Abstract
The red seaweed Laurencia dendroidea belongs to the Rhodophyta, a phylum of eukaryotic algae that is widely distributed across the oceans and that constitute an important source of bioactive specialized metabolites. Laurencia species have been studied since 1950 and were found to contain a plethora of specialized metabolites, mainly halogenated sesquiterpenes, diterpenes and triterpenes that possess a broad spectrum of pharmacological and ecological activities. The first committed step in the biosynthesis of triterpenes is the cyclization of 2,3-oxidosqualene, an enzymatic reaction carried out by oxidosqualene cyclases (OSCs), giving rise to a broad range of different compounds, such as the sterol precursors cycloartenol and lanosterol, or triterpene precursors such as cucurbitadienol and β-amyrin. Here, we cloned and characterized the first OSC from a red seaweed. The OSC gene was identified through mining of a L. dendroidea transcriptome dataset and subsequently cloned and heterologously expressed in yeast for functional characterization, which indicated that the corresponding enzyme cyclizes 2,3-oxidosqualene to the sterol precursor cycloartenol. Accordingly, the gene was named L. dendroidea cycloartenol synthase (LdCAS). A phylogenetic analysis using OSCs genes from plants, fungi and algae revealed that LdCAS grouped together with OSCs from other red algae, suggesting that cycloartenol could be the common product of the OSC in red seaweeds. Furthermore, profiling of L. dendroidea revealed cholesterol as the major sterol accumulating in this species, implicating red seaweeds contain a ‘hybrid’ sterol synthesis pathway in which the phytosterol precursor cycloartenol is converted into the major animal sterol cholesterol.
Introduction
The genus Laurencia belongs to the phylum Rhodophyta (red algae) and is widely distributed across the oceans [1]. Laurencia species are found between the intertidal and subtidal zones at a depth of 3m and are recognized as a crucial source of specialized (secondary) metabolites [2–5]. The specialized metabolites that are stored in Laurencia species are mainly halogenated compounds that play important ecological roles, such as chemical defence against bacterial colonization and infection [6–9]. Most of the described specialized metabolites isolated from red seaweeds are terpenes, especially triterpenes, sesquiterpenes and diterpenes [10, 11]. Besides ecological roles, these compounds have a wide array of activities and valuable medical properties [9]. Triterpenoids isolated from Laurencia species with important pharmacological properties include the squalenoid-derived triterpenoids, thyrsiferol and venustatriol (both isolated from L. viridis) and laurenmariannol and (21α)-21-hydroxythyrsiferol (isolated from L. mariannensis) with cytotoxic activity against P-388 cells [12, 13]. Furthermore three squalene-derived brominated triterpenes dehydrothyrsiferol, 10-epidehydrothyrisiferol and isodehydrothyrsiferol isolated from L. viridis also had cytotoxic activities against cancer cell lines [14].
All terpenes are synthesized from the universal building blocks isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) that can be generated via either the mevalonate (MVA) or the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway. Most bacteria, including the photosynthetic cyanobacteria, use the MEP pathway to synthesize terpenes. In contrast, animals, fungi, archaea and some Gram-positive bacteria exclusively rely on the MVA pathway for terpene synthesis [15]. It is suggested that plants acquired the MEP pathway through cyanobacterial endosymbiosis and consequently use both pathways for terpene biosynthesis [16]. In plants, the IPP and DMAPP generated through the cytosolic MVA pathway is mostly used for the synthesis of sesquiterpenes and triterpenes, whereas IPP and DMAPP generated through the plastidial MEP pathway are mostly used for the synthesis of all other types of terpenes [17, 18]. Green algae do not possess the genes for the MVA pathway and their sesquiterpene and triterpene building blocks are consequently derived from the MEP pathway [19–21]. However, red algae such as Laurencia species retained both IPP pathways during their evolution and use both biosynthetic routes [22].
A key step in the biosynthesis of triterpenes and sesquiterpenes is the sequential condensation of two units of IPP with DMAPP to yield farnesyl pyrophosphate (FPP). For the synthesis of triterpenes, two units of FPP are fused head-to-tail to form squalene, which is subsequently epoxidized to 2,3-oxidosqualene, the direct triterpene precursor [21, 23]. In most organisms, the epoxidation of squalene is carried out by squalene epoxidase [24], however, several species, including the diatom Phaeodactylum tricornutum, lack a conventional squalene epoxidase [21].
The great diversity of triterpene structures, with more than 100 different carbon skeletons known in the kingdom of life, is due to different oxidosqualene cyclases (OSCs) and, in particular, to the number of rearrangement steps that the different OSC enzymes can catalyse in the third stage of the cyclization reaction [25–27]. The OSCs belong to a gene superfamily divided into 10 groups according to their product specificity and higher rank phylogeny [28]. They are used both in primary metabolism (e.g. lanosterol/cycloartenol) or specialized metabolism [29, 30].
Some OSC enzymes have more than one final product, e.g. an OSC from Citrullus colocynthis produces cucurbitadienol and lanosterol [31]. Furthermore, OSC enzymes can produce precursors for both non-sterol triterpenes, like lupeol, and sterol triterpenes, like cycloartenol [32]. To characterize the enzymatic product, OSC genes are often ectopically expressed in yeast [21, 30, 33–35] or in plants via transient expression in Nicotiana benthamiana [36, 37].
