Abstract
Purpose
MicroRNA (miRNAs) have been previously implicated in scleral remodeling in normal eye growth. They have the potential to be therapeutic targets for prevention/retardation of exaggerated eye growth in myopia by modulating scleral matrix remodeling. To explore this potential, genome-wide miRNA and messenger RNA (mRNA) scleral profiles in myopic and control eyes from mice were studied.
Methods
C57BL/6J mice (n = 7; P28) reared under a 12L:12D cycle were form-deprived (FD) unilaterally for 2 weeks. Refractive error and axial length changes were measured using photorefraction and 1310-nm spectral-domain optical coherence tomography, respectively. Scleral RNA samples from FD and fellow control eyes were processed for microarray assay. Statistical analyses were performed using National Institute of Aging array analysis tool; group comparisons were made using ANOVA, and gene ontologies were identified using software available on the Web. Findings were confirmed using quantitative PCR in a separate group of mice (n = 7).
Results
Form-deprived eyes showed myopic shifts in refractive error (−2.02 ± 0.47 D; P < 0.01). Comparison of the scleral RNA profiles of test eyes with those of control eyes revealed 54 differentially expressed miRNAs and 261 mRNAs fold-change >1.25 (maximum fold change = 1.63 and 2.7 for miRNAs and mRNAs, respectively) (P < 0.05; minimum, P = 0.0001). Significant ontologies showing gene over-representation (P < 0.05) included intermediate filament organization, scaffold protein binding, detection of stimuli, calcium ion, G protein, and phototransduction. Significant differential expression of Let-7a and miR-16-2, and Smok4a, Prph2, and Gnat1 were confirmed.
Conclusions
Scleral mi- and mRNAs showed differential expression linked to myopia, supporting the involvement of miRNAs in eye growth regulation. The observed general trend of relatively small fold-changes suggests a tightly controlled, regulatory mechanism for scleral gene expression.
Keywords: microarray, microRNA, mouse, myopia, sclera
MicroRNAs (miRNAs) are small noncoding molecules that play pivotal roles in cell signaling through regulation of gene expression, specifically, by pairing with complementary messenger RNA (mRNA) sequences to either suppress translation or degrade mRNA.1–3 It is now recognized that they play crucial roles in normal physiological and pathologic processes, both in nonocular and in ocular tissues.4,5 Previous studies that focused on the human ocular sclera implicated miRNAs in normal ocular growth (axial elongation), with samples from very young, rapidly growing eyes, showing differential expression compared to those from adult (assumed) stable eyes. Some of the differentially expressed miRNAs could be linked to extracellular matrix remodeling pathways, making them potential targets for preventing or slowing the progression of myopia, which is largely a product of active scleral extracellular remodeling and thinning.6 As the first step in exploring the potential of miRNAs as therapeutic targets for myopia control, this study sought to understand their role in the scleral changes underlying myopia. We hypothesized that myopia development, when ocular axial elongation (“growth”) is exaggerated, is directly linked to differential regulation of scleral miRNAs.
This study used a mouse model of myopia, which has distinct advantages over other animal models of myopia in that the mouse possesses a well-characterized genome and a short gestational period with large litter sizes.7 In the context of myopia, visual FD has been shown to induce myopic shifts in refractive error by a number of different research laboratories.7–15 The mouse also has a fibrous sclera, like other mammals and primates.16,17 In this study, myopia was induced in young mice with short-term FD, and after the expected refractive error and ocular dimensional changes were confirmed, genome-wide expression profiling of mi- and mRNAs of scleral tissue was undertaken using microarray analyses, and gene ontology/bioinformatic analyses coupled with validation experiments, using quantitative PCR. Briefly, this study revealed differentially expressed mi- and mRNAs linked to myopia, implicating miRNAs and specific signaling pathways in scleral extracellular matrix remodeling and eye growth regulation.
Methods
Animals and Biometric Measurements
The C57BL/6J mice used in this study were reared under a 12L:12D cycle in an Atlanta VA-approved facility, with food and water provided ad libitum. All procedures were conducted according to the ARVO statement for the use of animals in ophthalmic and vision research and were approved by Institutional Animal Care and Use Committee. All mice (n = 14), 7 in each group for microarray and quantitative PCR (qPCR) analyses were unilaterally form-deprived (FD) for 2 weeks, starting at age P28, using white plastic diffusers on right eyes as previously described.8 Animals were anesthetized with ketamine (80 mg/kg) and xylazine (16 mg/kg) for the procedure and also for measuring refractive error, using photorefraction,11 and ocular biometry, using a 1310-nm spectral-domain optical coherence tomography (Bioptigen, Inc., Durham, NC, USA), as described previously.18
Tissue Collection and RNA Extraction
Mice were euthanized by cervical dislocation. Eyes were then enucleated and hemisected, and posterior eye cups were flat mounted, and whole-sclera samples were isolated under physiological saline, with the removal of the retina, retinal pigment epithelium, and choroid layers as well as the optic nerve head, using a dissecting microscope (EZ4 model; Leica, Buffalo Grove, IL, USA). Following homogenization of the samples using a BeadRuptor 24 (Omni, Inc., Kennesaw, GA, USA), RNA was extracted using miRVANA miRNA isolation kits (Life Technologies, Bengaluru, India) and then quantified and checked for purity using a NanoDrop (Thermo Scientific, Waltham, MA, USA) as well as a Bio-analyzer (Agilent Technologies, Santa Clara, CA, USA).
miRNA Microarrays
Scleral RNA samples from FD and fellow control eyes were analyzed commercially (Ocean Ridge Biosciences, Palm Beach Gardens, FL, USA) using custom multispecies microarrays (Microarrays, Inc. Huntsville, AL, USA), covering the 1279 mouse mature miRNAs in addition to human (2040) and rat (723) miRNAs from mirBASE version 19, consisting of epoxide glass substrates that had been spotted in triplicate with each probe. The samples were digested using DNase, and low-molecular weight RNA was first isolated by ultrafiltration through YM-100 columns (Merck Millipore, Darmstadt, Germany) and subsequently purified using the RNeasy MinElute clean-up kit (Qiagen, Hilden, Germany). The low-molecular weight RNA samples (50 ng) were 3′-end labeled with Oyster-550 fluorescent dye, using Flash Taq RNA labeling kit (Genisphere, Hatfield, PA, USA). Labeled low-molecular weight RNA samples were hybridized to the miRNA microarrays according to the recommended procedures. The microarrays were then scanned using a microarray scanner (Axon Genepix 4000B model; Molecular Devices, Sunnyvale, CA, USA), and data were extracted from images using GenePix version 4.1 software.
