Abstract
The karyotype is shaped by different chromosome rearrangements during species evolution. However, determining which rearrangements are responsible for karyotype changes is a challenging task and the combination of a robust phylogeny with refined karyotype characterization, GS measurements and bioinformatic modelling is necessary. Here, this approach was applied in Heterotaxis to determine what chromosome rearrangements were responsible for the dysploidy variation. We used two datasets (nrDNA and cpDNA, both under MP and BI) to infer the phylogenetic relationships among Heterotaxis species and the closely related genera Nitidobulbon and Ornithidium. Such phylogenies were used as framework to infer how karyotype evolution occurred using statistical methods. The nrDNA recovered Ornithidium, Nitidobulbon and Heterotaxis as monophyletic under both MP and BI; while cpDNA could not completely separate the three genera under both methods. Based on the GS, we recovered two groups within Heterotaxis: (1) "small GS", corresponding to the Sessilis grade, composed of plants with smaller genomes and smaller morphological structure, and (2) "large GS", corresponding to the Discolor clade, composed of plants with large genomes and robust morphological structures. The robust karyotype modeling, using both nrDNA phylogenies, allowed us to infer that the ancestral Heterotaxis karyotype presented 2n = 40, probably with a proximal 45S rDNA on a metacentric chromosome pair. The chromosome number variation was caused by ascending dysploidy (chromosome fission involving the proximal 45S rDNA site resulting in two acrocentric chromosome pairs holding a terminal 45S rDNA), with subsequent descending dysploidy (fusion) in two species, H. maleolens and H. sessilis. However, besides dysploidy, our analysis detected another important chromosome rearrangement in the Orchidaceae: chromosome inversion, that promoted 5S rDNA site duplication and relocation.
Introduction
The karyotype, i.e., the complete eukaryotic chromosome complement, was shaped during species evolution through chromosome rearrangements [1–8]. Fusion and fission are two of the most important chromosome rearrangements causing dysploidy; i.e., the variation in chromosome number due to rearrangements without any gain or loss of genetic material [9–11]. Some hypotheses have been proposed regarding the importance of fusion and fission in karyotype evolution: whereas some authors claim fusion as the most important type of rearrangement, since truly telocentric chromosomes either do not exist or are very rare [12]; others have suggested centric fission as the main process, as it minimizes the genetic risks due to deleterious reciprocal translocations, as postulated by the Minimal Interaction Theory [13–15]. So far, these hypotheses have rarely been tested in a phylogenetic framework (but see [11, 16]).
Elucidating karyotype evolutionary history is often challenging because the successive accumulation of chromosome rearrangements can obscure the order of events that have occurred across a lineage [17]. However, the use of chromosome number and other karyotype traits, such as chromosome morphology and the localization of heterochromatic bands and rDNA sites, within a phylogenetic framework, can help to reveal karyotype modifications that occurred during species evolution.
Orchids are good models for testing the fusion/fission hypotheses due to the frequent dysploidy variation documented in different genera. Among them, Heterotaxis Lindl., which comprises 13 species (Fig 1) [18], presents dysploid variation, between 2n = 40 and 42 among the six species with known chromosome numbers (Table 1). However, only one species–H. discolor [cited as Maxillaria discolor (Lodd. ex Lindl.) Rchb. f.]–has been analysed for additional chromosome markers, such as heterochromatic bands and rDNA sites [19].
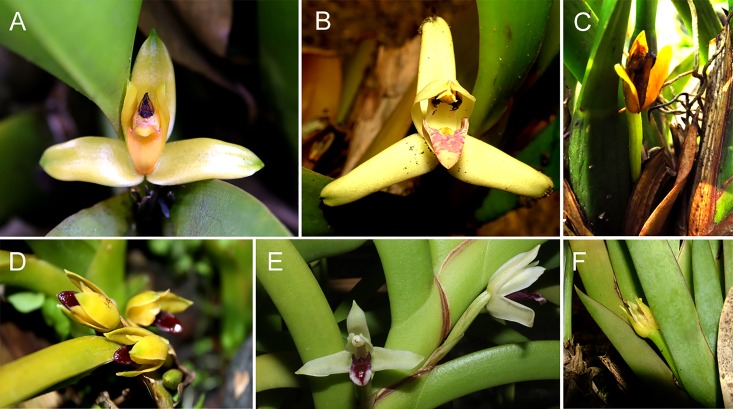
Fig 1. Heterotaxis flowers.
A, H. brasiliensis; B, H. violaceopunctata; C, H. villosa; D, H. superflua; E, H. equitans; F, H. valenzuelana. Photos by A. P. Moraes.
Table 1. Heterotaxis chromosome numbers.
| Species | 2n | n | Reference |
|---|---|---|---|
| Heterotaxis discolor (Lodd. ex Lindl.) Ojeda & Carnevali | 42 | [18][19] | |
| H. maleolens (Schltr.) Ojeda & Carnevali | 40 | [20] | |
| H. valenzuelana (A. Rich.) Ojeda & Carnevali | 40 | [20] | |
| H. villosa Barb. Rodr. | 20 | [21] | |
| H. sessilis (Lindl.) F. Barros | 20 | [21] | |
| H. violaceopunctata (Rchb. f.) F. Barros | 42 | [18] |
Morphologically, Heterotaxis is composed mainly of sympodial species with short rhizomes and laterally compressed, oblong unifoliate pseudobulbs, subtended by various leaf-bearing sheaths [18]. The only exceptions are H. equitans [= Maxillaria equitans (Schltr.) Garay] (Fig 1E) and H. valenzuelana [= Maxillaria valenzuelana (A. Rich.) Nash] (Fig 1F), which have a pseudomonopodial growth habit without pseudobulbs [22]. These two species were originally placed in Marsupiaria Hoehne, but they are currently classified as Heterotaxis, along with four other Brazilian Maxillaria Ruiz & Pav. species transferred to Heterotaxis: H. sessilis, H. superflua, H. villosa and H. violaceopunctata [23].
Traditionally, cytogenetic data have been superimposed onto phylogenetic trees to identify chromosome rearrangements throughout the evolution of the karyotype [24–25]. However, statistical approaches, as ancestral state reconstruction based on maximum likelihood, have recently been applied to infer karyotype evolution in a phylogenetic framework [11, 16, 26–28]. In such approaches, the phylogenetical proposals should present all species with karyotype data available and, when nuclear and chloroplast phylogenetic proposals are conflicting, both should be tested independently to reflect the more robust answer about karyotype evolution.
In order to determine which chromosome rearrangement is responsible by the dysploidy variation detected in Heterotaxis we aim:
(1st) amplify the knowledge about karyotype differences among Heterotaxis species based on chromosome number, heterochromatic blocks (number, distribution and type, i.e., CG-rich or AT-rich), rDNA (number and distribution of loci) and genome size (GS);
(2nd) build a phylogenetic framework based on Heterotaxis and close genera, Nitidobulbon and Ornithidium, using DNA sequence data (nuclear and chloroplast);
(3rd) implement model-based phylogenetic approaches to infer the chromosomal rearrangements responsible by chromosome number changes among Heterotaxis species. Aiming to get a robust answer about chromosome evolution, the two datasets, i.e., nuclear DNA and chloroplast DNA (nrDNA and cpDNA, respectively), were analysed separately and used comparatively as phylogenetic framework to model the karyotype evolution in Heterotaxis.
Materials and Methods
Taxon sampling
Efforts were made to sample the largest possible number of species for each analysis. A list of voucher information for all methodologies is provided in Table 2. In the subsection “Phylogenetic analyses”, 11 out the 13 Heterotaxis species, the three species of Nitidobulbon and four out the 35 of Ornithidium species were analysed. In the subsections “Chromosome analyses” and “Genome size estimation”, we used the six available Brazilian species of Heterotaxis, plus Mapinguari desvauxiana (Rchb.f.) Carnevali & R.B.Singer.
Table 2. Taxon sampling for all performed analyses.