To date, approximately 50 OSC genes have been cloned from various plant species, e.g. Arabidopsis thaliana, Artemisia annua, Lotus japonicus and Oryza sativa (S1 Table). So far, such studies have not been performed for OSC genes from red seaweeds. In this context, our aim was to clone and functionally characterize an OSC from the red seaweed L. dendroidea by expression in a sterol-engineered yeast strain.
Material and Methods
Identification and Cloning of LdCAS
To screen for potential OSCs, we performed a TBLASTX search in the transcriptome assembly of L. dendroidea [38] using the nucleotide sequence of the characterized A. annua cycloartenol synthase (CAS) (GenBank accession KM670093 [39]) as query. The domain composition of the sequence coding for the OSC from L. dendroidea was obtained through search for conserved domains using the National Center for Biotechnology Information (NCBI) Conserved Domain Database (CDD) [40].
Afterwards, L. dendroidea specimens obtained from an unialgal culture (see 38 for details) were frozen in liquid nitrogen and ground to a fine powder using a mortar and pestle. Total RNA was extracted with the Qiagen RNeasy® Mini Kit (Hilden, Germany) and cDNA was prepared with the Bio-Rad iScriptTM cDNA Synthesis Kit (Hercules, United States). The full-length coding sequence of LdCAS was amplified from this L. dendroidea cDNA using the primers LdCAS_Fw and LdCAS_RV (Table 1). The obtained PCR fragment was GatewayTM recombined into the GatewayTM vector pDONR207 and the resulting entry clone was sequence verified and further recombined into the in-house generated destination vector pESC-URA-tHMG1-DEST [41] to yield pESC-URA-tHMG1-DEST[GAL1/LdCAS].
Table 1. Primer sequences used in this study.
| Oligo | Sequence | Description |
|---|---|---|
| combi1715 | 5’-TAATACGACTCACTATAGGG-3’ | T7 sequencing primer |
| combi2287 | 5’-GGAATAAGGGCGACACGG-3’ | bla internal Rv |
| combi3244 | 5’-GTTAACCGGCCGCAAATTAAAGCC-3’ | HpaI-CYC1t Rv |
| combi3245 | 5’-ggggacaagtttgtacaaaaaagcaggcttaAAGGGAACAAAAGCTGGAGC-3’ | attB1-SNR52p Fw |
| combi3246 | 5’-ggggaccactttgtacaagaaagctgggtaAAAGCCTTCGAGCGTCCC-3’ | attB2-CYC1t Rv |
| combi3247 | 5’-GTTAAC GCTAGCGAGGGAACAAAAGCTGGAGC-3’ | HpaI-NheI-TEFp Fw |
| crispr014 | 5’-AGAGTTCCTCGGTTTGCCGATCATTTATCTTTCACTGCGGAGAAG-3’ | TRP1 gRNA left Rv |
| crispr031 | 5’-GGCAAACCGAGGAACTCTGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGG-3’ | TRP1 gRNA right fw |
| crispr059 | 5’-AACTGCATGGAGATGAGTCGTGGCATTAATAACAGAGTTCCTCGGTTTGCCAGTTATT-3’ | TRP1 HR donor Fw |
| crispr060 | 5’-AATAACTGGCAAACCGAGGAACTCTGTTATTAATGCCACGACTCATCTCCATGCAGTT-3’ | TRP1 HR donor Rv |
| crispr119 | 5’-AGGTAGTTCTGGTCCATTGG-3’ | TRP1 RFLP fw |
| crispr120 | 5’-ACACCATTTGTCTCCACACC-3’ | TRP1 RFLP rev |
| LdCAS_Fw | 5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGTTGTTTGGCGACTCAATG-3’ | LdCAS fw |
| LdCAS_RV | 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTATTATTGCTGCTGGCACGCTTTC-3’ | LdCAS rev |
Generation and Culturing of Saccharomyces cerevisiae (Yeast) Strains
All primers used to create novel yeast strains and all yeast strains created and/or used in this study are listed in Table 1 and Table 2, respectively. Yeast strains PA14, TM097, and TM122 were derived from strain TM1 in the S288c BY4742 background (26).
Table 2. Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| S288c BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Moses et al., 2014 |
| TM1 | S288c BY4742; Perg7::PMET3-ERG7 | Moses et al., 2014 |
| PA14 | TM1; trp1Δ0 | This study |
| TM097 | PA14; pESC-URA-tHMG1-DEST | This study |
| TM122 | PA14; pESC-URA-tHMG1-DEST[GAL1/LdCAS] | This study |
Yeast strain PA14 was derived from strain TM1 [21] by knocking out the TRP1 gene using CRISPR/Cas9. A plasmid for CRISPR/Cas9 in yeast that contains both Cas9 and a gRNA cassette was generated according to the system reported by DiCarlo and co-workers [42]. To this end, the Cas9 expression cassette was PCR-amplified from p414-TEF1p-Cas9-CYC1t (Addgene plasmid 43802) using primers combi3244 and combi3247, each containing a HpaI restriction site at the 5’ terminus. The resulting fragment was cloned into pJET1.2 and sequence verified. Subsequently, the Cas9 cassette was cut out using HpaI, gel-purified and cloned into the vector backbone of a PvuII-treated, dephosphorylated, and gel-purified pESC-URA plasmid (Agilent). The resulting plasmid was named pCAS1. Next, a GatewayTM cassette was PCR-amplified from pDEST14 (Invitrogen, Carlsbad, United States) using primers combi1715 and combi2287. The PCR fragment was treated with XbaI and NheI, gel purified and cloned into the NheI-linearized and dephosphorylated pCAS1, yielding pCAS-ccdB. The TRP1 knock-out construct was generated by PCR amplification of SNR52p and sgRNA-CYC1t from p426-SNR52p-gRNA.CAN1.Y-SUP4t (Addgene plasmid 43803) using primers combi3245 and crispr014 and combi3246 and crispr031, respectively. The individual fragments were fused by overlap extension PCR, subcloned into pDONR221, sequence verified and finally GatewayTM recombined into pCAS-ccdB, yielding pCAS-TRP1. Subsequently, 200 ng of pCAS-TRP1 and 10 μmol of double-stranded DNA (prepared by annealing the single-stranded oligonucleotides crispr059 and crispr060) as homologous recombination donor were co-transformed in yeast strain TM1. The resulting colonies were analyzed for positive CRISPR events by replica plating on SD medium (Clontech, Mountain View, United States) with or without tryptophan. Tryptophan auxotrophs were further confirmed by Sanger sequencing. Auxotrophic strains were cured of pCAS-TRP1 by counter-selection on plates containing 1 mg/mL 5-fluoroorotic acid (Zymo Research, Irvine, United States) and the resulting TM1-derived trp1 strain was named PA14.
