in all fifteen of the full-length Tp0624 spirochetal homologs (S2 Fig). These structural observations are consistent with our inability to demonstrate binding between DAP and Tp0624 in either DAP soaked crystals or by isothermal titration calorimetry.
Fig 3. Comparative Tp0624 domain analyses.
(A) Left: overlay of Tp0624 D3 (purple) on the OmpA-like domain of Acinetobacter baumannii (grey, PDB ID 3TD5) in complex with a peptidoglycan derivative (light grey ball-and-stick colored by element). Right: Structure-based sequence alignment of Tp0624 with the peptidoglycan binding residues of A. baumanii OmpA-like domain. Critical residues are bolded and displayed in the inset panels. Inset: Zoomed-in view of the peptidoglycan binding site on A. baumanii OmpA-like domain (grey, left), with hydrogen bonds to the diaminopimelic acid residue shown as dotted lines. Star indicates the loop that comprises the outer side of the pocket. Contrast with the same view of Tp0624 D3, which despite conservation of the essential Arg, shows the loss of a critical Asp to Ala, the displaced position of the outer loop (star), and the large residues (Leu/His) in the lower loop that project into the binding site. (B) Electrostatic surface of Tp0624 D3 in same orientation as (A) shows a clear basic patch comprising the putative ligand binding site. (C) Overlay of Tp0624 D1 (green) on the E. coli trigger factor (grey, PDB ID 1P9Y). Grey bracket, region of the trigger factor that has been functionalized for ribosome binding and is not conserved in Tp0624 D1. Green bracket, region of Tp0624 D1 that has been extended to form the interfaces with D2 and D3. (D) Tp0624 D2 is a previously uncharacterized domain showing no significant structural similarity to any domain in the PDB. Tp0624 consists of a four-stranded parallel top sheet (cyan), four-stranded mixed bottom sheet (orange), and flanking helices (yellow). Left: orthogonal views of Tp0624 D2 tertiary structure. Right: Topology diagram of Tp0624 D2.
Intriguingly, biochemical analyses of spirochete peptidoglycan, including that found in T. pallidum, have shown that DAP is not a detectable constituent for most spirochetes, with a few noted exceptions such as Leptospira spp. [39, 40]. The unique peptidoglycan of treponemes and most spirochetes is comprised of muramic acid, glucosamine, L-ornithine (DAP replacement in most spirochetes), D-glutamic acid, D/L-alanine, and sometimes glycine [39, 41, 42]. Inspection of the electrostatic surface of the putative binding site on Tp0624 D3 reveals a strongly basic region, consistent with the conservation of the critical Arg, but loss of the Asp to Ala (Fig 3B). The presence of an exposed pocket at the base of the domain that has been modified primarily by the loss of an acidic charge (responsible for coordinating the amine of the DAP headgroup; Fig 3A, left inset), makes it tempting to speculate that Tp0624 D3 might indeed anchor to the thin periplasmic peptidoglycan layer of T. pallidum, but that its divergently functionalized OmpA-like domain coordinates D-glutamic acid.
The N-terminus of Tp0624 reveals functionally repurposed (D1) and previously uncharacterized (D2) domain architectures
In contrast to D3, D1 and D2 do not show significant sequence identity to any proteins of known structure or function. Structural analysis of D1 (Fig 3C) using the DALI server showed moderate alignment with the ribosome binding domain of E. coli trigger factor (e.g. PDB ID 1P9Y); despite a sequence identity of only 4%, these domains overlay with an rmsd of 3.3 Å over 79 positions, corresponding to a Z-score of 5.4. While the central four-stranded beta-sheet and primary helix are well conserved, both ends are structurally divergent with apparent functional consequences. Specifically, the presence of helices at the base of Tp0624 D1 uniquely enable the domain to pack against D2 and D3, while the opposite end of D1 is truncated compared to the trigger factor where it is used to engage the ribosome [43]. These differences suggest that, while Tp0624 has retained a conserved core scaffold, it is repurposed for a yet to be determined function. Notably, differential functionalization of this scaffold in bacteria is common, with modified versions forming part of proteins ranging from a modular lytic transglycosylase thought to be important for the assembly and maturation of the bacterial cell wall (PDB ID 4CFO, Z-score 5.1) [44], to a plasmid relaxase domain important for conjugative plasmid transfer (PDB ID 3L6T, Z-score 4.7) [45].