The total number of species in each genus is presented in parenthesis after the genus identification. Voucher and origin are supplied for each specimen analysed for molecular psequences (nrDNA and cpDNA), karyotype and genome size approach.
| Genus | Species | Molecular1 | Karyotype | Genome Size | |||||
|---|---|---|---|---|---|---|---|---|---|
| Voucher | Origin | Voucher | Origin | Voucher | Origin | ||||
| Heterotaxis Lindl. (13 species) | |||||||||
| H. brasiliensis (Brieger & Illg) F.Barros | |||||||||
| Koehler 0150 | Brazil | AP 17 | Ubatuba, Brazil | AP 17 | Ubatuba, Brazil | ||||
| IBt 3244 | Paraty, Brazil | IBt 3244 | Paraty, Brazil | ||||||
| FZB 774 | Cultivated | IBt 322 | Brazil | ||||||
| IBt 13159 | Brazil | ||||||||
| IBt 4107 | Brazil | ||||||||
| H. discolor (Lodd. ex Lindl.) Ojeda & Carnevali | |||||||||
| Koehler 0311 | Brazil | - | - | - | - | ||||
| H. equitans (Schltr.) Ojeda & Carnevali | |||||||||
| Koehler 0141 | Brazil | - | - | ||||||
| IBt 979P | Brazil | IBt 979P | Brazil | IBt 979P | Brazil | ||||
| IBt 3931P | Belém, Brazil | IBt 3931P | Belém, Brazil | IBt 3931P | Belém, Brazil | ||||
| H. fritzii Ojeda & Carnevali | |||||||||
| Whitten 2672 | Colombia | - | - | - | - | ||||
| H. maleolens (Schltr.) Ojeda & Carnevali | |||||||||
| Atwood & Whitten 5055 | Honduras | - | - | - | - | ||||
| H. santanae (Carnevali & I. Ramírez) Ojeda & Carnevali | |||||||||
| Whitten 6725 | Ecuador | - | - | - | - | ||||
| H. sessilis (Lindl.) F. Barros | |||||||||
| Atwood & Whitten 5065 | Jamaica | - | - | - | - | ||||
| H. superflua (Rchb.f.) F. Barros | |||||||||
| Koehler 0153 | Brazil | IBt 2336 | Juruena, Brazil | IBt 2336 | Juruena, Brazil | ||||
| AP 76 | Manaus, Brazil | AP 76 | Manaus, Brazil | ||||||
| H. valenzuelana (A.Rich.) Ojeda & Carnevali | |||||||||
| Koehler 0263 | Brazil | IBt 3177 | Serra dos Órgãos, Brazil | IBt 3177 | Serra dos Órgãos, Brazil | ||||
| IBt A457 | Camanducaia, Brazil | IBt A457 | Camanducaia, Brazil | ||||||
| IBt A843 | Cananéia, Brazil | IBt A843 | Cananéia, Brazil | ||||||
| H. villosa (Barb. Rodr.) F. Barros | |||||||||
| Koehler 0367 | Brazil | IBt 3934P | Belém, Brazil | IBt 3934P | Belém, Brazil | ||||
| H. violaceopunctata (Rchb.f.) F.Barros | |||||||||
| Koehler 0129 | Brazil | IBt 10110 | Rondonia, Brazil | IBt 10111 | Rondonia, Brazil | ||||
| IBt 11518 | Cultivated | IBt 11518 | Cultivated | ||||||
| IBt 11519 | Cultivated | ||||||||
| IBt 1713 | Brazil | ||||||||
| Nitidobulbon Ojeda, Carnevali & GARomero (3 species) | |||||||||
| N. cymbidiodes (Dodson, J.T. Atwood & Carnevali) Ojeda & G.A. Romero | |||||||||
| Atwood & Whitten 5067 | Ecuador | - | - | - | - | ||||
| N. nasutum (Rchb. f.) Ojeda & Carnevali | |||||||||
| Koehler 0261 | Brazil | - | - | - | - | ||||
| N. proboscideum (Rchb. f.) Ojeda & Carnevali | |||||||||
| Atwood & Whitten 5056 | Venezuela | - | - | - | - | ||||
| Ornithidium Salisb. ex. R.Br. (35 species) | |||||||||
| O. adendrobium (Rchb. f.) M.A. Blanco & Ojeda | |||||||||
| Dressler 4231 | Panama | - | - | - | - | ||||
| O. coccinea (Jacq.) Salisb. ex R. Br. | |||||||||
| Atwood & Whitten 5092 | Puerto Rico | - | - | - | - | ||||
| O. conduplicatum Ames & C. Schweinf. | |||||||||
| Blanco 1660 | Costa Rica | - | - | - | - | ||||
| O. fulgens Rchb. f. | |||||||||
| Whitten 2630 | Panama | - | - | - | - | ||||
| Outgroup | |||||||||
| Brasiliorchis picta (Hook.) R.B. Singer, S. Koehler & Carnevali | |||||||||
| Koehler 0337 | Brazil | - | - | - | - | ||||
| Cryptocentrum latifolium Schltr. | |||||||||
| Whitten 2349 | Ecuador | - | - | - | - | ||||
| Inti bicallosa (Rchb.f.) M.A.Blanco | |||||||||
| Whitten 2748 | Ecuador | - | - | - | - | ||||
| Mapinguari desvauxiana (Rchb. f.) Carnevali & R.B. Singer | |||||||||
| Koehler 1585 | Brazil | - | - | IBt 2367 | Paraty, Brazil | ||||
| IBt 3961 | Jeriquara, Brazil | ||||||||
| IBt 4119 | Peruíbe, Brazil | ||||||||
| IBt 807 | Cananéia, Brazil | ||||||||
| Xylobium zarumense Dodson | |||||||||
| Whitten 1881 | Ecuador | - | - | - | - | ||||
1 –All data used in Phylogeny analysis were published by [29] and downloaded from GenBank (S2 Table), except the sequences for H. equitans IBt 979P and IBt3931P that were obtained here. Collection: FZB—Fundação ZooBotânica de Porto Alegre/RS, Brazil; IBt—Instituto de Botânica de São Paulo/SP, Brazil; AP–plants collected by Ana Paula Moraes with field study authorization by SISBIO/Brazil (37013–1 and 37417–1).
All specimens, but AP16 and AP46, were held in two living orchid collections available in Brazil (São Paulo Botany Institute—IBt—and Botanical Garden of Porto Alegre—FZB) (Table 2). The two specimens sampled from the field were collected in unprotected area and the authorization was emitted by SISBIO/Brazil (37013–1 and 37417–1).
Phylogenetic analyses
Phylogenetic analyses based on ITS, matK + trnK and atpB—rbcL spacer used sequences published by [29] and new sequences obtained for H. equitans (S1 Table). The species Xylobium zarumense Dodson, Inti bicalosa (Rchb.f.) M.A.Blanco and Cryptocentrum latifolium Schltr. were used as outgroup, following Ojeda et al. [30], plus Brasiliorchis picta (Hook.) R.B.Singer, S.Koehler & Carnevali and Mapinguari desvauxiana—both species previously considered Maxillaria.
The analyses were performed using the maximum parsimony (MP) criterion implemented in PAUP 4.0 [31] and Bayesian inference (BI) with MrBayes v.3.1.2 [32]. Both analyses were conducted on two separate matrices: (1) nrDNA and (2) cpDNA. All characters were considered unordered and equally weighted.
For the MP analysis, a heuristic search for the most parsimonious trees (MPT) included: (1) an initial round of tree searches with 1000 random addition sequence replicates (RASR), holding 10 trees at each step, and (2) tree bisection-reconnection (TBR) branch swapping with MULTREES, with steepest descent option in effect, saving a maximum of 50 trees at each replicate. All the shortest trees retained in memory were then included in a second round of searches involving exhaustive TBR branch swapping. Bootstrap support [33] was performed on each analysis using the program TreeRot v.2 [34]. Bootstrap values (BS) were evaluated as providing either moderate (50–74%) or strongly supported (75–100%).
