Abstract
Measuring the chemical composition of individual cells in mammalian organs can provide critical insights towards understanding the mechanisms leading to their normal and pathological function. In this work, single cell heterogeneity of islets of Langerhans is characterized with high throughput by microscopy-guided single cell matrix-assisted laser desorption/ionization mass spectrometry. Two levels of chemical heterogeneity were observed from the analysis of more than 3000 individual cells. Within a single islet, cellular heterogeneity was evident from the exclusive expression of the canonical biomarkers glucagon, insulin, pancreatic polypeptide (PP), and somatostatin within α-, β-, γ-, and δ-cells, respectively. We localized the neuropeptide WE-14, a known cell-to-cell signaling molecule, to individual δ-cells. Moreover, several unreported endogenous peptides generated by dibasic site cleavages of PP were detected within individual γ-cells. Of these, PP(27–36) was previously reported to activate the human Y4 receptor, suggesting it has a signaling role in vivo. Heterogeneity in cell composition was also observed between islets as evidenced by a 50-fold larger α–cell population in islets of the dorsal pancreas compared to the ventral-derived pancreatic islets. Finally, PP(27–36) was more abundant in γ-cells from the ventral region of the pancreas, indicating differences in the extent of PP-prohormone processing in the two regions of the pancreas.
Graphical abstract

The heterogeneity and variability of individual cells is critical for the survival and propagation of life. Cellular heterogeneity is thought to be necessary for higher-level systems, suggesting that “variation is function”, where nuanced variation across single cells provides a means of graded response.1 Greater phenotypic heterogeneity is also vital; for example, the cellular heterogeneity of islets of Langerhans, which are composed of four major cell types. Each type of endocrine cell is functionally interconnected, yet chemically distinct, providing plastic responses of an organism to fluctuating environments. Cellular heterogeneity facilitates the endocrine functions of islets of Langerhans that are essential for glucose homeostatis. 2,3
Due to the importance of islets of Langerhans in normal and pathological glucose utilization, the cellular heterogeneity and variability of this microorgan are well studied using approaches such as immunohistochemical profiling,4–7 single cell transcriptomics analysis,1,8,9 and metabolomic and proteomic profiling. 10,11 Mass spectrometry (MS) has become an important tool for the investigation of peptide content in organelles, cells, and tissues, such as the endocrine pancreas. MS has been efficiently used to probe the secretome, metabolome, peptidome, and proteome of whole islets.12–16 MS investigations of the peptide content of islets have revealed unknown cell-to-cell signaling molecules and indicated novel prohormone processing. However, most prior studies utilized individual or pooled islets, consisting of thousands of cells. Hence, these ensemble measurements conceal the biological variability and functional heterogeneity of individual cells. Matrix-assisted laser desorption/ionization (MALDI) MS is uniquely suitable for the detection and characterization of peptides in small samples, including single cells, due to its high sensitivity and low sample consumption. Beginning with the MALDI MS analysis of single neurons from Lymnaea stagnalis and Aplysia californica more than 20 years ago,17,18 the technique has evolved into a powerful tool for measuring peptide content in minute samples such as cells and even individual organelles.19–24
Microscopy-guided, single cell MALDI MS greatly increases sample throughput, facilitating the measurement of peptide and metabolite content for thousands of cells.10,11 Such rapid analysis of cell populations is key to successfully assessing cellular heterogeneity. The technique is amenable to peptide detection from virtually any type of cell. Non-targeted, label-free analysis of large populations coupled with multivariate statistical analysis allows detection of rare cellular phenotypes and enables the study of differential peptide expression in cellular subtypes. Here, using microscopy-guided, single cell MALDI MS we examined the cellular heterogeneity of islets of Langerhans located in the dorsal and ventral regions of the rat pancreas. By classifying each spectrum based on canonical and cell-specific peptide expression, the cellular composition of individual islets was directly measured and the proportion of α-, β-, γ-, and δ-cells was found to differ between the pancreatic regions. Furthermore, the dataset revealed novel prohormone processing products as well as their location-specific abundance.
RESULTS AND DISCUSSION
Intra- and Inter-islet Cellular Heterogeneity
High-throughput single cell MALDI MS profiling was used for phenotyping cells from islets of Langerhans to uncover the chemical cellular heterogeneity of islets in anatomically and developmentally distinct parts of the pancreas. Isolated from 48 individual islets (6 dorsal and 6 ventral islets each from a total of 4 animals), cells were deposited on indium tin oxide (ITO)-coated glass slides and examined with single cell MALDI MS. Single cell dispersions of individual islets were deposited on separated areas of the ITO slides to keep track of the source islet. Acquired mass spectra were classified and counted. Each successful measurement revealed the presence of chemically distinct hormone profiles in individual cells (Figures 1 and 2). A number of peptides characteristic for islet cell types, including glucagon (α-cells), insulin (β-cells), PP (γ-cells), and somatostatin-14 (δ-cells) were observed (Figure 1). The cellular composition of individual islets from the same pancreatic lobe were similar. In contrast, significant differences were found between cell populations of the islets of the dorsal and ventral pancreas. Our findings show that α-cells are more abundant in dorsal pancreas islets and γ-cells are more prevalent in ventral pancreas islets (Figure 2, Table 1, and Figure S1). A χ2-test for independence on the contingency table showed significant differences between islet cell populations in the two regions (nDtot = 1768, nVtot = 1738, p < 0.00001, n = 24 dorsal islets, n = 24 ventral islets). Importantly, these findings are in agreement with previously published histological reports focused only on major biomarkers due to the use of affinity probes.4–7 Here, our non-targeted and multiplex analytical technology generated a dataset to test for differential processing of islet prohormones between anatomical regions of the rat pancreas.
Figure 1.
Representative mass spectra acquired from individual α-, β-, γ-, and δ-cells, as well as an intact islet of Langerhans collected from the dorsal pancreas. Peptide cell-type qualifiers/biomarkers: glucagon m/z 3481.5 (α-cells), insulin m/z 5799.9 (β-cells), PP m/z 4397.2 (γ-cells), and somatostatin-14 m/z 1637.7 (δ-cells). These signals are highlighted with asterisks. Many other characteristic signals were observed in the cells as described herein.
Figure 2.