In striking contrast to the more than 1200 structural similarity hits for Tp0624 D1, no structural analogues of D2 were identified. Thus, the Tp0624 D2 fold can be described as a previously uncharacterized domain. The defining feature of this domain is its orthogonally packed four-stranded beta-sheets flanked on three sides by four alpha-helices (Fig 3D). The orthogonal orientation clearly distinguishes Tp0624 D2 from the beta-sandwiches of well-characterized pathogen proteins such as the Plasmodium 6-Cys and Toxoplasma SRS domains [46–49]. This new domain is assembled with a notably circuitous topology as the connectivity for the top and bottom sheets is 4-8-1-5 and 3-2-7-6, respectively, with the central two beta-strands of the top sheet derived from the N- and C-terminal sequence. No obvious pockets or charged patches are present on the surface, making it difficult to predict a specific function for Tp0624 D2. However, a possible functional interdependence of D1 and D2 is indicated by assessing the mobility of D2, which is tightly organized through the core beta-sandwich as well as the regions at the interface with D1, primarily helix 1 and a portion of the extended loop between beta-strands 1 and 2. In contrast, helix 2 positioned atop the beta-sandwich and the regions distal to the D1 interface are notably more flexible (Fig 2C).
The modular assembly of Tp0624 is positively correlated with treponeme pathogenicity
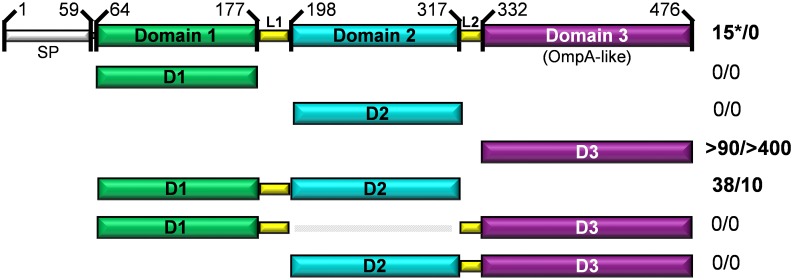
The intriguing modularity and domain architecture of Tp0624 indicate the possibility for functional interdependence, which led us to ask whether all three domains are reliant on each other for function or if specific domains are likely to act independently. To address these questions, we analyzed bacterial sequence databases using the domain boundaries established from the high-resolution structure (Fig 4). We found no examples of a sequence homologous to either D1 existing in isolation from D2 and D3, or D2 existing in isolation from D1 and D3. In contrast, sequences homologous to Tp0624 D3 were found in numerous proteins from both spirochetes and other bacteria that did not harbor domains homologous to D1 or D2. While the broad distribution of OmpA-like domains is not initially surprising, structural and binding studies reported here show clearly that Tp0624 D3 is unable to function as a canonical OmpA domain in binding conventional peptidoglycan.
Fig 4. Tp0624 amino acid sequence-based modularity analysis.
Orthologs of Tp0624 were identified using BLASTp. Tp0624 signal peptide (SP), domains (D1, green; D2, cyan; D3, purple), and linkers (L1 and L2, yellow) are shown. Indicated on the right are the numbers of orthologs with different domain combinations from spirochetes (left number) or other bacteria (right number). Sequences homologous to D1 and D2 only occur together, while D3 homologs are ubiquitous, and all three domains only occur together in treponemes (shown by an asterisk).