Data Preprocessing (miRNA Microarrays).
Spot intensities were obtained for the 10,330 total features (including triplicates) on each microarray by subtracting the median local background from the median local foreground for each spot. Detection thresholds (T) for each array were determined by calculating the 10% trim mean intensity of the negative controls spots and adding 5 times the SD of the background (nonspot area). The spot intensities and thresholds were transformed by taking the log (base 2) of each value. The normalization factor (N) for each microarray was determined by obtaining the 20% trim mean of spot intensities above threshold in all samples. The log2-transformed spot intensities for all 10,330 features were then normalized by subtracting N from each of the spot intensities and scaled by adding the grand mean of N across all microarrays. The mean probe intensities for each of the 3,431 mammalian and control probes on each of the arrays represented the average of the triplicate spot intensities. Spots flagged as poor quality during data extraction were omitted prior to averaging. The 1,257 mouse noncontrol log2-transformed, normalized, and averaged probe intensities were then filtered to obtain a final list of 526 mouse miRNA probes, meeting the requirement of intensities above T for at least 25% of all samples.
Quality Control (miRNA Microarrays).
Each array contained probes targeting 11 different synthetic miRNAs, each of which was added at a mass of 200 attomoles to each RNA sample prior to labeling and hybridization. The sensitivity of the microarray hybridization was confirmed by detection of hybridization signals well above the detection threshold for all 11 spiking controls. The array also contained a set of specificity control probes complementary to 3 different miRNAs. Each specificity control included a perfect match, a single mismatch, a double mismatch, and a shuffled version of the probe. The specificity of hybridization was confirmed by detection of hybridization signals corresponding to perfect matches and not modified versions of the probes. The reproducibility of the arrays was determined by comparing hybridization intensity within the triplicates on each array.
Differential Expression Analysis and Hierarchical Clustering (miRNA Microarrays).
There were 526 probes that met all requirements for inclusion in data analysis. The log2-transformed and normalized spot intensities for these probes were examined for differences between groups (i.e., treated versus fellow control eyes), by 1-way ANOVA, using National Institute of Aging array analysis software19 and applying the Bayesian error model with 20° of freedom. Statistical significance was determined using the false discovery rate method proposed by Benjamini and Hochberg.20 Principal component analysis was also performed in detectable probes using National Institute of Aging software. Data for the detectable miRNA probes were clustered using Cluster 3.0 software.21 Three rounds of gene median centering and gene median normalization were used to preprocess the data. Hierarchical clustering was conducted using centered correlation as the similarity metric and average linkage as the clustering method. Data from 1 control sample, which showed a relatively lower percentage detection of miRNAs, was excluded from statistical analyses.
mRNA Microarrays
Total RNA (50 ng) was converted to complementary DNA (cDNA) and amplified using a microarray system (Ovation Pico WTA system; NuGen, San Carlos, CA, USA) and analyzed using GeneChip Gene 2.0 ST microarrays (Affymetrix, Santa Clara, CA, USA; >28000 mRNAs; Genome Reference Consortium Mouse Build 38; transcript coverage and gene count derived from RefSeq download as of February 2012). Briefly, cDNA was fragmented and end-labeled with biotin, using GeneChip WT terminal labeling kit (Affymetrix). Labeled cDNA was then hybridized to GeneST arrays (Affymetrix), according to the manufacturer's instructions, after which microarrays were washed and stained using the Fluidics Station 450 and scanned with the Scanner 3000 7G (both from Affymetrix). Scanned images (CEL format files) were analyzed using the RMA algorithm to export the data CHP files using the Expression Console version 1.3.1 software (Affymetrix).
Data Preprocessing (mRNA Microarrays).
Intensity data from the probes were adjusted for background, quantile normalized, summarized, and log2-transformed using software and exported to obtain the log2-transformed and normalized probe set intensities. Briefly, spot intensities for each probe were calculated by subtracting median local background from median local foreground for each spot; these values were then transformed by taking the base 2 logarithm of each, which were finally normalized by subtracting the 70th percentile of the spot intensities of probes relative to mouse constitutive exons and adding back a scaling factor (grand mean of 70th percentile). Data were filtered for mouse noncontrol probe sets. After we removed data corresponding to low quality spots and control sequences, 33,793 mouse probes remained and were filtered further to identify all probes with intensity above the normalized threshold determined as follows: [log2 (±3 SD of raw local background) + mean of log2-transformed negative controls]. After filtering, 20,547 probes were above threshold in at least 25% of all samples.
Differential Expression Analysis and Hierarchical Clustering (mRNA Microarrays).
The log2-transformed and normalized spot intensities for the above-derived 20,547 probes were examined for significant differences between treated and fellow control eyes, as described earlier for miRNA differential expression analysis. Data from 1 experimental (goggled) sample and 1 control sample were not included in ANOVA analyses because they showed relatively lower percentages of detection of mRNAs.
Gene Ontologies (mRNA Microarrays) and miRNA-mRNA Interactions.