MALDI MS characterization of intact islets of Langerhans and corresponding single cell populations obtained from individual islets. (a) Intact islets have different peptide profiles in the dorsal (filled circles) and ventral (crosses) pancreas. Distributions of total ion current (TIC)-normalized intensities of insulin, glucagon, and PP signals acquired from intact islets. (b and c) Results of k-means clustering of single cell data shown in color: α-cells (red), β-cells (green), γ-cells (blue), δ-cells (purple). K-means clustering was used on the full data set to classify and count the different cell types observed in single cell preparations from islets of Langerhans based on TIC-normalized peptide signal intensities. The classification highlights differences in cell composition between islets from dorsal and ventral pancreata, reflected in the relative abundance of α- (red) and γ- (blue) cells. The data were dimension-reduced with principal component analysis for visualization purposes only, here shown with principal components (PC) 1, 2, and 4. (b) Dorsal pancreas islet cells (n = 1768). (c) Ventral pancreas islet cells (n = 1738).
Table 1.
Population fractions of cell types for dorsal (n = 24) and ventral (n = 24) rat pancreatic islets of Langerhans measured with MALDI MS analysis of single cell populations.
| Cell type | Dorsal | Ventral |
|---|---|---|
| α | 0.43 | 0.081 |
| β | 0.51 | 0.67 |
| γ | 0.045 | 0.24 |
| δ | 0.0090 | 0.0086 |
| ncells | 1768 | 1738 |
Discovery of Endogenous Pancreatic Prohormone-Originated Peptides and their Different Abundances in γ-Cells of Dorsal and Ventral Islets
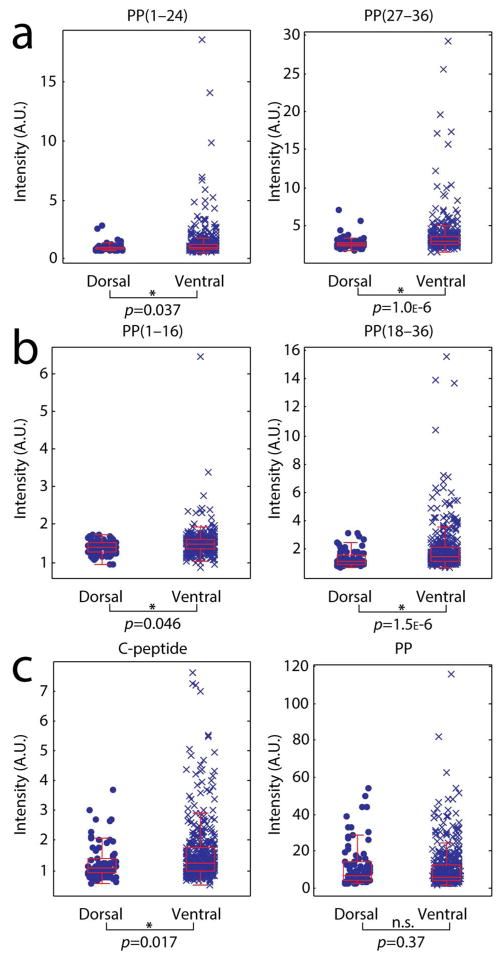
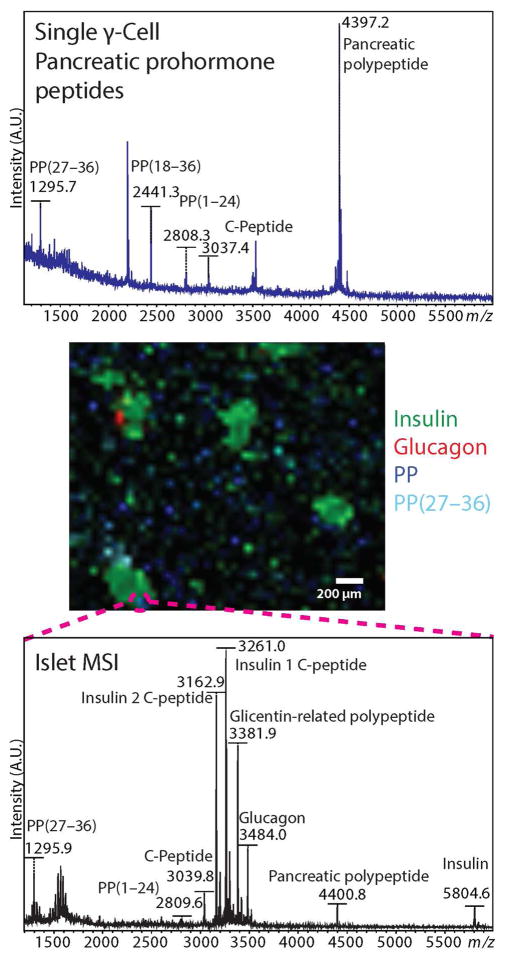
A statistically significant increase in pancreatic prohormone-related peptide signal intensities was observed in γ-cells from ventral islets when compared to dorsal islets collected from four animals (Figures 3 and 4). These include PP(1–24) +34%, PP(27–36) +32%, PP(1–16) +7%, and PP(18–36) +44%. In addition, correlation plots of signal intensities for PP against signal intensities for the other peptide products within single γ-cells indicate heterogeneity of the chemical content in cells from the ventral lobe, whereas cells from the dorsal lobe appear more clustered together (Figure S2). Parent ion m/z values of peptides detected with single cell MALDI MS were matched to m/z values of pancreatic prohormone peptides identified with LC–MS sequencing (Tables S1 and S2). We hypothesize that the peptides are formed by endogenous, enzymatic cleavage of the pancreatic prohormone at the dibasic or monobasic sites of full-length PP (Figures 3 and 4). This hypothesis is supported by MALDI mass spectrometry imaging (MSI) of fast-frozen pancreas sections from which we detected molecular signals corresponding to the PP(27–36) peptide, concomitant with full-length PP (Figure 5), adding confidence to the notion that PP(27–36) is synthesized endogenously. Interestingly, the relative signal intensity of the pancreatic prohormone C-terminal peptide (aa69–98) was 25% higher in cells of the ventral pancreas islets compared to those from the dorsal pancreas. No significant differences in the relative intensities of full-length PP were found in the same cellular populations.
Figure 3.