The pattern of domain combinations is similarly intriguing as D1 and D2 were found together independently of D3 in nearly 50 protein sequences from a variety of bacteria consistent with the integrated nature of these domains in the crystal structure (Fig 2B and 2C) and further supporting that the D1-D2 pair is a functional unit. In contrast, the pairings of D2-D3 or D1-D3 were not observed in isolation (Fig 4). Perhaps most notably, the combination of all three domains together is only observed in treponemes, primarily in pathogenic species but also in three of the six non-pathogenic species (T. primitia, T. azotonutricium, and T. caldaria), suggesting that D3 can augment a treponeme-specific function of the D1-D2 pair. Next, we assessed the evolutionary relationship between the Tp0624 treponemal orthologs using full-length sequences or the three individual domains (S3–S6 Figs). Strikingly, an evolutionary separation emerged where the full-length Tp0624 appeared to be more closely related to orthologs from pathogenic treponemes relative to non-pathogenic treponemes (S3 Fig). However, when each of the three domains was analyzed individually, this clear phylogenetic division was lost (S4–S6 Figs) indicating that inclusion of all three Tp0624 domains in one protein positively correlates with treponemal pathogenicity.
Putative mechanistic roles for Tp0624
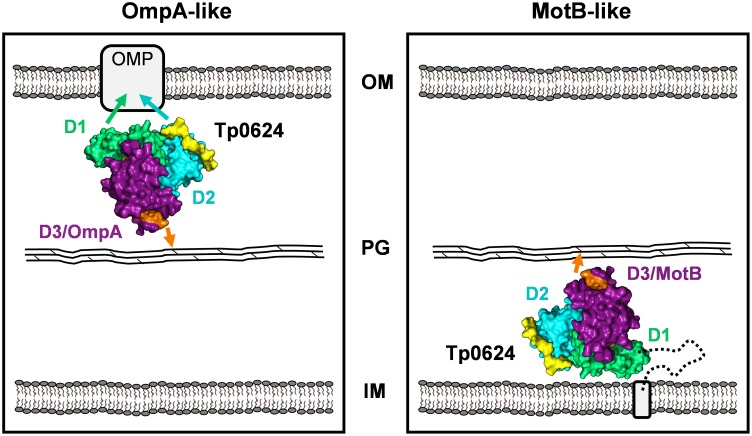
Based on the data reported here, we propose two models describing potential subcellular locations and corresponding functional peptidoglycan binding roles for Tp0624 (Fig 5). In the first model, Tp0624 is exported across the inner membrane via the Sec-mediated protein translocation pathway. Following cleavage of the signal peptide, Tp0624 coordinates the thin peptidoglycan layer via the OmpA-like domain (D3) resulting in the D1/D2 tandem unit being positioned for interactions with outer membrane-localized proteins (Fig 5, left panel [OmpA-like]). In this model, Tp0624 performs a similar function to the Neisseria meningitides OmpA-like domain containing protein, RmpM (Reduction-modifiable protein M) [8, 9] to promote stabilization of the outer membrane. In the second model, Tp0624 is also exported from the cytoplasm, but is retained in the inner membrane through its non-cleavable, hydrophobic N-terminal signal peptide with its functional head group directed into the periplasm (Fig 5, right panel [MotB-like]). Similar to other flagellar motor complex proteins, such as MotB, Tp0624 would be positioned to form a structural bridge between the peptidoglycan layer and the inner membrane. Comparable to the class of OmpA domain-like-containing proteins such as MotB and PomB, Tp0624 may further help to stabilize the flagellar motor complex within the cell envelope by functioning as another immobilized structure against which the flagellar rotor and rod rotate. Since T. pallidum possesses a MotB homolog (Tp0724) within a flagellar operon [5] it is perhaps more likely that an inner membrane location for Tp0624 would allow the protein to function as a scaffolding structure to maintain correct peptidoglycan positioning within the cell envelope and this could be facilitated by the D1/D2 tandem unit [5]. Given that MotB is a stabilizing component of the flagellar motor complex, and is only located at the ends of the treponeme, other periplasm-directed, inner membrane-anchored proteins may be involved in maintaining correct peptidoglycan positioning and providing structural support along the length of the treponeme [5]; Tp0624 could be one example of a protein that functions in this manner.
Fig 5. Model of the potential roles of Tp0624 in T. pallidum peptidoglycan binding and cell envelope structure stabilization.