Using WebGestalt software,22,23 we determined the distribution of differentially regulated genes among cellular, molecular, and biological processes, querying the Gene Ontology database and comparing the relative distribution of genes that met specific significance criteria to the distribution of all detected genes. The hypergeometric method was used, and significance was set at an adjusted P value of <0.05 (Benjamini and Hochberg method),20 with a minimum requirement of 2 or more genes being involved, using detectable genes with valid gene symbols (Genome Reference Consortium Mouse Build 38). Predicted gene targets for mouse miRNAs were downloaded from TargetScan,24 EiMMo,25 and Microcosm.26 Three statistical tests of enrichment were used to determine whether the predicted targets of any miRNAs exhibited altered expression levels, which included miRNA Target Enrichment Analysis/miTEA,27,28 permutation, and hypergeometric distribution tests. For miTEA, the list of differentially expressed genes ranked by fold-change was used as the input, and the tool searched for any miRNAs whose predicted targets (by any of the three prediction methods) were enriched at the top or bottom of the ranked list. For the hypergeometric test, the number of genes identified as being differentially expressed predicted target(s) of a given miRNA was compared to the expected number of such genes according to the hypergeometric distribution. A similar test was performed using a random selection of genes to build the distribution of expected gene numbers.
Quantitative PCR
Validation experiments using select miRNAs and mRNAs, chosen based on fold-change and/or statistically significant microarray findings, were conducted using TaqMan miRNA and gene expression assays using a separate set of FD and control samples (n = 7). Working within constraints imposed by assay and sample availability, we deliberately set a bias toward selecting targets toward those with potential involvement in matrix remodeling. TaqMan miRNA assays contained a stem-looped primer for reverse transcription and a sequence-specific TaqMan assay (containing forward and reverse primers along with a TaqMan MGB probe). Reverse transcription reactions specific to the tested miRNAs were performed using individual samples, and qPCRs were performed in triplicate (95°C for 10 minutes; 95°C for 15 seconds; 60°C for 1 minute for 40 cycles). Gene expression assays contained both unlabeled PCR primers and the TaqMan MGB (FAM dye-labeled) probe. Housekeeping genes snoRNA202 and snoRNA234 were used for miRNA assays, and GAPDH and POLR2A for the mRNA assays, based on previous studies demonstrating lack of association with myopia development (assay information is listed in Supplementary Table S1).29–32 The comparative CT threshold method was used to analyze qPCR data; the amount of target was normalized to endogenous reference(s), cycle threshold (2−ΔΔCt) values were determined, and fold-change differences between FD and control eyes were calculated.33 The geometric mean of the expression levels of housekeeping genes was used as the normalization factor.34 Two-tailed Student's t-tests were performed, assuming equal variance, and P values were thus obtained.
Results
Myopia Induction
In response to visual FD for 2 weeks, treated eyes showed statistically significant relative myopic shifts in refractive error (mean interocular differences: −2.90 ± 0.86 D; P < 0.05; and −1.25 ± 0.28 D; P < 0.01 for groups used in microarray and qPCR experiments, respectively) although interocular differences in axial length for the above groups did not reach statistical significance (−10.7 ± 9.9 and +3.2 ± 5.0 μm, respectively; P > 0.05). The interocular axial length difference data covering all experimental animals also did not reach statistical significance (−3.3 ± 5.4 μm; P > 0.05), whereas the refractive error differences remained significant (−2.02 ± 0.47 D; P < 0.01). Refractive error and axial length data for individual animals are summarized in Supplementary Table S2.
Differential Expression, Gene Ontologies, and miRNA-mRNA Interactions (miRNA and mRNA Microarrays)
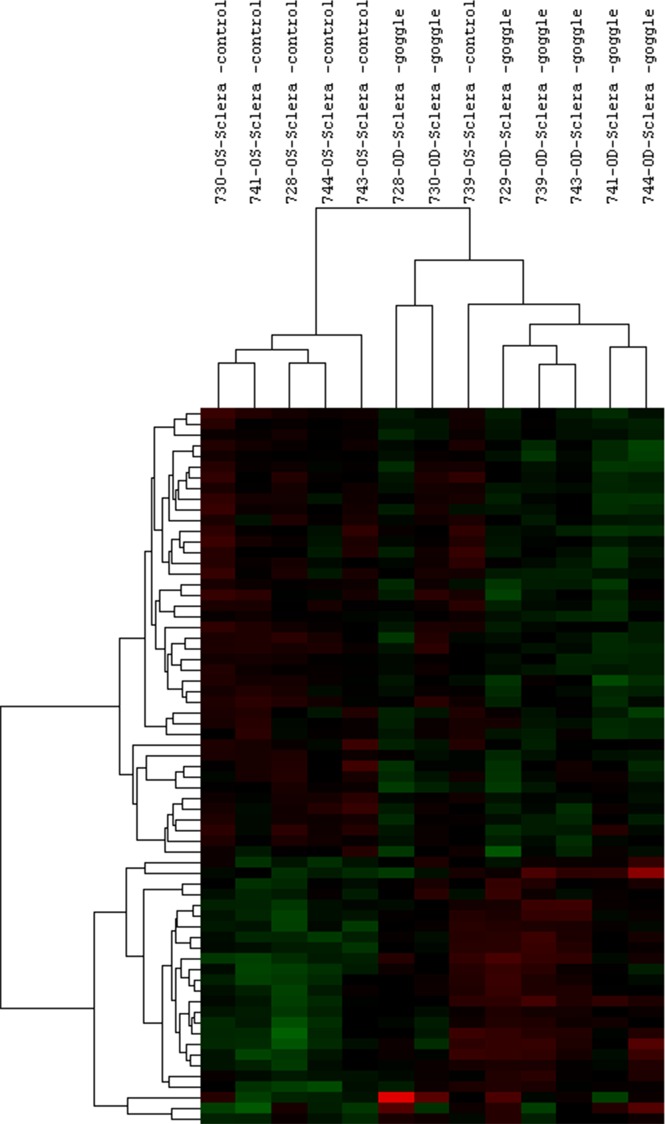
To compare expression levels between treated and control eyes, fold differences were calculated. Figures 1 and 2 show the cluster analyses performed on significant log2-transformed miRNA and mRNA probes, respectively. A total of 54 miRNAs showed significant differential expression, with 24 miRNAs being upregulated and 30 miRNAs downregulated, despite overall FD-induced changes in the levels of miRNA expression being low enough to moderate in size (maximum fold change [FC] = 1.63 in either direction; P < 0.05; minimum P = 0.0001). A total of 261 mRNAs showed significantly altered expression, with 177 showing upregulation and 84 showing downregulation (maximum FC = 2.7, P < 0.05; minimum P = 0.0001). We attribute the observed nondefinitive clustering to both biological variability and the relatively small proportion of probes showing statistically significant differential expression in relation to the total number of probes used for clustering. We summarized details of the main 20 significant miRNAs and mRNAs in Tables 1 and 2, respectively. All significant results showing fold-changes of 1.25 and above are also summarized in Supplementary Tables S3 and S4. All data presented here represent raw P values, because none of the targets met statistical significance based on the false discovery rate correction method, which is also consistent with the small size of our study.35
Figure 1.