For the BI analysis, the optimal model of sequence evolution for each molecular dataset was selected using jModeltest v.2.1.1 [35]. The Bayesian information criterion (BIC) implemented in jModeltest was used to choose the best-fitting evolutionary model for each sequence partition. Starting model parameters were assigned as uniform prior probabilities and further estimated during the analysis by allowing them to vary independently among data partitions. Twenty million generations were run using one cold and three incrementally heated Markov Chain Monte Carlo (MCMC) (Temp = 0.2), with parameters sampled every 2,000 generations. Two independent runs (Nruns = 2), starting from different random trees, were performed to ensure that the individual runs had converged to the same result. Log files were analysed with Tracer v.1.5 [36] to assess convergence and ensure that the MCMC had run long enough to obtain a valid estimate of the parameters. Based on inspection of the likelihood scores for each generation, the first 2,500 sampled generations were considered as burn-in and discarded from subsequent analyses. The post burn-in trees were imported into Tree Annotator v.1.5.4 [37], and a 50% majority-rule consensus tree was then reconstructed to obtain posterior probabilities of the clades. The majority-rule consensus tree was then analysed and edited into FigTree v.1.3.1. [38]. Posterior probabilities (PP) were considered strongly supported when equal to or higher than 0.95.
Chromosome analysis
Root tips were pre-treated in 8-hydroxyquinoline (0.002 M) for 24 h at 10°C, fixed in absolute ethanol:glacial acetic acid (3:1, v/v) for 24 h at room temperature, and stored at -20°C. The meristems were washed in distilled water and digested in 2% (w/v) cellulase (Onozuka) / 20% (v/v) pectinase (Sigma) / 1% macerozyme (Sigma) solution and squashed in a drop of 45% acetic acid. The cover slip was removed in liquid nitrogen. For chromosome banding, preparations aged for three days were stained with CMA (0.5 mg ml-1) for 1 h and counterstained with DAPI (1 mg ml-1) for 30 min. The slides were examined using a Leica DMRA2 epifluorescence microscope, photographed with a Leica camera, and analysed using Leica LAS 3.6 software. The best slides were distained in alcohol and stored for FISH analysis. Images were processed uniformly for colour balance, contrast and brightness using Adobe Photoshop CS5 (Adobe Systems, Inc.).
For in situ hybridization, a D2 probe from Lotus japonicus (Regel) K. Larsen [39] and an R2 probe from Arabidopsis thaliana (L.) Heynh. [40] were used to localize 5S and 45S rDNA, respectively. The 5S rDNA probe was labelled with digoxigenin-11-dUTP and the 45S rDNA probe with biotin-14-dUPT. In both cases, nick translation (Roche Biochemicals) was performed. The in situ hybridization mixture was composed of 50% (v/v) formamide, 10% (w/v) dextran sulphate and 0.1% (w/v) sodium dodecyl sulphate in 2 × saline-sodium citrate buffer (SSC) with 3–5 ng ml-1 of each probe. The 5S rDNA probe was detected with anti-digoxigenin conjugated to rhodamine (Roche Biochemicals), and the 45S rDNA probe was detected using an avidin-FITC conjugate (Roche Biochemicals). All slides were counterstained with 2 μg ml-1 DAPI in Vectashield mounting medium (Vector Laboratories). Metaphase images were obtained and processed as described above under "Chromosome banding".
Genome size estimation
To determine the DNA content of Heterotaxis species, approximately 25 mg of leaf tissue of each species was macerated with the same mass of the internal reference standard Zea mays L. cv. CE-777 (2C = 5.43 pg) [41]. The material was macerated in 1 ml of cold Tris buffer, using a scalpel blade to release the nuclei into suspension [42]. Nuclei were stained by adding 25 uL of a 1 mg ml-1 solution of propidium iodide (PI, Sigma®, USA). Additionally, 12.5 uL of RNase (2 μg ml-1) was added to each sample. The analysis was performed using the FACSCanto II cytometer (Becton Dickinson, San Jose, CA, USA), kindly made available by the Microbiology and Immunology Department of IBB-UNESP/Botucatu, Brazil. The histograms were obtained with FACSDiva software based on 20,000 events. A statistical evaluation was performed using the Flowing Software 2.5.1 (http://www.flowingsoftware.com/). One to five samples from each species were analysed twice, according to collection availability (Table 2). The GS obtained from each species were compared statistically by ANOVA followed by Tukey’s test using BioEstat v.5.3 [43].
Ancestral state reconstruction of chromosome number
Ancestral state reconstruction for base chromosome number was performed with ChromEvol v.2.0 [44–45] to identify which chromosome rearrangement–fission or fusion–was responsible for chromosome number variation in Heterotaxis. We considered the basic chromosome number (x) as the haploid chromosome number that most parsimoniously explain the chromosomal variability in the group and shows a clear relationship with the basic number of the closest related groups [7, 46]. We are aware of Peruzzi (2013) [47] who, after an extensive revision about the concept of base chromosome number, suggested that the inferred ancestral base number should be indicated by the symbol ‘ρ‘ to clearly differ from ‘x’. When appropriated, we cited the symbol ‘ρ‘ along with the ‘x’, for the sake of clarity.
The ChromEvol software (http://www.tau.ac.il/~itaymay/cp/chromEvol/) uses a likelihood method based on eight types of chromosome number changes along phylogenies. We ran all available models for each phylogenetic proposal (nrDNA and cpDNA under MP and BI) and used the Akaike information criterion (AIC) to select the best model for our dataset. The gain/loss of expected numbers of polyploidy events and the gains and losses of single chromosomes along each branch of the phylogeny were recorded based on the best-fitting model. Chromosome numbers were taken from the literature as well as obtained in the present study. The input data are presented in S2 Table.
Results
Molecular data information
The aligned nrDNA dataset consisted of 780 bp with 88 informative characters, and the aligned complete cpDNA dataset consisted of 3014 bp (1813 from the matK + trnK locus and 1201 from the atpB—rbcL intergenic region) with 96 informative characters.
Phylogenetic analyses: Maximum Parsimony
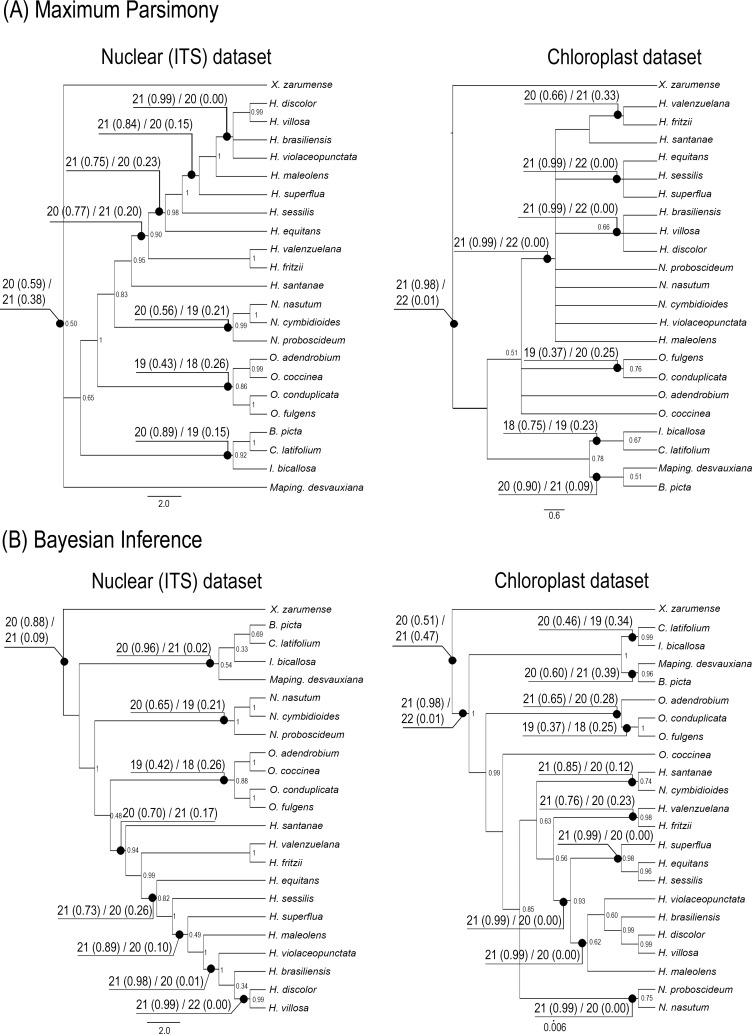
Based on the most parsimonious trees (MPTs) obtained, some incongruent clades were found between the nrDNA and cpDNA trees, mainly due to the H. equitans. Additional sequences were obtained for this species to avoid taxonomic errors, but the incongruence was maintained. Only nrDNA recovered the three genera, Heterotaxis, Nitidobulbon and Ornithidium, as monophyletic (Fig 2A). The cpDNA dataset recovered Nitidobulbon nested in a comb with Heterotaxis and Ornithidium as sister of Nitidobulbon + Heterotaxis. The CI and RI for the individual datasets were CI = 0.571 and RI = 0.734 for nrDNA and CI = 0.518 and RI = 0.718 for cpDNA.