Subsequently, the PA14 strain was transformed with the LdCAS expression construct or the empty pESC-URA-tHMG1-DEST vector to yield strain TM122 and the control strain TM097, respectively.
The yeast strains TM097 and TM122 were cultivated in the presence of methyl β-cyclodextrin (MβCD) as described [30]. On day 1, for each strain five individual yeast colonies were used to inoculate 5 mL of synthetic defined (SD) medium containing glucose with the–Ura dropout (DO) supplement (Clontech). The pre-cultures were grown for 24 h at 30°C with agitation (200 rpm). To induce heterologous gene expression, the pre-cultures were washed with 1 mL of sterile water and used to inoculate 15 mL of SD Gal/Raf medium containing galactose and raffinose with the–Ura DO supplement (Clontech). The induced cultures were incubated for 24 h and on day 3, methionine and MβCD were added to 1 mM and 5 mM, respectively. After a further 24 h incubation, MβCD was added once again to 5 mM. After a final 24 h incubation, the yeast cultures were stored at 4°C for three days and on day 8, 1 mL of each culture was extracted thrice with 500 μL of hexane. The pooled organic fractions were evaporated and derivatized with 10 μL of pyridine (Sigma-Aldrich, St. Louis, United States) and 50 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (Sigma-Aldrich) prior to gas chromatography-mass spectrometry (GC-MS) analysis. The authentic cycloartenol standard (Sigma-Aldrich) was derivatized following the same method.
Sterol extraction from L. dendroidea
Dried L. dendroidea material was ground to a fine powder under liquid nitrogen. Five milligram of the obtained powder was saponified by boiling it for 2 hours in 500 μL of 40% KOH and 500 μL of 50% EtOH. The resulting mixture was extracted thrice with 500 μL of hexane. The organic fractions were pooled, evaporated in vacuo and derivatized with 10 μL of pyridine (Sigma-Aldrich, St. Louis, United States) and 50 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (Sigma-Aldrich, St. Louis, United States) prior to GC-MS analysis. An authentic cholesterol standard (Sigma-Aldrich, St. Louis, United States) was derivatized following the same method.
GC-MS analysis
GC-MS analysis was performed using a GC model 6890 and MS model 5973 (Agilent, Santa Clara, United States). A VF-5ms capillary column (Varian CP9013, Agilent) was operated at a constant helium flow of 1 mL/min and 1 μL of the sample was injected in splitless mode. The oven was initially held at 80°C for 1 min, ramped to 280°C at a rate of 20°C/min, held at 280°C for 45 min, ramped to 320°C at a rate of 20°C/min, held at 320°C for 1 min, and at the end of the run cooled to 80°C at a rate of 50°C/min. Throughout the analysis, the injector was set to 280°C, the MS transfer line to 250°C, the MS ion source to 230°C, and the quadrupole to 150°C. A full electron ionization (EI) mass spectrum was generated by scanning the m/z range of 60–800 with a solvent delay of 7.8 min.
Phylogenetic analysis
All OSC sequences reported in [26] were downloaded from the Phytozome database (A. thaliana) and NCBI (other species). Additional OSC sequences released after this study were screened in literature and included in our phylogenetic analysis. Subsequently, a Python script [43, 44] was used to obtain only the coding sequences to facilitate the alignment. The coding sequences were translated into amino acids and aligned using the software SeaView [45] employing the Clustal Omega algorithm [46]. After this, we constructed a neighbour joining tree using the Kimura 2 parameter model [47] with 1,000 bootstrap replicates. In the end, the tree was edited using FigTree.
Results and Discussion
Identification of LdCAS
Mining of L. dendroidea transcriptome data [38] revealed only one full-length OSC with an open reading frame of 739 amino acids. This OSC was cloned from L. dendroidea cDNA and named LdCAS. The LdCAS sequence was submitted to GenBank (accession number KX343073). The search for LdCAS conserved domains returned one hit, the squalene cyclase (SQCY) domain subgroup 1 (NCBI CDDS ID: cd02892), which has an alpha 6 –alpha 6 barrel fold and belongs to the Isopren C2 like superfamily (NCBI CDDS ID: cl08267). Further, using the CAS1 from A. thaliana (UniProtKB ID: P38605), we were able to identify all five farnesyltransferase B subunit (PFTB) repeats in LdCAS. The mutagenesis sites, where the change of residues leads to the production of distinct compounds, and the active sites of AtCAS1 and LdCAS were all the same, suggesting that our LdCAS is indeed a cycloartenol synthase, as AtCAS1 is the cycloartenol synthase from A. thaliana, previously isolated and characterized [29].