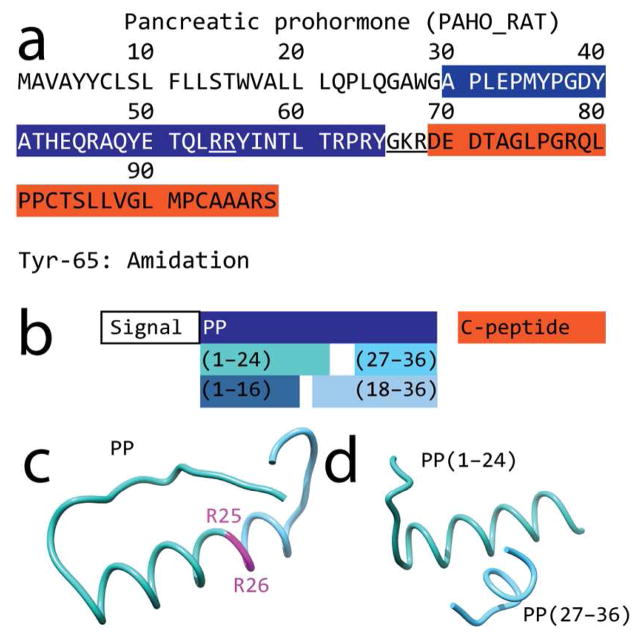
Sequence and predicted structure of peptides originating from the rat pancreatic prohormone. (a) Sequence of rat pancreatic prohormone (UNIPROT ID: P06303, PAHO_RAT); the dibasic sites are underlined. (b) Observed peptide products resulting from processing of the pancreatic prohormone (filled color boxes). (c) Ab initio-calculated structure of PP; the dibasic site is outlined in magenta. (d) Molecular modeling shows that the N- and C-terminal products (sea green and sky blue, respectively) resulting from cleavage at the dibasic site retain the α-helix folding of the parent peptide.
Figure 4.
Comparison of pancreatic prohormone peptide signals acquired from γ-cells of the dorsal and ventral islets (nDγ = 79 cells, nVγ = 418 cells). Box and whisker plots of TIC-normalized signal intensities are shown for (a) peptide products from internal dibasic cleavage of PP, left: PP(1–24), m/z 2808.2; right: PP(27–36), m/z 1295.7. (b) Peptide products from internal monobasic cleavage of PP, left: PP(1–16), m/z 1818.8; right: PP(18–36), m/z 2441.3. (c) Canonical peptide products from the pancreatic prohormone, left: C-terminal peptide, m/z 3037.4; right: PP, m/z 4397.2.
Figure 5.
Comparison of mass spectra acquired from a single γ-cell (top) and a pixel on a pancreatic islet cross-section (bottom). Color-coded molecular ion distributions for four peptides are shown on the MALDI MS image (center).
Although the internal dibasic site of PP has been recognized as a possible processing site,47 the resulting products have only been reported in CA-77 cells transfected with full-length pancreatic prohormone,26 and have not been described in previous studies using MS.12–16 Bioinformatic analysis of the pancreatic prohormone using the NeuroPred tool48 predicts that the dibasic RR site in the PP sequence is a potential processing site with a probability of 0.53 ± 0.10 and a 95% confidence interval (CI). The probability for processing at the C-terminal site resulting in full-length PP is 0.91 ± 0.04 (95% CI). This modeling suggests that the prohormone convertase processing of full-length PP at its dibasic site may form PP(27–36).
Molecular modeling using the PEP-FOLD tool49 indicates that PP(1–24) and PP(27–36) retain the α-helix secondary structure.40,41,49 Hence, the dibasic cleavage does not disrupt the shape of the C-terminus of PP. The region has an exact match to the C-terminal of the TRPRY-NH2 motif, suggesting that PP(27–36) may possess some affinity to NPY receptors. Other fragments of NPY, which exhibit the C-terminal motif TRPRY-NH2, display some bioactivity and binding affinity to NPY receptors, albeit more weakly than the full-length peptide. 38–45 Previous studies of N-terminal truncation of PP demonstrated the binding affinity of PP(27–36) to be 3 orders of magnitude weaker than PP. In functional assays, the maximum inhibition of cAMP accumulation of PP(27–36) was 50% of the full-length PP with an EC50 of 3.5 μM for PP(27–36) compared to 0.09 nM of PP.36,37 This suggests that PP(27–36) acts as a partial agonist to the Y4 receptor. The studies discussed above utilized simplified models that cannot be used to uncover all of the spatiotemporal activities in which PP(27–36) may participate in vivo.
We observed that the C-terminal peptide of pancreatic prohormone, as well as PP(1–24), PP(27–36), PP(1–18), and PP(20–36), displayed higher signal intensities in ventral islet-derived γ-cells. This finding, in conjunction with equal signal intensities of full-length PP, could be explained by heterogeneous expression or regulation of prohormone convertases for the shortened PP products, whereas the convertases yielding full-length PP remained the same in each set of islets. MALDI MSI and LC–MS also detected products of PP at its monobasic site, resulting in the formation of PP(1–16) and PP(18–36). Processing of the pancreatic prohormone at a single arginyl residue has previously been reported to occur,50 however, the exact processing pathway remains unclear. Our findings indicate these peptides are generated endogenously from the pancreatic prohormone and display a specific, heterogeneous distribution. Though the peptides display weaker binding affinity and less efficacy to G protein-coupled receptors compared to full-length PP, these molecules may operate within the feedback mechanism of Y4 receptors as partial agonists. It has been reported that the pancreatic prohormone is less amenable to processing with prohormone convertases than other NPY-family peptides.26 Although less common, prohormone processing may be tissue-specific, such that the final peptide products vary between anatomical regions, in agreement with our findings.26–35
Other Neuropeptides in Islets of Langerhans
WE-14 peptide (m/z 1677.8) from the chromogranin A prohormone was detected with LC–tandem MS analysis of extracts of homogenized islets of Langerhans (Table S1); however, due to homogenization, its cellular origin was uncertain. Further leveraging detection of rare phenotypes, single cell MALDI MS analysis of cellular populations obtained by islet dissociation revealed that WE-14 is selectively localized to δ-cells. WE-14 was accounted for among the top 50 most-intense analyte signals in δ-cells, but in no other cell types. Furthermore, WE-14 had a similar abundance between dorsal- and ventral-derived islet cells (2.50 ± 0.64, nDδ = 16; 3.2 ± 2.7, nVδ = 15; p = 0.94, n.s.). Rat islets of Langerhans have a complex spherical microarchitecture consisting of a core occupied by β-cells, with the other secretory cell types lining the periphery of the islet.6 A previous peptidomic study using MS revealed the presence of WE-14 in analyte extracts of homogenized islets of Langerhans, 15 and an immunohistochemical study localized WE-14 to the outer edge of rat islets of Langerhans where δ-cells are located.25 The results presented here are in good agreement with these previous findings but provide additional important details, including the localization of WE-14 in δ-cells within the islets of Langerhans. By isolating the rare δ-cells from the thousands of neighboring endocrine cells, their peptidome could be selectively investigated. Other neuropeptides were also detected in analyte extracts from islets of Langerhans using LC–MS, including aa513–532 (N- and C-terminal dibasic site processing) from the secretogranin-1 prohormone, and secretoneurin from the secretogranin-2 prohormone (Table S1). These peptides were not observed in our single cell data obtained with MALDI MS, which may reflect differences in analyte extraction due to sample preparation, ionization efficiency, and/or their lower abundance.