Fig 2.
The strict consensus trees generated using (A) Maximum Parsimony and (B) Bayesian Inference based on nrDNA and cpDNA datasets. Selected bootstrap values above 0.49 are shown below the branches. For each consensus tree, the results for ancestral base chromosome number evolution estimated by MLE is shown, presenting the two most likely base chromosome numbers (haploid) on selected nodes, followed by the probability in parenthesis.
Phylogenetic analyses: Bayesian inference
Three models were selected for each molecular marker: TiM3 + G for nrDNA, TPM1uf + G and TIM1 + G for cpDNA (atpB and matK + trnK, respectively). The tree recovered from the nrDNA dataset contained the three major clades, with strong support for Nitidobulbon (PP = 1), placed as sister of Ornithidium (moderate support—PP = 0.88) + Heterotaxis (marginal strong support—PP = 0.94) (Fig 2B—Nuclear dataset). The separation between Ornithidium and Heterotaxis received a low support (PP = 0.48). Based on cpDNA, N. cymbidioides was nested in Heterotaxis and O. coccinea was sister of Nitidobulbon + Heterotaxis (Fig 2B—Chloroplast dataset). Marsupiaria, as previously circumscribed, was nested within Heterotaxis, and neither the nrDNA nor cpDNA dataset placed H. valenzuelana and H. equitans close to each other. Again, incongruences between nrDNA and chloroplast datasets do not allowed to join both datasets.
Chromosome number and genome size
The 2n = 42 was observed in H. brasiliensis, H. villosa, H. violaceopunctata, H. equitans and H. superflua and 2n = 40 in H. valenzuelana (Table 3, Fig 3). Regarding the genome size (2C value; see S1 Fig), the six Heterotaxis species were divided into two groups (F = 29.7; p < 0.0001): (1) larger genomes, found in H. brasiliensis (8.64 pg), H. villosa (8.75 pg) and H. violaceopunctata (8.51 pg); and (2) smaller genomes, found in H. equitans (7.70 pg), H. superflua (7.67 pg) and H. valenzuelana (7.46 pg) (Table 3). Mapinguari desvauxiana, used as an outgroup in the phylogeny, had a 2C = 4.49 pg.
Table 3. Karyotype and genome size (2C) data for Heterotaxis.
| Species | Karyotype1 | Genome Size | |||||
|---|---|---|---|---|---|---|---|
| 2n | DAPI+ | CMA+ | 45S | 5S | 2C2 | CV3 | |
| Heterotaxis brasiliensis | 42 | 12–16, pr. | 4, ter. | 4, ter. | 2, ter. | 8.64 | 2.76 |
| 2, int. | |||||||
| Heterotaxis violaceopunctata | 8.51 | 2.43 | |||||
| Heterotaxis villosa | 8.75 | 2.57 | |||||
| Heterotaxis superflua | 7.67 | 2.65 | |||||
| Heterotaxis equitans | 4, (2/2—ter/int). | 7.70 | 3.15 | ||||
| Heterotaxis valenzuelana | 40 | - | 2, pr. | 2, pr. | 2, int. | 7.46 | 2.50 |
| Mapinguari desvauxiana* | 40 | - | 2, ter. | 2, ter. | 2, int. & 2, ter. | 4.49 | 4.26 |
1 pr. = proximal position on the chromosome; int. = intertitial position on the chromosome; ter. = terminal position on the chromosome.
2 2C values are given in picograms.
3CV = Coefficient of variation.
*Karyotype data were obtained by [19].
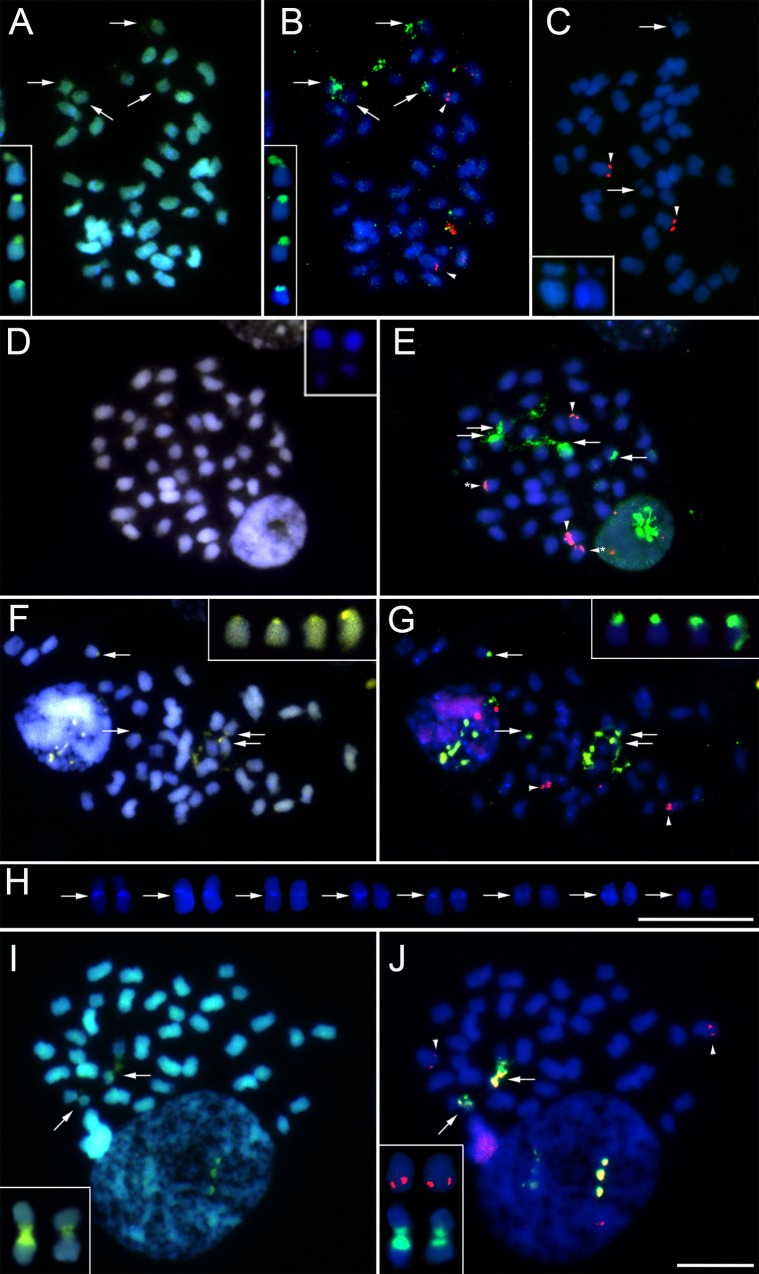
Fig 3. Chromosome analysis of Heterotaxis.