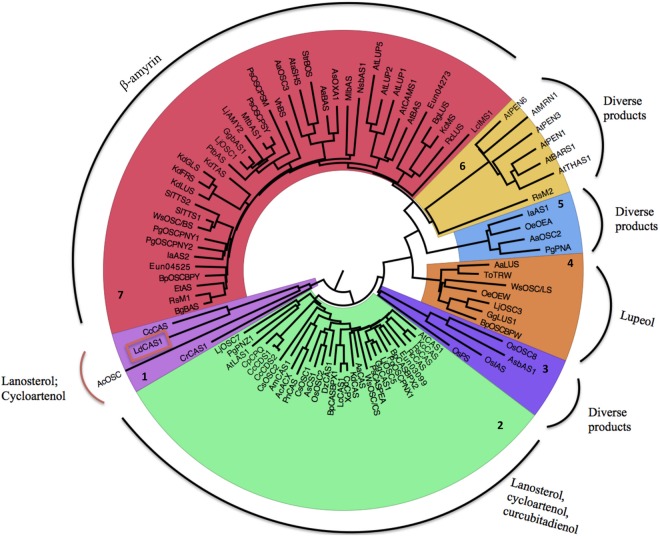
A phylogenetic tree using the amino acid sequences from putative OSCs from other species formed seven major groups (Fig 1). LdCAS clustered together with the OSCs from Chondrus crispus, a red seaweed, and with fungi and green algae OSCs (Fig 1, cluster 1). It is postulated that ergosterol is the final product of the CAS-dependent sterol biosynthesis pathway found in the genome of the green algae Chlamydomonas reinhardtii, as observed in fungi, despite the absence of the MVA pathway in green algae [48]. Since there is no other study that functionally characterized an OSC from red seaweeds, we could suggest that cycloartenol is the product of the OSC in red seaweeds.
Fig 1. Neighbour-joining tree of OSCs from several organisms.
OSC amino acid sequences (S1 Table) were aligned using Clustal Omega [59] with default parameters as implemented in the program SeaView [45] and all gaps were eliminated. The molecular evolutionary model chosen was Kimura 2 parameter [47]. The tree was reconstructed in SeaView with 1,000 bootstrap replicates. The name of each tip is the OSC name (source on S1 Table). Usually the first two characters represent the name of the species and the last three the product.
The genes coding for cycloartenol, lanosterol and cucurbitadienol synthases were all grouped together (Fig 1, cluster 2). These products are generated from the same precursor–the protesterol cation. The modification of the protesterol cation by carbon removal leads to lanosterol and cycloartenol (see [49, 50] for further details). The modification of this molecule with a second methyl and hydrogen migration leads to curcubitadienol [51]. A third group of OSCs (Fig 1, cluster 7) consisted mainly of β-amyrin synthases. Other enzymes found within this group produce lupeol and mixed products. The phylogenetic tree showed four other minor groups. One group (Fig 1, cluster 4) was formed by some lupeol synthases and other enzymes, while another group (Fig 1, cluster 6) is composed mainly by A. thaliana enzymes. The last two groups (Fig 1, clusters 3 and 5) are mainly constituted by a broad group of multifunctional OSCs, which make a range of cyclization products.
The division of the OSC family according their product and higher rank phylogeny is similar to that observed in other studies. Xue et al. [28] divided the OSC family of plants into ten groups based on this pattern: pentacyclic triterpenes, lanosterol, cucurbitadienol, cycloartenol and unknown function within dicots and pentacyclic triterpene, isoarborinol, parkeol, cycloartenol and unknown function within monocots. Gas-Pascual et al. [52] included species from other kingdoms and found seven distinct OSC groups: lupeol, β-amyrin, promiscuous β-amyrin, plant lanosterol and cycloartenol from plants, protists and algae plus planctomycete. Together, these studies indicate that functional characterization is necessary to properly define the product specificity of an OSC since some phylogenetic groups have more than one product or because they are composed of multifunctional OSCs.
Functional characterization of LdCAS in S. cerevisiae
To determine the enzymatic activity of LdCAS, we carried out functional characterization by means of heterologous expression in the yeast S. cerevisiae. Therefore, we first generated a S. cerevisiae strain capable of inducing terpene hyperproduction, as described previously [30, 53]. The yeast strain that we used was PA14, which is a modified version of the previously generated TM1 strain [30], containing an additional auxotrophic selection marker (see Material and Methods) and generated using the CRISPR/Cas9 genome editing tool [42]. As in the TM1 strain, the lanosterol synthase (ERG7) promoter in strain PA14 was replaced by a methionine repressible promoter. Addition of methionine to the yeast cultivation medium reduces the biosynthetic flux towards the sterols, leading to the accumulation of 2,3-oxidosqualene that can be used as a substrate for heterologously expressed OSCs. Additionally, a truncated feedback-insensitive version of the S. cerevisiae 3-hydroxy-3-methylglutaryl-CoA reductase 1 (tHMG1) enzyme was expressed from the high-copy-number plasmid pESC-URA to further increase the flux through the MVA pathway and thus further enhance the 2,3-oxidosqualene pool available for the heterologously expressed OSC.