CONCLUSIONS
Determining the mechanisms involved in modulating the synthesis and processing of endogenous peptides, and regulation mediated through ligand–receptor interactions, is integral to understanding normal physiology as well as diseased states. From pancreatic islets, we have detected cell type-specific expression of known peptide hormones displaying anatomical heterogeneity. Peptides resulting from the processing of PP were detected endogenously and found to be enriched in γ-cells from ventral islets. Furthermore, we located the neuropeptide WE-14 with high abundance in δ-cells. The physiological roles and mechanisms of action of both PP and WE-14 are less understood compared to many other peptide hormones such as insulin. The detection of such molecules and determination of their specific localization are initial steps towards determining signaling mechanisms and physiological effects. A more complete understanding of pancreatic cell-to-cell signaling hormones may help reveal the mechanisms of metabolic diseases, in particular, the development of type 2 diabetes mellitus — a disease affecting a growing number of individuals worldwide.
MALDI MS of individual islet cells allowed detection and colocalization of previously unreported peptides with well-studied pancreatic hormones. The non-targeted, single cell MALDI MS profiling facilitated measurements of the peptide content of rare cells (γ-cells in dorsal islets) and tests for significant differences in abundance for the same cell type derived from a more abundant source of cells (γ-cells in ventral islets). The method is label-free and capable of detecting hundreds of compounds in thousands of cells within an hour, enabling experiments that would not be feasible with traditional flow cytometry. Moreover, the sampling procedure for comparative analysis of endocrine cells is suitable for many other types of peptidergic system studies. Examples include investigating temporal changes of tissue microarchitecture and pathological or drug-induced changes in peptide production.
METHODS
Chemicals
Collagenase P (from Clostridium histolyticum) used in the enzymatic isolation of islets of Langerhans was purchased from Roche Diagnostics (Indianapolis, IN). Mass spectrometer calibration was performed using a Peptide Calibration Standard Kit II (angiotensin II, angiotensin I, substance P, bombesin, ACTH clip 1–17, ACTH clip 18–39, somatostatin 28, bradykinin fragment 1–7, renin substrate tetradecapeptide porcine with added bovine insulin) obtained from Bruker Daltonics (Billerica, MA). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Isolation of Islets of Langerhans and Single Cell Preparation
Male, four-month old Sprague-Dawley rats from Harlan Laboratories (Indianapolis, IN) were euthanized by decapitation. The vertebrate animal use protocol was approved by the Institutional Animal Care and Use Committee at the University of Illinois at Urbana–Champaign. Islet isolation was performed as described elsewhere,52 with minor modifications. Modified Gey’s balanced salt solution (mGBSS) was prepared, containing 1.5 mM CaCl2, 4.9 mM KCl, 0.2 mM KH2PO4, 11 mM MgCl2, 0.3 mM MgSO4, 138 mM NaCl, 27.7 mM NaHCO3, 0.8 mM NaH2PO4, and 25 mM HEPES dissolved in Milli-Q water (Millipore, Billerica, MA), with the pH adjusted to 7.2 using NaOH in Milli-Q water. For islet isolation, the mGBSS was supplemented to a final concentration of 5 mM D-glucose and 1% (w/v) bovine serum albumin (buffer 1). Each pancreas was injected through the bile duct with 2 mL of 1.4 mg/mL collagenase P solution dissolved in buffer 1. Next, the ventral and dorsal regions were surgically dissected following morphological landmarks along the lower duodenum as described elsewhere.5 The two resulting tissues were placed in separate glass vials, each containing 1 mL of the collagenase P solution. The pancreatic tissues were incubated in a recirculating water bath for 20–30 min at 37 °C to digest exocrine tissue while leaving islets primarily intact. The resulting suspension was washed twice with buffer 1 and centrifuged for 3 min at 300 × g. The resulting pellet was resuspended in 10 mL buffer 1 and islets were manually isolated with a micropipette under visual control using an inverted microscope.
To stabilize the cells and label their nuclei for fluorescent targeting, each islet was transferred to 20 μL of a staining and cell stabilization solution consisting of 40% (v/v) glycerol in buffer 1 with 0.1 mg/mL Hoechst 33342.53 Islets were incubated for 2 h at 15 °C before trituration of individual islets into single cells onto ITO-coated glass microscopy slides (Delta Technologies, Loveland, CO). Studies of intact islets were performed as above but without trituration. To decrease bias, single cell dispersions from individual islets were randomly placed onto subdivided areas of the slides, such that each slide contained cells from at least two different animals. These efforts ensured that significant differences in abundance were not due to batch effects between slides.
For the LC–MS experiments, about 100 islets were collected and transferred to an Eppendorf tube containing 200 μL acidified methanol (90% MeOH, 9% formic acid (FA), 1% H2O) for peptide extraction, sonicated for 5 min, and incubated on ice for 1 h. The sample was centrifuged for 20 min (20 000 × g, 4 °C), the supernatant was then dried down in a Savant SpeedVac vacuum concentrator (Thermo Scientific, Waltham, MA) and reconstituted in 50 μL 5% MeOH, 0.1% FA. Sample clean-up was performed with a C18 spin-column (Thermo Scientific) pre-equilibrated with 5% MeOH, 0.1% FA. After sample loading and analyte retention, the column was washed twice with 1 mL 5% MeOH, 0.1% FA. The retained peptides were eluted twice using 50 μL 70% MeOH, 0.1% FA. The final sample used for LC–MS analysis was prepared by lyophilizing the eluent and reconstituting it in 10 μL 5% MeOH, 0.1% FA.