Differences among patterns of miRNA expression examined by cluster analysis. Cluster analyses were performed on 67 log2-transformed miRNA probes with P < 0.05, using Gene Cluster 3.0. Data were adjusted by centering median, and centered correlation was used as distance measurement (n = 7).
Figure 2.
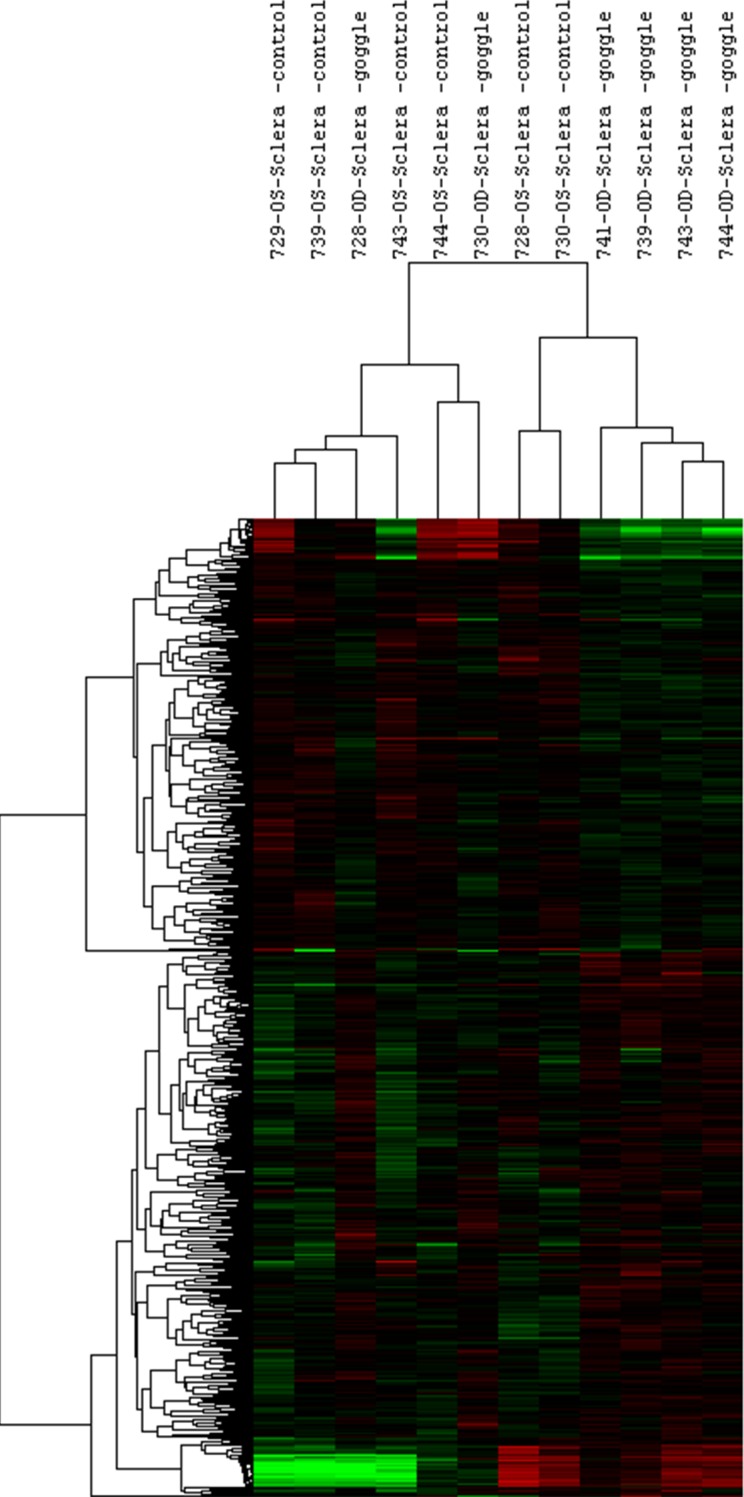
Patterns of mRNA expression differences examined by cluster analysis. Cluster analyses were performed on 691 log2-transformed mRNA probes with P < 0.05, using Gene Cluster 3.0. Data were adjusted by centering median, and centered correlation was used as distance measurement (n = 7).
Table 1.
Main Significant Differentially Expressed Scleral miRNAs During Myopia Development*
Table 2.