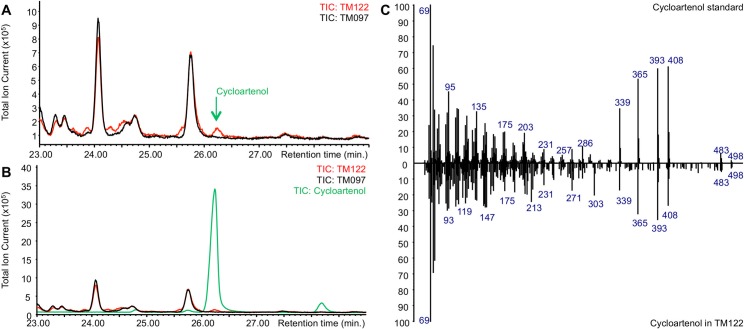
Two yeast strains were generated from strain PA14. Strain TM122 expressed both tHMG1 and LdCAS from the pESC-URA-tHMG1-DEST plasmid, whereas the control strain TM097 only expressed tHMG1 from the pESC-URA-tHMG1-DEST plasmid. Both yeast strains were cultivated in the presence of MβCD as described [30]. After yeast cultivation, the extract of the spent medium of the TM122 strain was checked by GC-MS and compared to the extract of the spent medium of the control strain TM097. This analysis revealed the presence of a chromatographic peak unique to strain TM122 at 26.2 min with the same retention time and electron ionization (EI) spectrum as an authentic cycloartenol standard (Fig 2). The finding that LdCAS encodes a cycloartenol synthase is further underscored by previous studies suggesting cycloartenol to be the precursor of all sterols in marine seaweeds [54].
Fig 2. Expression of LdCAS in S. cerevisiae leads to the production of cycloartenol.
(A) Overlay of GC-MS chromatograms from spent medium of the control yeast strain TM097 (black) and yeast strain TM122 expressing LdCAS (red). A peak unique to strain TM122 was observed with a retention time of 26.2 minutes. (B) Overlay of GC-MS chromatograms from spent medium of the control yeast strain TM097 (black) and yeast strain TM122 expressing LdCAS (red) and the GC-MS chromatogram of an authentic cycloartenol standard (green). The peak unique to strain TM122 has the same retention time as the authentic cycloartenol standard. (C) Comparison of the EI-MS spectra of the authentic cycloartenol standard (top) and the cycloartenol produced in strain TM122 (bottom).
Sterol profile of L. dendroidea and implications on sterol synthesis in red seaweeds
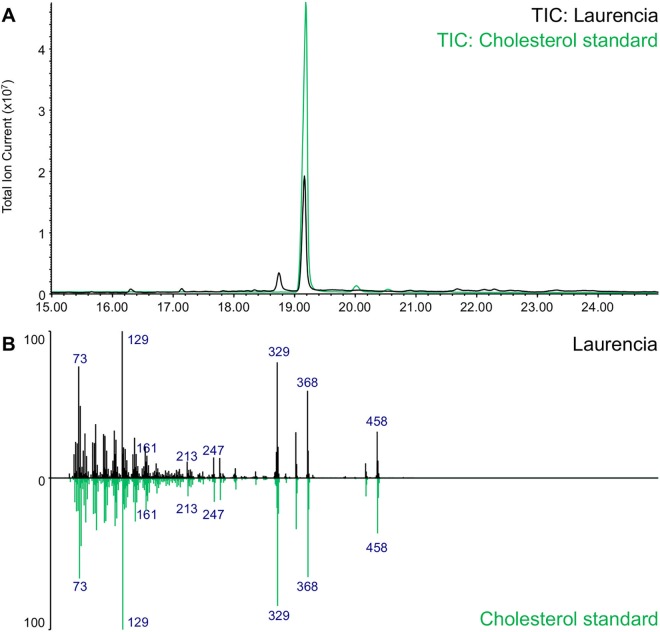
As primary metabolites, sterols are essential structural components of cell membranes. Cholesterol, the main animal sterol, provides structural integrity and fluidity to the cell membrane. In fungi, the major sterol is erogsterol. Campesterol, stigmasterol and sitosterol are the most abundant phytosterols. Like for animals, the main sterol in red seaweeds was reported to be cholesterol [55]. To validate the sterol composition of the red seaweed L. dendroidea, sterol profiling was carried out by GC-MS (Fig 3). This analysis confirmed that, like other Rhodophyta, L. dendroidea accumulates cholesterol as its major sterol.
Fig 3. Sterol profiling of Laurencia dendroidea using GC-MS.
(A) Total Ion Current chromatogram of the sterol extract of L. dendroidea (black) compared to an authentic cholesterol standard (green). (B) Comparison of the EI-MS spectra of cholesterol detected in L. dendroidea (black) and the authentic cholesterol standard (green).
As the phytosterol precursor cycloartenol is the cyclization product of 2,3-oxidosqualene, and the major accumulating sterol in red seaweeds is the ‘animal’ sterol cholesterol, the sterol biosynthesis in red seaweeds may be a hybrid pathway between the plant and animal sterol synthesis pathways. Notably, unlike other plants, Solanaceae plant species also accumulate cholesterol as a major sterol [56, 57]. The biosynthesis of cholesterol in Solanaceae has not been elucidated yet, but was shown to involve a sterol side chain reductase enzyme (SSR) 2 that catalyzes the conversion of cycloartenol into cycloartanol by reduction of the C-24(25) double bond. This reduction step forms the branch point between the synthesis of phytosterols and cholesterol in Solanaceae [56] and it could thus be postulated that a similar reduction step takes place in the cholesterol synthesis of red seaweeds. Notably, this SSR2 enzyme seems to have arisen from DWF1 [56], a reductase enzyme involved in phytosterol synthesis that catalyzes the reduction of C-24(28) double bonds in C-24 alkylsterols and that is homologous to 24-dehydrocholesterol reductase (DHCR24), the animal C-24(25) reductase that catalyzes the conversion of desmosterol into cholesterol [58]. Hence, a key step in the synthesis of cholesterol in red algae may be the reduction of the C-24(25) double bond by a DHCR24-like enzyme.