(Left) OmpA-like model: Tp0624 is exported across the inner membrane (IM) followed by cleavage and removal of the signal peptide. Tp0624 binds peptidoglycan (PG) via non-covalent interactions (orange arrow) involving the OmpA-like D3 while D1 and/or D2 interact with outer membrane-localized proteins (OMP). Residues from the treponemal conserved TAD(x)6E/QxxLSxxRA motif (putative non-canonical peptidoglycan binding motif) and surrounding pocket located within D3 are shown in orange (Right) MotB-like model: Tp0624 is translocated across the inner membrane, however the signal peptide is not cleaved and the protein remains anchored to the inner membrane, but with the functional head group directed into the periplasm. Tp0624 binds peptidoglycan via its OmpA-like domain, forming a structural bridge with the inner membrane. In this subcellular location, Tp0624 could function in cell envelope and/or flagellar stabilization.
Conclusions
T. pallidum has a strategically minimalized complement of proteins, yet is one of the most invasive pathogens. Key virulence traits of T. pallidum include its unusual peptidoglycan structure, fragile cell envelope and internal endoflagella, yet the molecular strategies to functionalize and stabilize these ultrastructural features are largely unknown. In this study, we determined the high-resolution structure of Tp0624, and showed it to be comprised of a modular, three-domain architecture incorporating an OmpA-like domain and a tandem pair of previously uncharacterized domains. Intriguingly, the OmpA-like domain (D3) is sufficiently divergent that it is unable to bind the conventional DAP peptidoglycan component and may be uniquely adapted to coordinate the novel peptidoglycan structures of T. pallidum and potentially rigidify and stabilize the cell envelope. While we have not been able to establish a function for the previously uncharacterized domain pair (D1-D2), its association with D3 appears to correlate positively with treponeme pathogenicity and thus may play an important role in supporting the function of D3. Overall, our data suggests that Tp0624 may coordinate peptidoglycan through an unconventional mechanism to stabilize the inner- or outer-membranes of the highly divergent human pathogen, T. pallidum.
Supporting Information
Superdex 75 HiLoad size exclusion chromatogram showing that Tp0624 (46 kDa) elutes as a monomer (elution peak of 44 kDa globular standard denoted by hash mark).
(PDF)
Orthologs of Tp0624 were identified using BLASTP (PSI-BLAST algorithm) and aligned in Clustal Omega and BioEdit sequence alignment editor. The predicted Tp0624 signal peptide cleavage site at T61-Q62 (SP), domains 1 (D1), 2 (D2), and 3 (D3), known OmpA domain peptidoglycan binding residues (TD(x)7NxxLSxxRA) and corresponding treponemal residues (red rectangles) are shown. Identical residues (black background), similar residues (gray background), and dissimilar residues (white background) are highlighted. At each position of the consensus sequence, an asterisk indicates full conservation among the 18 treponemes, a colon indicates strong conservation, and a dot indicates lower conservation.
(PDF)
A phylogenetic tree of 18 full-length Tp0624 ortholog sequences from treponemes was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was calculated. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method and were in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) 6 software. The tree was rooted with the Tp0624 ortholog from Spirochaeta thermophila (Accession number: WP_013314620). The dashed line indicates the phylogenetic divide between pathogens (top) and non-pathogens (bottom).
(PDF)
A phylogenetic tree of Tp0624 ortholog sequences corresponding to domain 1 from 18 treponemes was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was calculated. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method and were in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) 6 software. The tree was rooted with the Tp0624 ortholog from Spirochaeta thermophila (Accession number: WP_013314620). Pathogens (P) and non-pathogens (NP) are indicated.
(PDF)
A phylogenetic tree of Tp0624 ortholog sequences corresponding to domain 2 from 18 treponemes was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was calculated. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method and were in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) 6 software. The tree was rooted with the Tp0624 ortholog from Spirochaeta thermophila (Accession number: WP_013314620). Pathogens (P) and non-pathogens (NP) are indicated.