Main Significant Differentially Expressed Scleral mRNAs During Myopia Development*

Querying the Gene Ontology database using WebGestalt software revealed several significant Gene Ontology processes. Notably, genes showing differential expression are over-represented in processes related to intermediate filament organization, scaffold protein binding, detection of stimuli, visual perception, eye development, phototransduction, calcium ion homeostasis, G protein-coupled receptor, cell projection, and structural molecule activity. Complete lists of significant processes and networks are provided in Supplementary Tables S5 and S6 and shown graphically in Supplementary Figures S1 and S2. In analyses exploring potential miRNA-mRNA interactions, we found no enrichment of predicted mRNA targets (based on miRNA differential expression) in the list of differentially expressed mRNAs, for any of the methods used.
Validation qPCR
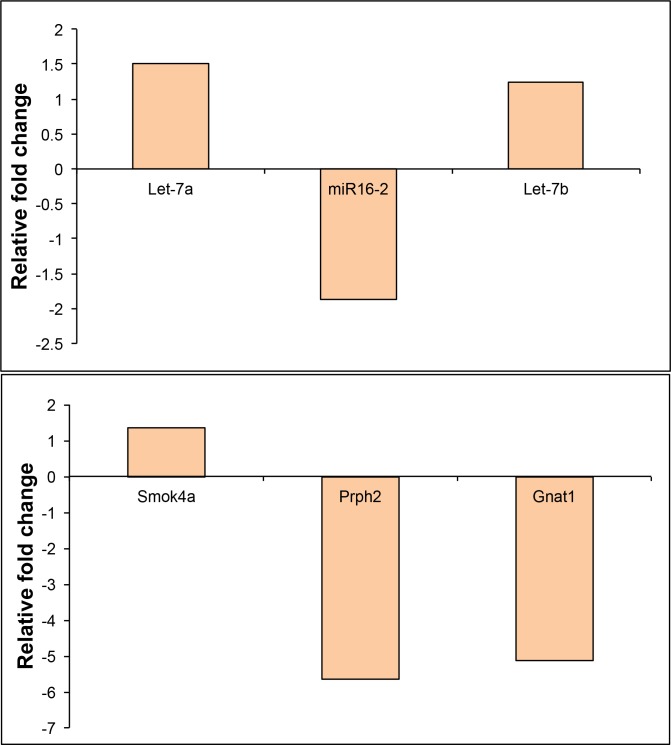
The results for miRNAs and mRNAs showing differential expression in microarray analyses based on their fold-change and/or statistical significance were validated using qPCR with an independent set of samples. For miRNAs, PCR fold-changes for Let-7a and miR-16 but not Let-7b reached statistical significance (Fig. 3, top). PCR fold-changes for the sperm motility kinase 4A (Smok4a), peripherin 2 (Prph2), and guanine nucleotide binding protein, alpha transducing 1 (Gnat1) genes also reached statistical significance (Fig. 3, bottom). Table 3 summarizes the 2−ΔΔCt values and standard deviations, as well as corresponding fold differences for all 3 miRNAs and all 3 mRNAs, along with related microarray findings.
Figure 3.
Quantitative PCR validation of microarray analysis results are shown for 3 miRNAs (top) and 3 mRNAs (bottom) were revealed to be differentially expressed in mouse sclera. Scleras from 2-week form deprived eyes and fellow control eyes were compared.
Table 3.
Mean ± SD Average 2−ΔΔCt Values for qPCR Analyses of Mouse Scleral Samples From FD and Control Eyes and Equivalent Microarray Data

Discussion
We report for the first time, genome-wide scleral expression profiles of both mi- and mRNAs in a mouse model of FD-induced myopia. Overall, several scleral mi- and mRNAs showed differential regulation after 2 weeks of FD myopia, although the magnitude of the myopia-related changes in miRNA expression was relatively small in all cases. Nonetheless, these findings are in line with the notion that miRNAs are “fine-tuners” of gene expression 36 and perhaps indicative of a tightly controlled, scleral gene regulatory nexus driven by miRNA regulation. Because this is the first study to investigate the genome-wide miRNA and mRNA profiles in the mouse sclera after FD, there are no available expression profiles for direct comparison. However, the magnitude of observed scleral mRNA expression changes are comparable to differential gene expression changes documented in the tree shrew model of myopia,29,37,38 although curiously, there is no overlap in genes showing differential regulation in these 2 species. This may reflect species differences and/or differences in the sampling time points used and magnitude of ocular growth responses.
The over-riding goal of this research was to identify scleral miRNAs that are differentially regulated in myopia and explore their potential to alter the course of scleral remodeling as a treatment strategy to prevent or slow ocular elongation. Short-term myopia induction treatments allow the identification of genes critical to active “myopic” growth, whereas later sampling from much larger, more myopic eyes may yield results that are confounded by associated biochemical and biomechanical changes in sclera. To this end, we specifically limited our treatments to 2 weeks, when changes in axial ocular dimensions were still small, and thus, our findings reflect relatively early events, although extracellular matrix loss and/or scleral thinning cannot be ruled out.