A-B, H. brasiliensis (population from Paraty, Brazil); C, H. brasiliensis (population from Ubatuba, Brazil); D-E, H. equitans; F-G, H. villosa; H, Heterotaxis chromosomes showing pericentromeric DAPI+ bands; I-J, H. valenzuelana. A, D, F and I: CMA (yellow)/DAPI (blue) banding. H: DAPI+ bands. B, E, G and J: in situ hybridization using 45S rDNA (green) and 5S rDNA (red). C: 5S rDNA (red). Arrows in A, F and I indicate CMA0/DAPI−or CMA+/DAPI−bands. Arrows and arrowheads in B, E, G and J show 45S rDNA and 5S rDNA, respectively. Arrowheads in C show 5S rDNA and arrows indicated the CMA–/DAPI−chromosome gap. Detail in A, F and I indicate chromosomes with CMA+/DAPI−bands and in B, G and J, the same chromosomes with 45S rDNA sites (green). Detail in J shows also the chromosome pair with 5S rDNA sites (red). Chromosomes in the inserts in A, B, I and J could be selected from an alternative metaphase. Insets in C and D show the chromosome with CMA–/DAPI−gap. Bars in H and J represent 10 m.
Karyotype characterization
CMA/DAPI banding
The chromosome banding showed four band types: CMA0/DAPI−(neutral on CMA and dull on DAPI; see arrows in Fig 3A and 3I), CMA+/DAPI−(bright on CMA and dull on DAPI; see arrows in Fig 3F and inserts in 3A, 3F and 3I), CMA–/DAPI+ (dull on CMA and bright on DAPI; see arrows in Fig 3H) and CMA–/DAPI−(dull on both fluorochromes; see arrows and detail in Fig 3C and detail in Fig 3D). Punctual CMA–/DAPI+ bands were observed in all species in the proximal region of 6–8 chromosome pairs (Fig 3H), which became more evident after in situ hybridization (Fig 3B and 3G). However, H. valenzuelana did not have any DAPI+ bands (Fig 3I and 3J).
The four terminal CMA+/DAPI−bands (see details in Fig 3A and 3F), sometimes were detected as CMA0/DAPI−bands (Fig 3A) and could be hardly seen in some metaphases (Fig 3D). The bands were observed in the terminal position on two acrocentric chromosome pairs in all species (see details in Fig 3A and 3F), except by H. valenzuelana, which had two CMA+/DAPI−bands in the proximal region on a metacentric chromosome pair (Fig 3I). A chromosome pair could be identified by an uncommon CMA–/DAPI−band in the proximal region (insets in Fig 3C and 3D). The absence of staining with both fluorochromes formed a gap, which was frequently distended, sometimes placing the short arm distant from the long arm.
In situ hybridization
The 45S rDNA sites were always co-localized with CMA+ bands. All species had four terminal 45S rDNA sites on acrocentric chromosomes (Fig 3B, 3E and 3G) with the exception of H. valenzuelana, which had two proximal 45S rDNA sites on a metacentric chromosome pair (see detail in Fig 3J). We observed two interstitial 5S rDNA sites in most species (Fig 3B, 3G and 3J and detail in 3J); however, H. brasiliensis from the Ubatuba population (São Paulo State, Brazil) had the two 5S rDNA sites in a terminal position (Fig 3C). Moreover, H. equitans had four sites: two interstitial sites in one chromosome pair and two terminal sites in another (Fig 3E).
Reconstruction of ancestral chromosome number
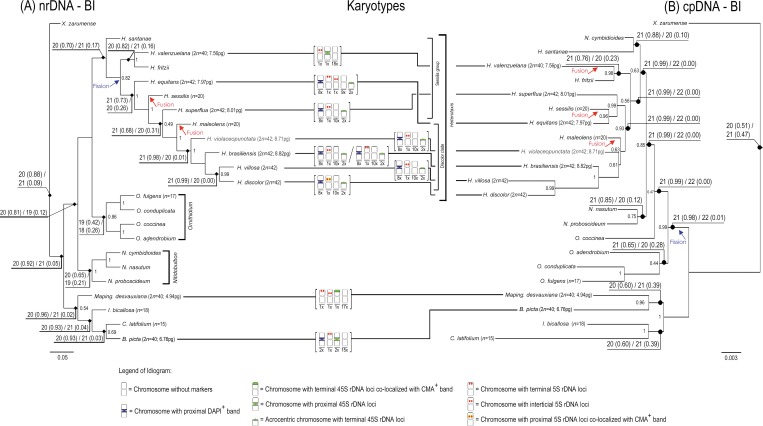
Due to the incongruences between nrDNA and cpDNA datasets we used the four phylogenetic proposals independently for ancestral reconstruction of the basic chromosome number (Fig 2). The results for nrDNA and cpDNA under BI are compared in Fig 4 with idiograms for all species analysed. For the four phylogenetical proposals, the best-fitting ML model was the combination of constant gain and loss without duplication (i.e., just dysploidy events) for the four phylogenetic hypotheses used (Table 4).
Fig 4.
Majority rule consensus tree generated using Bayesian Inference based on the (A) nrDNA and (B) cpDNA datasets, presenting also the ancestral base chromosome number evolution estimated by MLE and karyotypes obtained. Blue arrow indicate a probably point of chromosome gain (supposed fission), while red arrow indicate a probably point of chromosome loss (supposed fusion). The two most likely base chromosome numbers (haploid) are indicated on selected nodes, followed by the probability in parenthesis. Genome sizes are indicated at the terminal, after the species name. An idiogram for species with karyotype data is shown after the terminal. Data for B. picta were determined by [25]. Selected PP values above 0.49 are shown on the nodes.
Table 4. Likelihood estimates and AIC scores for the four phylogenetical proposals tested using the ChromEvol software.
| Maximum Parcimony | Bayesian Inference | |||
|---|---|---|---|---|
| MODEL | Log-likelihood | AIC | Log-likelihood | AIC |
| nrDNA | ||||
| CONST_RATE | -14.92 | 35.84 | -15.73 | 37.46 |
| CONST_RATE_DEMI | -14.92 | 35.84 | -15.73 | 37.46 |
| CONST_RATE_DEMI_EST | -14.92 | 37.84 | -15.73 | 39.46 |
| CONST_RATE_NO_DUPL | -14.92 | 33.84 | -15.73 | 35.46 |
| LINEAR_RATE | -15.08 | 40.15 | -15.89 | 41.77 |
| LINEAR_RATE_DEMI | -15.08 | 40.15 | -15.89 | 41.77 |
| LINEAR_RATE_DEMI_EST | -15.08 | 42.15 | -15.89 | 43.77 |
| LINEAR_RATE_NO_DUPL | -15.08 | 38.15 | -15.89 | 39.66 |
| cpDNA | ||||
| CONST_RATE | -22.41 | 50.81 | -19.46 | 44.92 |
| CONST_RATE_DEMI | -22.41 | 50.81 | -19.46 | 44.92 |
| CONST_RATE_DEMI_EST | -22.41 | 52.81 | -19.46 | 46.92 |
| CONST_RATE_NO_DUPL | -22.41 | 48.81 | -19.46 | 42.92 |
| LINEAR_RATE | -22.77 | 55.55 | -19.75 | 49.50 |
| LINEAR_RATE_DEMI | -22.77 | 55.55 | -19.75 | 49.50 |
| LINEAR_RATE_DEMI_EST | -22.77 | 57.61 | -19.75 | 51.6 |
| LINEAR_RATE_NO_DUPL | -22.77 | 53.55 | -19.75 | 47.5 |
When using phylogenetic hypotheses based on nrDNA, the best-fitting model suggested x = 20 (or ρ = 20, if following nomenclature suggested in [45]) as the inferred ancestral basic chromosome number of Heterotaxis. However, x = 21 (or ρ = 21) was suggested when using phylogenetic hypotheses based on cpDNA (Figs 2 and 4). In three out four tested phylogenetic hypothesis, x = 20 (or ρ = 20) was the inferred as the ancestral state for the whole group of species used in the phylogeny. Besides that just nrDNA datasets recovered the three genera as monophyletics, the difference among nrDNA x cpDNA phylogenetic hypothesis is when the first fission event occurred: (1) if in the beginning of Heterotaxis diversification, with two subsequent fusions in H. maleolens and H. sessilis, as suggested by nrDNA datatsets; or (2) before Heterotaxis diversification and the three fusions events occurred inside Heterotaxis genus, reducing the chromosome number from 2n = 42 to 2n = 40 in H. maleolens, H. sessilis and H. valenzuelana, as suggested by cpDNA datasets.