Conclusion
We were able to clone and characterize for the first time an OSC enzyme from a red seaweed. Through phylogenetic analysis we postulated that the product generated by LdCAS of L. dendroidea is cycloartenol. This finding was confirmed by the functional characterization of LdCAS in yeast and literature reports suggesting that cycloartenol is the sterol precursor of seaweed sterols. Furthermore, sterol profiling revealed that cholesterol is the major L. dendroidea sterol, implicating sterol synthesis in red seaweeds may be carried out via a hybrid of the plant and animal sterol synthesis pathways.
Supporting Information
(DOCX)
Acknowledgments
We thank CNPq, CAPES and FAPERJ for financial support. This work is part of G.C. PhD thesis in the graduate course of Dinâmica dos Oceanos e da Terra, supported by CAPES scholarship. J.P. is postdoctoral fellow of the Research Foundation Flanders. P.A. is indebted to the VIB International Fellowship Program for a predoctoral fellowship. RCP and FLT thank CNPq by Productivity Fellowships.
Data Availability
All data used in this study are specified in Table 1 (primer sequences), Table 2 (yeast strain sources) and S1 Table (GenBank accession number).
Funding Statement
We thank CNPq, CAPES and FAPERJ for financial support. This work is part of GC's PhD thesis in the graduate course of Dinâmica dos Oceanos e da Terra, supported by CAPES scholarship. JP is postdoctoral fellow of the Research Foundation Flanders. PA is indebted to the VIB International Fellowship Program for a predoctoral fellowship. RCP and FLT thank CNPq by Productivity Fellowships. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Guiry MD, Guiry GM. AlgaeBase: National University of Ireland, Galway; 2016 [04 January 2016]. Available from: http://www.algaebase.org.
- 2.Irie T, Suzuki M, Kurosawa E, Masamune T. Laurinterol, debromolaurinterol and isolaurinterol, constituents of Laurencia intermedia Yamada. Tetrahedron. 1970;26(13):3271–7. [Google Scholar]
- 3.Irie T, Suzuki M, Masamune T. Laurencin, a constituent from Laurencia species. Tetrah Let. 1965;6(16):1091–9. [Google Scholar]
- 4.Wang BG, Gloer JB, Ji NY, Zhao JC. Halogenated organic molecules of Rhodomelaceae origin: chemistry and biology. Chem Rev. 2013;113(5):3632–85. 10.1021/cr9002215 . [DOI] [PubMed] [Google Scholar]
- 5.Ji N-Y, Wang B-G. Nonhalogenated organic molecules from Laurencia algae. Phytoc Rev. 2013;13(3):653–70. 10.1007/s11101-013-9326-0 [DOI] [Google Scholar]
- 6.Vairappan CS, Anangdan SP, Tan KL, Matsunaga S. Role of secondary metabolites as defense chemicals against ice-ice disease bacteria in biofouler at carrageenophyte farms. J Appl Phyc. 2009;22(3):305–11. 10.1007/s10811-009-9460-7 [DOI] [Google Scholar]
- 7.Vairappan CS, Daitoh M, Suzuki M, Abe T, Masuda M. Antibacterial halogenated metabolites from the Malaysian Laurencia species. Phytochemistry. 2001;58(2):291–7. [DOI] [PubMed] [Google Scholar]
- 8.Vairappan CS, Kawamoto T, Miwa H, Suzuki M. Potent Antibacterial Activity of Halogenated Compounds against Antibiotic-Resistant Bacteria. Planta Med. 2004;70(11):1087–90. 10.1055/s-2004-832653 [DOI] [PubMed] [Google Scholar]
- 9.Pereira RC, Da Gama BA, Teixeira VL, Yoneshigue-Valentin Y. Ecological roles of natural products of the brazilian red seaweed Laurencia obtusa. Braz J Biol. 2003;63(4):665–72. [DOI] [PubMed] [Google Scholar]
- 10.Iliopoulou D, Roussis V, Pannecouque C, De Clercq E, Vagias C. Halogenated sesquiterpenes from the red alga Laurencia obtusa. Tetrahedron. 2002;58:6749–55. [Google Scholar]
- 11.Cabrita MT, Vale C, Rauter AP. Halogenated compounds from marine algae. Mar Drugs. 2010;8(8):2301–17. 10.3390/md8082301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernández J, Souto ML, Norte M. Evaluation of the cytotoxic activity of polyethers isolated from Laurencia. Bio Med Chem. 1998;6(12):2237–43. 10.1016/s0968-0896(98)80004-7 [DOI] [PubMed] [Google Scholar]
- 13.Ji NY, Li XM, Xie H, Ding J, Li K, Ding LP, et al. Highly Oxygenated Triterpenoids from the Marine Red Alga Laurencia mariannensis (Rhodomelaceae). Helv Chim Acta. 2008;91:1940–6. [Google Scholar]
- 14.Li YX, Himaya SW, Kim SK. Triterpenoids of marine origin as anti-cancer agents. Molecules. 2013;18(7):7886–909. 10.3390/molecules18077886 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arendt P, Pollier J, Callewaert N, Goossens A. Synthetic biology for production of natural and new-to-nature terpenoids in photosynthetic organisms. Plant J. 2016;87(1):16–37. 10.1111/tpj.13138 [DOI] [PubMed] [Google Scholar]