(PDF)
A phylogenetic tree of Tp0624 ortholog sequences corresponding to domain 3 from 15 treponemes was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was calculated. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method and were in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) 6 software. The tree was rooted with the Tp0624 ortholog from Spirochaeta alkalica (Accession number: WP_018525408). Pathogens (P) and non-pathogens (NP) are indicated.
(PDF)
Acknowledgments
The authors gratefully acknowledge the staff at the Canadian Light Source (CLS) and the Stanford Synchrotron Radiation Lightsource (SSRL).
Data Availability
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 5JIR.
Funding Statement
This work was supported by Natural Sciences and Engineering Research Council of Canada grants to MJB and CEC. MJB and CEC gratefully acknowledge the Canada Chairs Research Program for salary support.
References
- 1.Organization WH. Prevalence and incidence of sexually transmitted infections Chlamydia trachomatis, Neisseria gonorrhoeae, syphilis and Trichomonas vaginalis: methods and results used by WHO to generate 2005 estimates. Geneva: World Health Organization; 2011. [Google Scholar]
- 2.Newman L, Kamb M, Hawkes S, Gomez G, Say L, Seuc A, et al. Global estimates of syphilis in pregnancy and associated adverse outcomes: analysis of multinational antenatal surveillance data. PLoS Med. 2013;10(2):e1001396 10.1371/journal.pmed.1001396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nusbaum MR, Wallace RR, Slatt LM, Kondrad EC. Sexually transmitted infections and increased risk of co-infection with human immunodeficiency virus. J Am Osteopath Assoc. 2004;104(12):527–35. . [PubMed] [Google Scholar]
- 4.Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281(5375):375–88. . [DOI] [PubMed] [Google Scholar]
- 5.Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La Vake C, et al. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol. 2009;191(24):7566–80. 10.1128/JB.01031-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Howell JK, Bradley SD, Zheng Y, Zhou ZH, Norris SJ. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J Mol Biol. 2010;403(4):546–61. 10.1016/j.jmb.2010.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Confer AW, Ayalew S. The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet Microbiol. 2013;163(3–4):207–22. 10.1016/j.vetmic.2012.08.019 . [DOI] [PubMed] [Google Scholar]
- 8.Grizot S, Buchanan SK. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol Microbiol. 2004;51(4):1027–37. . [DOI] [PubMed] [Google Scholar]
- 9.Maharjan S, Saleem M, Feavers IM, Wheeler JX, Care R, Derrick JP. Dissection of the function of the RmpM periplasmic protein from Neisseria meningitidis. Microbiology. 2016;162(2):364–75. 10.1099/mic.0.000227 . [DOI] [PubMed] [Google Scholar]
- 10.Volokhina EB, Beckers F, Tommassen J, Bos MP. The beta-barrel outer membrane protein assembly complex of Neisseria meningitidis. J Bacteriol. 2009;191(22):7074–85. 10.1128/JB.00737-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JS, Lee WC, Yeo KJ, Ryu KS, Kumarasiri M, Hesek D, et al. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J. 2012;26(1):219–28. 10.1096/fj.11-188425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouveret E, Benedetti H, Rigal A, Loret E, Lazdunski C. In vitro characterization of peptidoglycan-associated lipoprotein (PAL)-peptidoglycan and PAL-TolB interactions. J Bacteriol. 1999;181(20):6306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima S, Imada K, Sakuma M, Sudo Y, Kojima C, Minamino T, et al. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol Microbiol. 2009;73(4):710–8. 10.1111/j.1365-2958.2009.06802.x . [DOI] [PubMed] [Google Scholar]
- 14.Reboul CF, Andrews DA, Nahar MF, Buckle AM, Roujeinikova A. Crystallographic and molecular dynamics analysis of loop motions unmasking the peptidoglycan-binding site in stator protein MotB of flagellar motor. PLoS One. 2011;6(4):e18981 10.1371/journal.pone.0018981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roujeinikova A. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: implications for peptidoglycan recognition. Proc Natl Acad Sci U S A. 2008;105(30):10348–53. 10.1073/pnas.0803039105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charon NW, Goldstein SF. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu Rev Genet. 2002;36:47–73. 10.1146/annurev.genet.36.041602.134359 . [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191(16):5026–36. 10.1128/JB.00340-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy GE, Leadbetter JR, Jensen GJ. In situ structure of the complete Treponema primitia flagellar motor. Nature. 2006;442(7106):1062–4. 10.1038/nature05015 . [DOI] [PubMed] [Google Scholar]