Discussion
The interpretation of the pattern of chromosome evolution detected here is supported by a set of phylogeny hypothesis suggesting the occurrence of chromosome fission and subsequent chromosome fusion. The chromosome data obtained from species of Heterotaxis are consistent with the variation observed in other Orchidaceae genera: frequent dysploidy (caused by chromosome fission and fusion), 5S rDNA position/number changes and a diversity of heterochromatic bands revealed by CMA/DAPI banding.
Phylogenetic relationships
Genus Heterotaxis
Traditionally, the Heterotaxis species were organized into two major morphological groups–Sessilis and Discolor [18, 29, 30]. Both groups were recovered here, Sessilis as a grade and Discolor as a clade.
Sessilis grade: This group of species comprised H. santanae, H. valenzuelana, H. fritzii, H. equitans, H. sessilis and H. superflua (Fig 4), all of which are conspicuously succulent with small vegetative, floral organs and small GSs, as well.
Discolor clade: This clade is well supported (see both nrDNA phylogeny trees in Fig 2) and morphologically characterized by robust vegetative and floral parts and the presence of a three lobed lip with an ovate apex. This clade contains five robust species–H. maleolens, H. violaceopunctata, H. brasiliensis, H. villosa and H. discolor which present also a large GS (Fig 4).
Genera Ornithidium and Nitidobulbon
The organization of cpDNA trees probably reflect previous hybridization event between Heterotaxis and Ornithidium and Nitidobulbon species. The proximity of Nitidobulbon and Heterotaxis is reflected by morphological similarities between genera and the three species currently grouped in Nitidobulbon were traditionally included in Heterotaxis and, even nowadays, are sometimes misidentified [23, 48, 49].
Karyotypes in Heterotaxis
In this study, we report new chromosome numbers for H. brasiliensis, H. superflua and H. equitans and also confirm previous reports for H. violaceopunctata and H. valenzuelana. However, we found discrepancies between the chromosome number previously reported for H. villosa (n = 20) (Table 1, [21]) and that obtained from our analysis (2n = 42; Table 3). Such intraspecific karyotype variation suggests either the occurrence of counting errors/misidentifications or occurrence of different cytotypes; i.e., populations with divergent karyotypes. Such difference could be due to polyploidy, aneuploidy or dysploidy rearrangements [1–3], what could be the case of H. villosa.
The presence of multiple cytotypes, specially dysploidy cytotypes, seems to be neglected in taxonomic and ecological studies [4, 5, 8, 46, 50]. However, reports of such variation among populations are common [51–52], even in taxonomic groups in which dysploidy is considered rare, such as in subfamily Mimosoideae (Leguminosae; 1.46% of species show dysploidy) [53–54]. Unfortunately, Blumenschein & Paker [21] did not deposit any vouchers of the analysed material; therefore, the possibility of misidentification should not be ruled out, especially considering the challenging taxonomy of this genus [18, 23, 30, 55].
Karyotype and GS evolution in Heterotaxis
Recently, Escudero et al. [11] analysed chromosome gains and losses in 15 angiosperms clades, including the subtribe Orchidinae (Orchideae: Orchidaceae). The authors proposed dysploidy as a predominant mechanism in Orchidinae, as previously suggested for other subfamilies of Orchidaceae [56, 57]. Dysploidy is traditionally suggested as the cause of chromosome number variation in subfamily Cypripedioideae [58], in the genus Paphiopedilum Pfitzer [59] and in tribe Neottieae, including Cephalanthera Rich. [60, 61], Epipactis Zinn and Neottia Guett. [62]. These studies suggest that dysploidy plays a key role in the chromosome evolution of Orchidaceae [56].
The variation in chromosome number detected in Heterotaxis also appears to be caused by dysploidy. However, the separation between taxonomic groups, grade Sessilis and clade Discolor, is more likely caused by repetitive DNA variation, increasing the GS in the clade Discolor. It is traditionally assumed that plants with large GS present large morphological traits [63]. However, this hypothesis could be confirmed just in small groups of related species and, when using higher phenotypic scales, this relationship is often reduced [64, 65].
Our findings support the inference that the dysploidy variation was primarily caused by chromosome fission in an ancestral presenting 2n = 40 and a proximal 45S rDNA site on a metacentric chromosome pair. Such species, after a fission in the 45S rDNA site, originated species with 2n = 42 and two acrocentric chromosome pairs with terminal 45S rDNA sites.
Despite some doubt about when the chromosome fission occurred, it is certainly that a fission event happened just before Heterotaxis diversification (cpDNA phylogeny) or at the beginning of Heterotaxis diversification, after separation of H. santanae, H. fritzii and H. valenzuelana (nrDNA phylogeny). However, considering that nrDNA dataset provides a more robust phylogeny, we can assume x = 20 (or ρ = 20) to Heterotaxis. The occurence of one fission originated the 2n = 42 and the two subsequent fusion, in H. sessilis and H. maleolens, restored the 2n = 40 in these two species. The hypothesis of chromosome evolution presented here diverge of both White’s hypothesis [12] and Minimal Interaction Theory [13–15], but proposed a more dynamic karyotype evolution with both event occurring repeated times.
Actually, some chromosome characteristics facilitated the occurrence of repeatedly fusion-fission chromosome events. For example, the chromosome bouquet configuration during meiosis, i.e. telomere clustered together at one side and centromeres clustered at the opposite side during chromosome pairing in meiosis [66], facilitates chromosome rearrangements [67]. Such configuration facilitate chromosome centromeres fission, as well as, fusion of chromosome terminals. Moreover, the 45S rDNA is a fragile site in the chromosome, susceptible to breaks and unions, and after a chromosome rearrangement, the unbound terminals can join, facilitating chromosome fusion [66]. These small breaks and chromosome fusion events are common and can occur many times in the same chromosome site. Therefore, such rearrangements could be responsible for a large proportion of the chromosome number variation observed in the Orchidaceae.
However, rearrangements other than fusion/fission events are also responsible for modelling the karyotype. Here, inversions seem to play an important role in chromosome evolution. It is generally accepted that 5S rDNA sites vary less in number and position than do 45S rDNA sites [68]. However, the Orchidaceae seems to be an exception, with their 5S rDNA sites being highly variable in number, position and sequence [25, 69, 70]. The duplication of the 5S rDNA site in H. equitans and the site position changes detected in one population of H. brasiliensis support inversion as the second more important chromosome rearrangement in Heterotaxis karyotype evolution.
The variation in 5S rDNA position observed in H. brasiliensis is likely the consequence of a paracentric inversion, moving the sites from an interstitial position to a terminal position. In H. equitans, one of the points of chromosome breakage (allowing the chromosome inversion to occur) was probably inside the 5S rDNA site and, after inversion, the rearrangement originated a second site. In this sense, inversion happened twice during Heterotaxis evolution and seems to be frequent in the Orchidaceae, as observed in Cephalanthera [61] and Christensonella subulata (Lindl.) Szlach., Mytnik, Górniak & Śmiszek [25]. In addition to dysploidy, inversion is probably a recurrent chromosome rearrangement modelling Orchidaceae karyotypes.
Conclusion
The refined karyotype characterization analysed under a phylogenetic context reinforces the dysploidy importance in the chromosome evolution and the GS importance in the separation of groups of species. If in one hand, larger GS coincides with larger morphological structures; in the other hand the chromosome number variation seems to be a very dynamic rearrangement not related with groups separation in Heterotaxis. Following the well resolved phylogenetic hypothesis, nrDNA under MP and BI, it is likely that 2n = 40 is an ancestral state, while 2n = 42, observed in the majority of the species, is likely to be a derived condition. However, chromosome fusions restored the ancestral condition in H. maleolens and H. sessilis. The cpDNA suggested three fusion event in Heterotaxis with 2n = 42 as ancestral, but cpDNA has a lower phylogenetic resolution when compared with nrDNA hypothesis.