- 16.Lange BM, Rujan T, Martin W, Croteau R. Isoprenoid biosynthesis: The evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci U S A. 2000;97(24):13172–7. 10.1073/pnas.240454797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong Y, Morris-Natschke SL, Lee KH. Biosynthesis, total syntheses, and antitumor activity of tanshinones and their analogs as potential therapeutic agents. Nat Prod Rep. 2011;28(3):529–42. 10.1039/c0np00035c . [DOI] [PubMed] [Google Scholar]
- 18.Yang D, Du X, Liang X, Han R, Liang Z, Liu Y, et al. Different roles of the mevalonate and methylerythritol phosphate pathways in cell growth and tanshinone production of Salvia miltiorrhiza hairy roots. PLoS One. 2012;7(11):e46797 10.1371/journal.pone.0046797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massé G, Belt S, Rowland S, Rohmer M. Isoprenoid biosynthesis in the diatoms Rhizosolenia setigera (Brightwell) and Haslea ostrearia (Simonsen). Proc Natl Acad Sci U S A. 2004;101(13):4413–8. 10.1073/pnas.0400902101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohr M, Schwender J, Polle JE. Isoprenoid biosynthesis in eukaryotic phototrophs: a spotlight on algae. Plant Sci. 2012;185–186:9–22. 10.1016/j.plantsci.2011.07.018 . [DOI] [PubMed] [Google Scholar]
- 21.Fabris M, Matthijs M, Carbonelle S, Moses T, Pollier J, Dasseville R, et al. Tracking the sterol biosynthesis pathway of the diatom Phaeodactylum tricornutum. New Phytol. 2014;204(3):521–35. 10.1111/nph.12917 . [DOI] [PubMed] [Google Scholar]
- 22.Schwender J, Zeidler J, Gröner R, Müller C, Focke M, Braun S, et al. Incorporation of 1-deoxy-d-xylulose into isoprene and phytol by higher plants and algae. FEBS Letters. 1997;414(1):129–34. 10.1016/s0014-5793(97)01002-8 [DOI] [PubMed] [Google Scholar]
- 23.Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66:212–29. 10.1111/j.1365-313X.2011.04520.x [DOI] [PubMed] [Google Scholar]
- 24.Summons RE, Bradley AS, Jahnke LL, Waldbauer JR. Steroids, triterpenoids and molecular oxygen. Philos Trans R Soc Lond B Biol Sci. 2006;361(1470):951–68. 10.1098/rstb.2006.1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu R, Fazio GC, Matsuda SPT. On the origins of triterpenoid skeletal diversity. Phytochemistry. 2004;65(3):261–91. 10.1016/j.phytochem.2003.11.014 [DOI] [PubMed] [Google Scholar]
- 26.Thimmappa R, Geisler K, Louveau T, O'Maille P, Osbourn A. Triterpene biosynthesis in plants. Annu Rev Plant Biol. 2014;65:225–57. 10.1146/annurev-arplant-050312-120229 . [DOI] [PubMed] [Google Scholar]
- 27.Abe I, Liu W, Oehlschlager A, Prestwich G. Mechanism-Based Active Site Modification of Oxidosqualene Cyclase by Tritium-Labeled 18-Thia-2,3-Oxidosqualene. J Am Chem Soc. 1996;118(38):9180–1. [Google Scholar]
- 28.Xue Z, Duan L, Liu D, Guo J, Ge S, Dicks J, et al. Divergent evolution of oxidosqualene cyclases in plants. New Phytol. 2012;193(4):1022–38. 10.1111/j.1469-8137.2011.03997.x . [DOI] [PubMed] [Google Scholar]
- 29.Corey E, Matsuda S, Bartel B. Isolation of an Arabidopsis thaliana gene encoding cycloartenol synthase by functional expression in a yeast mutant lacking lanosterol synthase by the use of a chromatographic screen. Proc Natl Acad Sci U S A. 1993;90(24):11628–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moses T, Pollier J, Almagro L, Buyst D, Van Montagu M, Pedreno MA, et al. Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16alpha hydroxylase from Bupleurum falcatum. Proc Natl Acad Sci U S A. 2014;111(4):1634–9. 10.1073/pnas.1323369111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidovich-Rikanati R, Shalev L, Baranes N, Meir A, Itkin M, Cohen S, et al. Recombinant yeast as a functional tool for understanding bitterness and cucurbitacin biosynthesis in watermelon (Citrullus spp.). Yeast. 2015;32(1):103–14. 10.1002/yea.3049 . [DOI] [PubMed] [Google Scholar]
- 32.Babiychuk E, Bouvier-Navé P, Compagnon V, Suzuki M, Muranaka T, Montagu MV, et al. Allelic mutant series reveal distinct functions for Arabidopsis cycloartenol synthase 1 in cell viability and plastid biogenesis. Proc Natl Acad Sci U S A. 2008;105(8):3163–8. 10.1073/pnas.0712190105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhar N, Rana S, Razdan S, Bhat WW, Hussain A, Dhar RS, et al. Cloning and functional characterization of three branch point oxidosqualene cyclases from Withania somnifera (L.) Dunal. J Biol Chem. 2014;289(24):17249–67. 10.1074/jbc.M114.571919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abe I, Prestwich G. Molecular cloning, characterization, and functional expression of rat oxidosqualene cyclase cDNA. Proc Natl Acad Sci U S A. 1995;92(20):9274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin YL, Lee YR, Tsao NW, Wang SY, Shaw JF, Chu FH. Characterization of the 2,3-Oxidosqualene Cyclase Gene from Antrodia cinnamomea and Enhancement of Cytotoxic Triterpenoid Compound Production. J Nat Prod. 2015;78(7):1556–62. 10.1021/acs.jnatprod.5b00020 . [DOI] [PubMed] [Google Scholar]