- 19.Nguyen CC, Saier MH Jr, Structural and phylogenetic analysis of the MotA and MotB families of bacterial flagellar motor proteins. Res Microbiol. 1996;147(5):317–32. . [DOI] [PubMed] [Google Scholar]
- 20.Petrosova H, Pospisilova P, Strouhal M, Cejkova D, Zobanikova M, Mikalova L, et al. Resequencing of Treponema pallidum ssp. pallidum strains Nichols and SS14: correction of sequencing errors resulted in increased separation of syphilis treponeme subclusters. PLoS One. 2013;8(9):e74319 10.1371/journal.pone.0074319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelkar YD, Ochman H. Genome reduction promotes increase in protein functional complexity in bacteria. Genetics. 2013;193(1):303–7. 10.1534/genetics.112.145656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson B, Martin A. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun. 2011;79(9):3476–91. 10.1128/IAI.00179-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smajs D, McKevitt M, Howell JK, Norris SJ, Cai WW, Palzkill T, et al. Transcriptome of Treponema pallidum: gene expression profile during experimental rabbit infection. J Bacteriol. 2005;187(5):1866–74. 10.1128/JB.187.5.1866-1874.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osbak KK, Kenyon C, Lithgow K, Houston S, Meehan C, Strouhat M, et al. Characterizing the Syphilis-Causing Treponema pallidum ssp. pallidum proteome using complementary mass spectrometry. PLoS Negl Trop Dis. 2016;Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukehart SA, Marra CM. Isolation and laboratory maintenance of Treponema pallidum. Curr Protoc Microbiol. 2007;Chapter 12:Unit 12A 1. 10.1002/9780471729259.mc12a01s7 . [DOI] [PubMed] [Google Scholar]
- 26.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–81. Epub 2011/04/05. 10.1107/S0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1204–14. 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):479–85. Epub 2010/04/13. 10.1107/S0907444909038360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40(Pt 4):658–74. Epub 2007/08/01. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–21. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 1):32–41. Epub 2006/12/14. 10.1107/S0907444906045975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–67. 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. Epub 2010/01/09. 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8(10):410–5. . [DOI] [PubMed] [Google Scholar]
- 36.Tuteja R. Type I signal peptidase: an overview. Arch Biochem Biophys. 2005;441(2):107–11. 10.1016/j.abb.2005.07.013 . [DOI] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–97. Epub 2007/08/08. 10.1016/j.jmb.2007.05.022 . [DOI] [PubMed] [Google Scholar]
- 38.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–9. 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holt SC. Anatomy and chemistry of spirochetes. Microbiol Rev. 1978;42(1):114–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanagihara Y, Kamisango K, Yasuda S, Kobayashi S, Mifuchi I, Azuma I, et al. Chemical compositions of cell walls and polysaccharide fractions of spirochetes. Microbiol Immunol. 1984;28(5):535–44. . [DOI] [PubMed] [Google Scholar]
- 41.Joseph R, Holt SC, Canale-Parola E. Peptidoglycan of free-living anaerobic spirochetes. J Bacteriol. 1973;115(1):426–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umemoto T, Ota T, Sagawa H, Kato K, Takada H, Tsujimoto M, et al. Chemical and biological properties of a peptidoglycan isolated from Treponema pallidum kazan. Infect Immun. 1981;31(2):767–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kristensen O, Gajhede M. Chaperone binding at the ribosomal exit tunnel. Structure. 2003;11(12):1547–56. . [DOI] [PubMed] [Google Scholar]