In addition to dysploidy, inversions appear to take part in modelling Orchidaceae karyotypes, moving 5S rDNA sites and sometimes duplicating them. The identification of chromosome rearrangements presented here reinforces the importance of a phylogenetical framework and statistical methods for ancestral state reconstruction, shedding light on chromosome rearrangements throughout species diversification.
Supporting Information
Representative flow histograms of relative fluorescence (X–axis) obtained after isolation of nuclei from Heterotaxis and M. desvauxiana with internal calibration standards, Zea mays and Vicia fava, respectively. Peaks are identified in each Fig A, Zea mays; B, H. brasiliensis; C, H. villosa; D, H. violaceopunctata; E, H. equitans; F, H. superflua; G, H. valenzuelana; H, Vicia fava; I, Mapinguari desvauxiana.
(TIF)
All sequences were published by [29].
(DOC)
Missing data are represented as "x".
(DOC)
Acknowledgments
The authors are grateful to Dra. Samantha Koehler for all comments on the text.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the São Paulo Research Foundation (http://www.fapesp.br/en/; 2008/03673-8 and 2011/22215-3 to APM), CONACyT (http://www.conacyt.mx/; to DIOA), and the National Council of Scientific and Technological Research (http://cnpq.br/CNPq; Research Productivity Grant 304506/2013-3 to FB and 06142/2011-2 to EFM).
References
- 1.Stebbins GL (1971) Chromosomal Evolution in Higher Plants. United States of America: Addison-Wesley Publishing. [Google Scholar]
- 2.King M (1993) Species Evolution: the role of chromosome change United Kingdom: Cambridge University Press. [Google Scholar]
- 3.Levin DA (2002) The role of chromosomal change in plant evolution United Kingdom: Oxford University Press. [Google Scholar]
- 4.Schubert I (2007) Chromosome evolution. Cur Opin Plant Biol 10: 109–115. [DOI] [PubMed] [Google Scholar]
- 5.Faria R, Navarro A (2010) Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol 25: 660–669. 10.1016/j.tree.2010.07.008 [DOI] [PubMed] [Google Scholar]
- 6.Rieseberg LH (2001) Chromosomal rearrangements and speciation. Trends Ecol Evol 16: 351–358. [DOI] [PubMed] [Google Scholar]
- 7.Guerra M (2008) Chromosome numbers in plant cytotaxonomy. Cytog Genome Res 350: 339–350. [DOI] [PubMed] [Google Scholar]
- 8.Schubert I, Vu GTH (2016) Genome stability and evolution: attempting a holistic view. Trends Plant Sci 21:749–757. 10.1016/j.tplants.2016.06.003 [DOI] [PubMed] [Google Scholar]
- 9.Jones K (1998) Robertsonian fusion and centric fission in karyotype evolution of higher plants. The Bot Rev 64: 273–289. [Google Scholar]
- 10.Perry J, Slater HR, Choo KHA (2004) Centric fission—simple and complex mechanisms. Chrom Resear 12: 627–640. [DOI] [PubMed] [Google Scholar]
- 11.Escudero M, Martín-Bravo S, Mayrose I, Fernández-Mazuecos M, Fiz-Palacios O, Hipp AL et al. 2014. Karyotypic changes through dysploidy persist longer over evolutionary time than polyploid changes. PloS One 9: e85266 10.1371/journal.pone.0085266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White MJD (1973) Animal Cytology and Evolution. United Kingdom: Cambridge University Press. [Google Scholar]
- 13.Imai HT, Maruyama T, Gojobori T, Inoue Y, Crozier RH (1986) Theoretical basis for karyotype evolution. 1. The minimum-interaction hypothesis. Am Nat 128: 900–920. [Google Scholar]
- 14.Imai HT, Taylor RW, Crozier RH (1988) Modes of spontaneous chromosomal mutation and karyotype evolution in ants with reference to the minimum interaction hypothesis. Jpn J Genet 63: 159–185. [DOI] [PubMed] [Google Scholar]
- 15.Imai HT, Satta Y, Takahata N (2001) Integrative study on chromosome evolution of mammals, ants and wasps based on the minimum interaction theory. J Theor Biol 210: 475–497. 10.1006/jtbi.2001.2327 [DOI] [PubMed] [Google Scholar]
- 16.Moraes AP, Souza-Chies TT, Stiehl-Alves EM, Piccolli P, Eggers L, Siljak-Yakovlev S et al. (2015) Evolutionary trends in Iridaceae: new cytogenetic findings from the New World. Bot J Linn Soc 177: 27–49. [Google Scholar]
- 17.Schubert I, Lysak MA (2011) Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet 27: 207–216. 10.1016/j.tig.2011.03.004 [DOI] [PubMed] [Google Scholar]
- 18.Ojeda I, Carnevalli C (2009) Heterotaxis In: Pridgeon AM, Cribb P J, Chase MW, Rasmussen FN eds. 2009. Genera Orchidacearum, Volume 5 (Part 2) (1° ed., p. 147–152). Oxford University Press Inc. [Google Scholar]
- 19.Cabral JS, Felix LP, Guerra M (2006). Heterochromatin diversity and its co-localization with 5S and 45S rDNA sites in chromosomes of four Maxillaria species (Orchidaceae). Gen Mol Biol 29: 659–664. [Google Scholar]
- 20.Carnevali G (1991) The cytology and the pollinaria in the Maxillariinae (Orchidaceae). Master’s thesis, University of Missouri, St. Louis, Missouri, USA.
- 21.Blumenschein A, Paker IU (1963) Número de cromossomas nas Maxillariinae (Orchidaceae). Ciên Cult 15: 255. [Google Scholar]
- 22.Hoehne FC (1947) Reajustamento de algumas espécies de Maxillarias do Brasil, com a criação de dois novos gêneros para elas. Arq. Bot. Est. São Paulo 2: 65–73. [Google Scholar]
- 23.Barros F (2002) Notas taxonômicas para espécies brasileiras dos gêneros Epidendrum e Heterotaxis (Orchidaceae). Hoehnea 29: 109–113. [Google Scholar]
- 24.Koehler S, Cabral JS, Whitten WM, Williams NH, Singer RB, Neubig KM, et al. (2008) Molecular phylogeny of the neotropical genus Christensonella (Orchidaceae, Maxillariinae): species delimitation and insights into chromosome evolution. Ann Bot 102: 491–507. 10.1093/aob/mcn128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moraes AP, Leitch IJ, Leitch AR (2012) Chromosome studies in Orchidaceae: karyotype divergence in Neotropical genera in subtribe Maxillariinae. Bot J Linn Soc 170: 29–39. [Google Scholar]
- 26.Sousa A, Cusimano N, Renner SS (2014) Combining FISH and model-based predictions to understand chromosome evolution in Typhonium (Araceae). Ann Bot 113:669–80. 10.1093/aob/mct302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellicer J, Kelly LJ, Magdalena C, Leitch IJ (2013) Insights into the dynamics of genome size and chromosome evolution in the early diverging angiosperm lineage Nymphaeales (water lilies). Genome 13: 1–13. [DOI] [PubMed] [Google Scholar]
- 28.Pellicer J, Kelly LJ, Leitch IJ, Zomlefer WB, Fay MF (2014) A universe of dwarfs and giants: genome size and chromosome evolution in the monocot family Melanthiaceae. New Phytol 201: 1484–97. 10.1111/nph.12617 [DOI] [PubMed] [Google Scholar]
- 29.Whitten WM, Blanco MA, Williams NH, Koehler S, Carnevali G, Singer RB, et al. (2007) Molecular phylogenetics of Maxillaria and related genera (Orchidaceae:Cymbidieae) based on combined molecular data sets. Amer J Bot 94: 1860–1889. [DOI] [PubMed] [Google Scholar]
- 30.Ojeda I, Carnevali G, Williams NH, Whitten WM (2003) Phylogeny of the Heterotaxis Lindley complex (Maxillariinae): evolution of the vegetative architecture and pollination syndromes. Lankesteriana 7: 45–47. [Google Scholar]
- 31.Swofford DL (2002) PAUP*: Phylogenetic analysis using parsimony. Version 4 Sinauer Associates. [Google Scholar]
- 32.Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 33.Bremer K (1988) The limits of aminoacid sequence data in angiosperm phylogenetic reconstruction. Evolution 42: 795–803. [DOI] [PubMed] [Google Scholar]
- 34.Sorensen MD (1999) TreeRot, version 2 Boston University. [Google Scholar]
- 35.Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rambaut A, Drummond AJ (2007) Tracer v1.5. http://beast.bio.ed.ac/Tracer.