Optical Imaging for Registration of Fiducial Marks and Single Cell Locations
To create a system of spatial coordinates, a set of fiducial marks were made on conductive ITO slides. Cross marks were made prior to single cell suspension deposition with a diamond pen on 10–15 locations spread across each slide. Glycerol-stabilized cells or individual islets were deposited onto the slides. After overnight incubation in a minimal volume of glycerol-containing solution at ambient conditions, the slides were quickly rinsed with 150 mM ammonium acetate, pH 10, and dried with a gentle stream of nitrogen gas.54 Cells and fiducial markers were located using a Nanozoomer digital slide-scanner system (Hamamatsu, Middlesex, NJ). Silver paint applied with a marking pen surrounding the dispersed cells was targeted for autofocusing to acquire fluorescent and bright-field images of the suspension area. The images were processed and analyzed to determine the relative coordinates of cells, islets, and fiducial markers.
Single Cell MALDI MS Profiling
To provide a higher throughput and more reproducible coating than via a typical airbrush, an automatic sprayer system was developed using low-cost electric motors and linear actuators to coat up to four slides simultaneously (Figure S3). Pumping matrix solution through a fused silica capillary inserted into a stainless steel tube, similar to prototype desorption electrospray ionization sources, generated the nebulizing spray. The slides were affixed to a rotating plate with the nebulizer oscillating radially over them. By rapidly rotating the samples and performing numerous oscillating passes, uncertainty in sprayer position was averaged and an even MALDI matrix coating was achieved. A solution of 50 mg/mL 2,5-dihydroxybenzoic acid (DHB) in an acetone/H2O mixture, 1/1 (v/v) with 0.05% trifluoroacetic acid (TFA), delivered at 0.5 mL/min, was used in sample preparation for MALDI MS. For single cell profiling, the nebulizer was placed 1 cm above the surface and oscillated over the samples 25 times with a nitrogen gas pressure of 50 psi, resulting in a DHB coating of ~0.2 mg/cm2. Intact islets required a thicker, dryer coating, with the sprayer operated at a 7 cm distance from the sample with nebulizing gas pressure at 100 psi and 100 passes.
A point-based similarity registration algorithm55 was utilized to align the relative coordinates of cells, islets, and fiducial points on the ITO slides with the stage positions of a Bruker ultrafleXtreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics). Scored crosshairs, acting as fiducial markers on the slides, were located in the bright-field image and in the ultrafleXtreme camera system. Python scripts written in-house (see Supporting Information and Supplementary Software) were utilized to mark fiducial locations and input corresponding stage locations to the registration model (Figure S4). Implementing a simple threshold and group algorithm, the same software was then used to find cells in the fluorescent and bright-field images (Figure S5), allowing selective recognition of the biological structure by user-defined levels of fluorescence signal intensity, cell size, and cell circularity. Pixel positions in the microscopy image were transformed to fractional distances, which are required to generate custom geometry files with the registration parameters. In addition to saving information on the registration points and cell-finding parameters, the software generated a custom geometry file for direct import to the flexControl software (Bruker Daltonics) that operated the MALDI MS automatic acquisition. The Additional Methods found in the Supporting Information include further details regarding cell localization accuracy and details of the Python script.
Spectra were acquired with a Bruker ultrafleXtreme MALDI-TOF/TOF mass spectrometer equipped with a frequency tripled Nd-YAG solid state laser (Bruker Daltonics). The mass scan window was set to m/z 400–6000 and the laser set to the “Ultra” footprint setting at an ~100-μm footprint diameter. The Bruker ultrafleXtreme AutoXecute feature was utilized with the custom geometry file as previously reported. 24 Each spectrum represents the summed signals acquired during 1000 laser shots fired at 1000 Hz. From the 48 dispersed islets analyzed, approximately 32,000 spots were profiled based on fluorescence microscopy identification of the locations of single cells. A 100-μm distance filter removed half of the spectra to ensure each profile corresponded to a single cell (Figure S6). The dataset was then imported into ClinProTools software (Bruker Daltonics) with a 16-fold data reduction to perform null spectra exclusion, baseline subtraction, and total ion current normalization. Initial examination of the principal component analysis (PCA) loading plots suggested the major contributors to sample variance were classical biochemical markers for each cell type (glucagon, insulin, somatostatin, and PP). The mass accuracy for peptides observed with MALDI MS are listed in Supporting Table 2. To simplify comparison with previous histological reports, the intensities of somatostatin, glucagon, PP, and insulin were exported from ClinProTools for further analysis in MATLAB (Mathworks, Natick, MA). After parsing the xml files, empty spectra were excluded by removing samples with peptide intensities of less than 3.5 times the median value of each peptide. Cell phenotyping was performed by k-means clustering using a cosine distance, and then further validated with the same threshold based on the median intensity. To visualize the classes on a single plot, the dimension of the data was reduced with PCA as shown in Figure 2. The cell counts from each cluster were then matched to their corresponding cellular populations, shown in Table 1.
Next, the inter-anatomical differences between pancreatic islet and cellular subtypes were evaluated. Mass spectra were classified based on their peptide content and the dorsal and ventral islet-derived cells were considered as two separate classes in the ClinProTools analyses. The 50 most intense signals between m/z 1000–6000 were evaluated for statistically significant differences for each of the four cell types. The dataset was tested for univariate normality with the Anderson–Darling test, which showed the data not to be normally distributed. Hence, statistical tests of differences were performed with the Wilcoxon test applying the Benjamini–Hochberg procedure for false discovery rate correction, with p < 0.05 considered to be statistically significant.
MALDI MSI
MSI was performed on sections of rat pancreas. The tissue was fast frozen after dissection and sectioned in 5-μm thick slices without chemical fixation at −25 °C using a Leica CM 3050 S cryostat (Leica Microsystems, Bannockburn, IL). Pancreas sections were deposited on ITO-coated glass slides at room temperature (23–25 °C). Specimens for MSI were spray coated with 5 mg/mL 2-(4-hydroxyphenylazo) benzoic acid in a methanol/H2O mixture (20/80 v/v) containing 0.1% FA and 0.01% TFA with an artist’s airbrush. MSI was performed with an utrafleXtreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) operating in reflector mode at positive polarity. The laser beam was set to “ultra”, corresponding to an ~100 μm footprint. The mass spectra were acquired in 25- or 50-μm-spaced arrays, with some expected oversampling. The MS calibration standards were deposited onto slide locations nearby the tissue sections. The peptide ions, and in some cases known endogenous ions, such as several of the most common lipids, as well as MALDI matrix ions, were used in the post-processing step for data recalibration. MSI data acquisition and processing was performed with flexControl, flexImaging, and ClinPro-Tools software (all Bruker Daltonics).