- 44.Artola-Recolons C, Lee M, Bernardo-Garcia N, Blazquez B, Hesek D, Bartual SG, et al. Structure and cell wall cleavage by modular lytic transglycosylase MltC of Escherichia coli. ACS Chem Biol. 2014;9(9):2058–66. 10.1021/cb500439c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nash RP, Habibi S, Cheng Y, Lujan SA, Redinbo MR. The mechanism and control of DNA transfer by the conjugative relaxase of resistance plasmid pCU1. Nucleic Acids Res. 2010;38(17):5929–43. 10.1093/nar/gkq303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford J, Grujic O, Bruic E, Czjzek M, Grigg ME, Boulanger MJ. Structural characterization of the bradyzoite surface antigen (BSR4) from Toxoplasma gondii, a unique addition to the surface antigen glycoprotein 1-related superfamily. J Biol Chem. 2009;284(14):9192–8. Epub 2009/01/22. 10.1074/jbc.M808714200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crawford J, Lamb E, Wasmuth J, Grujic O, Grigg ME, Boulanger MJ. Structural and functional characterization of SporoSAG: a SAG2-related surface antigen from Toxoplasma gondii. J Biol Chem. 2010;285(16):12063–70. Epub 2010/02/19. 10.1074/jbc.M109.054866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tonkin ML, Arredondo SA, Loveless BC, Serpa JJ, Makepeace KA, Sundar N, et al. Structural and biochemical characterization of Plasmodium falciparum 12 (Pf12) reveals a unique interdomain organization and the potential for an antiparallel arrangement with Pf41. J Biol Chem. 2013;288(18):12805–17. Epub 2013/03/21. 10.1074/jbc.M113.455667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker ML, Peng F, Boulanger MJ. The Structure of Plasmodium falciparum Blood-Stage 6-Cys Protein Pf41 Reveals an Unexpected Intra-Domain Insertion Required for Pf12 Coordination. PLoS One. 2015;10(9):e0139407 10.1371/journal.pone.0139407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Superdex 75 HiLoad size exclusion chromatogram showing that Tp0624 (46 kDa) elutes as a monomer (elution peak of 44 kDa globular standard denoted by hash mark).
(PDF)
Orthologs of Tp0624 were identified using BLASTP (PSI-BLAST algorithm) and aligned in Clustal Omega and BioEdit sequence alignment editor. The predicted Tp0624 signal peptide cleavage site at T61-Q62 (SP), domains 1 (D1), 2 (D2), and 3 (D3), known OmpA domain peptidoglycan binding residues (TD(x)7NxxLSxxRA) and corresponding treponemal residues (red rectangles) are shown. Identical residues (black background), similar residues (gray background), and dissimilar residues (white background) are highlighted. At each position of the consensus sequence, an asterisk indicates full conservation among the 18 treponemes, a colon indicates strong conservation, and a dot indicates lower conservation.
(PDF)
A phylogenetic tree of 18 full-length Tp0624 ortholog sequences from treponemes was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was calculated. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method and were in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) 6 software. The tree was rooted with the Tp0624 ortholog from Spirochaeta thermophila (Accession number: WP_013314620). The dashed line indicates the phylogenetic divide between pathogens (top) and non-pathogens (bottom).
(PDF)
A phylogenetic tree of Tp0624 ortholog sequences corresponding to domain 1 from 18 treponemes was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was calculated. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method and were in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) 6 software. The tree was rooted with the Tp0624 ortholog from Spirochaeta thermophila (Accession number: WP_013314620). Pathogens (P) and non-pathogens (NP) are indicated.
(PDF)
A phylogenetic tree of Tp0624 ortholog sequences corresponding to domain 2 from 18 treponemes was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was calculated. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method and were in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) 6 software. The tree was rooted with the Tp0624 ortholog from Spirochaeta thermophila (Accession number: WP_013314620). Pathogens (P) and non-pathogens (NP) are indicated.
(PDF)
A phylogenetic tree of Tp0624 ortholog sequences corresponding to domain 3 from 15 treponemes was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was calculated. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method and were in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) 6 software. The tree was rooted with the Tp0624 ortholog from Spirochaeta alkalica (Accession number: WP_018525408). Pathogens (P) and non-pathogens (NP) are indicated.
(PDF)
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 5JIR.