Of the candidate miRNAs validated using qPCR, Let-7a has been shown to take part in NF-κβ signaling and in the regulation of type I collagen.39,40 Although NF-κβ signaling has not been studied in the context of scleral remodeling and myopia, regulation of type I collagen is directly relevant because it is the main component of the scleral matrix, and myopic scleras show reduced collagen levels.41 Other members of the Let-7 family also showed differential expression in our study, including Let-7b, previously shown to be involved in COL1A1 and COL1A2 regulation in skin fibroblasts.42 However, the microarray finding for this miRNA failed the qPCR validation test, reiterating the need for validation studies to rule out false positives from high-throughput assays. We also cannot rule out the possibility that other members of the Let-7 family (e.g., Let-7a) play compensatory roles in the absence of changes in expression of Let-7b. Only further studies can address this possibility. miR-16, another miRNA showing significant differential expression, has been previously implicated in cell proliferation, TNF/NF-kB signaling, and apoptosis.43–46 Interestingly, there is some evidence from previous studies that scleral fibroblast proliferation is reduced in myopia, as reflected in decreased DNA production, although neither the rate of turnover of scleral fibroblasts nor the role of apoptosis are well understood.47,48
Three candidate mRNAs among the main significant genes from our microarray findings were validated using qPCR, Gnat1, Prph2, and Smok4a. The roles in scleral growth regulation of all 3 of these genes are at best poorly understood. Mutations in Gnat1 are associated with congenital stationary night blindness.49 Gnat1−/− mice lacking functional rods have also been shown to exhibit disrupted refractive development and a lack of response to FD.50 Although these studies argue for a role of retinal Gnat1 in this abnormal growth pattern, Gnat1 has also been reported to be expressed in other tissues such as skin, heart, and liver.51 Prph2 is a cell surface glycoprotein belonging to the tetraspanin family, in which mutations result in retinal degenerations.52,53 It has not been reported in the sclera before, and we can only speculate, based on known functions of tetraspanins and what is known about scleral structure, that Prph2 has the potential to take part in signal transduction and the maintenance of scleral extracellular matrix. To the best of our knowledge, there are no published studies describing the function of Smok4a. Given the limited scope of the validation experiments, they can be viewed only largely as technical confirmations and cannot substitute for detailed functional studies, required to reveal specific roles.
Among other main candidate mRNAs that were not chosen for validation are several members of the keratin family (Krt4, −5, −14, −15, and −17), all of which were upregulated in the myopic eyes of our mice. Although not widely studied in sclera, some keratins have been previously reported in normal human sclera (see supplemental information in Young et al.54). They also hold the potential to influence scleral extracellular matrix because they are part of important intermediate filament network machinery that connects the cell periphery/extracellular matrix to the nuclear matrix via the actin microfilaments.55 Surprisingly, we also found differentially expressed in our scleral samples, genes linked to retinal pathologies, but not linked previously to scleral remodeling. Given that the sclera is separated from the retina by the intervening choroid and the ease of collection of scleral tissue samples, we are inclined to rule out retinal contamination as the explanation for these results and instead argue for the presence of the above genes in the sclera, whose functions are yet to be characterized.
Highlighting the complex nature of scleral growth regulation, our investigations identified a wide array of cellular processes that may be involved in myopia development in the mouse model. Roles in scleral matrix remodeling mechanisms are suggested by some of the identified processes, for example, intermediate filament organization, scaffold protein binding, and structural molecule activity. However, we also have identified other processes generally deemed retinal and linked to detection of stimuli, visual perception and phototransduction. Could the rhodopsin gene be serving an unknown role in the sclera, given that it has been reported in pigmented skin cells and that the sclera harbors melanocytes?56–58 As another example, gene enrichment in the visual perception process could reflect the activity of G protein-coupled receptors, given that the most effective off-label drug treatment for myopia, atropine, is an antagonist of G protein-coupled muscarinic receptors, with the sclera being one potential site of action.59 Unfortunately similar enrichment analysis of miRNAs was not possible because of the low number achieving statistical significance.
Our miRNA-mRNA interaction analyses did not reveal any enrichment of predicted mRNA targets in our differentially expressed mRNA list. Nonetheless, as the same samples were processed for miRNA and mRNA profiling, our findings must be considered representative of actual biological events. Possible explanations for the lack of overlap between predicted targets and the differentially expressed mRNAs could include a relatively small number of differentially expressed mi- and mRNAs during scleral remodeling, resulting in only weak enrichment signals (no more were present than would be expected by a random draw of genes in a high-throughput experiment); or a high number of false positives among predicted interactions, given that they are based on several factors including sequence information, expression levels, physical chemistry properties and complex algorithms; or redundancy in regulation and small effects (several miRNAs acting at once to bring about a subtle change in gene expression).
There are limitations to our study. First, the sample size was small. However, given the lack of any published myopia-related genome-wide scleral mi- and mRNA expression data, we consider this study an important first step toward expanding our knowledge of the molecular landscape of the sclera. The extremely low amounts of RNA available from mice sclera also limited qPCR validation experiments to just a small number of mi- and mRNAs. Additional validation studies, as well as studies exploring the specific roles of miRNAs in mRNA regulation, are critical. The redundancy in miRNA regulation also calls for the application of unbiased bioinformatic approaches to provide initial rationale in selecting targets for such studies. Nonetheless, our decision to investigate miRNA and mRNA changes simultaneously rather than limiting our study to the differentially expressed miRNAs alone, avoided the need to rely exclusively on predicted mRNAs, which can only be speculative in nature. This decision seems also validated by our results, which reinforce the need for healthy skepticism about functional predictions and call for careful modeling of our own data prior to functional validations.
In conclusion, this is the first study, albeit on a small scale, to document genome-wide miRNA and mRNA expression changes in the sclera in an animal model of myopia. Our study revealed myopia-related changes in the expression of a number of miRNAs and genes, some of which have not been previously associated with scleral matrix remodeling. These findings provide a platform for further investigations aimed at understanding the roles of miRNAs in scleral mRNA regulation and matrix remodeling and potentially uncovering novel therapeutic scleral targets for preventing or slowing myopia.
Supplementary Material
Acknowledgments
Supported by US National Institutes of Health Grants NIH-NEI K12 EY017269 (RM) (Berkeley Clinical Scientist Development Program), NIH-NEI K08 EY022670 (RM), NIH-NEI R01 EY016435 (MTP), NIH-NEI R01 EY012392 (CFW).