NanoLC–Fourier Transform (FT)-Ion Cyclotron Resonance (ICR) MS
Peptide identification was performed via an FT-ICR mass spectrometer (LTQ-FT Ultra, Thermo Scientific) coupled with a nanoLC system (Eksigent 1D Plus, Eksigent, Dublin, CA). Peptide extract (2 μL) from islets was mixed with 8 μL of loading solvent (5% acetonitrile (ACN), 0.2% FA), injected onto a peptide trap column (150 μm inner diameter (i.d.) × 2 cm length, 5 μm Magic AQ particles, 100 Å pore size, New Objective, Inc., Woburn, MA) and desalted with the loading solvent. The column was flushed with loading solvent. The trap column was then placed in line with the analytical column (PicoFrit column, 75 μm i.d. × 15 cm length, 5 μm Magic AQ particles, 100 Å pore size, New Objective). Mixtures of ACN/water with 0.2% FA were used as chromatographic solvents A (5/95 v/v) and B (95/5 v/v). The analytes were separated with a flow rate of 300 nL/min over a gradient with a solvent A and B mixture as follows: 0–10 min, 0–20% solvent B; 10–65 min, 20–55% solvent B. For MS acquisition, the mass scan window was set to m/z 300–2000, data-dependent precursor selection was restricted to the top five most intense ions, dynamic exclusion was enabled with a repeat count of 2, and an exclusion duration of 180 s.
Peptide Sequencing
Native Thermo LC–MS data in raw file format were processed with PEAKS 7 (Bioinformatics Solutions Inc., ON, Canada) for peptide sequencing. The data were searched against a UniprotKB/SwissProt rat database of canonical sequences (June 2015 release, Rattus norvegicus, 7923 entries). The parent mass error tolerance was set to 50 ppm, the fragment mass error tolerance was set to 0.01 Da. Enzymatic digestion was set to “none”. Acetylation (N-term and K), amidation, oxidation (M), Pyro-glu (E and Q), half of a disulfide bridge, and phosphorylation were allowed as variable modifications. The filtering conditions used (peptide −10 log P ≥ 15, protein −10 log P ≥ 20, proteins unique peptide ≥ 0) resulted in a false discovery rate of 1.5% for peptide spectrum matches.
Bioinformatic Analysis
Probabilities for processing of dibasic sites in the pancreatic prohormone (PAHO_RAT) were calculated using the NeuroPred tool (available online at http://neuroproteomics.scs.illinois.edu), with mammalian as the selected model option.
Ab initio calculations of PP, PP(1–24), and PP(27–36) folding were performed using the PEP-FOLD tool (available online at http://mobyle.rpbs.univ-paris-diderot.fr), with the sequence for full-length PP obtained from Uniprot KB (PAHO_RAT).
Supplementary Material
Acknowledgments
The project described was supported by the National Institutes of Health, Award Number P30 DA018310 from the National Institute on Drug Abuse, and Award Number U01 MH109062 from the National Institute of Mental Health, and fellowships to T.J.C. from the Training Program at Chemistry-Interface with Biology (T32 GM070421), the National Science Foundation Graduate Research Fellowship Program, and the Springborn Fellowship. The Core Facilities at the Carl R. Woese Institute for Genomic Biology, UIUC, provided access to the digital pathology scanner. We thank Ning Yang, UIUC, for technical assistance with the FT-ICR MS.
Footnotes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website.
Additional Methods: description of the single cell MALDI MS workflow and Python script details; supporting figures S1–S6 and Supporting Table 1 (PDF).
Supporting Table 2, LC–MS data from islet peptide extractions (Microsoft Excel spreadsheet).
Supplementary software, used for generation of the Bruker ultrafleXtreme MALDI-TOF/TOF mass spectrometer coordinates for single cell locations (ZIP file archive containing Python scripts).
References
- 1.Dueck H, Eberwine J, Kim J. Variation is function: Are single cell differences functionally important? Bioessays. 2015;38:172–180. doi: 10.1002/bies.201500124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wills QF, Boothe T, Asadi A, Ao Z, Warnock GL, Kieffer TJ, Johnson JD. Statistical approaches and software for clustering islet cell functional heterogeneity. Islets. 2016;8:48–56. doi: 10.1080/19382014.2016.1150664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benninger RP, Hutchens T, Head W, McCaughey M, Zhang M, Le Marchand S, Satin L, Piston D. Intrinsic islet heterogeneity and gap junction coupling determine spatiotemporal Ca2+ wave dynamics. Biophys J. 2014;107:2723–2733. doi: 10.1016/j.bpj.2014.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baetens D, Malaisse-Lagae F, Perrelet A, Orci L. Endocrine pancreas: three-dimensional reconstruction shows two types of islets of langerhans. Science. 1979;206:1323–1325. doi: 10.1126/science.390711. [DOI] [PubMed] [Google Scholar]
- 5.Elayat AA, el-Naggar MM, Tahir M. An immunocytochemical and morphometric study of the rat pancreatic islets. J Anat. 1995;186:629–637. [PMC free article] [PubMed] [Google Scholar]
- 6.Suckale J, Solimena M. Pancreas islets in metabolic signaling–focus on the beta-cell. Front Biosci. 2008;13:7156–7171. doi: 10.2741/3218. [DOI] [PubMed] [Google Scholar]
- 7.Merkwitz C, Blaschuk OW, Schulz A, Lochhead P, Meister J, Ehrlich A, Ricken AM. The ductal origin of structural and functional heterogeneity between pancreatic islets. Prog Histochem Cytochem. 2013;48:103–140. doi: 10.1016/j.proghi.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Battich N, Stoeger T, Pelkmans L. Control of transcript variability in single mammalian cells. Cell. 2015;163:1596–1610. doi: 10.1016/j.cell.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Klughammer J, Farlik M, Penz T, Spittler A, Barbieux C, Berishvili E, Bock C, Kubicek S. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2015;17:178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubakhin SS, Romanova EV, Nemes P, Sweedler JV. Profiling metabolites and peptides in single cells. Nat Meth. 2011;8:S20–S29. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science. 2013;342 doi: 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- 12.Edwards JL, Kennedy RT. Metabolomic analysis of eukaryotic tissue and prokaryotes using negative mode MALDI time-of-flight mass spectrometry. Anal Chem. 2005;77:2201–2209. doi: 10.1021/ac048323r. [DOI] [PubMed] [Google Scholar]