- 37.Rambaut A, Drummond AJ (2007) TreeAnnotator, v1.5.4. http://beast.bio.ed.ac.uk/
- 38.Rambaut A (2009) FigTree v1.3.1. http://tree.bio.ed.ac.uk/software/figtree/.
- 39.Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161: 1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wanzeböck EM, Schofer C, Schweizer D, Bachmair A (1997) Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana. Plant J 11: 1007–1016. [DOI] [PubMed] [Google Scholar]
- 41.Lysak MA, Dolezel J (1998) Estimation of nuclear DNA content in Sesleria (Poaceae). Caryologia 52: 123–132. [Google Scholar]
- 42.Dolezel J, Binarova P, Lucretti S (1989) Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plant 31: 113–120. [Google Scholar]
- 43.Ayres M, Ayres M Jr, Ayres DL, Santos AA (2007) BioEstat—Aplicações estatísticas nas áreas das ciências bio-médicas, version 5.3 Instituto de Desenvolvimento Sustentável Mamirauá. [Google Scholar]
- 44.Mayrose I, Barker MS, Otto SP (2010) Probabilistic models of chromosome number evolution and the inference of polyploidy. Syst Biol 59: 132–44. 10.1093/sysbio/syp083 [DOI] [PubMed] [Google Scholar]
- 45.Glick L, Mayrose I (2014) ChromEvol: Assessing the pattern of chromosome number evolution and the inference of polyploidy along a phylogeny. Mol Biol Evol 31: 1914–1922. 10.1093/molbev/msu122 [DOI] [PubMed] [Google Scholar]
- 46.Guerra M (2012) Cytotaxonomy: The end of childhood. Plant Biosyst 146: 703–710. [Google Scholar]
- 47.Peruzzi L (2013) “x” is not a bias, but a number with real biological significance. Plant Biosyst 147: 1238–1241. [Google Scholar]
- 48.Christenson EA (1999) Maxillaria, an overview. In: Proceedings of the 16th world orchid conference, USA, 279–292.
- 49.Atwood JT, Mora de Retana DE (1999) Flora Costaricensis. Tribe Maxillarieae: Subtribes Maxillariinae and Oncidiinae. Fieldiana 40: 1–84. [Google Scholar]
- 50.Severns PM, Liston A (2008) Intraspecific chromosome number variation: a neglected threat to the conservation of rare plants. Conserv Biol 22:1641–1647. 10.1111/j.1523-1739.2008.01058.x [DOI] [PubMed] [Google Scholar]
- 51.Nakata M, Hashimoto T (1983) Karyomorphological studies on species of Pleurothallis, Orchidaceae. Ann Tsukuba Bot Gard 2: 11–322. [Google Scholar]
- 52.Pellicer J, Clermont S, Houston L, Rich TCG, Fay MF (2012) Cytotype diversity in the Sorbus complex (Rosaceae) in Britain: sorting out the puzzle. Ann Bot 110: 1185–1193. 10.1093/aob/mcs185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biondo E, Miotto STS, Schifino-Wittmann MT (2005) Números cromossômicos e implicações sistemáticas em espécies da subfamília Caesalpinioideae (Leguminosae) ocorrentes na região sul do Brasil. Rev Bras Bot 28: 797–808. [Google Scholar]
- 54.Dahmer N, Simon MF, Schifino-Wittmann MT, Hughes CE, Miotto STS, Giuliani JC (2011) Chromosome numbers in the genus Mimosa L.: cytotaxonomic and evolutionary implications. Plant Syst Evo 291:211–220. [Google Scholar]
- 55.Ojeda I, Fernández-Concha GC, Romero-González GA (2005) New species and combinations in Heterotaxis Lindley (Orchidaceae: Maxillariinae). Novon 15: 572–582. [Google Scholar]
- 56.Felix LP, Guerra M (2000) Cytogenetics and cytotaxonomy of some Brazilian species of Cymbidioid orchids. Genet Mol Biol 23: 957–978. [Google Scholar]
- 57.Felix LP, Guerra M (2010) Variation in chromosome number and the basic number of subfamily Epidendroideae (Orchidaceae). Bot J Linn Soc 163: 234–278. [Google Scholar]
- 58.Cox AV, Abdelnour GJ, Bennett MD, Leitch IJ (1998) Genome size and karyotype evolution in the slipper orchids (Cypripedioideae: Orchidaceae). Amer J Bot 85: 681–687. [PubMed] [Google Scholar]
- 59.Karasawa K. & Tanaka R. 1980. C-banding study on centric fission in the chromosomes of Paphiopedilum. Cytologia 45: 97–102. [Google Scholar]
- 60.Schwarzacher T, Schweizer D. 1982. Karyotype analysis and heterochromatin differentiation with Giemsa C-banding and fluorescent counterstaining in Cephalanthera (Orchidaceae). Plant Syst Evol 141: 91–113. [Google Scholar]
- 61.Moscone EA, Samuel R, Schwarzacher T, Schweizer D, Pedrosa-Harand A (2007) Complex rearrangements are involved in Cephalanthera (Orchidaceae) chromosome evolution. Chrom Resear 15: 931–945. [DOI] [PubMed] [Google Scholar]
- 62.Giuseppina B, Brullo C, Pulvirenti S, Scrugli A, Terrasi MC. D’Emerico S (2010) Advances in chromosomal studies in Neottieae (Orchidaceae): constitutive heterochromatin, chromosomal rearrangements and speciation. Caryologia 63: 184–191. [Google Scholar]
- 63.Bennett MD (1987) Variation in genomic form in plants and its ecological implications. New Phytologist 106: 177–200 [Google Scholar]
- 64.Beaulieu JM, Leitch IJ, Patel S, Pendharkar A, Knight CA (2005) Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytologist 179:975–986. [DOI] [PubMed] [Google Scholar]
- 65.Knight CA, Beaulieu JM (2008) Genome size scaling through phenotype space. Annals of Botany 101:759–766. 10.1093/aob/mcm321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Apples R, Morris R, Gill BS, May CE. 1998. Chromosome Biology. Boston: Kluwer Academic Publishers. [Google Scholar]
- 67.Wang X, Jin D, Wang Z, Guo H, Zhang L, Wang L, Li J, Paterson AH (2015) Telomere-centric genome repatterning determines recurring chromosome number reductions during the evolution of eukaryotes. New Phytologist 205: 378–389. 10.1111/nph.12985 [DOI] [PubMed] [Google Scholar]
- 68.Roa F, Guerra M (2012) Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol Biol 12: 225–238. 10.1186/1471-2148-12-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lan T, Albert VA (2011) Dynamic distribution patterns of ribosomal DNA and chromosomal evolution in Paphiopedilum, a lady’s slipper orchid. BMC Evol Biol 11: 126–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Souza BCQ (2011) Citogenética da subtribo Laeliinae (Orchidaceae:Epidendroideae): Regiões heterocromáticas e localização do DNA ribossomal. Dissertation, Universidade Federal da Paraíba.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative flow histograms of relative fluorescence (X–axis) obtained after isolation of nuclei from Heterotaxis and M. desvauxiana with internal calibration standards, Zea mays and Vicia fava, respectively. Peaks are identified in each Fig A, Zea mays; B, H. brasiliensis; C, H. villosa; D, H. violaceopunctata; E, H. equitans; F, H. superflua; G, H. valenzuelana; H, Vicia fava; I, Mapinguari desvauxiana.
(TIF)
All sequences were published by [29].
(DOC)
Missing data are represented as "x".
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.