Disclosure: R. Metlapally, None; H.N. Park, None; R. Chakraborty, None; K.K. Wang, None; C.C. Tan, None; J.G. Light, None; M.T. Pardue, None; C.F. Wildsoet, None
References
- 1. Gregory RI,, Chendrimada TP,, Cooch N,, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005; 123: 631–640. [DOI] [PubMed] [Google Scholar]
- 2. Hutvagner G. Small RNA asymmetry in RNAi: function in RISC assembly and gene regulation. FEBS Lett. 2005; 579: 5850–5857. [DOI] [PubMed] [Google Scholar]
- 3. Pattanayak D,, Agarwal S,, Sumathi S,, Chakrabarti SK,, Naik PS,, Khurana SM. Small but mighty RNA-mediated interference in plants. Indian J Exp Biol. 2005; 43: 7–24. [PubMed] [Google Scholar]
- 4. Sampson VB,, Yoo S,, Kumar A,, Vetter NS,, Kolb EA. MicroRNAs and potential targets in osteosarcoma: review. Front Pediatr. 2015; 3: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu S. microRNAs and inherited retinal dystrophies. Proc Natl Acad Sci U S A. 2015; 112: 8805–8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Metlapally R,, Gonzalez P,, Hawthorne FA,, Tran-Viet KN,, Wildsoet CF,, Young TL. Scleral micro-RNA signatures in adult and fetal eyes. PLoS One. 2013; 8: e78984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pardue MT,, Stone RA,, Iuvone PM. Investigating mechanisms of myopia in mice. Exp Eye Res. 2013; 114: 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Faulkner AE,, Kim MK,, Iuvone PM,, Pardue MT. Head-mounted goggles for murine form deprivation myopia. J Neurosci Methods. 2007; 161: 96–100. [DOI] [PubMed] [Google Scholar]
- 9. Tejedor J,, de la Villa P. Refractive changes induced by form deprivation in the mouse eye. Invest Ophthalmol Vis Sci. 2003; 44: 32–36. [DOI] [PubMed] [Google Scholar]
- 10. Schmucker C,, Schaeffel F. In vivo biometry in the mouse eye with low coherence interferometry. Vision Res. 2004; 44: 2445–2456. [DOI] [PubMed] [Google Scholar]
- 11. Schaeffel F,, Burkhardt E,, Howland HC,, Williams RW. Measurement of refractive state and deprivation myopia in two strains of mice. Optom Vis Sci. 2004; 81: 99–110. [DOI] [PubMed] [Google Scholar]
- 12. Barathi VA,, Boopathi VG,, Yap EP,, Beuerman RW. Two models of experimental myopia in the mouse. Vision Res. 2008; 48: 904–916. [DOI] [PubMed] [Google Scholar]
- 13. Tkatchenko TV,, Shen Y,, Tkatchenko AV. Mouse experimental myopia has features of primate myopia. Invest Ophthalmol Vis Sci. 2010; 51: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pardue MT,, Faulkner AE,, Fernandes A,, et al. High susceptibility to experimental myopia in a mouse model with a retinal on pathway defect. Invest Ophthalmol Vis Sci. 2008; 49: 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang F,, Yan T,, Shi F,, et al. Activation of dopamine D2 receptor is critical for the development of form-deprivation myopia in the C57BL/6 mouse. Invest Ophthalmol Vis Sci. 2014; 55: 5537–5544. [DOI] [PubMed] [Google Scholar]
- 16. Zhou X,, Huang Q,, An J,, et al. Genetic deletion of the adenosine A2A receptor confers postnatal development of relative myopia in mice. Invest Ophthalmol Vis Sci. 2010; 51: 4362–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chakravarti S,, Paul J,, Roberts L,, Chervoneva I,, Oldberg A,, Birk DE. Ocular and scleral alterations in gene-targeted lumican-fibromodulin double-null mice. Invest Ophthalmol Vis Sci. 2003; 44: 2422–2432. [DOI] [PubMed] [Google Scholar]
- 18. Park H,, Qazi Y,, Tan C,, et al. Assessment of axial length measurements in mouse eyes. Optom Vis Sci. 2012; 89: 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sharov AA,, Dudekula DB,, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005; 21: 2548–2549. [DOI] [PubMed] [Google Scholar]
- 20. Benjamini Y,, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc. Series B (Methodological). 1995: 57; 289–300. [Google Scholar]
- 21. de Hoon MJ,, Imoto S,, Nolan J,, Miyano S. Open source clustering software. Bioinformatics. 2004; 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- 22. Wang J,, Duncan D,, Shi Z,, Zhang B. WEB-based GEne SeT AnaLysis toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013; 41: W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang B,, Kirov S,, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005; 33: W741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lewis BP,, Burge CB,, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 25. Gaidatzis D,, van Nimwegen E,, Hausser J,, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007; 8: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Griffiths-Jones S,, Saini HK,, van Dongen S,, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008; 36: D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eden E,, Navon R,, Steinfeld I,, Lipson D,, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009; 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Steinfeld I,, Navon R,, Ach R,, Yakhini Z. miRNA target enrichment analysis reveals directly active miRNAs in health and disease. Nucleic Acids Res. 2013; 41: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo L,, Frost MR,, He L,, Siegwart JT, Jr,, Norton TT. Gene expression signatures in tree shrew sclera in response to three myopiagenic conditions. Invest Ophthalmol Vis Sci. 2013; 54: 6806–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao H,, Frost MR,, Siegwart JT, Jr,, Norton TT. Patterns of mRNA and protein expression during minus-lens compensation and recovery in tree shrew sclera. Mol Vis. 2011; 17: 903–919. [PMC free article] [PubMed] [Google Scholar]