- 13.Schmudlach A, Felton J, Cipolla C, Sun L, Kennedy RT, Dovichi NJ. Sample preparation protocol for bottom-up proteomic analysis of the secretome of the islets of Langerhans. Analyst. 2016;141:1700–1706. doi: 10.1039/c5an02265g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M. Quantitative proteomic analysis of single pancreatic islets. Proc Natl Acad Sci USA. 2009;106:18902–18907. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boonen K, Baggerman G, D’Hertog W, Husson SJ, Overbergh L, Mathieu C, Schoofs L. Neuropeptides of the islets of Langerhans: a peptidomics study. Gen Comp Endocrinol. 2007;152:231–241. doi: 10.1016/j.ygcen.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Stewart KW, Phillips ARJ, Whiting L, Jüllig M, Middleditch MJ, Cooper GJS. A simple and rapid method for identifying and semi-quantifying peptide hormones in isolated pancreatic islets by direct-tissue matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2011;25:3387–3395. doi: 10.1002/rcm.5239. [DOI] [PubMed] [Google Scholar]
- 17.Jimenez CR, van Veelen PA, Li KW, Wildering WC, Geraerts WPM, Tjaden UR, van der Greef J. Rapid communication: neuropeptide expression and processing as revealed by direct matrix-assisted laser desorption ionization mass spectrometry of single neurons. J Neurochem. 1994;62:404–407. doi: 10.1046/j.1471-4159.1994.62010404.x. [DOI] [PubMed] [Google Scholar]
- 18.Garden RW, Moroz LL, Moroz TP, Shippy SA, Sweedler JV. Excess salt removal with matrix rinsing: Direct peptide profiling of neurons from marine invertebrates using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Mass Spectrom. 1996;31:1126–1130. doi: 10.1002/(SICI)1096-9888(199610)31:10<1126::AID-JMS403>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 19.Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI MS: distribution of neuropeptides within single neurons. Anal Chem. 2003;75:5374–5380. doi: 10.1021/ac034498+. [DOI] [PubMed] [Google Scholar]
- 20.Neupert S, Predel R. Mass spectrometric analysis of single identified neurons of an insect. Biochem Biophys Res Commun. 2005;327:640–645. doi: 10.1016/j.bbrc.2004.12.086. [DOI] [PubMed] [Google Scholar]
- 21.Altelaar AFM, Taban IM, McDonnell LA, Verhaert PDEM, de Lange RPJ, Adan RAH, Mooi WJ, Heeren RMA, Piersma SR. High-resolution MALDI imaging mass spectrometry allows localization of peptide distributions at cellular length scales in pituitary tissue sections. Int J Mass Spectrom. 2007;260:203–211. [Google Scholar]
- 22.Jarecki JL, Andersen K, Konop CJ, Knickelbine JJ, Vestling MM, Stretton AO. Mapping neuropeptide expression by mass spectrometry in single dissected identified neurons from the dorsal ganglion of the nematode Ascaris suum. ACS Chem Neurosci. 2010;1:505–519. doi: 10.1021/cn1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen R, Ouyang C, Xiao M, Li L. In situ identification and mapping of neuropeptides from the stomatogastric nervous system of Cancer borealis. Rapid Commun Mass Spectrom. 2014;28:2437–2444. doi: 10.1002/rcm.7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ong T, Kissick DJ, Jansson ET, Comi TJ, Romanova EV, Rubakhin SS, Sweedler JV. Classification of large cellular populations and discovery of rare cells using single cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 2015;87:7036–7042. doi: 10.1021/acs.analchem.5b01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barkatullah SC, Curry WJ, Johnston CF, Hutton JC, Buchanan KD. Ontogenetic expression of chromogranin A and its derived peptides, WE-14 and pancreastatin, in the rat neuroendocrine system. Histochem Cell Biol. 1997;107:251–257. doi: 10.1007/s004180050110. [DOI] [PubMed] [Google Scholar]
- 26.Wulff BS, Johansen TE, Dalbøge HH, O’Hare MMT, Schwartz TW. Processing of two homologous precursors, pro-neuropeptide Y and pro-pancreatic polypeptide, in transfected cell lines expressing different precursor convertases. J Biol Chem. 1993;268:13327–13335. [PubMed] [Google Scholar]
- 27.Brakch N, Galanopoulou AS, Patel YC, Boileau G, Seidah NG. Comparative proteolytic processing of rat prosomatostatin by the convertases PC1, PC2, furin, PACE4 and PC5 in constitutive and regulated secretory pathways. FEBS Lett. 1995;362:143–146. doi: 10.1016/0014-5793(95)00229-3. [DOI] [PubMed] [Google Scholar]
- 28.Itoh Y, Tanaka S, Takekoshi S, Itoh J, Osamura RY. Prohormone convertases (PC1/3 and PC2) in rat and human pancreas and islet cell tumors: subcellular immunohistochemical analysis. Pathol Int. 1996;46:726–737. doi: 10.1111/j.1440-1827.1996.tb03541.x. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka S, Kurabuchi S, Mochida H, Kato T, Takahashi S, Watanabe T, Nakayama K. Immunocytochemical localization of prohormone convertases PC1/PC3 and PC2 in rat pancreatic islets. Arch Histol Cytol. 1996;59:261–271. doi: 10.1679/aohc.59.261. [DOI] [PubMed] [Google Scholar]
- 30.Rawdon BB, Larsson LI. Development of hormonal peptides and processing enzymes in the embryonic avian pancreas with special reference to co-localisation. Histochem Cell Biol. 2000;114:105–112. doi: 10.1007/s004180000169. [DOI] [PubMed] [Google Scholar]
- 31.Portela-Gomes GM, Stridsberg M. Selective processing of chromogranin A in the different islet cells in human pancreas. J Histochem Cytochem. 2001;49:483–490. doi: 10.1177/002215540104900408. [DOI] [PubMed] [Google Scholar]
- 32.Webb GC, Dey A, Wang J, Stein J, Milewski M, Steiner DF. Altered proglucagon processing in an alpha-cell line derived from prohormone convertase 2 null mouse islets. J Biol Chem. 2004;279:31068–31075. doi: 10.1074/jbc.M404110200. [DOI] [PubMed] [Google Scholar]