- 36.Khakimov B, Kuzina V, Erthmann PO, Fukushima EO, Augustin JM, Olsen CE, et al. Identification and genome organization of saponin pathway genes from a wild crucifer, and their use for transient production of saponins in Nicotiana benthamiana. Plant J. 2015;84(3):478–90. 10.1111/tpj.13012 . [DOI] [PubMed] [Google Scholar]
- 37.Andre CM, Legay S, Deleruelle A, Nieuwenhuizen N, Punter M, Brendolise C, et al. Multifunctional oxidosqualene cyclases and cytochrome P450 involved in the biosynthesis of apple fruit triterpenic acids. New Phytol. 2016;211(4):1279–94. 10.1111/nph.13996 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Oliveira LS, Tschoeke DA, de Oliveira AS, Hill LJ, Paradas WC, Salgado LT, et al. New Insights on the terpenome of the red seaweed Laurencia dendroidea (Florideophyceae, Rhodophyta). Mar Drugs. 2015;13(2):879–902. 10.3390/md13020879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moses T, Pollier J, Shen Q, Soetaert S, Reed J, Erffelinck M-L, et al. OSC2 and CYP716A14v2 Catalyze the Biosynthesis of Triterpenoids for the Cuticle of Aerial Organs of Artemisia annua. Plant Cell. 2015;27(1):286–301. 10.1105/tpc.114.134486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–6. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiallos-Jurado J, Pollier J, Moses T, Arendt P, Barriga-Medina N, Morillo E, et al. Saponin determination, expression analysis and functional characterization of saponin biosynthetic genes in Chenopodium quinoa leaves. Plant Sci. 2016;250:188–97. 10.1016/j.plantsci.2016.05.015 [DOI] [PubMed] [Google Scholar]
- 42.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41(7):4336–43. 10.1093/nar/gkt135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–3. 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cock PJ, Gruning BA, Paszkiewicz K, Pritchard L. Galaxy tools and workflows for sequence analysis with applications in molecular plant pathology. PeerJ. 2013;1:e167 10.7717/peerj.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–4. 10.1093/molbev/msp259 . [DOI] [PubMed] [Google Scholar]
- 46.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:1–6. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–20. [DOI] [PubMed] [Google Scholar]
- 48.Schwender J, Gemünder C, Lichtenthaler H. Chlorophyta exclusively use the 1-deoxyxylulose 5-phosphate/2-C-methylerythritol 4-phosphate pathway for the biosynthesis of isoprenoids. Planta. 2001;212:416–23. 10.1007/s004250000409 [DOI] [PubMed] [Google Scholar]
- 49.Wendt K, Schulz G, Corey E, Liu D. Enzyme mechanisms for polycyclic triterpene formation. Angew Chem Int Ed. 2000;39:2812–33. [PubMed] [Google Scholar]
- 50.Acimovic J, Rozman D. Steroidal triterpenes of cholesterol synthesis. Molecules. 2013;18(4):4002–17. 10.3390/molecules18044002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cattel L, Ceruti M, Viola F, Delprino L, Balliano G, Duriatti A, et al. The Squalene-2,3-Epoxide Cyclase as a Model for the Development of New Drugs Lipids. 1986;21(1):31–8. [DOI] [PubMed] [Google Scholar]
- 52.Gas-Pascual E, Berna A, Bach TJ, Schaller H. Plant oxidosqualene metabolism: cycloartenol synthase-dependent sterol biosynthesis in Nicotiana benthamiana. PLoS One. 2014;9(10):e109156 10.1371/journal.pone.0109156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirby J, Keasling JD. Metabolic engineering of microorganisms for isoprenoid production. Nat Prod Rep. 2008;25(4):656–61. 10.1039/b802939c . [DOI] [PubMed] [Google Scholar]
- 54.Sambamurty AVSS. A Textbook of Algae: I. K. International Pvt. Ltd.; 2005.
- 55.Nasir M, Saeidnia S, Mashinchian-Moradi A, Gohari AR. Sterols from the red algae, Gracilaria salicornia and Hypnea flagelliformis, from Persian Gulf. Pharmacogn Mag. 2011;7(26):97–100. 10.4103/0973-1296.80663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawai S, Ohyama K, Yasumoto S, Seki H, Sakuma T, Yamamoto T, et al. Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell. 2014;26(9):3763–74. 10.1105/tpc.114.130096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cardenas PD, Sonawane PD, Pollier J, Vanden Bossche R, Dewangan V, Weithorn E, et al. GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat Commun. 2016;7:10654 10.1038/ncomms10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh P, Saxena R, Srinivas G, Pande G, Chattopadhyay A. Cholesterol biosynthesis and homeostasis in regulation of the cell cycle. PLoS One. 2013;8(3):e58833 10.1371/journal.pone.0058833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38(Web Server issue):W695–9. 10.1093/nar/gkq313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All data used in this study are specified in Table 1 (primer sequences), Table 2 (yeast strain sources) and S1 Table (GenBank accession number).