- 31. Barathi VA,, Kwan JL,, Tan QS,, et al. Muscarinic cholinergic receptor (M2) plays a crucial role in the development of myopia in mice. Dis Model Mech. 2013; 6: 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lim W,, Kwan JL,, Goh LK,, Beuerman RW,, Barathi VA. Evaluation of gene expression profiles and pathways underlying postnatal development in mouse sclera. Mol Vis. 2012; 18: 1436–1448. [PMC free article] [PubMed] [Google Scholar]
- 33. Schmittgen TD,, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008; 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 34. Vandesompele J,, De Preter K,, Pattyn F,, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002; 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pawitan Y,, Michiels S,, Koscielny S,, Gusnanto A,, Ploner A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics. 2005; 21: 3017–3024. [DOI] [PubMed] [Google Scholar]
- 36. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 37. Guo L,, Frost MR,, Siegwart JT, Jr,, Norton TT. Scleral gene expression during recovery from myopia compared with expression during myopia development in tree shrew. Mol Vis. 2014; 20: 1643–1659. [PMC free article] [PubMed] [Google Scholar]
- 38. McBrien NA,, Gentle A. Role of the sclera in the development and pathological complications of myopia. Prog Retin Eye Res. 2003; 22: 307–338. [DOI] [PubMed] [Google Scholar]
- 39. Makino K,, Jinnin M,, Hirano A,, et al. The downregulation of microRNA let-7a contributes to the excessive expression of type I collagen in systemic and localized scleroderma. J Immunol. 2013; 190: 3905–3915. [DOI] [PubMed] [Google Scholar]
- 40. Liu C,, Wang L,, Chen W,, et al. USP35 activated by miR let-7a inhibits cell proliferation and NF-kappaB activation through stabilization of ABIN-2. Oncotarget. 2015; 6: 27891–27906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Norton TT,, Miller EJ. Collagen and protein levels in sclera during normal development, induced myopia and recovery in tree shrews. Invest Ophthalmol Vis Sci. 1995; 36: S740. [Google Scholar]
- 42. Liu J,, Luo C,, Yin Z,, et al. Downregulation of let-7b promotes COL1A1 and COL1A2 expression in dermis and skin fibroblasts during heat wound repair. Mol Med Rep. 2016; 13: 2683–2688. [DOI] [PubMed] [Google Scholar]
- 43. Shin VY,, Jin H,, Ng EK,, et al. NF-kappaB targets miR-16 and miR-21 in gastric cancer: involvement of prostaglandin E receptors. Carcinogenesis. 2011; 32: 240–245. [DOI] [PubMed] [Google Scholar]
- 44. Lezina L,, Purmessur N,, Antonov AV,, et al. miR-16 and miR-26a target checkpoint kinases Wee1 and Chk1 in response to p53 activation by genotoxic stress. Cell Death Dis. 2013; 4: e953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Balakrishnan A,, Stearns AT,, Park PJ,, et al. MicroRNA mir-16 is anti-proliferative in enterocytes and exhibits diurnal rhythmicity in intestinal crypts. Exp Cell Res. 2010; 316: 3512–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. An F,, Gong B,, Wang H,, et al. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis. 2012; 17: 702–716. [DOI] [PubMed] [Google Scholar]
- 47. Norton TT,, Rada JA. Reduced extracellular-matrix in mammalian sclera with induced myopia. Vis Res. 1995; 35: 1271–1281. [DOI] [PubMed] [Google Scholar]
- 48. Gentle A,, McBrien NA. Modulation of scleral DNA synthesis in development of and recovery from induced axial myopia in the tree shrew. Exp Eye Res. 1999; 68: 155–163. [DOI] [PubMed] [Google Scholar]
- 49. Naeem MA,, Chavali VR,, Ali S,, et al. GNAT1 associated with autosomal recessive congenital stationary night blindness. Invest Ophthalmol Vis Sci. 2012; 53: 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Park H,, Jabbar SB,, Tan CC,, et al. Visually-driven ocular growth in mice requires functional rod photoreceptors. Invest Ophthalmol Vis Sci. 2014; 55: 6272–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Su AI,, Wiltshire T,, Batalov S,, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004; 101: 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Manes G,, Guillaumie T,, Vos WL,, et al. High prevalence of PRPH2 in autosomal dominant retinitis pigmentosa in france and characterization of biochemical and clinical features. Am J Ophthalmol. 2015; 159: 302–314. [DOI] [PubMed] [Google Scholar]
- 53. Shankar SP,, Birch DG,, Ruiz RS,, et al. Founder Effect of a c.828+3A>T splice site mutation in peripherin 2 (PRPH2) causing autosomal dominant retinal dystrophies. JAMA Ophthalmol. 2015; 133: 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Young TL,, Hawthorne F,, Feng S,, et al. Whole genome expression profiling of normal human fetal and adult ocular tissues. Exp Eye Res. 2013; 116: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Loschke F,, Seltmann K,, Bouameur JE,, Magin TM. Regulation of keratin network organization. Curr Opin Cell Biol. 2015; 32: 56–64. [DOI] [PubMed] [Google Scholar]
- 56. Miyashita Y,, Moriya T,, Kubota T,, Yamada K,, Asami K. Expression of opsin molecule in cultured murine melanocyte. J Invest Dermatol. 2001; 6: 54–57. [DOI] [PubMed] [Google Scholar]
- 57. van der Werf F,, Baljet B,, Otto AJ. Pigment-containing cells in extraocular tissues of the primate. Doc Ophthalmol. 1992; 81: 357–368. [DOI] [PubMed] [Google Scholar]
- 58. Bron AJ,, Tripathi RC,, Tripathi B. The cornea and sclera. : Wolffs Anatomy of the Eye and Orbit. London: Chapman and Hall Medical; 1997: 233–278. [Google Scholar]
- 59. Barathi VA,, Beuerman RW. Molecular mechanisms of muscarinic receptors in mouse scleral fibroblasts: prior to and after induction of experimental myopia with atropine treatment. Mol Vis. 2011; 17: 680–692. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.