- 33.Rholam M, Fahy C. Processing of peptide and hormone precursors at the dibasic cleavage sites. Cell Mol Life Sci. 2009;66:2075–2091. doi: 10.1007/s00018-009-0007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozawa S, Katsuta H, Suzuki K, Takahashi K, Tanaka T, Sumitani Y, Nishida S, Yoshimoto K, Ishida H. Estimated proinsulin processing activity of prohormone convertase (PC) 1/3 rather than PC2 is decreased in pancreatic beta-cells of type 2 diabetic patients. Endocr J. 2014;61:607–614. doi: 10.1507/endocrj.ej13-0506. [DOI] [PubMed] [Google Scholar]
- 35.Katsuta H, Ozawa S, Suzuki K, Takahashi K, Tanaka T, Sumitani Y, Nishida S, Kondo T, Hosaka T, Inukai K, Ishida H. The association between impaired proinsulin processing and type 2 diabetes mellitus in non-obese Japanese individuals. Endocr J. 2015;62:485–492. doi: 10.1507/endocrj.EJ14-0611. [DOI] [PubMed] [Google Scholar]
- 36.Walker MW, Smith KE, Bard J, Vaysse PJJ, Gerald C, Daouti S, Weinshank RL, Branchek TA. A Structure-activity analysis of the cloned rat and human Y4 receptors for pancreatic Polypeptide1. Peptides. 1997;18:609–612. doi: 10.1016/s0196-9781(97)00070-3. [DOI] [PubMed] [Google Scholar]
- 37.Bard JA, Walker MW, Branchek T, Weinshank RL. 5,976,814. US Patent. 1999 Nov 2;
- 38.Gehlert DR, Schober DA, Beavers L, Gadski R, Hoffman JA, Smiley DL, Chance RE, Lundell I, Larhammar D. Characterization of the peptide binding requirements for the cloned human pancreatic polypeptide-preferring receptor. Mol Pharmacol. 1996;50:112–118. [PubMed] [Google Scholar]
- 39.Cerdá-Reverter JM, Larhammar D. Neuropeptide Y family of peptides: structure, anatomical expression, function, and molecular evolution. Biochem Cell Biol. 2000;78:371–392. [PubMed] [Google Scholar]
- 40.Berglund MM, Lundell I, Eriksson H, Söll R, Beck-Sickinger AG, Larhammar D. Studies of the human, rat, and guinea pig Y4 receptors using neuropeptide Y analogues and two distinct radioligands. Peptides. 2001;22:351–356. doi: 10.1016/s0196-9781(01)00337-0. [DOI] [PubMed] [Google Scholar]
- 41.Keire DA, Bowers CW, Solomon TE, Reeve JR., Jr Structure and receptor binding of PYY analogs. Peptides. 2002;23:305–321. doi: 10.1016/s0196-9781(01)00602-7. [DOI] [PubMed] [Google Scholar]
- 42.Balasubramaniam A, Mullins DE, Lin S, Zhai W, Tao Z, Dhawan VC, Guzzi M, Knittel JJ, Slack K, Herzog H, Parker EM. Neuropeptide Y (NPY) Y4 receptor selective agonists based on NPY(32–36): development of an anorectic Y4 receptor selective agonist with picomolar affinity. J Med Chem. 2006;49:2661–2665. doi: 10.1021/jm050907d. [DOI] [PubMed] [Google Scholar]
- 43.Parker MS, Sah R, Sheriff S, Balasubramaniam A, Parker SL. Internalization of cloned pancreatic polypeptide receptors is accelerated by all types of Y4 agonists. Regul Pept. 2005;132:91–101. doi: 10.1016/j.regpep.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Walther C, Mörl K, Beck-Sickinger AG. Neuropeptide Y receptors: ligand binding and trafficking suggest novel approaches in drug development. J Pept Sci. 2011;17:233–246. doi: 10.1002/psc.1357. [DOI] [PubMed] [Google Scholar]
- 45.Domin H, Pięketa E, Piergies N, Święch D, Kim Y, Proniewicz LM, Proniewicz E. Neuropeptide Y and its C-terminal fragments acting on Y2 receptor: Raman and SERS spectroscopy studies. J Colloid Interface Sci. 2015;437:111–118. doi: 10.1016/j.jcis.2014.09.053. [DOI] [PubMed] [Google Scholar]
- 46.Li J, Tian Y, Wu A. Neuropeptide Y receptors: a promising target for cancer imaging and therapy. Regen Biomater. 2015;2:215–219. doi: 10.1093/rb/rbv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boel E, Schwartz TW, Norris KE, Fiil NP. A cDNA encoding a small common precursor for human pancreatic polypeptide and pancreatic icosapeptide. EMBO J. 1984;3:909–912. doi: 10.1002/j.1460-2075.1984.tb01904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Southey BR, Amare A, Zimmerman TA, Rodriguez-Zas SL, Sweedler JV. NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 2006;34:W267–W272. doi: 10.1093/nar/gkl161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Y, Maupetit J, Derreumaux P, Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J Chem Theory Comput. 2014;10:4745–4758. doi: 10.1021/ct500592m. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz TW. Cellular peptide processing after a single arginyl residue. Studies on the common precursor for pancreatic polypeptide and pancreatic icosapeptide. J Biol Chem. 1987;262:5093–5099. [PubMed] [Google Scholar]
- 51.Atkins N, Jr, Mitchell JW, Romanova EV, Morgan DJ, Cominski TP, Ecker JL, Pintar JE, Sweedler JV, Gillette MU. Circadian integration of glutamatergic signals by little SAAS in novel suprachiasmatic circuits. PLoS ONE. 2010;5:1–13. doi: 10.1371/journal.pone.0012612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carter JD, Dula SB, Corbin KL, Wu R, Nunemaker CS. A practical guide to rodent islet isolation and assessment. Biol Proced Online. 2009;11:3–31. doi: 10.1007/s12575-009-9021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tucker KR, Li Z, Rubakhin SS, Sweedler JV. Secondary ion mass spectrometry imaging of molecular distributions in cultured neurons and their processes: comparative analysis of sample preparation. J Am Soc Mass Spectrom. 2012;23:1931–1938. doi: 10.1007/s13361-012-0472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berman ESF, Fortson SL, Checchi KD, Wu L, Felton JS, Wu KJJ, Kulp KS. Preparation of single cells for imaging/profiling mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1230–1236. doi: 10.1016/j.jasms.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 55.Fitzpatrick JM, Hill DLG, Maurer CR., Jr . Handbook of Medical Imaging. In: Fitzpatrick JM, Sonka M, editors. Handbook of medical imaging, volume 2. Medical image processing and analysis. Vol. 2. SPIE Press; Bellingham WA, USA: 2009. pp. 447–513. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.